Abstract
Toll-like receptors (TLRs) and cytoplasmic RNA sensors have been reported to be involved in the regulation of hepatitis B virus (HBV) replication, but remain controversial due to the lack of a natural infectious model. Our current study sets out to characterize aspects of the role of the innate immune system in eliminating HBV using hydrodynamic-based injection of HBV replicative plasmid and knockout mice deficient in specific pathways of the innate system. The evidence indicated that viral replication was not affected by MAVS or TICAM-1 knockout, but absence of interferon regulatory factor 3 (IRF-3) and IRF-7 transcription factors, as well as the interferon (IFN) receptor, had an adverse effect on the inhibition of HBV replication, demonstrating the dispensability of MAVS and TICAM-1 pathways in the early innate response against HBV. Myd88-/- mice did not have a significant increase in the initial viremia, but substantial viral antigen persisted in the mice sera, a response similar to Rag2-/- mice, suggesting that the MyD88-dependent pathway participated in evoking an adaptive immune response against the clearance of intrahepatic HBV. Taken together, we show that the RNA-sensing pathways do not participate in the regulation of HBV replication in a mouse model; meanwhile MyD88 is implicated in the HBV clearance.
Key Words: Type I interferon, Hepatitis B virus regulation, Toll/IL-1R homology domain-containing adaptor molecule 1, Mitochondrial antiviral signaling protein, Pathogen-associated molecular patterns
Introduction
Hepatitis B virus (HBV) is a noncytopathic human DNA (hepadna) virus that infects hepatocytes causing acute and chronic hepatitis [1]. More than 360 million people are chronically infected with HBV worldwide. Although less than 5% of HBV-infected patients develop persistent infections that progress to liver cirrhosis and hepatocellular carcinoma, HBV causes about 20% of hepatocellular carcinoma deaths [2]. The adaptive immune response is widely acknowledged as pivotal in the defense against HBV. However, the role of innate immunity during HBV infection remains controversial since analysis in patients at the early stage of infection is unfeasible. In addition, no reliable cell-based in vitro infection system or convenient animal model is available.
During HBV infection, the HBV genome is delivered into the nucleus. Infection is defined by the formation of covalently closed circular DNA. Following formation of covalently closed circular DNA, viral mRNA and pregenomic RNA are transcribed [3, 4]. The pregenomic RNA is subsequently converted to a partially double-stranded genome by the viral DNA polymerase. Unlike other DNA viruses, HBV uses an RNA proviral intermediate that must be copied back into DNA for replication. Although these replication steps are sequestered in the nucleus of infected cells, cytoplasmic DNA/RNA sensors are reported to affect the efficacy of HBV replication [5, 6]. The association between cytoplasmic pattern recognition receptors and the dynamics of the HBV life cycle in HBV-infected cells needs to be clarified.
Viral RNA is sensed by the innate immune system by either Toll-like receptor 3 (TLR3) or cytoplasmic sensors such as retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5). RIG-I and MDA5 mainly participate in type I interferon (IFN) induction in conjunction with the adaptor molecule, mitochondrial antiviral signaling protein (MAVS; also called IPS-1, Cardif, or VISA) [7, 8, 9]. The Toll/IL-1R homology domain-containing adaptor molecule 1 (TICAM-1; also called TRIF) is the adaptor of TLR3, which senses viral RNA on the endosomal membrane [8, 9, 10]. Several DNA sensors, most of which signal through STING for type I IFN induction, have been reported in recent years [11]. A few reports have also mentioned that MAVS participates in DNA sensing in certain human cells whereby poly-dA/dT DNA is found to signal via RIG-I. Later, it was also shown that poly-dA/dT serves as a template for RNA polymerase III to make RIG-I ligands [12, 13, 14]. Nevertheless, this hypothesis is unresolved in mouse cells. Once stimulated by the viral DNA/RNA, these adaptor proteins activate IFN regulatory factor (IRF)-3 and IRF-7, which induce type I IFN production [7, 8, 9]. These pattern recognition receptor-mediated early innate immune responses are crucial in controlling viral replication and spread before the initiation of more specific and powerful adaptive immune responses [8, 9, 15].
Despite numerous studies on HBV pathogenesis, the putative molecular patterns of HBV that trigger cellular responses remain unknown. A few reports have suggested that the antiviral response against HBV is mediated by the RIG-I/MAVS pathway in the cytosol and its activation is blocked by HBV polymerase in infected cells [16, 17, 18]. However, no definitive evidence in vivo is available because analysis on the gene expression and effectors required for elimination of the replicative template has been especially difficult. Since viral clearance is a multifaceted process, we hydrodynamically injected a naked HBV plasmid DNA into wild-type (WT) and gene-disrupted mice deficient in TICAM-1, MAVS, TICAM-1/MAVS, IRF-3/IRF-7, IFNAR, MyD88, or RAG2 to identify and characterize the immunological events for HBV clearance. With the availability of various gene-disrupted mice, our study allows the identification of pathways crucial for the clearance of HBV.
Materials and Methods
Animal Studies
All mice were backcrossed with C57BL/6 mice more than seven times before use. Ticam-1−/-[19] and Mavs−/-[20] mice were generated in our laboratory as described previously, while Ticam-1−/-Mavs−/- mice were generated by crossing Ticam-1−/- mice with Mavs−/- mice. Irf-3−/−/Irf-7−/− and Ifnar−/− mice were kindly provided by T. Taniguchi (University of Tokyo, Tokyo, Japan). Myd88−/− mice were provided by Drs. K. Takeda and S. Akira (Osaka University, Osaka, Japan). Rag2−/− mice were kindly provided by Dr. N. Ishii (Tohoku University, Sendai, Japan). Female C57BL/6J mice were purchased from CLEA Japan (Tokyo) and used at 7–9 weeks of age. All mice were maintained under specific pathogen-free conditions in the animal facility at Hokkaido University Graduate School of Medicine (Sapporo, Japan). Animal experiments were performed according to the guidelines set by the Animal Safety Center, Japan.
Hydrodynamic Transfection of Mice with HBV1.4 Plasmid
The pTER1.4xHBV plasmid containing 1.4-genome length sequences of HBV that were previously shown to produce a similar sedimentation in sucrose density gradient centrifugation to HBV extracted from the serum of carriers [21] was used in this study. A total of 50 μg of the plasmid was injected into the tail vein of 7- to 9-week-old mice in a volume of 2.0 ml of TransIT-QR hydrodynamic delivery solution (Mirus, USA). The total volume was delivered within 3–8 s. Plasmid DNA was prepared by using an EndoFree plasmid system (Qiagen, Germany) according to the manufacturer's instructions.
