Abstract
Neural Message Passing for graphs is a promising and relatively recent approach for applying Machine Learning to networked data. As molecules can be described intrinsically as a molecular graph, it makes sense to apply these techniques to improve molecular property prediction in the field of cheminformatics. We introduce Attention and Edge Memory schemes to the existing message passing neural network framework, and benchmark our approaches against eight different physical–chemical and bioactivity datasets from the literature. We remove the need to introduce a priori knowledge of the task and chemical descriptor calculation by using only fundamental graph-derived properties. Our results consistently perform on-par with other state-of-the-art machine learning approaches, and set a new standard on sparse multi-task virtual screening targets. We also investigate model performance as a function of dataset preprocessing, and make some suggestions regarding hyperparameter selection.
Keywords: Message passing neural network, Graph convolution, Virtual screening, Machine learning, Deep learning
Introduction
QSAR (Quantitative Structure Activity Relationships) have been applied for decades in the development of relationships between physicochemical properties of chemical substances and their biological activities to obtain a reliable mathematical and statistical model for prediction of the activities of new chemical entities. The major aim of QSAR study is to reduce the number of compounds synthesized during the drug development, a notoriously long and costly process, hence the desire to improve its efficiency from a drug discovery perspective. After Hansch proposed the QSAR concept [1], engineering molecular descriptors to build accurate models for the prediction of various properties has become the standard approach to QSAR modelling. Researchers [2–6] have proposed numerous descriptors to represent molecular 2D and 3D structures, aiming to correlate these descriptors with predicted endpoints. Approaches to generating representations using the graph representation of a molecule include graph kernels [7], and perhaps most importantly in the present context, ECFP (Extended Connectivity Circular Fingerprints) [8]. Once a descriptor set has been defined, various modelling methods, including linear mapping methods like linear regression, partial least square and non-linear methods like support vector machine, random forest etc., are applied to building models. Recently, deep neural network methods have become the latest weapon in a Cheminformatician’s arsenal for doing QSAR.
Over the past decade, deep learning has become a staple in the machine learning toolbox of many fields and research areas [9, 10]. Notably in the pharmaceutical area, in recent years AI has shown incredible growth, and is being used now not just for bioactivity and physical–chemical property prediction, but also for de novo design, image analysis, and synthesis prediction, to name a few. This rapid growth is due in part to the substantial increase in available biochemical data thanks to the rise of techniques such as High Throughput Screening (HTS) and parallel synthesis, and also to the recent surge in parallel computational power that can be feasibly attained by harnessing General Purpose computing on Graphics Processing Units (GPGPU).
Efforts have also been taken to enable neural networks to do representation learning, i.e. the neural network is able to learn descriptors itself instead of relying on predefined molecular descriptors. Among these, the graph convolution network (GCN) is gaining popularity and various architectures have been proposed in data science community. The first Graph Neural Networks (GNNs) was put forward by Gori et al. in 2005 [11], presenting an architecture for learning node representations using recurrent neural networks capable of acting on directed, undirected, labelled, and cyclic graphs. This work was later expanded upon by Micheli [12] and Scarselli et al. [13] In 2013, the Graph Convolutional Network (GCN) was presented by Bruna et al. [14] using the principles of spectral graph theory. Many other forms of GNN have been presented since then, including, but not limited to, Graph Attention Networks [15], Graph Autoencoders [16–19], and Graph Spatial–Temporal Networks [20–23].
In GCNs and some other forms of GNNs, information is propagated through a graph in a manner similar to how conventional convolutional neural networks (CNNs) treat grid data (e.g. image data). However, whilst graph-based deep learning shares some connection with CNNs with respect to local connectivity of the component data, CNNs exploit the properties of regular connectivity, shift-invariance, and compositionality to achieve their noteworthy performance. In order to cope with the irregularity of graph data, alternative approaches must be designed, most notably to circumvent the issue of irregular non-Euclidean data, and to be invariant to the graph representation.
Whilst many implementations are designed for use on a single large graph, such as social networks or citation graphs, approaches designed for use on multiple smaller graphs such as graphs of small molecule are also desired for their potential use in, amongst other things, drug design. Duvenaud [24] proposed the neural fingerprint method, describing it as an analogue of ECFP, as one of the first efforts in applying graph convolution model on chemistry related problems. The notable advancement embodied in the neural fingerprint approach with regards to predecessing concepts such as graph kernels and ECFP, is that the generation of descriptors is adapted—learned—during training. Other molecular graph convolution methods were reported by Kearnes et al. [25] and Coley [26] as extensions to Duvenaud’s method. Recently researchers from Google [27] put forward an new NN architecture called as message passing neural networks (MPNNs) and used the MPNNs to predict quantum chemical properties. The MPNN framework contains three common steps: (1) message passing step, where, for each atom, features (atom or bond features) from its neighbours are propagated, based on the graph structure, into a so called a message vector; (2) update step, where embedded atom features are updated by the message vector; (3) aggregation step, where the atomic features in the molecule are aggregated into the molecule feature vector. These molecule feature vector can then be used in a dense layer to correlate with the endpoint property. It has been shown that the MPNN framework has a high generalizability such that several popular graph neural network algorithms [24–26, 28, 29] can be translated into the MPNN framework. Several research groups have made various extensions to the MPNN framework to augment it for work on cheminformatic problems [30].
Like GCN methods, MPNN model learns task specific molecule features from the graph structure and avoid feature engineering in the pre-processing stage. This type of method also presents an approach for the secure sharing of chemical data, i.e. it is possible to disseminate trained models for activity predictions without the risk of reverse-engineering IP-sensitive structural information [31–33].
We introduce a selection of augmentations to known MPNN architectures, which we refer to as Attention MPNN (AMPNN) and Edge Memory Neural Network (EMNN) [34], and evaluate them against published benchmark results with a range of metrics. The EMNN network shares architectural similarities to the D-MPNN model published by Yang et al. [35] that was developed concurrently to this work [36], but the D-MPNN includes additional chemical descriptor information. We applied these two types of neural network to eight datasets from the MoleculeNet [30] benchmark and analyse the performances and offer chemical justification for these results with respect to both architecture and parameter selection.
Method
Concepts of graphs
A graph is a set of nodes and a set of edges, which are pairs of elements of . If the members of E are ordered pairs, the graph is said to be directed. In the graph representation of a molecule, atoms are viewed as nodes and indicates there is a bond between atoms and . This representation is an undirected graph: we do not consider a bond to have a direction, so we do not distinguish between .
In the given context, a graph comes together with a feature vector corresponding to each node and an edge feature vector corresponding to each edge .
Message passing neural network
The Message Passing Neural Network [27] is a deep learning architecture designed for implementation in chemical, pharmaceutical and material science contexts. They were introduced as a framework to generalise several proposed techniques [14, 24, 25, 28, 29, 37, 38], and have demonstrated state-of-the-art results on multiple related benchmarks. For the specific MPNN implementations used for experiments in this paper, the most important predecessor is the Gated Graph Sequence Neural Network (GGNN) [28].
In simplistic terms, MPNNs operate by the following mechanism: An initial set of states is constructed, one for each node in the graph. Then, each node is allowed to exchange information, to “message”, with its neighbours. After one such step, each node state will contain an awareness of its immediate neighbourhood. Repeating the step makes each node aware of its second order neighbourhood, and so forth. After a chosen number of “messaging rounds”, all these context-aware node states are collected and converted to a summary representing the whole graph. All the transformations in the steps above are carried out with neural networks, yielding a model that can be trained with known techniques to optimise the summary representation for the task at hand.
