Abstract
Macroautophagy/autophagy defines an evolutionarily conserved catabolic process that targets cytoplasmic components for lysosomal degradation. The process of autophagy from initiation to closure is tightly executed and controlled by the concerted action of autophagy-related (Atg) proteins. Although substantial progress has been made in characterizing transcriptional and post-translational regulation of ATG/Atg genes/proteins, little is known about the translational control of autophagy. Here we report that Psp2, an RGG motif protein, positively regulates autophagy through promoting the translation of Atg1 and Atg13, two proteins that are crucial in the initiation of autophagy. During nitrogen starvation conditions, Psp2 interacts with the 5′ UTR of ATG1 and ATG13 transcripts in an RGG motif-dependent manner and with eIF4E and eIF4G2, components of the translation initiation machinery, to regulate the translation of these transcripts. Deletion of the PSP2 gene leads to a decrease in the synthesis of Atg1 and Atg13, which correlates with reduced autophagy activity and cell survival. Furthermore, deactivation of the methyltransferase Hmt1 constitutes a molecular switch that regulates Psp2 arginine methylation status as well as its mRNA binding activity in response to starvation. These results reveal a novel mechanism by which Atg proteins become upregulated to fulfill the increased demands of autophagy activity as part of translational reprogramming during stress conditions, and help explain how ATG genes bypass the general block in protein translation that occurs during starvation.
Subject terms: Macroautophagy, Vacuole
Introduction
Cellular homeostasis requires a proper balance between synthesis and degradation. One major degradative pathway in eukaryotic organisms is autophagy (“self-eating”), by which cytosolic proteins, damaged or superfluous organelles and invading pathogens are targeted and delivered to the vacuole (in yeast) or lysosomes (in mammals) for recycling.1 Based on the inducing signals and temporal aspects of induction, the type of cargo and the mechanism of sequestration, autophagy can be divided into distinct types. Macroautophagy (hereafter autophagy) is the best characterized pathway, which involves the de novo formation of a phagophore, a transient double-membrane structure that carries out the sequestration of cytoplasm, and then seals to form an autophagosome.
As an evolutionarily conserved survival mechanism of all eukaryotic cells, autophagy primarily acts as an adaptive response to environmental adversity, especially nutrient limitation. Studies in yeast have pioneered our understanding of the molecular mechanisms of autophagy. More than 40 autophagy-related (ATG) genes in yeast have been identified to mediate this process, and these genes exhibit homology from yeast to human.2 With subsequent studies in various animal models, many additional physiological processes have been linked to autophagy including intracellular protein quality control, maintenance of tissue homeostasis, animal development, and innate and adaptive immunity.3 Given the important roles autophagy plays, it is not surprising that autophagy needs to be stringently regulated to avoid either excessive or insufficient activity. Dysregulated autophagy is related to many human diseases, including cancer, neurodegeneration, metabolic disorders, macular degeneration, and liver and heart diseases.4,5
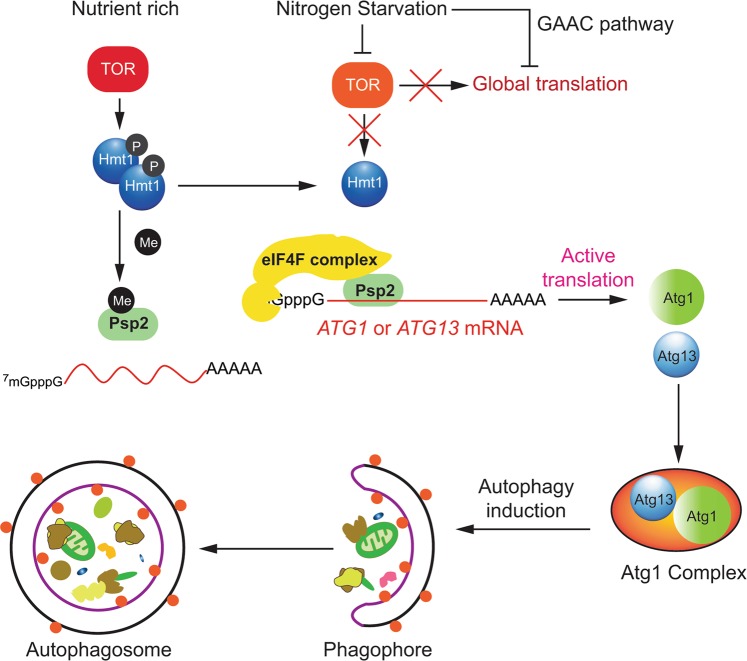
The level of autophagy is dynamic, allowing adaptation to intracellular cues and environmental changes. Autophagy is normally kept at basal levels to maintain cellular homeostasis, while it also can be rapidly augmented as a survival mechanism in response to stress signals such as starvation, growth factor depletion, and hypoxia. The regulation of autophagy is largely done through regulating the expression of ATG genes. Under nutrient-deprivation conditions, global translation is downregulated through two signaling pathways: the target of rapamycin (TOR) pathway and the general amino acid control (GAAC) pathway,6,7 to manage starvation stress with limited resources. However, at the same time, the expression of most ATG genes is upregulated to support the increased demands of autophagy activity.8,9 This upregulation is critical to keep autophagy at a proper amplitude.8,10,11 A tremendous amount of research has focused on transcriptional and post-translational regulation of ATG/Atg genes/proteins,12 but little is known about the translational control of autophagy. In fact, selective translation of mRNAs is a widespread mechanism of gene regulation, and contributes to diverse biological processes, most of which are sensitive to stress, cellular energy, and nutrient availability.13,14 Thus, we considered the following question: Is there a specific translational control for ATG mRNAs?
Previous translatome studies have reported increased translation efficiency for some ATG genes including ATG1, ATG3, ATG8 and ATG19 upon amino acid withdrawal, suggesting the existence of translational regulation.15,16 Two recent studies demonstrated that EIF5A (eukaryotic translation initiation factor 5A) is required for efficient translation of ATG3 through a DGG tripeptide motif, and the RNA binding protein ELAVL1/HuR (ELAV like RNA binding protein 1) enhances the translation of ATG5, ATG12, and ATG16L1 through their 3′ untranslated region (UTR), providing new insights into the translational control of autophagy.11,17 The translational regulation of other ATG genes still remains unexplored.