Quantification of HBV DNA by Real-Time PCR
To determine the HBV DNA in the serum, 30 μl of each serum sample was pretreated with 20 units of DNase I (Roche, Germany) at 37°C overnight. The encapsidated viral DNA was extracted with the SMITEST kit (Genome Science Laboratories, Tokyo, Japan) following the manufacturer's instructions and dissolved in 20 μl of TE-buffer. The purified viral genome was quantified by real-time PCR using the SYBR green master mix (Life Technologies, USA) and the HBV-DNA-F/R primer (see suppl. table 1 for primer sequences; for all online suppl. material, see www.karger.com/doi/10.1159/000365113). Amplification conditions included initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 5 s, and extension at 72°C for 6 s. The lower detection limit of this assay is 1,000 copies.
Immunohistochemical Staining for HBV Core Antigen
For immunohistochemical staining of the HBV core antigen (HBcAg), mouse livers were fixed with 4% paraformaldehyde overnight, cryoprotected in 30% sucrose, and sectioned at a thickness of 10 μm using Leica cryostat and mounted on Superfrost glass slides. Sections were incubated with the primary antibody (anticore polyclonal rabbit antibody, DAKO) overnight, followed by incubation with an immunoperoxidase technique involving avidin-biotin peroxidase complexes (Vectastain ABC kit; Vector Laboratories, Burlingame, Calif., USA) according to a method reported previously [22].
HBV Surface Antigen Antigenemia
Mice were bled on the days mentioned after injection of pTER-1.4xHBV and serum was isolated by centrifugation. Concentration of HBV surface antigen (HBsAg) in the serum was quantified by sandwich ELISA in commercial ELISA kits following the manufacturer's protocol (XpressBio, USA). The reporting unit is the signal/cutoff ratio of the 1,000-fold diluted serum at an O.D. of 450 nm.
Southern Blotting to Detect Intracapsid HBV DNA
Viral DNA was isolated from intracellular viral capsids and detected with a specific DIG-labeled probe as described previously [21]. In brief, to isolate the viral DNA, mouse livers were homogenized and subjected to overnight sodium dodecyl sulfate-proteinase K digestion followed by phenol extraction and ethanol precipitation. Twenty micrograms of the isolated DNA was separated in 1% agarose gel, transferred onto Immobilon-Ny+ charged nylon membrane (Milipore), and detected with a full-length HBV-DNA probe labeled by the DIG DNA labeling and detection kit (Roche Diagnostics, Basel, Switzerland) according to the instructions provided by the manufacturer.
Anti-HBs Antibody ELISA
IgG antibodies specific for HBsAg were detected by ELISA as described previously [23] with slight modification. A 96-well plate was coated with antigen of HBs in carbonate buffer and followed by blocking of 2% BSA. Plasma samples were diluted 5× and then incubated in the antigen-coated wells for 3 h at room temperature. A horseradish peroxidase-conjugated goat anti-mouse IgGγ (Southern Biotechnology, USA) and TMB were used to develop the signal. Plates were read at 450 nm. Normal mouse plasma was used to generate cutoff values. The antibody titers are reported as the reciprocal of A450 (sample)/A450 (2.1' normal mouse average) at which samples with a value >1 were considered to have scored positive.
Quantitative HBV or Cytokines mRNA in the Organs
Each organ was extracted from the mice on the days mentioned after hydrodynamic injection of the HBV plasmid. Total RNA of the organs was isolated with TRIzol according to the manufacturer's protocol. Using 0.5-1 µg of total RNA as a template, cDNA was obtained using a high-capacity cDNA transcription kit (Applied Biosystems) according to manufacturer's instructions. qPCR was performed using a Step One real-time PCR system (Applied Biosystems). The expression of cytokine mRNA was normalized to that of β-actin mRNA in each organ, and the fold increase was determined by dividing the expression in each sample by that of the mice receiving the control plasmid. The primer sequences are described in online supplementary table 1.
Quantitative cGAS, STING, and MAVS Expression in Cell Lines
Total RNA was isolated from L929 cells, RAW264.7 cells, immortalized mouse hepatocytes, Huh7 cells, and HepG2 cells with TRIzol according to the manufacturer's protocol. Using 0.5-1 µg of total RNA as a template, cDNA was obtained using a high-capacity cDNA transcription kit (Applied Biosystems) according to manufacturer's instruction. qPCR was performed using a Step One real-time PCR system (Applied Biosystems). The expression of each targeted mRNA was normalized to that of β-actin mRNA in each sample and shown as a relative expression. The primer sequences are described in online supplementary table S1.
Reporter Gene Assay
To prepare the HBV RNA, immortalized mouse hepatocytes previously established in our laboratory [24] were transfected with either control plasmid or pTER1.4xHBV. Total RNA containing the HBV RNA was isolated after 12 h and confirmed with RT-PCR, while the RNA transfected with only control plasmid was used as a control. The isolated RNA was later used as stimuli for the reporter gene assay of IFN-β. Briefly, the immortalized hepatocytes were again transfected with the reporter plasmids. After 16 h, the immortalized hepatocytes were transfected with the stimuli including PIC, a control plasmid, HBV RNA, and pTER1.4xHBV using FuGENE HD (Roche). Cells were lysed at the time point mentioned using a passive lysis buffer, and Firefly and Renilla luciferase activities were determined using a dual-luciferase reporter assay kit. Firefly luciferase activity was normalized by Renilla luciferase activity and was expressed as the fold stimulation relative to activity in nonstimulated cells.
Statistical Analysis
The statistical significance of the obtained data in this study was analyzed using a two-tail unpaired t test and p < 0.05 was regarded as statistically significant.