More formally, MPNNs contain three major operations: message passing, node update, and readout. Using a message passing neural network entails iteratively updating a hidden state of each node . This is done according to the following formulas:
| 1 |
| 2 |
where is the message function, is the node update function, is the set of neighbours of node in graph , is the hidden state of node at time , and is a corresponding message vector. For each atom , messages will be passed from its neighbours and aggregated as the message vector from its surrounding environment. Then the atom hidden state is updated by the message vector.
The formula for the readout function is shown in formula 3:
| 3 |
where is a resulting fixed-length feature vector generated for the graph, and is a readout function that is invariant to node ordering, an important feature that allows the MPNN framework to be invariant to graph isomorphism. The graph feature vector then is passed to a fully connected layer to give prediction. All functions , and are neural networks and their weights are learned during training. While details are given in the following sections, we provide summary differences between our presented architectures in Tables 1, 2, 3 and 4.
Table 1.
Core differences between model architectures
| Model | Hidden states | Denotion of neighbourhood | Message aggregation scheme |
|---|---|---|---|
| MPNN | |||
| AMPNN |
, where
|
||
| EMNN |
, where
|
Table 2.
Aggregation function special cases
| Model | Hidden states | Aggregation form |
|---|---|---|
| MPNN | ||
| AMPNN | ||
| EMNN |
|
Table 3.
Other model architecture differences
| Model | Pre-message passing | Update | Pre-readout |
|---|---|---|---|
|
MPNN AMPNN |
NA | NA | |
| EMNN |
Table 4.
Model readout function and post-readout function
| Model | Readout function | Post-readout |
|---|---|---|
| All | FFNN |
SELU message passing neural network (SELU-MPNN)
Our first architecture involved the basic MPNN framework, but with the use of the SELU activation function [39] instead of more traditional batch or layer norm functions. The SELU activation function is parameterised to converge towards a zero mean and unit variance, and removed the need to experiment with different normalisation approaches (batch, layer, tensor, etc.) explicitly. All other architectures we propose also use SELU as their activation functions. Whilst many of the graph neural network approaches presented by MolNet can be cast into the MPNN framework, we chose to use SELU-MPNN as our baseline for our implementation of the framework due to the increased convergence speed that SELU offers [40]. This affords us consistent results within our framework for a less biased comparison to more basic methods.
Apart from the different choice of activation function and hidden layers in the message function, the model we in our experiments denote SELU-MPNN shares great similarity with the original GGNN.
Attention message passing neural network (AMPNN)
Here we propose a further augmentation to the MPNN architecture by considering a more general form of the MPNN message summation step (Eq. 1). Using simple summation to convert an unknown cardinality set of vectors into a single vector is hypothetically an expressive bottleneck. Potential better ways to implement such aggregation functions are currently being researched [41–44]. In the current study we extend previous MPNN models for graph-level prediction by employing a straight forward aggregation function with an attention mechanism. The attention mechanism has been proposed on image recognition and language translation problems amongst others [41, 45, 46] and have achieved better performance compared with normal deep neural network algorithms. We denote our specific implementation of the extended framework an Attention Message Passing Neural Network (AMPNN). Its most important predecessor is, as for our SELU-MPNN, the GGNN [28].
As mentioned earlier, the non-weighted summation in message passing function (Eq. 1) of the original MPNN constitutes a potential limitation. In the AMPNN framework, a computationally heavier but potentially more expressive attention layer is proposed in the message passing stage to aggregate messages (Eq. 4). Equation 1 is replaced by the more general formula:
| 4 |
where is an aggregate function invariant to the ordering of set members at step t. Just as for the original MPNN, the message to node is computed based on its neighbours , but the method of aggregation is not restricted to being a simple summation. The here chosen to be able to investigate the architecture is that of the SELU-MPNN augmented with an attention mechanism. This is mainly inspired by [41] and essentially eliminates the cardinality dimension of the set of neighbours by taking weighted sums. Formally, our layer is
| 5 |
Two feed forward neural network (FFNN) and are used for each edge type and give output vectors with the same length. The and the fraction bar represent Hadamard multiplication and Hadamard division, respectively. Note that because of the output dimensionality of , the softmax-like operation embodied in the fraction of Eq. 5 uses a multitude of weightings rather than just one.
The network turns the hidden state of atom into an embedding vector, while the network embeds the atom hidden states into weight vectors which are turned into weight coefficients after the softmax operation. Notably, the softmax operation is done along the cardinality dimension of the set of weight vectors. Thus, the contribution of one element in the embedding vector depends on equivalent element of weight vectors in the set.
In the node update stage, similar to the GGNN, the node hidden states are updated via a gated recurrent unit, where the is treated as the input and the current node hidden state is used as the hidden state of the GRU
| 6 |
At the initial state (t = 0), is the predefined atom feature vector. After the message passing and node updating steps are iterated for K steps, a readout function is applied to aggregate the hidden state of all the nodes in the graph into a graph level feature vector using two FFNNs. More precisely we use the GGNN readout function,
| 7 |
where and are FFNNs, the denotes Hadamard multiplication, is the sigmoid function and the (,) of the right hand side denotes concatenation. The generated graph feature vector is then passed into the final FFNN layer to make prediction.
Edge Memory Neural Network (EMNN)
The message passing concept in the MPNN framework computes the message to a centre atom by aggregating information from its neighbourhood atoms in a symmetric fashion. Another MPNN-inspired model in our study has a hidden state in each directed edge (every bond has two directed edges in the directed graph) instead of in the nodes. In the directed graph, each bond (node–node connection) has two directed edges, thus two hidden states. The hidden state of a directed edge is updated based on hidden states of edges whose heads coincide with its tail (Fig. 1). We call this model an Edge Memory Neural Network (EMNN). In the resulting message passing step, the update of a hidden state has a corresponding direction.
Fig. 1.
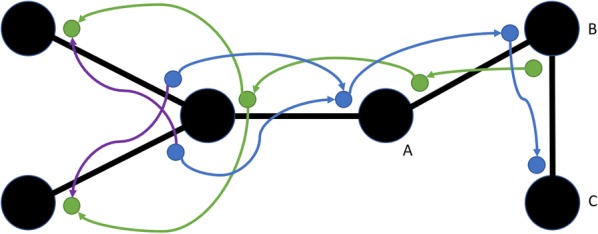
The message passing from directed neighbouring edges to another edge in EMNN. Blue and green dots represent each directed hidden state for edges. Each coloured arrow is used to represent a respective message pass within the graph—purple represents the transition from one arbitrary direction to the other when the graph branches
This model shares underlying principles with the D-MPNN architecture proposed by Yang et al. [35] which also uses directed edges to improve MPNN performance. Their proposed model also injects additional chemical descriptor information alongside the FFNN after the message passing stage. Another notable difference between these architectures is our implementation of the afore-mentioned attention mechanism in the aggregation function. We include the D-MPNN model in our result and discussion to compare implementations and contrast the performance benefits of additional descriptor information, as has been explored in other literature [47]. We refer to their manuscript for further details on their implementation and architecture.
One hypothetical advantage compared to MPNN is explained in the following. Consider a small graph of three nodes A, B and C connected as A–B–C, as illustrated on the right-hand side of Fig. 1. If information passage from A to C is relevant to the task, two message passes are necessary with conventional MPNN. In the first pass, information is passed from A to B, as desired. However, information is also passed from C to B, so that part of B’s memory is being occupied with information that C already has. This back-and-forth passing of information happening in an MPNN hypothetically dilutes the useful information content in the hidden state of node B. When hidden states instead reside in the directed edges as per EMNN, this cannot happen. The closest thing corresponding to a hidden state in B is the hidden states in the edges and . The update of uses information from , but not from .