Translational regulation can either target global mRNAs by inhibiting or activating general translational machinery or target specific mRNAs through trans-acting RNA binding factors including RNA-binding proteins (RBPs) and miRNAs.18 This type of regulation can take place at each of the three steps of translation: initiation, elongation, and termination, with the rate-limiting step, initiation, being the most common and effective target.19 RBPs, which can interfere with various steps of initiation in a transcript-specific manner, are the major regulators of translation. For the purpose of discovering yeast RBPs involved in promoting ATG mRNA translation in response to starvation, we performed an Atg1 expression screen using deletion mutants in genes encoding RGG motif-containing proteins. RGG motif proteins are a group of RBPs characterized by the presence of multiple Arg-Gly-Gly and Arg-Gly-X repeats, and they have been implicated in regulating transcription, pre-mRNA splicing and mRNA translation.20,21 The evolutionarily conserved RGG motif has an RNA-binding property and also mediates protein-protein interactions.22,23 Recently, a subset of RGG motif proteins in yeast were found to form a complex with eIF4E-eIF4G through their RGG motifs, and several RGG motif proteins in mammalian cells were reported to target specific mRNAs to regulate their translation.24–27
Our screen for RGG motif proteins that affect Atg1 protein levels led to the identification of Psp2, an RBP with four RGG motifs at its C terminus, as a positive translational regulator for autophagy. Under nitrogen-starvation conditions, Psp2 interacts with translation initiation factors and binds the 5′ UTR of ATG1 and ATG13 transcripts to promote their translation. The association of Psp2 with ATG mRNA is dependent on its RGG motif. Furthermore, arginine residues within the RGG-motif region are required for the function of Psp2. We found that Psp2 is arginine methylated by the methyltransferase Hmt1 during nutrient-rich conditions. Starvation abolishes the methyltransferase activity of Hmt1, preventing methylation of newly synthesized Psp2, and leaving the majority of Psp2 unmethylated. Unmethylated Psp2 shows higher binding affinity to ATG1 mRNA, consistent with the observation that cells expressing a Psp2 arginine-methylation-mimetic mutant have lower Atg1 expression and autophagy activity. Taken together, we show for the first time that expression of Atg1 and Atg13 is regulated at the translational level by the RBP Psp2 during nitrogen starvation. The switch of this regulation is arginine methylation controlled by starvation/TOR signaling.
Results
The RGG motif protein Psp2 promotes Atg1 expression under nitrogen-starvation conditions
We carried out a screen using a library of deletion mutants for genes encoding RGG motif-containing proteins with the goal of identifying yeast RBPs involved in promoting ATG mRNA translation in response to starvation. For two reasons we used changes in Atg1 protein level as the initial readout: First, Atg1 plays essential roles in the induction of autophagy; second, the expression of this protein is highly upregulated upon nutrient deprivation, whereas global translation is downregulated (Fig. 1a; Supplementary information, Fig. S1). Among all the mutants screened, we found that psp2∆ cells showed a consistently decreased level of Atg1 compared to wild-type (WT) cells during nitrogen starvation, with the greatest difference (more than a 2-fold decrease) following 1 day of starvation (Fig. 1a). This is similar to what we observed with another stress stimulus: amino acid starvation (Supplementary information, Fig. S1b). Furthermore, following treatment with the TOR inhibitor rapamycin, which normally results in an upregulation of autophagy and an increase in Atg1, the psp2∆ cells also showed markedly less Atg1 even shortly after autophagy induction (Supplementary information, Fig. S1c). This result suggests that Psp2 might function downstream of TOR.
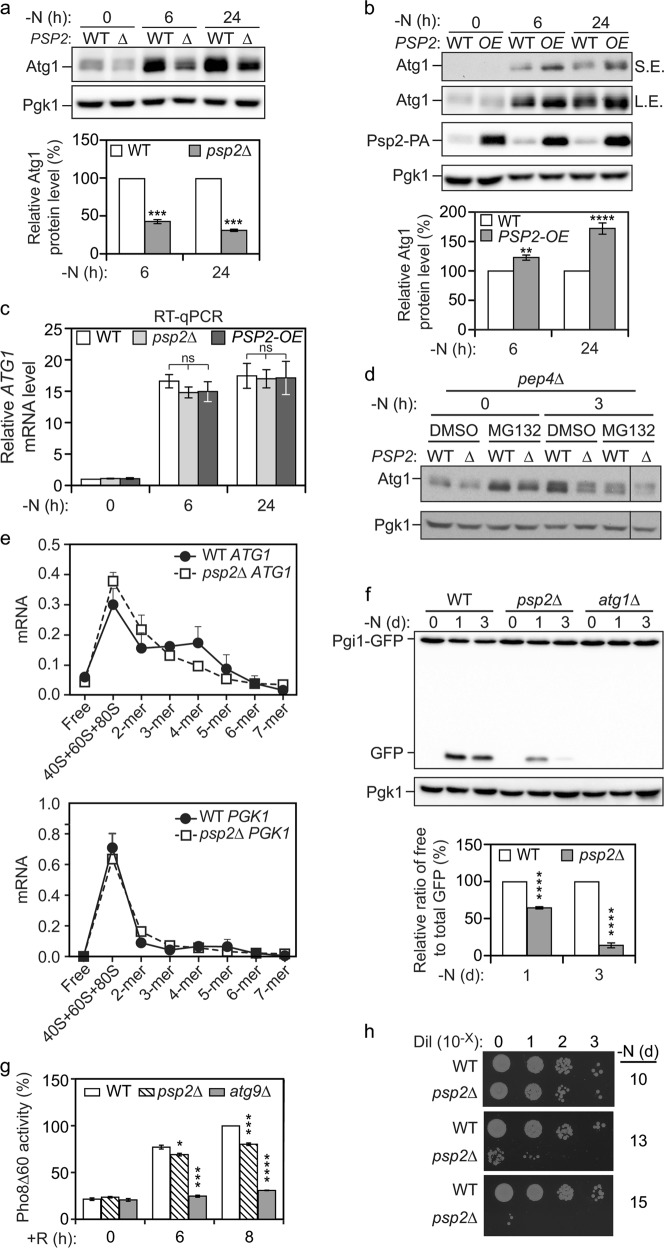
Fig. 1.