Results
MAVS and TICAM-1 Are Dispensable in Suppressing HBV Replication
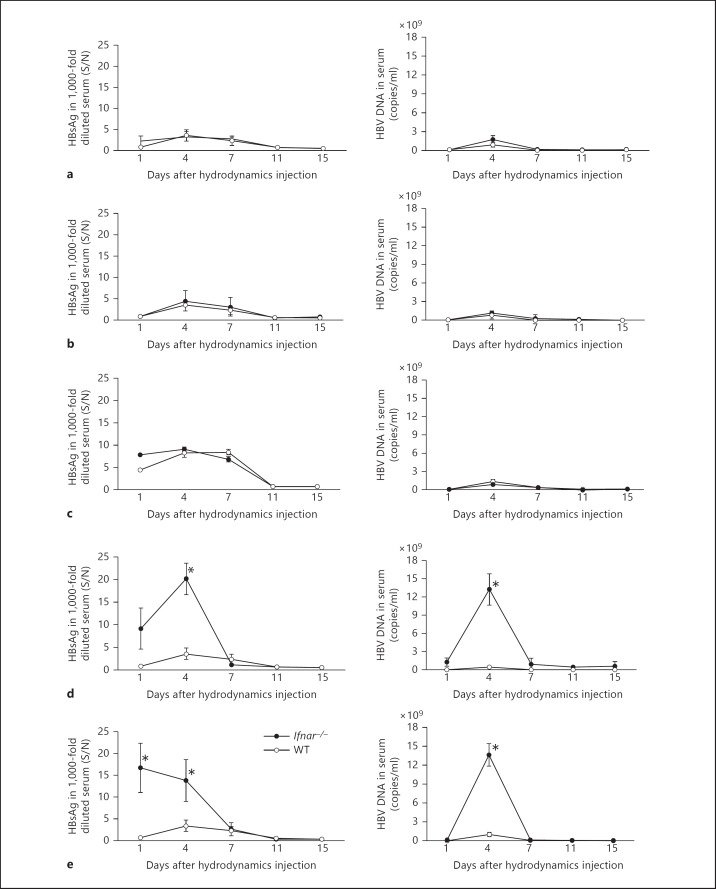
We hydrodynamically transfected replication-competent HBV DNA into Mavs-/- or Ticam-1-/- and Mavs-/-/ Ticam-1-/- mice to access the role of these viral RNA-sensing pathways in response to HBV. Serum HBsAg and HBV-DNA levels were monitored regularly as surrogate markers of HBV replication in vivo. WT mice displayed acute self-limiting hepatitis with peak HBs antigenemia on day 4 after DNA injection (fig. 1a-c). Subsequently, HBsAg in sera decreased and terminated by day 11. Mavs-/- and Ticam-1-/- mice displayed HBsAg clearance kinetics that closely paralleled the WT mice response (fig. 1a, b, left panels). Serum HBV-DNA levels were quantified using real-time PCR. The average titer of serum HBV DNA in 15 WT mice injected with HBV DNA was below 1 × 104 copies/ml 1 day after injection and reached 2 × 109 copies/ml 4 days after injection (fig. 1a-c, right panels). At later time points, most mice showed no detectable virus titer. Similar results were obtained with Mavs-/- and Ticam-1-/- mice (fig. 1a, b). The serum HBV-DNA and HBsAg results showed only a marginal effect for the absence of MAVS or TICAM-1 compared to WT mice. The results suggested that the pathways involving these two adaptor proteins were dispensable for triggering the immune responses that suppressed HBV replication.
Fig. 1.
IFNAR and IRF-3/IRF-7 are critically associated with regulation of HBV propagation in mice but not MAVS and/or TICAM-1. HBsAg or HBV DNA were measured with sera from Mavs-/- (n = 13) (a), Ticam-1-/- (n = 10) (b), Ticam-1-/-/Mavs-/- (n = 6) (c), Irf-3−/−/Irf-7−/− (n = 12) (d), and Ifnar-/- (n = 13) (e) mice compared to WT mice (n = 15). These mice were hydrodynamically injected with 50 μg of the pTER-1.4xHBV plasmid containing full-genome HBV DNA. Mouse sera were isolated at the time points indicated. The HBsAg titers in the 1,000-fold diluted serum (left) and HBV DNA (right) in the knockout mice (⚫) were compared to the WT mice (⚪). Serum HBsAg titers were determined with an enzyme immunoassay at O.D. 450 nm [calculated as signal-over-noise ratios (S/N)]. Sera HBV DNA were determined by Q-PCR and indicated as copies per milliliter. Error bars indicate SD. The statistical p values were analyzed and no significant differences were observed in a-c. * p < 0.01 in d and e are time points statistically different between WT and transgenic mice.
To determine whether the RIG-I/MDA5-MAVS and TLR3-TICAM-1 RNA-sensing pathways were dispensable for suppressing the HBV replication, similar studies were performed in mice lacking both the MAVS and TICAM-1 adaptor proteins (fig. 1c). No notable differences were observed between WT and MAVS/TICAM-1 double-knockout mice in serum HBsAg and HBV-DNA levels, consistent with other data obtained. In addition, similar kinetics of intrahepatic clearance of the HBV template as well as HBV replication was observed in WT, Mavs-/-, and Ticam-1-/- mice as revealed by Southern blotting using HBV-specific probes (online suppl. fig. 1).
To ensure the efficiency of delivery of the HBV transcriptional template into the mouse liver, a plasmid harboring the lacZ gene was used to transfect the liver cells using the hydrodynamic injection method. X-gal (a substrate for lacZ) staining showed that nearly the entire liver of injected mice has successfully received the injected plasmid (online suppl. fig. 2). An independent determination of transfection efficiency was carried out using a plasmid harboring the GFP fragment. The comparable transfection efficiencies observed did not differ significantly among the different mouse strains (data not shown). Furthermore, quantification of HBV mRNA in the organs of WT and knockout mice on day 3 after hydrodynamic injection revealed that HBV mRNA was amplified mainly in the liver but not in other organs, including kidney, lung, heart, spleen, and thymus (online suppl. fig. 3). Only weak HBV signals were detected in other organs in some types of knockout mice. These results demonstrated that HBV replication in vivo using the injection method was efficient and liver specific.