As shown in Fig. 1, the flow of messages in each edge is directional where the message flows from a node (tail node) to another node (head node). Formally, the set of edge hidden states taken into account when updating edge of the directed graph is
In the EMNN, before message passing takes place, the two node features are embedded into an edge feature by feeding a concatenation of the original edge and node feature vectors through a FFNN ,
At the initial state , are the raw bond feature vector and atom feature vector respectively and (,) refers to the concatenation operation.
The edge hidden state of at time is updated according to Eqs. 8–10:
| 8 |
Note that each directed edge has both a static edge feature and the time-mutated edge state contributing. is instantiated as a vector of zeros. One choice of aggregation function is
| 9 |
| 10 |
is the message for edge at iteration . is an attention based aggregation function similar to the one used in the AMPNN. means all the edges involving node including the edge itself. Equation 10 is the update of edge using a GRU unit.
After message passing iterations, a node hidden state for each node is taken as the sum of the edge hidden state of edges that the node is end to,
This is done to be able to utilize the same readout functions as seen effective for the MPNNs. The readout function for EMNN is the same as in AMPNN (Eq. 7).
Summary of architectural differences
All models we present are available from our git repository as abstract classes, and have been designed from the ground-up in the Pytorch [48] framework to allow modification at all points, and have been tested using CUDA libraries for GPU acceleration.
Bayesian optimisation
Bayesian Optimisation is a method for returning the next best expected value of an N-dimensional surface by utilising all available information, in contrast to local gradient or Hessian approximation techniques. Gaussian processes are fit around datapoints as they become available, and by using suitable evaluator types, estimates of the next datapoints to be evaluated can be obtained, and a balance between surface exploration and locality optimisation can be struck. We used Expected Improvement as the acquisition function, and Local Penalisation [49] as the evaluator type in order to make batch predictions and hence explore our hyperparameter surface in parallel. The hyperparameters used in the NN were tuned using the Bayesian optimization package GPyOpt [50].
The hyperparameters searched in Bayesian optimization and their constrained ranges are listed in Table 5. Due to architectural differences and an increased number of parameters, the optimisation range for the EMNN was slightly tightened.
Table 5.
A list of hyperparameters optimised for each architecture type, and the domains over which they were optimised
| Hyperparameter | SELU-MPNN | AMPNN | EMNN |
|---|---|---|---|
| Learn-rate | {} | {} | {} |
| Message-size | [10,16,25,40] | [10,16,25,40] | NA |
| Message-passes | [1–10] | [1–10] | [1–8] |
| Msg-hidden-dim | [50,85,150] | [50,85,150] | [50,85,150] |
| Gather-width | [30,45,70,100] | [30,45,70,100] | [30,45,70,100] |
| Gather-emb-hidden-dim | [15,26,45,80] | [15,26,45,80] | [15, 26, 45] |
| Gather-att-hidden-dim | [15,26,45,80] | [15,26,45,80] | [15, 26, 45] |
| Out-hidden-dim | [360,450,560] | [360,450,560] | [360,450,560] |
| Out-dropout-p | {0.0–0.1} | {0.0–0.1} | {0.0–0.1} |
| Out-layer-shrinkage | {0.2–0.6} | {0.2–0.6} | {0.2–0.6} |
| Att-hidden-dim | NA | [50,85,150] | [50,85,150] |
| Edge-emb-hidden-dim | NA | NA | [60,105,180] |
| Edge-embedding-size | NA | NA | [30,50,80] |
Square brackets indicate discrete domains
NA not applicable
Datasets
We used a selection of 8 datasets presented in the MoleculeNet (MolNet) [30] benchmarking paper to evaluate the networks. Datasets (shown in Table 6) were split according to the methods described in the MolNet paper. Datasets were split either randomly, or by Bemis-Murcko scaffold [51]. In the case of randomly split sets, three sets were produced, split by fixed random seeds. Each dataset was split into train/test/validation sets in the ratio 80/10/10 as per the MolNet procedure. Optimal hyperparameters were determined based on their performance on the validation set of the primary split. Once optimal hyperparameters were selected three models were trained, one for each split, and the test scores for the best validation set epoch were averaged and the standard deviation calculated. In the case of scaffold splitting, test runs were still performed three times, and variation in the runs is the result of randomly initiated weights and biases. Each task in each dataset was normalised prior to training, and the results were transformed back after being passed through the model. Normalisation was done the same way as MolNet, with the notable exception of QM8.1 The node features generated from the datasets were: Atom Type, Atom Degree, Implicit Valence, Formal Charge, Number of Radical Electrons, Hybridization (SP, SP2, SP3, SP3D, SP3D2), Aromaticity, and Total Number of Hydrogens. These features were generated as per the MolNet Deepchem functions. For edge features, the bond types were limited to single bonds, double bonds, triple bonds and aromatic bonds.
Table 6.
The selection of datasets on which models were trained, and details pertaining to these sets
| Dataset | Tasks | Type | Compounds | Split | Metric |
|---|---|---|---|---|---|
| MUV | 17 | Classification | 93,127 | Random | PRC-AUC |
| HIV | 1 | Classification | 41,913 | Scaffold | ROC-AUC |
| BBBP | 1 | Classification | 2053 | Scaffold | ROC-AUC |
| Tox21 | 12 | Classification | 8014 | Random | ROC-AUC |
| SIDER | 27 | Classification | 1427 | Random | ROC-AUC |
| QM8 | 12 | Regression | 21,786 | Random | MAE |
| ESOL | 1 | Regression | 1128 | Random | RMSE |
| LIPO | 1 | Regression | 4200 | Random | RMSE |
The QM8 dataset [52] contains electronic spectra calculated from coupled-cluster (CC2) and TD-DFT data on synthetically feasible small organic molecules. The ESOL [53] dataset comprises aqueous solubility values for small molecules, “medium” pesticide molecules, and large proprietary compounds from in-house Syngenta measurements. The LIPO dataset comprises lipophilicity data. The MUV dataset [54] contains PubChem bioactivity data specially selected and arranged by refined nearest-neighbour analysis for benchmarking virtual screening approaches. The HIV dataset [55] comprises classification data for compound anti-HIV activity. The BBBP dataset [56] contains data regarding compound ability to penetrate the blood–brain barrier. The Tox21 dataset [57] was released as a data analysis challenge to predict compound toxicity against 12 biochemical pathways. The SIDER set [58] is a collection of drugs and corresponding potential adverse reactions grouped following MedDRA classifications [59] according to previous usage [60].
Preprocessing
Datasets were used both directly as provided from the MolNet repository without any preprocessing, and with some preprocessing procedure. Dataset preprocessing constituted transformation of the given SMILES string to that of the standardised charge-parent molecule, and reintroduction of ‘missing value’ labels where appropriate in multitask sets, which we refer to as SMD (Standardised Missing Data) preprocessing (Fig. 2). Charge-parent fragmentation was performed using the MolVS standardizer [61], which returned the uncharged version of the largest organic covalent unit in the molecule or complex. In the original datasets, these values were imputed as inactive as per previous literature. The reintroduction of ‘missing value’ labels allows the use of a masking loss function that operates over the set [Active, Inactive, Missing] and does not include missing data in the loss calculation. This prevents backpropagation of molecule-target information in multitask datasets when it is not available.
Fig. 2.
Examples of ionic complexes found in the datasets, and their charge-parent standardized counterparts, as used in the SMD datasets
Results
We present our results as a comparison against the MoleculeNet paper [30], showing test set performances and relative test set errors to the best reported graph-based MoleculeNet architecture, as well as other classical machine learning models. We show our architectures (SELU-MPNN, AMPNN and EMNN models) for both the unaltered and for the SMD preprocessed data, compared against the literature values for the original datasets to allow for fair benchmarking comparison for both the methods and for the preprocessing approaches. Complete tables are available in Additional file 1, alongside model performance information and statistical tests. The results from the literature for other machine learning methods were also reported to have hyperparameters optimised by the authors, using Bayesian Optimisation where applicable, so should present a fair comparison. Some techniques are missing for some larger datasets; this is because they were not reported in the original publications, presumably due to computational limits. Our runs were performed only for the models we present, and these are compared against values taken from literature benchmark studies for other models.