Psp2 is a positive regulator of Atg1 expression and autophagy activity during nitrogen starvation. a, b Psp2 is a positive regulator of Atg1 expression. WT (SEY6210 for 1a and YZY051 for 1b), psp2∆ (YZY050), or ZEO1p-PSP2 (XLY439) cells were grown in YPD medium until mid-log phase and then starved for nitrogen for 6 h or 1 day. Protein extracts were analyzed by western blot with anti-Atg1 and anti-Pgk1 (loading control) antisera. Representative images and quantification of the data are shown. Atg1 level was measured and first normalized to Pgk1 and then normalized to that of WT cells in the same condition (set to 100%). Mean ± SEM of n ≥ 3 independent experiments are indicated. Student’s t-test; **P < 0.01, ***P < 0.001, ****P < 0.0001. c Deletion or overexpression of PSP2 does not affect the ATG1 mRNA level. WT (SEY6210), psp2∆ (YZY050), and OE-PSP2 (XLY439) cells were grown in YPD until mid-log phase and then starved for nitrogen for 6 h or 1 day. Total RNA was extracted and the ATG1 mRNA level was quantified by RT-qPCR. The ATG1 mRNA level was normalized to WT cells in growing conditions (set to 1). Mean ± SEM, n = 3 independent experiments for 6 h starvation, n = 4 independent experiments for 1 day starvation. Student’s t-test; ns not significant. d Psp2 regulates the Atg1 level in a manner that is independent of Atg1 degradation. WT (TVY1) and psp2∆ (YZY092) cells that lack PEP4 were treated with control (DMSO) or a proteasome inhibitor (MG132; 75 µM) for 3 h in the indicated conditions (YPD or SD-N media). The Atg1 level was analyzed by western blot. The line indicates that the separated lane was run on the same gel, but was not contiguous with the lane next to it. e Deletion of PSP2 results in a redistribution of ATG1 mRNA to a lower polysome/monosome fraction during starvation conditions. Distribution of ATG1 or PGK1 mRNAs in sucrose gradient fractions from WT (SEY6210) or psp2∆ (YZY050) cells after nitrogen starvation as determined by qRT-PCR. Mean ± SEM are indicated. f Autophagy is reduced in psp2∆ cells. WT (XLY306), psp2∆ (XLY440), and atg1∆ (XLY307) cells in which Pgi1 was tagged with GFP were grown in YPD medium until mid-log phase and then starved for nitrogen for the indicated times. The ratio of free GFP to total GFP (free GFP plus Pgi1-GFP) was quantified. The ratio of processed GFP at each time point was then normalized to that of WT cells at the same time point (set to 100%). Mean ± SEM of n = 3 independent experiments are indicated. Student’s t-test; ****P < 0.0001. g Psp2 positively regulates autophagy. WT (WLY176), psp2∆ (YZY063), and atg9∆ (YKF527) cells were grown in YPD medium until the mid-log phase and then treated with 100 nM rapamycin for 6 or 8 h. Autophagy activity was measured using the Pho8∆60 assay. Pho8∆60 activity was normalized to WT cells with 8-h rapamycin treatment (set to 100%). Means ± SEM of n = 3 or 4 independent experiments are indicated. Student’s t-test; *P < 0.05, ***P < 0.001, ****P < 0.0001. h Loss of Psp2 leads to reduced cell survival. WT (YZY231) and psp2∆ (YZY232) cells were grown in YPD to mid-log phase and then starved for the indicated times. The indicated dilutions were grown on YPD plates for 2 days
Psp2 is a cytosolic protein that contains several RGG motifs at its C terminus. The Psp2 protein is mostly known as a high-copy suppressor of DNA polymerase mutations, and that it binds mRNA and promotes P-body assembly.28–30 A large-scale analysis indicates that Psp2 is associated with the translation initiation complex, but its role in translational regulation remains unknown.31 Because deletion of PSP2 had no effect on global gene expression in both growing and starvation conditions (Supplementary information, Fig. S1a), we ruled out the possibility that Psp2 affects general protein synthesis. To further investigate the role of Psp2 in ATG1 gene expression, we overexpressed Psp2 by replacing its endogenous promoter with that of ZEO1 at the chromosomal locus. Overexpression (OE) of Psp2 substantially increased Atg1 protein abundance compared to WT cells after nitrogen starvation (Fig. 1b). Together, these results identified Psp2 as a positive regulator of Atg1 expression.
The Atg1 protein level is controlled by transcriptional and post-transcriptional regulation in both yeast and mammalian cells.10,32,33 To exclude potential effects of Psp2 on ATG1 transcription or mRNA stability, we measured the level of ATG1 mRNA in WT, psp2∆ and PSP2-OE cells, by real-time quantitative RT-PCR (qRT-PCR). We found that the ATG1 mRNA level was highly upregulated upon nitrogen starvation as reported previously9 (Fig. 1c); however, there was no significant difference between WT cells and those either deleted for or overexpressing PSP2, indicating that the effect of Psp2 on ATG1 expression occurs most likely at the translational level.
The reduced Atg1 level in psp2∆ cells might be due to decreased protein synthesis or increased protein degradation. In eukaryotic cells, proteins are primarily degraded either through the ubiquitin-proteasome pathway or autophagy-lysosomal proteolysis. During nitrogen starvation, ULK1 (a mammalian homolog of yeast Atg1) is degraded in both autophagy-dependent and proteasome-dependent pathways,34 whereas yeast Atg1 can undergo autophagy-dependent degradation in vacuoles,35,36 but it is not clear whether and how it is targeted to the proteasome. To examine the effect of modulating Psp2 levels on the synthesis of Atg1, we blocked the two degradation pathways by deleting the gene encoding the major vacuolar protease, Pep4, which should stabilize vacuolar Atg1, and by treating the cells with the proteasome inhibitor MG132. Whether or not we blocked the proteasome pathway, strains lacking PSP2 consistently showed a significantly reduced level of Atg1 in starvation conditions (Fig. 1d). In agreement with this finding, overexpression of Psp2 in pep4∆ cells, where vacuolar degradation should no longer account for a difference in protein level, led to an elevated level of Atg1 compared to the WT (Supplementary information, Fig. S1d). These data indicated that Psp2 is regulating Atg1 synthesis rather than its degradation.
To further confirm and quantify the effect of PSP2 deletion on translation of the ATG1 mRNA during nitrogen starvation, we used polysome profiling followed by qRT-PCR to chart the distribution of mRNA in polysomes. Translation is typically regulated at the initiation step, which is reflected by ribosome loading of a particular mRNA. Thus, higher levels of translation are reflected by higher ribosome occupancy, which will shift the population of that mRNA deeper into the polysome fraction. In WT cells, the ATG1 mRNA was actively translated during nitrogen starvation, as shown by the fraction of the mRNA occupied by 4 or more ribosomes; however, this fraction was reduced in the absence of Psp2 (Fig. 1e). The ATG1 mRNA was redistributed toward the monosome fraction when PSP2 was deleted, leading to decreased translation units (Supplementary information, Fig. S1e). In contrast, the distribution of PGK1 mRNA, which served as a control, was unaffected (Fig. 1e; Supplementary information, Fig. S1e). Thus, under nitrogen-starvation conditions, Psp2 is specifically required for increasing the ribosome load on ATG1 mRNA.
Psp2 is a positive regulator of autophagy
Because Psp2 promotes the expression of ATG1 in starvation conditions, we speculated that it might be a positive regulator of autophagy. To test if autophagy activity is impaired in psp2∆ cells, we developed a processing assay based on Pgi1-GFP. Pgi1 is a long-lived cytosolic glycolytic enzyme,37 whose expression is stable even during prolonged nitrogen starvation (Fig. 1f). Upon starvation, Pgi1-GFP is expected to be randomly delivered to the vacuole for degradation though nonselective autophagy, similar to most other cytosolic proteins. In the vacuole, Pgi1 will be degraded, whereas the GFP moiety remains relatively stable38; accordingly, we observed the accumulation of free GFP in WT cells after starvation. In contrast, in atg1∆ cells, only full-length Pgi1-GFP was detected, indicating the degradation of the chimera is dependent on autophagy. Thus, the conversion of Pgi1-GFP into GFP can be used as a readout for autophagic flux. In the psp2∆ strain, we observed a markedly lower level of Pgi1-GFP processing relative to the control after 1 day of starvation, and this difference became more significant following 3 days of nitrogen starvation (Fig. 1f). We also observed similar results when we measured the processing of two additional long-lived cytosolic GFP fusion proteins, Fba1 (fructose-1,6-biphosphate aldolase 1)-GFP, and Pgk1 (3-phosphoglycerate kinase)-GFP,39,40 suggesting decreased autophagy flux in psp2∆ cells (Supplementary information, Fig. S1f, g).