To further assess the possibility of HBV RNA acting as pathogen-associated molecular patterns to trigger the induction of type I IFN in hepatocytes, we transfected the immortalized hepatocytes with a plasmid containing the full genome of HBV as well as RNA containing the HBV mRNA. Along with the synthetic analog of dsRNA, poly(I:C), as a control, we determined the activity of the IFN-β promoter upon the stimulation using reporter gene assay (online suppl. fig. 4). Unlike poly(I:C), neither the full genome of HBV nor RNA induced any activity of the type I IFN promoter in the immortalized hepatocytes. Furthermore, we quantified the endogenous expression of genes including cGas, Sting, and Mavs in the hepatocyte cell lines in order to access the intrinsic RNA or DNA-sensing pathways (online suppl. fig. 5). We found that the hepatocyte cell lines, including those originating from mice and humans, expressed extremely low amounts of Sting compared to the intrinsic Mavs. However, other cell lines, including RAW 264.7 (murine macrophage cell line) and L929 (murine fibrosarcoma cell line), have higher endogenous expression of Sting in comparison to Mavs.
IRF-3/IRF-7 and IFNAR Are Critical Factors for HBV Replication Regulation
To investigate the mechanisms underlying the rapid termination of HBV replication in WT mice, we examined HBV clearance in IRF-3-/IRF-7-deficient mice. Activation of transcription factors including IRF-3 or IRF-7 is essential for raising immune responses including IFN production [25]. Unlike Mavs-/-, Ticam-1-/-, or WT mice, mice lacking the transcription factors IRF-3/IRF-7 had markedly high amounts of HBsAg and HBV DNA in sera (fig. 1d). A sharp peak of HBsAg in sera occurred in Irf-3−/−/Irf-7−/− mice on day 4 after injection. However, in spite of the high virus titer at the early stage, HBsAg and DNA in sera were cleared with kinetics that paralleled the WT mice response, and viremia was eliminated by day 11. Hence, the substantial differences in the serum viremia between WT and Irf-3−/−/Irf-7−/− mice in the early stage after transfection presumably reflects the importance of the genes being expressed with these transcription factors in the suppression of the HBV replication. IRF-3 and IRF-7 are the key molecules in the suppression of HBV viremia in the early stage after HBV injection.
Since type I IFN stimulates the IFNAR pathway to amplify type I IFN production, we hydrodynamically transfected HBV plasmid into mice lacking the gene of the type I IFN receptor (Ifnar-/-) and assessed the suppression of HBV replication. Ifnar-/- mice showed markedly high titers of viral DNA and antigens in sera (fig. 1e) similar to Irf-3−/−/Irf-7−/− mice.
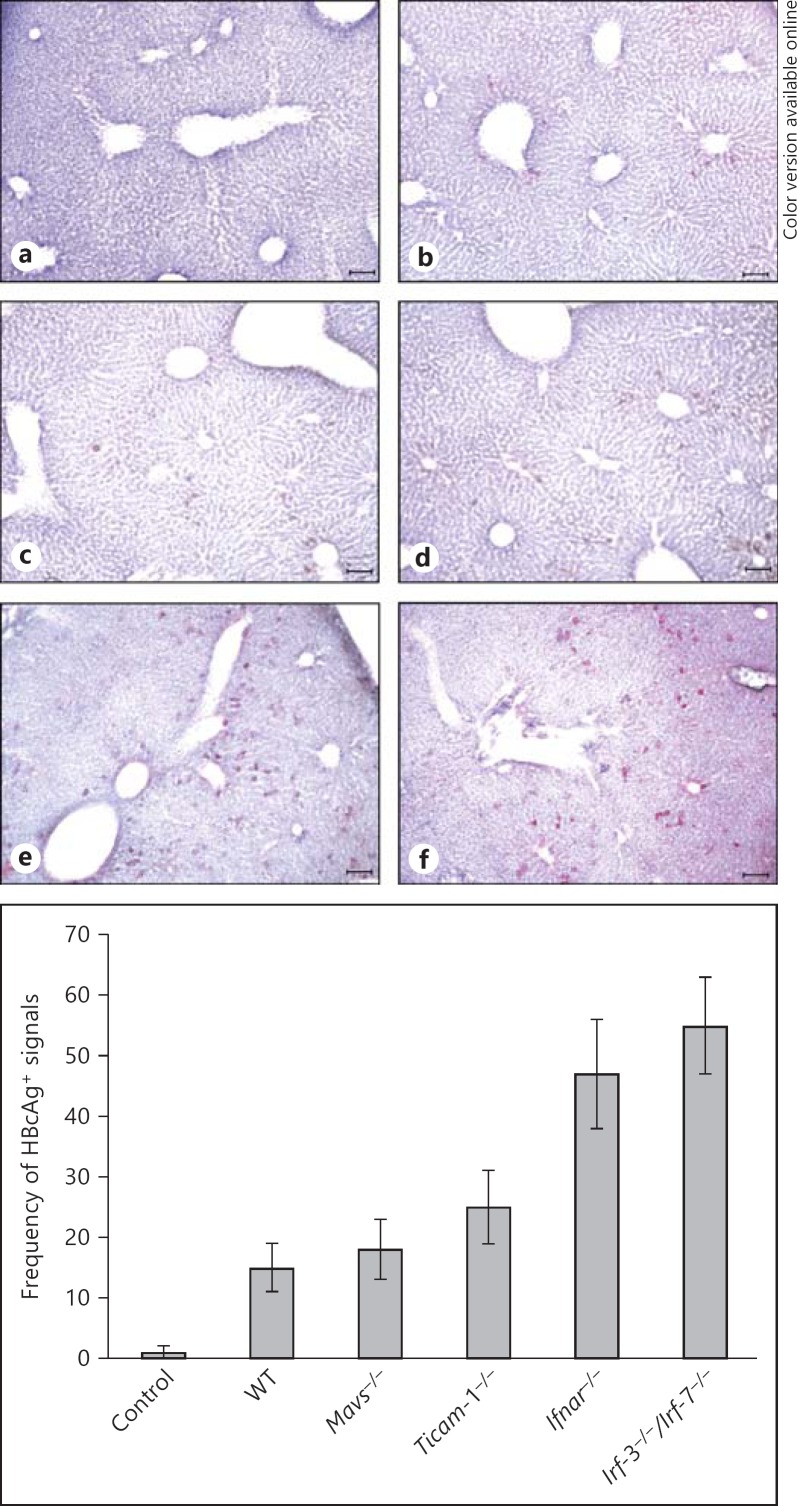
The presence of HBcAg-positive hepatocytes was also monitored by immunohistochemical staining of liver sections from mice of each strain at day 4 after the injections (fig. 2). Data from the observed HBcAg-positive hepatocytes were in good agreement with the results on sera HBsAg and HBV DNA: only deficiency of IRF-3/IRF-7 and IFNAR resulted in a sharp increase of viremia in mice in the early stage (earlier than day 4). Fewer HBcAg-positive hepatocytes were observed in Mavs-/- and Ticam1-/- as well as WT mice at day 4 after injection than in Irf-3−/−/Irf-7−/− or Ifnar-/- mice (fig. 2).