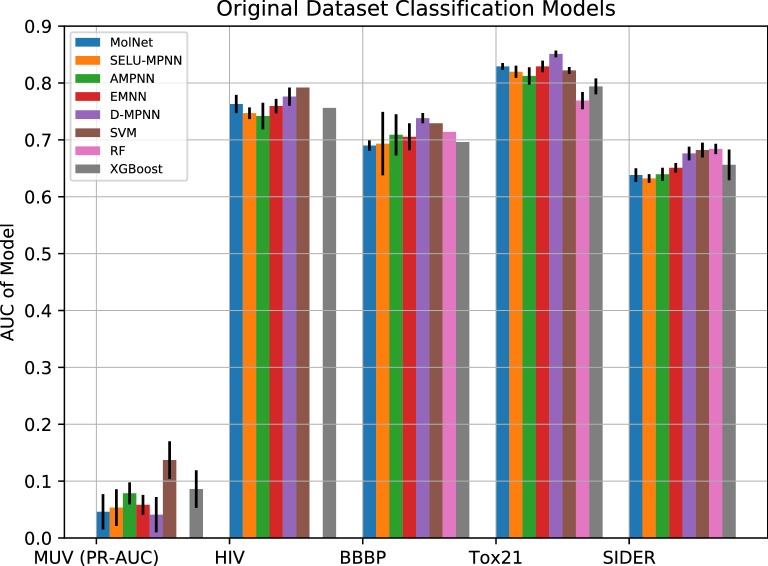
Performance in terms of AUC in classification on the original dataset was on par with state of the art for the majority of models, with the exception of the MUV set (Fig. 3), where a modest increase in performance was observed relative to MolNet. However, this increase was not significant compared to Support-Vector Machines, which had the highest performance by a large margin. The AMPNN architecture was the best of our presented approaches, with the third highest overall performance on the MUV dataset. The D-MPNN showed a mild performance increase over our architectures for sets other than MUV.
Fig. 3.
Predictive performances of machine-learning approaches relative to the best MolNet graph model. With the exception of MUV, the metric used is ROC-AUC. The higher the y-axis is, the better the model performs
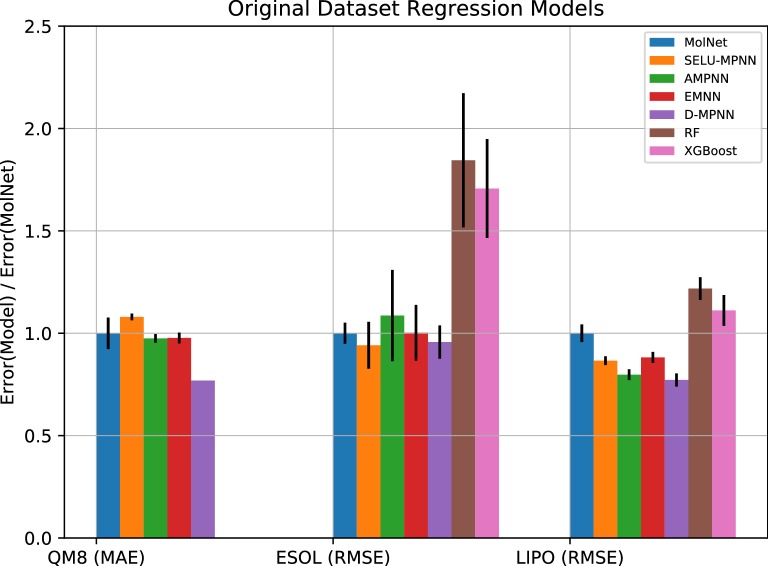
In terms of regression on the original datasets (Fig. 4), the AMPNN was also one of the best performing architectures we present, achieving the lowest error with smallest variance on two of the three sets, covering single and multi-task problems. Performance on the QM8 and ESOL datasets over our three presented architectures was more-or-less on par with MolNet, performing better than Random Forest and XGBoost models, and being beaten by the D-MPNN consistently. However, on the lipophilicity set, all our presented architectures achieved a lower error than all other presented approaches excepting the D-MPNN, which was rivalled by the AMPNN implementation. The Random Forest and XGBoost results are to be expected, as these approaches are much more suited to classification than regression.
Fig. 4.
Regression errors of machine-learning approaches relative to the best MolNet graph model. Metrics are specified for each dataset. The lower the y-axis is, the better the model performs
Performance in classification on the SMD preprocessed dataset was also on par with state of the art for the majority of models, again with the exception of the MUV set (Fig. 5). Little change was observed between the preprocessing techniques for the rest of the datasets, with minor improvement observed in the Tox21 models, a couple of the SIDER and HIV models, and one BBBP model. However, the MUV performance was considerably increased, with two of our architectures (SELU-MPNN and AMPNN) performing as well as SVM model, at three times the predictive power of the presented MolNet architecture. The EMNN network was the best performing architecture, beating SVM models and presenting a predictive power on average over four times higher than MoleculeNet original performance, with only a slightly higher variance.
Fig. 5.
Predictive performances of our machine-learning approaches on the SMD sets relative to MolNet and the respective original models. With the exception of MUV, the metric used is ROC-AUC. The higher the y-axis is, the better the model performs
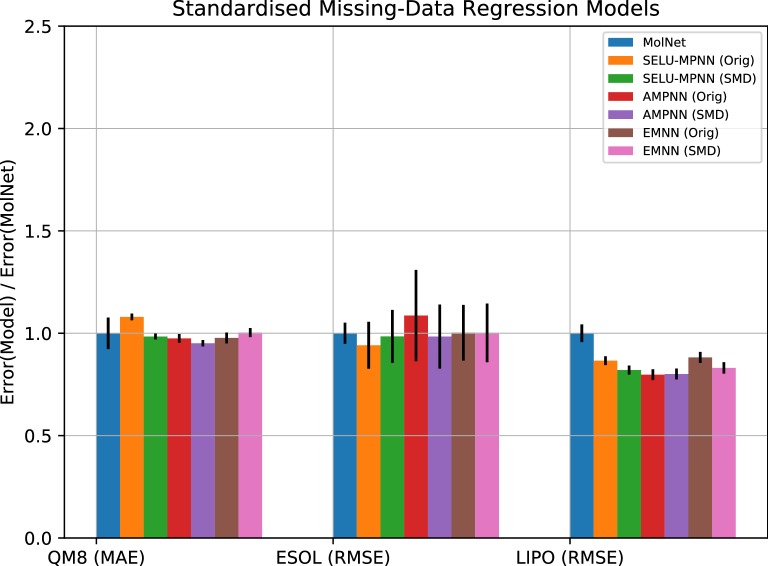
Regression on the SMD datasets (Fig. 6) also showed a little improvement overall versus the original datasets. The AMPNN was again one of the best performing architectures we present, achieving the lowest error with the smallest variance of the SMD models on the same two of the three sets as before, and showing a marked improvement on the ESOL dataset with this preprocessing approach. The lipophilicity set also showed lower overall error with these approaches, though the improvement is minor compared to the improved performance in classification.
Fig. 6.
Regression errors of our machine-learning approaches for the SMD sets relative to MolNet and the respective original models. Metrics are specified for each dataset. The lower the y-axis is, the better the model performs
Overall, we have demonstrated increased predictive power for some of our architectures dependent on task modelled. We have also demonstrated an improved dataset preprocessing technique that can increase the modelling capabilities of our networks under certain circumstances.