To extend our analysis, we used the quantitative Pho8∆60 assay.41 Pho8∆60 is a mutant form of the vacuolar phosphatase that is delivered to the vacuole through autophagy, allowing for the proteolytic activation of its phosphatase activity. To avoid the early saturation of Pho8∆60 activity in WT cells, we treated the cells with rapamycin instead of nitrogen starvation. After 6-h rapamycin treatment, WT cells showed a substantial induction of Pho8∆60 activity, which was blocked by the deletion of ATG9 (Fig. 1g). In agreement with the chimeric GFP processing assays, psp2∆ cells showed a significant decrease in autophagy activity compared to WT cells after 6-h and 8-h rapamycin treatment.
Defects in autophagy activity cause reduced life span and increased cell death, thus leading to a loss in viability during nutrient deprivation.42 To test the physiological importance of Psp2-dependent regulation of autophagy, we monitored the viability of yeast cells lacking Psp2 after prolonged nitrogen starvation. Compared to WT cells, psp2∆ cells showed a noticeable reduction in viability after 10 days of starvation, which was further exacerbated with longer starvation (Fig. 1h). Collectively, these data suggest that Psp2 positively regulates autophagy during nitrogen starvation conditions, possibly by promoting ATG1 translation.
Psp2 binds to ATG1 mRNA
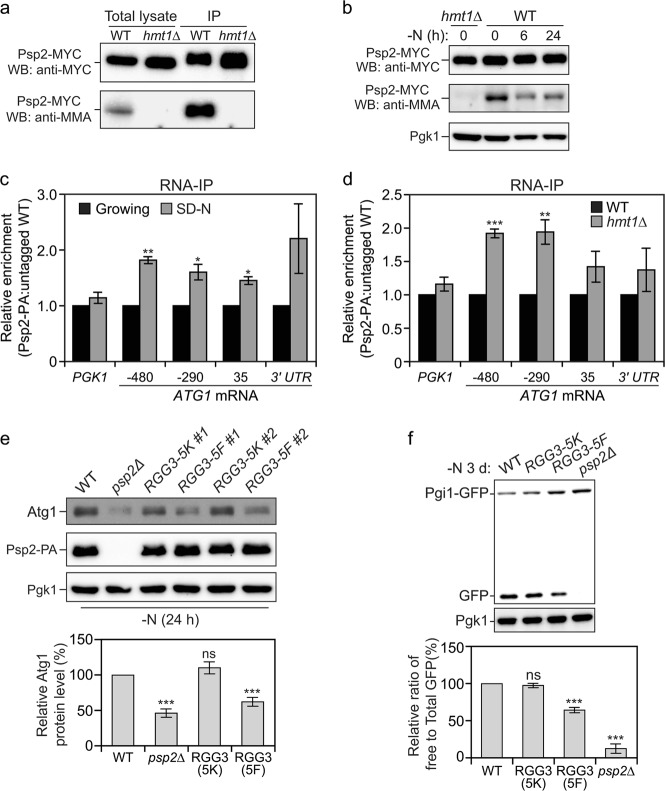
To address whether Psp2 directly targets ATG1 mRNA to promote its translation, we performed an RNA immunoprecipitation (RIP) assay.43 To carry out this analysis we generated a strain with a protein A (PA) tag integrated at the chromosomal PSP2 locus. We precipitated Psp2-PA with IgG, which binds to the protein A-tag, using cell lysates from the tagged strain and an untagged WT strain as a control. Cells were starved for nitrogen prior to lysis, and co-precipitated RNAs were quantified by qRT-PCR. We observed significant enrichment of ATG1 mRNA in the Psp2-PA RIP relative to the control (untagged WT) RIP (Fig. 2a). In addition, we used PGK1, whose expression is not affected by the depletion of Psp2, as a negative control. In this case, there was minimal (background) enrichment, which confirmed that Psp2 directly associates with ATG1 mRNA. Although many features of an mRNA can contribute to its interaction with RBPs, most binding sites for regulatory proteins are located within the 3′ or 5′ UTRs.44 To investigate which region on the ATG1 transcript serves as the binding site for Psp2, we used a set of probes that amplify different fragments along the transcript. The 5′ UTR and 3′ UTR of ATG1 mRNA are 549 nucleotides (nt) and 595 nt, respectively.45,46 All of the ATG1 mRNA fragments and PGK1 mRNA amplified during qRT-PCR are within 105–145 base pairs (bps), which are within the optimal amplicon length for qRT-PCR. We found a higher enrichment at both the 5′ UTR and 3′ UTR compared to the open reading frame (ORF) (Fig. 2a), suggesting that Psp2 might target those regions directly for binding.
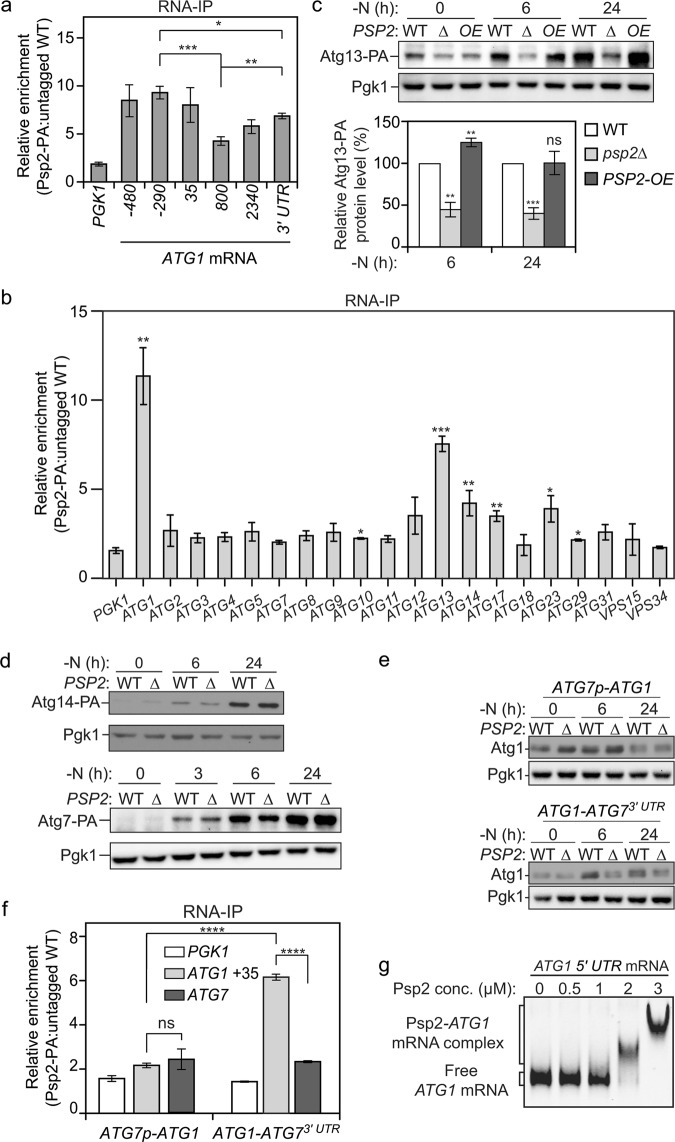
Fig. 2.