Fig. 2.
Lacking IFNAR and IRF-3/IRF-7 causes an increase of HBcAg in mouse liver injected with the HBV replicative plasmid. The HBc protein in the livers on day 3 after injection was visualized with immunohistochemical staining of the mice liver sections embedded in OCT using an anti-HBc antibody for HBcAg. Representative sections are shown. HBcAg-positive cells were absent in the WT mice that received only the control plasmid (a). Only marginal differences were observed in the frequency of HBcAg-positive cells between WT (b), Mavs-/- (c), and TICAM-1-/- (d) mice. Frequency of HBcAg-positive cells in the livers of the Ifnar-/- (e) and Irf-3−/−/Irf-7-/- (f) mice are more prevalent compared to the WT mice. The scale bars represent 10 μm. The images are displayed at 200× magnification. Frequency of HBcAg-positive signals between the different mouse strains shown is based on 3 images of each.
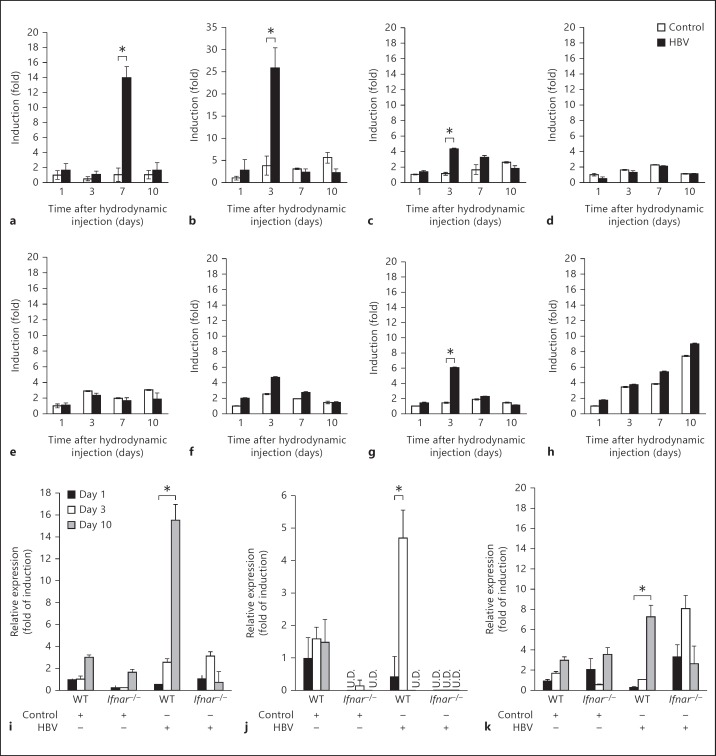
To gain insight into cytokine production in the liver in response to the HBV genome and its replication, we quantified the expression of type I IFN, IFN-γ, IL-7, IL-12p40, and chemokines including CXCL9, CXCL10, and CXCL11 mRNA in the livers of WT mice receiving either the control plasmid or plasmid carrying the HBV full genome on days 1, 3, 7, and 10 after hydrodynamic injection. Replication of HBV in the liver did not cause any significant changes in the expression of the cytokines or chemokines except the IFNs and CXCL-10 (fig. 3a-h). A similar study was carried out in WT and Ifnar-/- mice in order to further elaborate the type I IFN production. The IFNs increased in WT mice livers receiving the HBV full genome compared to the mouse livers receiving the control plasmid (fig. 3i-k). This increase was not observed in Ifnar-/- mice lacking the INF receptor. Although there appeared to be slight individual-to-individual differences in the apparent peaks of IFN-α induction, the result indicated that IFN-β was responsible for suppressing HBV replication early. However, the reason for the lag in the induction of IFN-γ between the WT and Ifnar-/- mice remains unclear.
Fig. 3.
Type I and II IFN expression is induced by HBV replication, and lacking the type I IFN receptor (IFNAR) causes failure of these inductions. WT mice were hydrodynamically injected with 50 μg of the pTER-1.4xHBV or control plasmid as described, and livers were isolated on days 1, 3, 7, and 10 after injection. The expression of IFN-α (a), IFN-β (b), IFN-γ (c), IL-7 (d), IL-12p40 (e), CXCL-9 (f), CXCL-10 (g), and CXCL-11 (h) mRNA was determined by reverse transcription followed by real-time PCR, and was expressed as the fold of induction relative to the WT mice receiving the control plasmid. Induction of IFNs and CXCL-10 was observed in the mice receiving the HBV plasmid. Similar studies were conducted in the WT and Ifnar-/- mice: IFN-α (i), IFN-β (j), and IFN-γ (k). Ifnar-/- mice show reduced expression of the IFNs compared to the WT. Data represent the mean of 3 mice on each strain and time point mentioned. * p < 0.05. U.D. = Undetected.
Taken together, these results suggest that type I IFN was indispensable for suppressing HBV replication in the early stage after viral genome entry. Type I IFN binds to its receptor to induce intracellular antiviral proteins to disrupt HBV replication. The results, however, infer that intrahepatic HBV clearance at the later stage is independent of IFN.