Discussion
Datasets
Classification
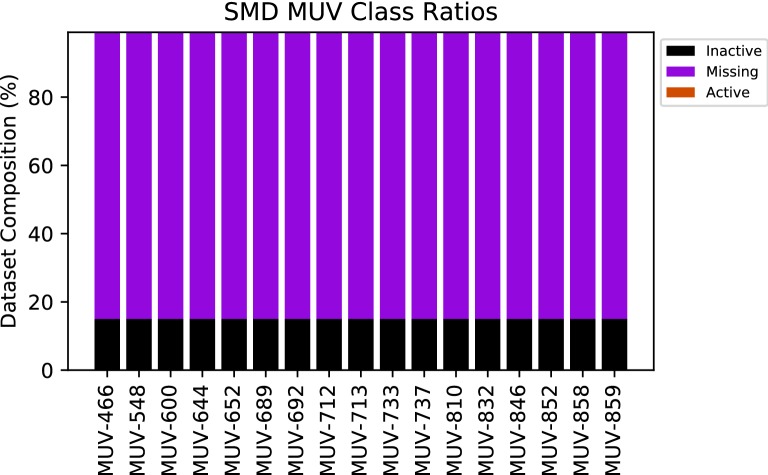
The reintroduction of missing data labels is likely the cause of the increased MUV performance over other methods. As shown in Table 7 and Fig. 7, approximately 84% of the data points in the MUV multitask set are unlabelled. In the original datasets, these points are imputed as inactives, which may introduce a large erroneous class imbalance to the dataset and affect performance.
Table 8.
Task Information for the MUV dataset
| Task label | Target | Mode of interaction | Target class | Assay type |
|---|---|---|---|---|
| MUV-466 | S1P1 rec. | Agonists | GPCR | Reporter Gene |
| MUV-548 | PKA | Inhibitors | Kinase | Enzyme |
| MUV-600 | SF1 | Inhibitors | Nuclear Receptor | Reporter Gene |
| MUV-644 | Rho-Kinase2 | Inhibitors | Kinase | Enzyme |
| MUV-652 | HIV RT-RNase | Inhibitors | RNase | Enzyme |
| MUV-689 | Eph rec. A4 | Inhibitors | Rec. Tyr. Kinase | Enzyme |
| MUV-692 | SF1 | Agonists | Nuclear Receptor | Reporter Gene |
| MUV-712 | HSP 90 | Inhibitors | Chaperone | Enzyme |
| MUV-713 | ER-a-coact. bind. | Inhibitors | PPIc | Enzyme |
| MUV-733 | ER-β-coact. bind. | Inhibitors | PPIc | Enzyme |
| MUV-737 | ER-a-coact. bind. | Potentiators | PPIc | Enzyme |
| MUV-810 | FAK | Inhibitors | Kinase | Enzyme |
| MUV-832 | Cathepsin G | Inhibitors | Protease | Enzyme |
| MUV-846 | FXIa | Inhibitors | Protease | Enzyme |
| MUV-852 | FXIIa | Inhibitors | Protease | Enzyme |
| MUV-858 | D1 rec. | Allosteric modulators | GPCR | Reporter Gene |
| MUV-859 | M1 rec. | Allosteric inhibitors | GPCR | Reporter Gene |
Table 7.
Number of Actives, inactives, and missing datapoints in the classification sets used in the study
| Classification set | Number of actives | Missing datapoints | Number of inactives |
|---|---|---|---|
| MUV | 398 | 1,066,216 | 199,359 |
| HIV | 1232 | 0 | 31,669 |
| BBBP | 1341 | 0 | 290 |
| Tox21 | 4617 | 12,821 | 57,730 |
| SIDER | 17,440 | 0 | 13,367 |
Fig. 7.
Ratio of actives, inactives, and missing data for each task in the MUV dataset. Actives represent such a small proportion that they are not visible in this diagram
When treating missing data as inactive in the original datasets, actives represent only 0.03% of the dataset, whereas ignoring missing data as with SMD sets the actives represent approximately 0.2% of the dataset, nearly an order of magnitude more. Heavily unbalanced datasets are notoriously tricky to train models on, and a reduction of this bias may explain the performance improvements of SMD processed data over the original MUV dataset.
As the SMD MUV dataset greatly outperformed other deep-learning approaches, we present a deeper analysis on this set. Per-task results (Fig. 8) ranged between minimal learned knowledge and well-learned knowledge when averaged across the three runs, and were on the whole very consistent between architectures. Tasks 548 and 644, and tasks 832, 846 and 852 are of particular note: These correspond to Kinase Inhibitors and Protease Inhibitors respectively, and are our highest-performing tasks with the exception of task 712.
Fig. 8.
Per-task results for the SMD MUV test set. Translations between task label and target information are available in Table 8
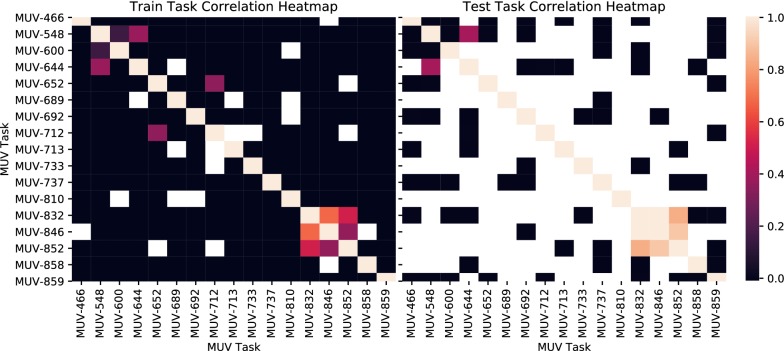
An analysis of these tasks gave a greater insight into one reason for the performance boost. As shown in Fig. 9, these tasks had a much greater activity correlation than others, i.e. ligands observed to be active or inactive for these tasks were likely to share similar activity with the others. This allows the network to much more effectively pick up on common structural features and learn them as reported in other studies [62, 63]. However, in the case where missing data is imputed as inactive, these correlations become more difficult to learn, as negative counterexamples examples are artificially introduced. Other tasks, such as the PPIc or GPCR tasks, are more challenging to learn; by the nature of the target, the structural diversity of the actives compounded with the sparsity of the data, the class imbalances and the lack of transfer learning examples, results in very low performance.
Fig. 9.
Correlation heatmaps between tasks for the training and test sets. These have been averaged across all splits. White indicates no data available for correlation (at least one missing datapoint for all pairs)
The other tasks display generally poor activity, or occasional performance peaks. Due to the extremely limited number of active compounds per task in the test-set, these performance peaks are expected to be sporadic and not true signal. Indeed, for task MUV-733, there were no active compounds in the test set for two of the three splits2 as split by MolNet procedure. As a method for improving performance, for future work we suggest encoding structural features of the target alongside the ligand may be one approach that could be used when correlated target information is not available.
The imputation of missing data as inactives in smaller sets with fewer missing labels has a much smaller impact. Tox21, with only approximately 17% missing data, has a barely perceptible change in active/inactive ratios when missing data is ignored—changing from 6.1% active to 7.4% (Additional file 1). The performance increase here is therefore more likely to be due to false imputation of inactives in the dataset disrupting the learning process and making learning molecular features harder, than it is to be from a confusion of transfer learning examples.
The SIDER (no missing labels) performance demonstrates our algorithms are remarkably resilient to multiple unbalanced sets in a multitask setting, performing on par with most other contemporary machine learning algorithms (Additional file 1). They maintain an advantage even against algorithms that must be trained as multiple single-task models instead of a singular multitask algorithm. The performance increase between the Original and SMD datasets was found to be negligible.