Psp2 promotes the translation of Atg1 and Atg13 by targeting their 5′ UTR. a, b Psp2 directly targets ATG1 mRNA. WT (SEY6210) and PSP2-PA (YZY051) cells were grown in YPD medium to mid-log phase and then starved for nitrogen for 2 h. Cells were subjected to RNA immunoprecipitation as described in experimental procedures. qRT-PCR experiments were performed to show the enrichment of ATG mRNAs based on the Psp2 RIP assay. “-” and without “-” mean the start of the amplicon is upstream or downstream of the start codon, respectively. The amplified 3′ UTR of the ATG1 mRNA refers to the sequence starting from 25 bps upstream of the stop codon and is 124 bps long. All of the fragments amplified during qRT-PCR are within 105–145 bps. Mean ratios ± SEM of n = 3 independent experiments of ATG mRNA levels in Psp2-PA:non-tag RIP are indicated. PGK1 mRNA served as a negative control. Enrichment of different regions in ATG1 mRNA and other ATG mRNAs are shown in a, b, respectively. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001; ns not significant. c, d Psp2 also positively regulates Atg13 expression. Cells of the indicated strains (for Atg13: ZYY202, YZY235 and YZY236; for Atg7: JMY322, YZY128; for Atg14: YZY059 and YZY060) were grown in YPD medium until the mid-log phase and then starved for nitrogen for the indicated times. Protein extracts were analyzed as in Fig. 1a. Representative images of western blots are shown in c, d, and quantification of the Atg13-PA level is shown in c. Mean ± SEM of n = 3 independent experiments are indicated. Student’s t-test; **P < 0.01, ***P < 0.001; ns not significant. The 3′ UTR and 5′ UTR of ATG13 was not altered in the tested strain. e The regulation of Atg1 translation by Psp2 is dependent on the 5′ UTR of ATG1 mRNA. Atg1 protein levels were measured in atg1∆ cells and atg1∆ psp2∆ cells expressing either ATG1 under the control of the ATG7 promoter (XLY442 and XLY443) or ATG1 with the ATG7 3′ UTR (XLY349 and XLY441) under the indicated conditions by western blot. Representative images are shown. f The enrichment of the ATG1 +35 mRNA fragment was measured by qRT-PCR experiments using RIP in atg1∆ cells and atg1∆ PSP2-PA cells expressing either ATG1 under the ATG7 promoter (YZY185) or ATG1 with the ATG7 3′ UTR (YZY184). The enrichment was quantified and shown as in Fig. 2a; PGK1 and ATG7 mRNAs served as negative controls. Student’s t-test; ****P < 0.0001; ns not significant. g Psp2 binds the 5′ UTR of ATG1 mRNA in vitro. A gel-shift assay was performed to analyze the binding of Psp2 to ATG1 mRNA. A 500-bp construct representing the 5′ UTR of ATG1 mRNA was incubated with increasing concentrations of purified Psp2. A representative image is shown out of 4 repeats
Psp2 also promotes the translation of Atg13
Atg1 is not the only Atg protein whose expression is highly upregulated during starvation, which prompted us to ask if there are any other ATG genes that are also regulated by Psp2. To answer this, we used the same method to examine the enrichment of other ATG mRNAs using specific primers amplifying an mRNA sequence of approximately 120 bps spanning from the 5′ UTR to the ORF. Among them, ATG13 and ATG14 mRNAs showed the highest enrichment, approximately 8 and 4 fold, respectively, in the Psp2-PA RIP versus the control RIP (Fig. 2b). Both Atg13 and Atg14 are in the core machinery of autophagy, and their expression is upregulated during starvation.9,47 Because the amounts of VPS30/ATG6 and ATG16 mRNAs were too low to be detected in RIP samples, we directly compared their protein levels between WT and psp2∆ cells. Deletion of PSP2 did not reduce either Vps30/Atg6 or Atg16 expression (Supplementary information, Fig. S2a).
Next, we sought to test if Psp2 indeed promotes the translation of Atg13 and Atg14 by measuring their protein levels in psp2∆ cells using western blot. Both proteins were induced upon starvation, whereas the Atg13 level in psp2∆ cells was significantly less than that in WT cells during prolonged nitrogen starvation (Fig. 2c). In agreement with this result, overexpression of Psp2 led to an increase in Atg13 expression after 6 h of nitrogen starvation. In contrast, Atg14 failed to show any significant difference between psp2∆ cells and WT cells (Fig. 2d). Furthermore, we measured the amount of Atg7, the mRNA of which was not pulled down by Psp2-PA RIP (Fig. 2b). Consistent with the RIP assay, deleting PSP2 had essentially no effect on the expression of Atg7 (Fig. 2d). Finally, we examined one additional protein, Atg9, which we have previously shown is regulated at the transcriptional level by Pho23.8 The protein level of Atg9 was not affected by Psp2 depletion in either the WT or pep4∆ background (Supplementary information, Fig. S2b). Taken together, these data identified ATG13 as another target of Psp2.
Psp2 targets the 5′ UTR of the ATG1 transcript
To extend our analysis, we decided to investigate whether it is the 3′ UTR or the 5′ UTR of ATG1 mRNA that is targeted by Psp2. We relied on the observation that Atg7 expression was not affected by PSP2 deletion and generated atg1∆ strains in which ATG1 was either expressed under the control of its endogenous promoter and contained the ATG7 3′ UTR, or expressed under the control of the ATG7 promoter with the endogenous ATG1 3′ UTR. When the 3′ UTR of ATG1 was switched with that of ATG7, the psp2∆ cells still showed a substantial decrease in Atg1 protein level compared to WT cells during starvation (Fig. 2e), to a similar extent as seen with the endogenous 3′ UTR (Fig. 1a), indicating that the 3′ UTR of ATG1 is not required for Psp2 function. This was further confirmed by examining cells where the ATG1 3′ UTR was replaced with that of ADH1 (Supplementary information, Fig. S2c), another gene that is not targeted by Psp2 (data not shown), which displayed a similar phenotype. In contrast, switching the ATG1 5′ UTR with that of ATG7 (ATG7 promoter [ATG7p]) prevented the decrease in Atg1 protein in psp2∆ cells (Fig. 2e), suggesting that the 5′ UTR of ATG1 mRNA contains regulatory elements for Psp2 targeting.