HBV Clearance in a Later Stage by Acquired Immunity
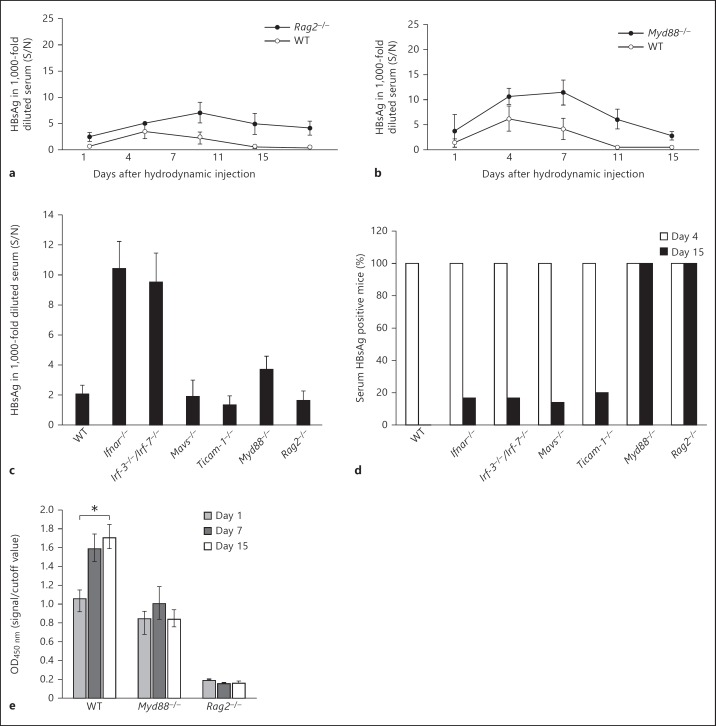
Previous studies by Yang et al. [23] and other groups showed that HBV replication persists indefinitely in globally immunodeficient mice such as NOD/Scid mice hydrodynamically injected with the replication-competent plasmid carrying the full genome of HBV. To investigate whether the elevated viral titer in Ifnar-/- and Irf-3−/−/Irf-7−/− mice on day 4 after hydrodynamic injection and intrahepatic HBV clearance were related to immune effectors including T and B cells, HBV clearance was examined in Rag-2-/- mice. The lack of V(D)J recombination in this strain resulted in failure to produce mature B or T lymphocytes. As shown in figure 4, the absence of mature T and B cells in the Rag-2-/- mice did not result in elevated viral titer immediately after transfection, unlike in Ifnar-/- and Irf-3−/−/Irf-7−/− mice. However, Rag-2-/- mice failed to clear the input plasmid and HBV products, as sera HBsAg and HBV DNA were detected up to day 15 (fig. 4a), by the time viral replication was terminated in all the other strains tested (fig. 4c, d). In other words, activation of the immune effectors such as the B and T cells is responsible for the intrahepatic HBV clearance, their activation being independent of IFN and IRF-3/IRF-7.
Fig. 4.
Mice lacking RAG2 and MyD88 show insufficient clearance of HBV. a, b The Rag2-/-, Myd88-/-, and WT mice were hydrodynamically injected with 50 μg of pTER-1.4xHBV and HBsAg in the mouse sera at the time points indicated and analyzed with ELISA as described. c HBsAg in 1,000-fold diluted serum from all the mice strains including WT, Ifnar-/-, Irf-3−/−/Irf-7-/-, Mavs-/-, Ticam-1-/-, Myd88-/-, and Rag2-/- at day 4 after the hydrodynamic injections. Only Ifnar-/- and Irf-3−/−/Irf-7-/- mice show a remarkable increase, while a moderate increase of sera HBsAg was seen in Myd88-/- mice. d HBsAg persistence rates in all the mice strains receiving pTER1.4HBV were determined by the percentage of serum HBsAg-positive mice on day 4 (□) and day 15 (■) after the hydrodynamic injections. Serum HBsAg was found to be persistent only in mice deficient in MyD88 and RAG2 on day 15 as 100% of the mice from these two strains were HBsAg positive (n = 8 for each mice strain). e Lacking MyD88 and RAG2 leads to the failure of the knockout mice to produce anti-HBs IgG compared to the WT mice on day 15 after injection as determined by ELISA using antigen of HBs (n = 3 for each mice strain). * p < 0.05. S/N = Signal-over-noise ratio.
MyD88 Deficiency Leads to Slower HBV Clearance
The MyD88-dependent pathway has been known to lead to the production of inflammatory cytokines and is common to all TLRs, except TLR3 [22]. To examine whether a MyD88-dependent pathway is required in the intrahepatic clearance of the HBV, we monitored the serum HBsAg in MyD88-deficient mice. As shown in figure 4b, an increase in sera HBsAg in Myd88−/− mice was observed, although without particular antigenemia peaks at the early stage of transfection in Ifnar-/- and Irf-3−/−/Irf-7−/− mice (fig. 4b, c). Instead, a delay in the elimination of the HBV was observed (fig. 4b, d). Typically, WT mice or other mouse strains lose serum HBsAg from day 11 after injection. However, serum antigen was detectable on day 15 in Myd88−/− mice. Delayed elimination of HBV plasmid and single-strand DNA in the liver was observed in Southern analysis of the liver from Myd88−/− mice compared with WT, Mavs-/-, and Ticam-1-/- mice (online suppl. fig. 1).
Additionally, ELISA to determine anti-HBsAg antibody production in mouse sera after hydrodynamic injection revealed that anti-HBs antibody was produced in WT mice from day 7 and peaked at day 15 (fig. 4e). RAG2-deficient mice lacking mature T and B cells failed to produce any antibody, and Myd88−/− mice also had lower or nearly undetectable anti-HBs antibody in serum in comparison to the typical response of WT mice at later transfection stages. These results suggested that MyD88 and RAG2 were crucial for triggering acquired immunity against HBV in vivo.
Discussion
In the present study, several different knockout mice were analyzed in an attempt to define the mechanism of innate immunity against HBV in vivo. The evidence we obtained indicated that viral replication was not affected by MAVS or TICAM-1 knockout, but absence of IRF-3 or IRF-7 transcription factors, as well as the IFN receptor, had an adverse effect on the inhibition of HBV replication. The results herein demonstrated that the TICAM-1 and MAVS pathways were not required in either suppressing the virus replication or intrahepatic clearance of HBV replicative plasmid in vivo.
Although a DNA virus, HBV has the unique feature of replicating via an RNA proviral intermediate that is copied into DNA. Thus, defining the virus component, either HBV DNA or RNA that triggers the antiviral response is crucial to understand the immune mechanisms that are responsible for eliminating HBV during infection. HBV RNA has been suggested as the putative pathogen-associated molecular pattern of HBV in a few reports [16, 17, 18, 26]. HBx or HBs inhibits IFN-β induction followed by activation of TLR3 or RIG-I pathways with poly(I:C) or SeV, respectively. However, these findings must be interpreted with caution, as poly(I:C) and SeV are heterologous inducers for evaluating either the TLR3 or RIG-I pathway [16, 17]. No definitive conclusion on activation of the TLR3 or RIG-I pathway by HBV RNA in vivo has been reported yet.