The networks perform on-par with other approaches for single-task classification—the HIV and BBBP classification sets. During the dataset analysis we observed that some compounds exist in counterionic forms in some datasets, which may not be optimal for ADMETox modelling: the charge-parent aspect of the SMD preprocessing was introduced to convert molecules to more pharmacologically-relevant forms as they may exist in the body. This was naïvely done by removing complexes from the datasets, notably ionic complexes such as those shown in Fig. 2, under the assumption that the largest fragment contributes the effect, and to ensure the consistency of charge representation. Further, there was an initial concern that, as ionic bonds are not modelled in the models’ edge types, information would not be able to propagate between the disjoint components of the complex, and smaller components such as the sodium ions would act as artefacts in the graph and introduce noise. However, the lack of performance difference between the two suggests that the readout function bridged these gaps successfully, and the network can be robust against multiple fragments. As well as HIV and BBBP, this is supported by the negligible performance difference between the SIDER models of the two sets.
Regression
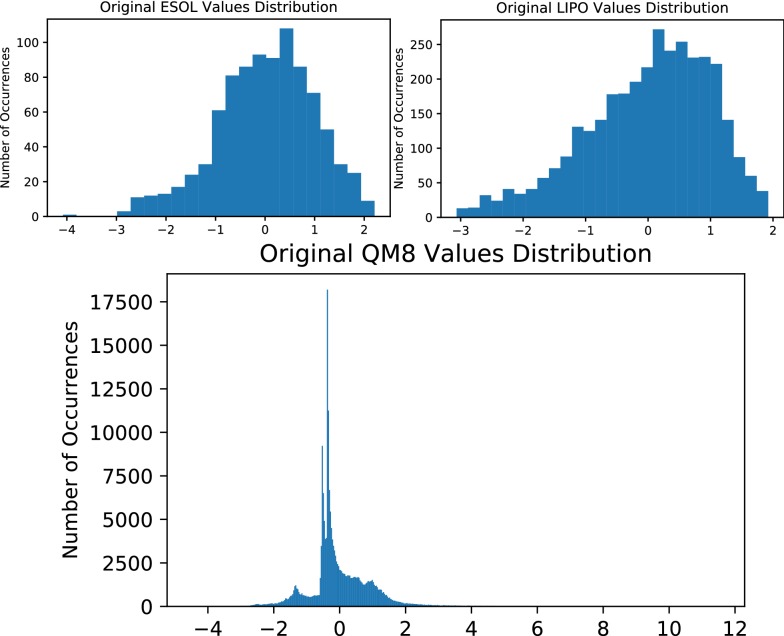
The models performed in general on-par with existing models in regression modelling, with a significant reduction in error when working on the LIPO dataset. The models seem robust against various distributions of values, with ESOL and LIPO datasets resembling skewed normal distributions and QM8 resembling a much more atypical distribution, with most values centred in a singular narrow range close to zero (Fig. 10).
Fig. 10.
Distribution of property values from the ESOL, LIPO and QM8 regression datasets after normalisation by mean and standard deviation
It is not known whether improvement can be further gained in some of these modelled tasks. The ESOL solubility models, for example, are close to the estimated experimental error of the original data. The estimated experimental error of drug-like compound solubility is usually cited as an RMSE around 0.6 logS units [64]. Simpler molecules nevertheless can be modelled with a much lower error around 0.3–0.4 log units [65]—this same study further suggests that the limit of ca. 0.6 log units for drug-like compounds may not be due to experimental or data curation issues, but a limit of QSPR modelling as applied to these databases. The creation of large datasets suitable for training complex models with lower experimental error is a nontrivial task, as solubility is a difficult property to measure correctly in a high throughput scenario: The ‘gold-standard’ measure for solubility—the shake-flask method, is a comparatively costly and time-consuming approach.
In contrast to the estimation of error for experimental physical chemical properties, other datasets can be difficult to give a lower bound of error, for example the QM8 dataset. DFT is in theory exact, however in practice a small but important energy component must be approximated. Though modern approximations provide useful accuracy for practical purposes, errors are not strictly variational, so systematic improvement is problematic. Compounding this, practical implementations introduce other errors (from e.g. choice of basis set, grid resolution), and as such quantifying the limit of how well neural networks can model these properties is difficult.
Hyperparameters
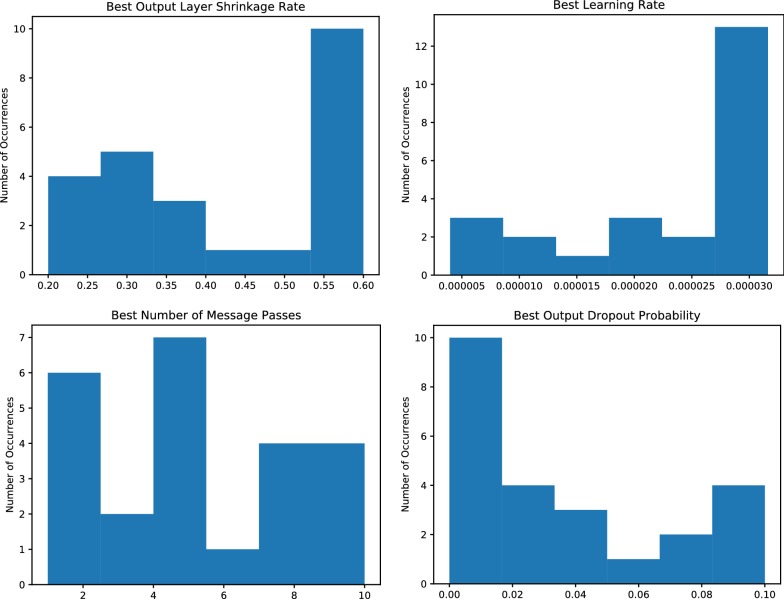
Due to the extensive hyperparameter optimisation that was performed during the training process, we analysed the distributions of hyperparameters to see if there were any tendencies towards optimal configurations for future work. Of the optimised hyperparameters (Table 5) we found that the shrinkage rate of the output fully-connected layer, the learning rate, the number of message passing iterations, and the output layer dropout rate were of note (Fig. 11). Other hyperparameters did not display any notable trends.
Fig. 11.
Aggregate distributions of hyperparameters observed over all tasks and architectures on the SMD datasets after optimisation
We found that generally a higher output layer shrinkage rate and a higher learning rate was more optimal for network performance. The learning rate was often hitting the maximum allowed value of the specified optimisation domain, which may indicate that performance could be further improved if this limit was expanded, pushing the distribution towards a more uniform coverage.
Conversely, dropout was observed to be generally lower in optimal hyperparameters across model training. Whilst this may generally be undesirable as it can lead to model overfitting, the evaluation of the model in a train/test/validation splitting approach should penalise any tendencies to overfit. This would imply that other aspects of the MPNN architecture act as feature regularisation and prevent this, though this cannot be stated conclusively. Figures supplied in the ESI suggest that no notable overfitting was observed during training, which may give the approach inherent advantages over machine learning methods that are traditionally more prone to overfitting. The number of message passes did not show any clear trend, and can be assumed to be heavily dependent on task and other hyperparameters. Some tasks such as ESOL and Tox21 however showed a small bias towards fewer message passing iterations, which makes sense as features such as hydrogen bond donors/acceptors, toxicophores etc. can be very localised and large contributing factors to these properties.