Next, we asked whether these phenotypes are due to a disrupted Psp2-ATG1 mRNA interaction. To this end, we performed RIP in strains where either the 5′ UTR or 3′ UTR of ATG1 was switched with that of ATG7 as above, using both PGK1 and ATG7 mRNA as negative controls. Consistent with the change in Atg1 protein level, we did not observe any enrichment of ATG1 mRNA in the Psp2-PA RIP when its endogenous 5′ UTR was replaced, whereas 3′ UTR switching still allowed substantial enrichment (Fig. 2f). This result further confirmed that Psp2 targets the 5′ UTR of ATG1 mRNA to promote its translation. The enrichment seen for the 3′ UTR with the Psp2 complex by RIP (Fig. 2a) might be due to the formation of circularized mRNA when ATG1 is actively translated.
To verify observations made in vivo, we decided to recapitulate the interaction between Psp2 and the 5′ UTR of the ATG1 transcript in vitro. To this end, Psp2 was recombinantly expressed and purified. The ATG1 mRNA was synthesized using T7 RNA polymerase, generating 500-bp transcripts bearing the nucleotide sequence upstream of the ATG1 ORF. The interaction between Psp2 and the RNA transcript was monitored using a gel-shift assay. Results verified that full-length Psp2 directly bound to the 5′ UTR of ATG1 mRNA (Fig. 2g). As a control, we also tested whether or not RNA transcripts representing the 5′ UTR of ATG7 could bind to Psp2 in vitro. This interaction was not observed using RNA-IP in yeast cells. The results, however, showed that Psp2 could associate with the 5′ UTR of ATG7 mRNA, but not as strongly as seen with ATG1 (Supplementary information, Fig. S2d, e). The observed 3-fold difference in binding affinities, coupled with the observation that the amount of ATG1 mRNA is estimated to be 2-fold higher than that of ATG7 (unpublished RNAseq data) after 2 h of nitrogen starvation, may partially account for the lack of ATG7 transcript that immunoprecipitated with Psp2-PA in vivo. Furthermore, there might be other potential RBPs in the cell that preferentially interact with the ATG7 mRNA, and these may compete with Psp2.
The RGG motif is essential for Psp2 to promote ATG1 expression
The RGG motif/box contains multiple clustered RGG/RG repeats interspersed with a varied length of spacers (usually 0–4 amino acids).20,21 Psp2 harbors thirteen RGG/RG repeats in four different RGG motifs at its C terminus (Fig. 3a). Because the RGG motif possesses an RNA-binding property and also mediates protein-protein interactions,21,22 we speculated that it may play important roles in promoting ATG gene translation. To define the function of the Psp2 RGG motif, we constructed truncation mutants in which the C-terminal 56, 156, or 175 residues were deleted, so that they only have two, one or no RGG motifs, respectively (Supplementary information, Fig. S3a). We first examined if removing RGG motifs affects ATG1 expression based on western blot. Compared to the WT, cells expressing all three truncation mutants showed lower Atg1 protein levels, to a similar extent as seen with the complete PSP2 deletion strain (Supplementary information, Fig. S3b), suggesting that the RGG motifs might be required for Psp2 function. Next, we focused on Psp2∆175, also named Psp2∆RGG, the variant lacking all the C-terminal RGG motifs. We found that the ability of Psp2 to bind ATG1 mRNA was completely abolished in Psp2∆RGG cells (Fig. 3b), suggesting that the RGG motifs are crucial to Psp2’s mRNA binding capability. To corroborate our result in vitro, we performed a gel-shift assay using purified recombinant Psp2∆RGG. With this mutant, we did not observe any shift in the ATG1 mRNA band (Fig. 3c), supporting our hypothesis that RGG motifs are required in order for Psp2 to interact with RNA.
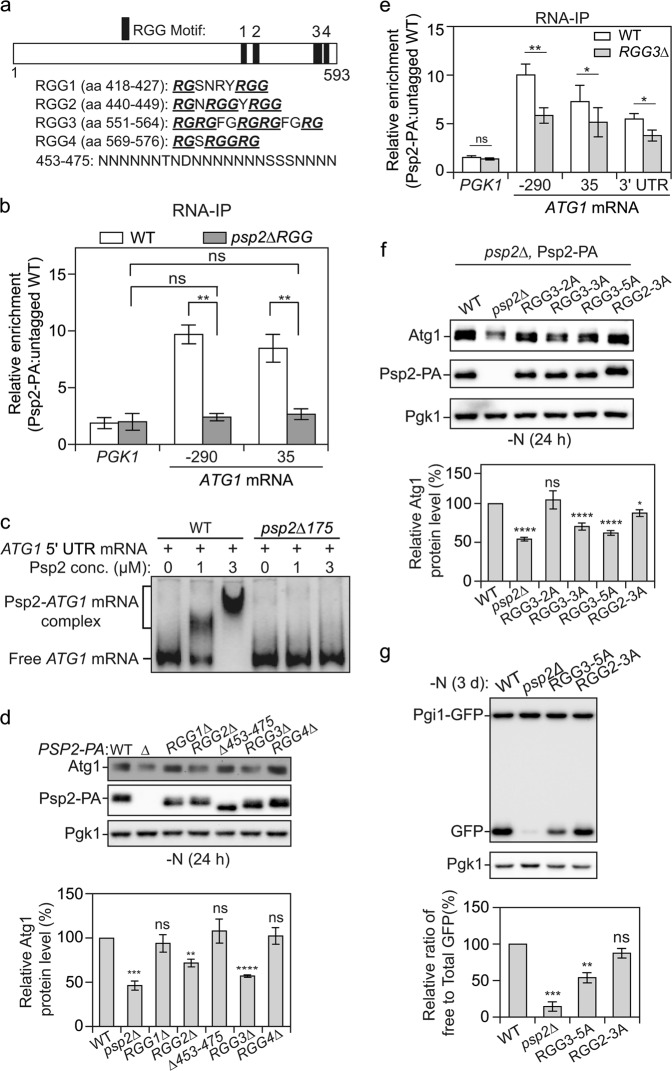
Fig. 3.