Viral RNA is recognized largely by RIG-I or MDA5 in the cytosol of infected cells [27, 28] and by TLR3 or TLR7/8 in the endosome of other noninfected cells [29, 30]. These RNA sensors require MAVS, TICAM-1, or MyD88 as adaptor proteins to induce type I IFN [28]. On the other hand, cytoplasmic DNA is recognized by DNA sensors including DAI, IFI16, RIG-I, DHX9 (helicase), and cGAS [31]. STING is the only adaptor for all IFN-inducing DNA sensors in mouse cells reported so far [30, 32, 33], although some of these sensors are reported to induce type I IFN via MAVS in human cells. These adaptors, TICAM-1, MAVS, and STING, are all linked to activation of IRF-3/IRF-7 which act as transcription factors that induce activation of the type I IFN promoter during viral infections. Involvement of different pathways in the induction of type I IFN is critically dependent on the virus species and cell type. Cell type-specific contributions of other sensors, including DEAD box helicases, might occur in some cases of infection. However, in hepatocytes, the control plasmid per se exhibited no IFN-inducing response, suggesting that the HBV replication is a critical step for IFN induction. Actually, no contribution of other sensors except RIG-I/MDA5 and TLR3 has been reported so far.
Using the murine hydrodynamic injection model, we found that mice deficient in IRF-3 and IRF-7 or IFNAR do not inhibit HBV replication as effectively as their WT counterparts and result in elevated HBV titers in mice sera and livers. These findings imply that type I IFN acting on IFNAR is indispensable for evoking anti-HBV protective responses although such a hypothesis is in disagreement with previous findings that HBV does not induce detectable changes in type I IFN expression during the early weeks of infection [34]. There are a few possibilities of how type I IFN is produced in mice receiving HBV template plasmid. One of them is that HBV could be recognized by pathways that do not link to MAVS or TICAM-1 and facilitate IFN production in the cytoplasm. For instance, STING-dependent signaling leads to type I IFN induction, and it has been shown that this can be MAVS and TICAM-1 independent. Notably, STING-dependent signaling is especially associated with DNA-mediated induction of type I IFN via IRF-3/IRF-7, and genomic DNA is an important part of HBV replication. It would be interesting to clarify such hypotheses using Sting-/- mice in the near future.
To elucidate the molecular pattern which triggers type I IFN induction, we transfected either HBV DNA or RNA into immortalized hepatocytes. To our surprise, we were unable to detect significant IFN-β induction with either HBV replicative DNA or HBV RNA. As we looked into the possible reasons to account for the lack of innate immune responses against HBV in hepatocytes, we found that the endogenous expression of STING in hepatocyte cell lines including HepG2 and immortalized mouse hepatocytes is extremely low compared to other cell lines like macrophages or dendritic cells, thus suggesting that STING-dependent signaling might play a crucial role in inducing type I IFN in response to HBV. The produced IFN in turn activates the IFNAR pathway. There are various cells populations in the liver that express IFNAR and therefore subsequently initiate a natural signaling cascade for amplification of IFN production via the Jak-STAT pathway.
Another possible way for HBV to induce IFN is via the HBV-stimulated nonparenchymal or resident myeloid cells. Even though there has been no report suggesting that HBV substantially infects pDCs, Isogawa et al. [5], demonstrated that freshly isolated CD11c+ cells of intrahepatic myeloid cells rather than the hepatocytes expressed TLRs including TLR2, 3, and 9. Therefore, resident myeloid cells might induce IFN to further prevent the spread of HBV by activating the IFNAR pathway in bystander cells or hepatocytes.
Although Myd88-/- mice receiving an HBV-DNA injection did not exhibit significantly high virus titers in the early phase unlike those observed in Ifnar-/- and Irf-3-/-/Irf-7-/- mice, interestingly MyD88 is required for the intrahepatic clearance of the HBV replicative template. The fact that the transcriptional template persists in the absence of MyD88 suggests that MyD88 may play a pivotal role in intrahepatic HBV clearance in the mouse model. Notably, MyD88 is the adaptor molecule for TLR7 and 9 in pDCs [35, 36]. Deficiency of MyD88 in pDCs may result in failure to induce acquired immunity for HBV. Our findings show that HBV-specific antibodies are efficiently produced in WT, but not in Myd88-/- mice. In addition, the number of pDCs has been previously reported to be reduced in vivo during several systemic viral infections including HBV [37]. In one of the most recent reports, Lv et al. [38], showed that HBV-derived CpG induces potent IFN-α production by human pDCs, which may partially explain how pDCs interact with HBV in infection. However, the cause of weak participation in the early response of IFN induction in Myd88-/- mice remains to be determined.
Recombinant IFN-α is a standard treatment for chronic HBV patients. Nevertheless, direct treatment with IFN yields only about 30% improvement in HBV patients and little is known about why most chronic HBV patients do not respond to IFN therapy [39]. As demonstrated in our study, virus persistency can be independent of the type I IFN-inducing system. This observation leads to the suggestion that type I IFN is indispensable for inducing antiviral molecules to control viral replication and spread before the onset of more specific and powerful adaptive immune responses. This appeared to be factual at least in our knockout mouse models as virus titers were highly elevated in Ifnar-/- mice in the initial days after injection. Conversely, type I IFN did not have any influential effects on clearance of the HBV template in the later stages. Such observations coincide with the latest study conducted in patients with chronic HBV infection by Tan et al. [40], in which IFN-α treatment was shown to modulate innate immune parameters in the patients, but without any detectable effect on HBV-specific adaptive immunity. The missing link between the induction of type I IFN and anti-HBV cellular effectors needs to be further investigated in mouse models, including the mechanism of MyD88 participation in activation of the cellular immune response during infection. Elucidating molecular mechanisms between innate pattern sensing and evoking cellular effectors may provide a reasonable explanation for the failure of IFN-treatment in HBV infection.
Collectively, our study validates the use of the hydrodynamic transfection method in mimicking acute HBV infection in mouse models and demonstrated the host-virus relationship during HBV infection in many aspects. Since HBV infectious models with immunologically well-defined laboratory animals do not exist, the result presented in this study herein provides an insight into the dispensability of RNA sensors for induction of IFN by HBV RNA and the complexity of innate and adaptive immunity during HBV clearance.