Conclusion
We have introduced two augmentations to the MPNN framework that have shown performance on-par or greater than existing benchmarking models. One is the Attention MPNN, and the other the Edge Memory NN, both of which performed competitively with state of the art machine learning techniques of both traditional and deep learning varieties. The introduction of the attention scheme to our baseline MPNN framework added minimal model overhead, and offers no disadvantages for its use compared to the baseline model, in situations where it is effective. The EMNN had computational cost disadvantages, however, its use may be justified in situations where it offers significant performance increases: We demonstrate that our algorithms can outperform state-of-the-art models in virtual screening settings, notably demonstrated on sparse multi-task datasets, even without the inclusion of target structural information. Further, the inclusion of an attention mechanism may aid in model interpretability, as explored in other literature [66]. We were fairly consistently outperformed by the analogous D-MPNN architecture on other tasks, however we noted generally comparable performance without the inclusion of additional chemical descriptor information, using only low-level chemical graph data. We have analysed different approaches to multitask modelling and dataset preprocessing that have demonstrated increased performance under specific conditions, most notably presenting that the graceful handling of missing data can contribute significantly to model performance in highly sparse datasets. Further, we have performed an extensive hyperparameter optimisation over many model parameters and provided a summary analysis of some more common hyperparameters, indicating potential starting values for future work.
Supplementary information
Additional file 1. Additional tables and figures.
Acknowledgements
We would like to thank Zhenqin Wu for assistance in reproducing the MolNet datasets for use in this publication, and Dr. Igor Tetko for comments that improved the manuscript.
Authors’ contributions
MW assisted with code development, experiment design and analysis, and wrote the manuscript. EL assisted heavily with the development of the codebase and the initial augmentations, and co-authored the manuscript. HC helped with project analysis and cosupervised the project with OE. All authors read and approved the final manuscript.
Funding
The project leading to this article received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska‐Curie Grant Agreement No. 676434, “Big Data in Chemistry” (“BIGCHEM”, http://bigchem.eu). The article reflects only the authors’ view, and neither the European Commission nor the Research Executive Agency are responsible for any use that may be made of the information it contains.
Availability of data and materials
The code we used in this paper is published and available at https://github.com/edvardlindelof/graph-neural-networks-for-drug-discovery.
Competing interests
The authors declare that they have no competing interests.
Footnotes
In other regression datasets, the dataset was normalised, and then split into train/test/validation splits, whereas QM8 was split and then each split normalised. We chose to normalise QM8 in the same manner as the other regression sets, splitting after normalising over the whole set.
In future work, to mitigate these issues in highly sparse, highly unbalanced datasets, we encourage the use of alternative splitting approaches such as stratified sampling. Alternatively, if random sampling is preferring, repeat selection of seeds until at least one active is available for each task is recommended.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
M. Withnall and E. Lindelöf contributed equally to this work
Contributor Information
M. Withnall, Email: followup@withnall.org.uk
E. Lindelöf, Email: edvardlindelof@gmail.com
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13321-019-0407-y.
References
- 1.Flynn GL. Substituent constants for correlation analysis in chemistry and biology. J Pharm Sci. 1980 doi: 10.1002/jps.2600690938. [DOI] [Google Scholar]
- 2.Ruecker G, Ruecker C. Counts of all walks as atomic and molecular descriptors. J Chem Inf Comput Sci. 1993;33(5):683–695. doi: 10.1021/ci00015a005. [DOI] [Google Scholar]
- 3.Mauri A, Consonni V, Pavan M, Todeschini R. Dragon software: an easy approach to molecular descriptor calculations. Match. 2006;56(2):237–248. [Google Scholar]
- 4.Nettles JH, et al. Flexible 3D pharmacophores as descriptors of dynamic biological space. J Mol Graph Model. 2007;26(3):622–633. doi: 10.1016/j.jmgm.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Todeschini R, Consonni V. Handbook of molecular descriptors. Weinheim: Wiley VCH; 2000. [Google Scholar]
- 6.Todeschini R, Lasagni M, Marengo E. New molecular descriptors for 2D and 3D structures. Theory. J Chemom. 1994;8(4):263–272. doi: 10.1002/cem.1180080405. [DOI] [Google Scholar]
- 7.Kriege NM, Johansson FD, Morris C (2019) A Survey on Graph Kernels. ArXiv190311835 Cs Stat
- 8.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23(6):1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 10.Baskin II, Winkler D, Tetko IV. A renaissance of neural networks in drug discovery. Expert Opin Drug Discov. 2016;11(8):785–795. doi: 10.1080/17460441.2016.1201262. [DOI] [PubMed] [Google Scholar]
- 11.Gori M, Monfardini G, Scarselli F (2005) A new model for learning in graph domains. In: Proceedings. 2005 IEEE international joint conference on neural networks, vol. 2, pp 729–734 10.1109/ijcnn.2005.1555942
- 12.Micheli A. Neural network for graphs: a contextual constructive approach. IEEE Trans Neural Netw. 2009;20(3):498–511. doi: 10.1109/TNN.2008.2010350. [DOI] [PubMed] [Google Scholar]
- 13.Scarselli F, Gori M, Tsoi AC, Hagenbuchner M, Monfardini G. The graph neural network model. IEEE Trans Neural Netw. 2009;20(1):61–80. doi: 10.1109/TNN.2008.2005605. [DOI] [PubMed] [Google Scholar]
- 14.Bruna J, Zaremba W, Szlam A, LeCun Y (2013) Spectral networks and locally connected networks on graphs. ArXiv13126203 Cs
- 15.Lee JB, Rossi RA, Kim S, Ahmed NK, Koh E (2018) Attention models in graphs: a survey. ArXiv180707984 Cs
- 16.Cao S, Lu W, Xu Q (2016) Deep neural networks for learning graph representations. In: Thirtieth AAAI conference on artificial intelligence
- 17.Wang D, Cui P, Zhu W (2016) Structural deep network embedding. In: Proceedings of the 22Nd ACM SIGKDD international conference on knowledge discovery and data mining, New York, pp 1225–1234. 10.1145/2939672.2939753
- 18.Pan S, Hu R, Long G, Jiang J, Yao L, Zhang C (2018) Adversarially regularized graph Autoencoder for graph embedding. ArXiv180204407 Cs Stat
- 19.Yu W, et al. (2018) Learning Deep Network Representations with Adversarially Regularized Autoencoders. In: Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining, New York, pp 2663–2671. 10.1145/3219819.3220000
- 20.Yan S, Xiong Y, Lin D. Spatial temporal graph convolutional networks for skeleton-based action recognition. In: Thirty-second AAAI conference on artificial intelligence
- 21.Li Y, Yu R, Shahabi C, Liu Y (2017) Diffusion convolutional recurrent neural network: data-driven traffic forecasting. ArXiv170701926 Cs Stat
- 22.Jain A, Zamir AR, Savarese S, Saxena A (2016) Structural-RNN: deep learning on spatio-temporal graphs presented at the Proceedings of the IEEE conference on computer vision and pattern recognition. pp 5308–5317
- 23.Yu B, Yin H, Zhu Z (2018) Spatio-temporal graph convolutional networks: a deep learning framework for traffic forecasting. In: Proc. Twenty-Seventh Int Jt Conf Artif Intell. pp 3634–3640. 10.24963/ijcai.2018/505