Psp2 promotes ATG gene expression in an RGG-motif-dependent manner. a The indicated positions and sequences of RGG motifs (1 to 4) in Psp2 are shown. RG/RGG repeats in the RGG motif are bolded and underlined. b The RGG motif in Psp2 is important for ATG1 mRNA binding in vivo. WT (SEY6210), PSP2-PA (YZY051) and psp2∆175-PA (YZY116) cells were subjected to RNA immunoprecipitation. The enrichment of the indicated ATG1 mRNA fragments was measured by qRT-PCR experiments, quantified and shown as in Fig. 2a; PGK1 mRNA served as a negative control. Student’s t-test; **P < 0.01; ns not significant. c The RGG motif in Psp2 is required for ATG1 mRNA binding in vitro. A 500-bp construct representing the 5′ UTR of the ATG1 transcript was incubated with increasing concentrations of purified recombinant full-length Psp2 and Psp2∆175. A representative gel is shown. d RGG2 and RGG3 in Psp2 are important for its function in regulating Atg1 expression. WT (YZY051), psp2∆ (YZY050), and cells with the indicated truncations were grown in YPD medium until mid-log phase and then starved for nitrogen for 1 day. The Atg1 level was analyzed by western blot. Representative images and quantification of the data are shown. Mean ± SEM of n = 3 independent experiments are indicated. Student’s t-test; **P < 0.01, ***P < 0.001, ****P < 0.0001; ns not significant. e The RGG3 motif in Psp2 is important for ATG1 mRNA binding. WT (SEY6210), PSP2-PA (YZY051), and PSP2[RGG3∆]-PA (YZY169) cells were subjected to RNA immunoprecipitation. The enrichment of the indicated ATG1 mRNA fragments was measured by qRT-PCR experiments, quantified and shown as in Fig. 2a). PGK1 mRNA served as a negative control. Student’s t-test; *P < 0.05, **P < 0.01; ns not significant. f Arginines in the RGG2 and RGG3 motifs are important for the function of Psp2. The Atg1 level was measured by western blot in the indicated strains after 1 day of nitrogen starvation. A representative image and quantification are shown. Mean ± SEM of n > = 4 independent experiments are indicated. Student’s t-test; *P < 0.05, ****P < 0.0001; ns not significant. g The Pgi1-GFP processing assay was performed in the indicated strains. Cells were grown in YPD medium until mid-log phase and then starved for nitrogen for 3 days. The processing of Pgi1-GFP was quantified as in Fig. 1f. Mean ± SEM of n = 3 independent experiments are indicated. Student’s t-test; **P < 0.01, ***P < 0.001; ns not significant
Because the 175 amino acids at the Psp2 C terminus might contain other functional structures, and large truncations might disrupt protein structure and integrity, to further examine whether the RGG motif is required for Psp2 function, we constructed a set of strains in which each individual RGG motif was deleted. We also included a ∆453–475 mutant, as 453–475 is a poly-N sequence, and asparagine-rich domains have been implicated in RNA recognition and RNA binding48,49 (Fig. 3a). In Psp2[RGG2∆] and Psp2[RGG3∆] cells, but not Psp2[RGG1∆], Psp2[RGG4∆] or Psp2[∆453–475] cells, the Atg1 protein level was significantly decreased compared to WT cells under nitrogen-starvation conditions (Fig. 3d). To determine if Atg1 reduction is due to decreased mRNA binding ability, we chose to examine whether Psp2[RGG3∆]-PA binds ATG1 mRNA similar to Psp2-PA as assessed by the RIP assay, because Psp2[RGG3∆] cells showed the highest decrease of Atg1 among all the RGG deletion strains tested. As expected, deleting the RGG3 motif led to markedly reduced ATG1 mRNA enrichment compared to the WT, especially for the RNA region 290 nucleotides upstream in the 5′ UTR (Fig. 3e). Nonetheless, we still observed some level of ATG1 mRNA associated with Psp2[RGG3∆]-PA, suggesting that apart from the RGG3 motif, other protein domains might be also involved in the Psp2-mRNA interaction, such as the RGG2 motif.
Arginine residues within the RGG motif are important for its binding with different RNAs.50–52 To test whether the arginine residues in the Psp2 RGG motif are functionally critical, we mutated the coding regions for these residues in RGG2 and RGG3 by replacing arginines with alanines in a plasmid stably expressing Psp2-PA. We observed that the expression of Psp2 in the Arg-to-Ala mutants was comparable to that in the WT (Fig. 3f), indicating that the mutations did not affect protein stability. Consistent with the results in RGG motif-deletion mutants, compared to psp2∆ cells with WT Psp2-PA, psp2∆ cells expressing Psp2R557,559,563A-PA (RGG3-3A) or Psp2R551,553,557,559,563A-PA (RGG3-5A) showed significantly decreased ATG1 expression, to a similar level as that seen in the Psp2[RGG3∆] strain (Fig. 3f). We observed that the number of arginine-to-alanine mutations had a dosage-dependent effect on the Atg1 protein level, with essentially no effect seen with Psp2R551,553A-PA (RGG3-2A). Furthermore, autophagy activity was also diminished when the five arginines in the RGG3 motif were substituted with alanines based on the Pgi1-GFP processing assay (Fig. 3g). In contrast, psp2∆ cells with Psp2R440,443,447A-PA (RGG2-3A) did not show a substantial reduction in Atg1 levels (Fig. 3f) or a significantly reduced Pgi1-GFP processing (Fig. 3g), indicating some difference in the extent of the phenotype dependent on the particular RGG motif. A possible explanation with regard to the autophagy activity is that the slight decrease in Atg1 expression may not compromise the ability of the RGG2-3A strain to induce autophagy. Taken together, these data indicate that arginine residues within the Psp2 RGG motif, and especially RGG3, are required for normal Atg protein expression and autophagy activity during nitrogen starvation.
Psp2 interacts with eIF4E and eIF4G2
Translation initiation is the most common target of translational control.53 During this process, eukaryotic translation initiation factor 4E (eIF4E) binds to the m7GpppG cap as part of the eIF4F complex, which also contains eIF4G, a central adaptor and scaffolding protein for other initiation factors.54 Previous large-scale affinity purification and protein-fragment complementation assays have shown that Psp2 can be co-purified with several proteins involved in translation initiation, including Cdc33 (eIF4E), Tif4632 (eIF4G2), Ded1 and Dbp1.31,55,56 Among these proteins, eIF4E and eIF4G are the most frequent targets of translational regulators.57,58 To further address how Psp2 promotes Atg1 and Atg13 translation, we examined the interaction between Psp2 and eIF4E or eIF4G by co-immunoprecipitation (IP).
Endogenous Cdc33-GFP was co-precipitated with Psp2-PA in both growing and starvation conditions (Fig. 4a), whereas in the control experiment in which Psp2 was not tagged with PA, neither protein was efficiently precipitated. Despite the fact that Psp2-PA in starvation samples was partially degraded during incubation in the IP/lysis buffer, we still observed that starvation significantly enhanced the interaction between Cdc33 and Psp2 (Fig. 4a). Although Psp2-PA failed to precipitate eIF4G1 by this approach (data not shown), we detected an endogenous interaction between Psp2 and Tif4632 (eIF4G2) (Supplementary information, Fig. S4a). Next, we asked if ATG1 or other mRNAs were involved in bridging this interaction. The addition of RNase A to lysates did not reduce the amount of Cdc33-GFP affinity isolated with Psp2-PA in either growing or starvation conditions (Fig. 4b; Supplementary information, Fig. S4b), suggesting that this interaction is RNA independent.