Disclosure Statement
The authors declare no financial or commercial conflict of interest.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, and Culture and the Ministry of Health, Labor, and Welfare of Japan, and the Yasuda Cancer Foundation (T.S.) and the Ono Foundation (T.S.). Financial support by a MEXT Grant-in-Project ‘The Carcinogenic Spiral’, ‘The National Cancer Center Research and Development Fund (23-A-44)’, and the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) are gratefully acknowledged. We are grateful to Drs. H. Shime, J. Kasamatsu, K. Funami, and M. Tatematsu in our laboratory for their fruitful discussions. We thank Dr. F.V. Chisari (Scripps Research Institute, La Jolla, Calif., USA) for providing us with HBV plasmid and sleeping beauty.
References
- 1.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 2.Zuckerman JN, Zuckerman AJ. Current topics in hepatitis B. J Infect. 2000;41:130–136. doi: 10.1053/jinf.2000.0720. [DOI] [PubMed] [Google Scholar]
- 3.Ganem D, Prince AM. Hepatitis B virus infection - natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 4.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol. 2005;79:7269–7272. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Lu M, Meng Z, Trippler M, Broering R, Szczeponek A, Krux F, Dittmer U, Roggendorf M, Gerken G, Schlaak JF. Toll-like receptor-mediated control of HBV replication by nonparenchymal liver cells in mice. Hepatology. 2007;46:1769–1778. doi: 10.1002/hep.21897. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 8.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci USA. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Kumar M, Jung SY, Hodgson AJ, Madden CR, Qin J, Slagle BL. Hepatitis B virus regulatory HBx protein binds to adaptor protein IPS-1 and inhibits the activation of beta interferon. J Virol. 2011;85:987–995. doi: 10.1128/JVI.01825-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Wang Y, Wen C, Wu Q, Shi W, Zhong H. The hepatitis B virus X protein disrupts innate immunity by down regulating mitochondrial antiviral signaling protein. J Immunol. 2010;185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 18.Yu S, Chen J, Wu M, Chen H, Kato N, Yuan Z. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKepsilon and DDX3. J Gen Virol. 2010;91:2080–2090. doi: 10.1099/vir.0.020552-0. [DOI] [PubMed] [Google Scholar]
- 19.Oshiumi H, Okamoto M, Fujii K, Kawanishi T, Matsumoto M, Koike S, Seya T. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J Immunol. 2011;187:5320–5327. doi: 10.4049/jimmunol.1101503. [DOI] [PubMed] [Google Scholar]
- 20.Akazawa T, Ebihara T, Okuno M, Okuda Y, Shingai M, Tsujimura K, Takahashi T, Ikawa M, Okabe M, Inoue N, et al. Antitumor NK activation induced by the Toll-like receptor 3-TICAM-1 (TRIF) pathway in myeloid dendritic cells. Proc Natl Acad Sci USA. 2007;104:252–257. doi: 10.1073/pnas.0605978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi C, Ishino H, Tsuge M, Fujimoto Y, Imamura M, Takahashi S, Chayama K. G to A hypermutation of hepatitis B virus. Hepatology. 2005;41:626–633. doi: 10.1002/hep.20580. [DOI] [PubMed] [Google Scholar]
- 22.Tian Y, Chen WL, Kuo CF, Ou JH. Viral-load-dependent effects of liver injury and regeneration on hepatitis B virus replication in mice. J Virol. 2012;86:9599–9605. doi: 10.1128/JVI.01087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aly HH, Oshiumi H, Shime H, Matsumoto M, Wakita T, Shimotohno K, Seya T. Development of mouse hepatocyte lines permissive for hepatitis C virus (HCV) PLoS One. 2011;6:e21284. doi: 10.1371/journal.pone.0021284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Li Y, Mao A, Li C, Tien P. Hepatitis B virus X protein suppresses virus-triggered IRF3 activation and IFN-beta induction by disrupting the VISA-associated complex. Cell Mol Immunol. 2010;7:341–348. doi: 10.1038/cmi.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 28.Onomoto K, Yoneyama M, Fujita T. Regulation of antiviral innate immune responses by RIG-I family of RNA helicases. Curr Top Microbiol Immunol. 2007;316:193–205. doi: 10.1007/978-3-540-71329-6_10. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011 doi: 10.1002/rmv.680. DOI: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 30.Diebold SS. Recognition of viral single-stranded RNA by Toll-like receptors. Adv Drug Deliv Rev. 2008;60:813–823. doi: 10.1016/j.addr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki T, Oshiumi H, Miyashita M, Aly HH, Matsumoto M, Seya T. Cell type-specific subcellular localization of phospho-TBK1 in response to cytoplasmic viral DNA. PLoS One. 2013;8:e83639. doi: 10.1371/journal.pone.0083639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87:1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino K, Sasaki I, Sugiyama T, Yano T, Yamazaki C, Yasui T, Kikutani H, Kaisho T. Critical role of IkappaB Kinase alpha in TLR7/9-induced type I IFN production by conventional dendritic cells. J Immunol. 2010;184:3341–3345. doi: 10.4049/jimmunol.0901648. [DOI] [PubMed] [Google Scholar]
- 36.Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440:949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 37.Swiecki M, Wang Y, Vermi W, Gilfillan S, Schreiber RD, Colonna M. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J Exp Med. 2011;208:2367–2374. doi: 10.1084/jem.20110654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv S, Wang J, Dou S, Yang X, Ni X, Sun R, Tian Z, Wei H. Nanoparticles encapsulating HBV-CpG induce therapeutic immunity against hepatitis B virus infection. Hepatology. 2014;59:385–394. doi: 10.1002/hep.26654. [DOI] [PubMed] [Google Scholar]
- 39.Ter Borg MJ, Hansen BE, Bigot G, Haagmans BL, Janssen HL. ALT and viral load decline during PEG-IFN alpha-2b treatment for HBeAg-positive chronic hepatitis B. J Clin Virol. 2008;42:160–164. doi: 10.1016/j.jcv.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Tan AT, Hoang LT, Chin D, Rasmussen E, Lopatin U, Hart S, Bitter H, Chu T, Gruenbaum L, Ravindran P, Zhong H, Gane E, Lim SG, Chow WC, Chen PJ, Petric R, Bertoletti A, Hibberd ML. Reduction of HBV replication prolongs the early immunological response to IFNα therapy. J Hepatol. 2014;60:54–61. doi: 10.1016/j.jhep.2013.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data