- 24.Duvenaud DK, et al. Convolutional networks on graphs for learning molecular fingerprints. In: Cortes C, Lawrence ND, Lee DD, Sugiyama M, Garnett R, et al., editors. Advances in neural information processing systems. New York: Curran Associates Inc; 2015. pp. 2224–2232. [Google Scholar]
- 25.Kearnes S, McCloskey K, Berndl M, Pande V, Riley P. Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des. 2016;30(8):595–608. doi: 10.1007/s10822-016-9938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coley CW, Barzilay R, Green WH, Jaakkola TS, Jensen KF. Convolutional embedding of attributed molecular graphs for physical property prediction. Journal of chemical information and modeling. 2017;57(8):1757–1772. doi: 10.1021/acs.jcim.6b00601. [DOI] [PubMed] [Google Scholar]
- 27.Gilmer J, Schoenholz SS, Riley PF, Vinyals O, Dahl GE (2017) Neural Message Passing for Quantum Chemistry. ArXiv170401212 Cs
- 28.Li Y, Tarlow D, Brockschmidt M, Zemel R (2015) Gated Graph Sequence Neural Networks. ArXiv151105493 Cs Stat
- 29.Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. ArXiv160902907 Cs Stat
- 30.Wu Z, et al. MoleculeNet: a benchmark for molecular machine learning. Chem Sci. 2018;9(2):513–530. doi: 10.1039/C7SC02664A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bologa C, Allu TK, Olah M, Kappler MA, Oprea TI. Descriptor collision and confusion: toward the design of descriptors to mask chemical structures. J Comput Aided Mol Des. 2005;19(9–10):625–635. doi: 10.1007/s10822-005-9020-4. [DOI] [PubMed] [Google Scholar]
- 32.Filimonov D, Poroikov V. Why relevant chemical information cannot be exchanged without disclosing structures. J Comput Aided Mol Des. 2005;19(9–10):705–713. doi: 10.1007/s10822-005-9014-2. [DOI] [PubMed] [Google Scholar]
- 33.Tetko IV, Abagyan R, Oprea TI. Surrogate data—a secure way to share corporate data. J Comput Aided Mol Des. 2005;19(9–10):749–764. doi: 10.1007/s10822-005-9013-3. [DOI] [PubMed] [Google Scholar]
- 34.Withnall Michael, Lindelöf Edvard, Engkvist Ola, Chen Hongming. Artificial Neural Networks and Machine Learning – ICANN 2019: Workshop and Special Sessions. Cham: Springer International Publishing; 2019. Attention and Edge Memory Convolution for Bioactivity Prediction; pp. 752–757. [Google Scholar]
- 35.Yang K, et al (2019) Are learned molecular representations ready for prime time?,” ArXiv190401561 Cs Stat
- 36.Lindelöf (2019) Deep Learning for Drug Discovery, Property Prediction with Neural Networks on Raw Molecular Graphs,” Masters Thesis, Chalmers
- 37.Defferrard M, Bresson X, Vandergheynst P. Convolutional neural networks on graphs with fast localized spectral filtering. In: Lee DD, Sugiyama M, Luxburg UV, Guyon I, Garnett R, editors. Advances in neural information processing systems. New York: Curran Associates Inc; 2016. pp. 3844–3852. [Google Scholar]
- 38.Schütt KT, Arbabzadah F, Chmiela S, Müller KR, Tkatchenko A. Quantum-chemical insights from deep tensor neural networks. Nat Commun. 2017;8:13890. doi: 10.1038/ncomms13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klambauer G, Unterthiner T, Mayr A, Hochreiter S. Self-normalizing neural networks. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, Garnett R, editors. Advances in neural information processing systems. New York: Curran Associates Inc; 2017. pp. 971–980. [Google Scholar]
- 40.“Deepchem/contrib/mpnn at master deepchem/deepchem GitHub.” https://github.com/deepchem/deepchem/tree/master/contrib/mpnn. Accessed 12 Aug 2019
- 41.Veličković P, Cucurull G, Casanova A, Romero A, Lio P, Bengio Y (2017) Graph attention networks. ArXiv171010903 Cs Stat
- 42.Rezatofighi SH, et al.(2018) Deep Perm-Set Net: Learn to predict sets with unknown permutation and cardinality using deep neural networks. ArXiv180500613 Cs
- 43.Zaheer M, Kottur S, Ravanbakhsh S, Poczos B, Salakhutdinov R, Smola A (2017) Deep Sets. ArXiv170306114 Cs Stat
- 44.Ilse M, Tomczak JM, Welling M (2018) Attention-based deep multiple instance learning. ArXiv180204712 Cs Stat
- 45.Liu Y, Sun C, Lin L, Wang X (2016) Learning natural language inference using bidirectional LSTM model and inner-attention. ArXiv160509090 Cs
- 46.Fu J, Zheng H, Mei T (2017) Look closer to see better: recurrent attention convolutional neural network for fine-grained image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. p. 4438–4446
- 47.Kimber TB, Engelke S, Tetko IV, Bruno E, Godin G (2018) Synergy effect between convolutional neural networks and the multiplicity of SMILES for improvement of molecular prediction. ArXiv181204439 Cs Stat
- 48.Paszke A, et al. (2017) Automatic differentiation in PyTorch
- 49.González J, Dai Z, Hennig P, Lawrence N (2015) Batch Bayesian optimization via local penalization. ArXiv150508052 Stat
- 50.González J (2016) Gpyopt: A bayesian optimization framework in python
- 51.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(15):2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 52.Ramakrishnan R, Hartmann M, Tapavicza E, von Lilienfeld OA. Electronic spectra from TDDFT and machine learning in chemical space. J Chem Phys. 2015;143(8):084111. doi: 10.1063/1.4928757. [DOI] [PubMed] [Google Scholar]
- 53.Delaney JS. ESOL: estimating aqueous solubility directly from molecular structure. J Chem Inf Comput Sci. 2004;44(3):1000–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- 54.Rohrer SG, Baumann K. Maximum unbiased validation (MUV) data sets for virtual screening based on PubChem bioactivity data. J Chem Inf Model. 2009;49(2):169–184. doi: 10.1021/ci8002649. [DOI] [PubMed] [Google Scholar]
- 55.AIDS Antiviral Screen Data - NCI DTP Data - National Cancer Institute - Confluence Wiki. https://wiki.nci.nih.gov/display/NCIDTPdata/AIDS+Antiviral+Screen+Data. Accessed 10 July 2019
- 56.Martins IF, Teixeira AL, Pinheiro L, Falcao AO. A Bayesian approach to in silico blood-brain barrier penetration modeling. J Chem Inf Model. 2012;52(6):1686–1697. doi: 10.1021/ci300124c. [DOI] [PubMed] [Google Scholar]
- 57.“Tox21.” https://tripod.nih.gov/tox21/challenge/index.jsp. Accessed 10 July 2019
- 58.Kuhn M, Letunic I, Jensen LJ, Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44(D1):D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.“MedDRA |.” https://www.meddra.org/. Accessed 10 July 2019
- 60.Altae-Tran H, Ramsundar B, Pappu AS, Pande V. Low data drug discovery with one-shot learning. ACS Cent Sci. 2017;3(4):283–293. doi: 10.1021/acscentsci.6b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swain M (2018) MolVS: molecule validation and standardization
- 62.Ishida F, Saji H, Maruya E, Furihata K. Human platelet-specific antigen, Siba, is associated with the molecular weight polymorphism of glycoprotein Ib alpha. Blood. 1991;78(7):1722–1729. doi: 10.1182/blood.V78.7.1722.1722. [DOI] [PubMed] [Google Scholar]
- 63.Sosnin S, Vashurina M, Withnall M, Karpov P, Fedorov M, Tetko IV. A survey of multi-task learning methods in chemoinformatics. Mol Inform. 2019;38(4):e1800108. doi: 10.1002/minf.201800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Adv Drug Deliv Rev. 2002;54(3):355–366. doi: 10.1016/S0169-409X(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 65.Palmer DS, Mitchell JBO. Is experimental data quality the limiting factor in predicting the aqueous solubility of druglike molecules? Mol Pharm. 2014;11(8):2962–2972. doi: 10.1021/mp500103r. [DOI] [PubMed] [Google Scholar]
- 66.Chen C, Hou J, Shi X, Yang H, Birchler JA, Cheng J (2019) Interpretable attention model in transcription factor binding site prediction with deep neural networks. bioRxiv, p 648691. 10.1101/648691
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional tables and figures.
Data Availability Statement
The code we used in this paper is published and available at https://github.com/edvardlindelof/graph-neural-networks-for-drug-discovery.