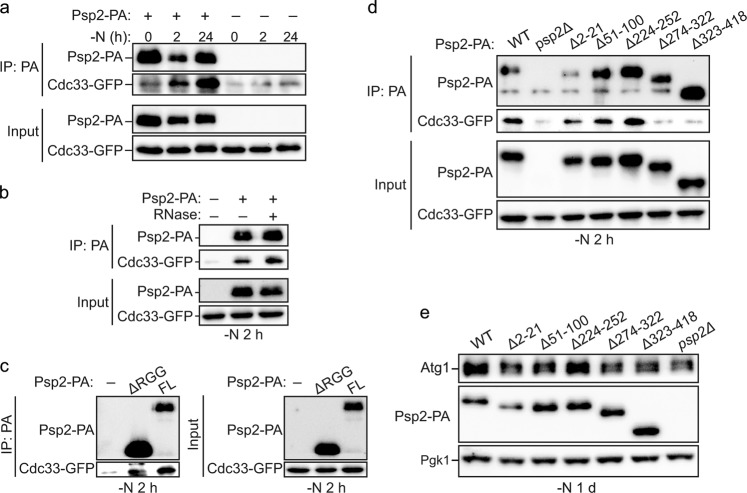
Fig. 4.
Psp2 interacts with eIF4E and eIF4G2. a Psp2 interacts with eIF4E. Cells expressing Psp2-PA and Cdc33-GFP (YZY131) or only Cdc33-GFP (untagged control; YZY132) were grown in YPD medium until mid-log phase, then starved for nitrogen for 2 h or 1 day. Cell lysates were prepared and then subjected to protein-A-immunoprecipitation (IP: PA) as described in Experimental Procedures. The samples were analyzed by western blot with anti-PA and anti-GFP antibodies. b The interaction between Psp2 and eIF4E is RNA independent. Cells were grown in YPD medium until mid-log phase, then starved for nitrogen for 2 h. Immunoprecipitation was performed as described in Fig. 4a and the RNase treatment during incubation with IgG beads was conducted as described in Experimental Procedures (see also Supplementary information, Fig. S4b). c The RGG motif is not required for Psp2-eIF4E interaction. Cdc33-GFP cells expressing Psp2-PA, Psp2∆175-PA (YZY139) or untagged Psp2 were grown in YPD medium until mid-log phase, then starved for nitrogen for 2 h. Immunoprecipitation was performed as described in Fig. 4a (see also Supplementary information, Fig. S4c). d Psp2 mutants lacking amino acids 274–322 (PSP2(∆274-322)) or amino acids 323–418 (PSP2(∆323-418)) were unable to interact with eIF4E. Cells (YZY050) expressing different Psp2 truncation variants were grown in YPD medium until mid-log phase, then starved for nitrogen for 2 h. Immunoprecipitation was performed as described in Fig. 4a. e PSP2(∆274–322) and PSP2(∆323-418) mutants showed a reduced Atg1 level during nitrogen starvation. The Atg1 protein level was measured by western blot in the indicated strains after 1 day of nitrogen starvation. A representative image is shown
To identify the region within Psp2 required for binding to eIF4E or eIF4G, we first tested Psp2∆RGG (Psp2∆175), as the RGG motif could mediate protein-protein interaction, and several other yeast RGG motif proteins interact with eIF4E or eIF4G in a manner dependent on these motifs.24 However, we found that Psp2∆RGG retained the ability to pull down eIF4E in an endogenous co-IP assay (Fig. 4c; Supplementary information, S4c). Considering that the protein level of Psp2∆RGG was much higher than Psp2, we speculated that RGG motifs might contribute to, but not be essential for, the interaction between Psp2 and eIF4E or eIF4G. This result is also consistent with the finding that the interaction is RNA-independent, because RGG motifs are RNA binding domains. Considering that Psp2 is a protein whose functions largely remain unknown, and it contains no known protein interaction domains other than the RGG motifs, we constructed a series of truncation mutants of Psp2 based on the prediction of its intrinsic disordered regions and protein binding regions (Supplementary information, Fig. S4d). We observed that a region within amino acids 274–418 was required for the interaction between Psp2 and Cdc33 (Fig. 4d). Cells expressing a Psp2 mutant lacking this region also had a decreased Atg1 level after starvation (Fig. 4e). These data demonstrate that the association between Psp2 and eIF4E and/or eIF4G is required for promoting Atg1 expression.
Arginines in Psp2 are methylated by Hmt1
Arginine methylation is a posttranslational modification most commonly found in RNA-binding proteins and it has been implicated in various aspects of RNA metabolism and translation.59 Arginine residues within RGG/RG motifs are preferred sites for methylation by protein arginine methyltransferases (PRMTs).21 Arginine methylation in the RGG motif could have either positive or negative effects on its direct interaction with RNA by either increasing the hydrophobicity to enhance binding affinity, or interrupting H-bond interaction and creating steric hindrance to prevent close interaction with mRNA.52,60–63 Because Psp2 has been reported to be arginine methylated in the context of the RGG motif under nutrient-rich conditions,64 we asked whether this methylation could regulate Psp2 function.
To address this question, we first verified that Psp2 is methylated in vivo. Accordingly, we performed MYC affinity isolation in yeast strains expressing MYC epitope-tagged Psp2 under nutrient-replete conditions. The immunoprecipitated samples were detected by western blot using anti-MYC antibody to confirm the precipitation, and MMA, a mono-methyl arginine-specific antibody, to assess the methylation status of Psp2.65 We also included Psp2∆RGG-MYC cells as a control because all of the arginines in the truncated region are within the RGG motifs; all of the remaining arginines are outside of these motifs. We observed that Psp2-MYC was indeed arginine methylated (Fig. 5a) and this occurred only at the arginines in the RGG motif (Supplementary information, Fig. S5a). Because Hmt1 is the predominant protein arginine methyltransferase in yeast,66 we examined if Psp2 is a substrate of Hmt1 in vivo using the same approach in HMT1 knockout cells. Psp2 was no longer arginine methylated in hmt1∆ cells (Fig. 5a), indicating that Psp2 is arginine methylated in an Hmt1-dependent manner.
Fig. 5.
Psp2 arginine methylation by Hmt1 controls its translational regulation activity. a Psp2 is arginine-methylated by Hmt1. Psp2-MYC was affinity isolated with anti-MYC-beads from PSP2-MYC (YZY133) and PSP2-MYC hmt1∆ (YZY149) cells. The samples were analyzed by western blot with anti-MYC and mono-methyl arginine (MMA)-specific antibody. b The amount of methylated Psp2 markedly decreased upon nitrogen starvation. PSP2-MYC cells were grown in YPD medium until mid-log phase and then starved for nitrogen for 6 h or 1 day. Protein extracts were analyzed by western blot with anti-MYC, anti-Pgk1 (loading control) and mono-methyl arginine (MMA)-specific antibodies. PSP2-MYC hmt1∆ cells collected during growing conditions (0 h -N) serve as a negative control. c Starvation enhanced the interaction between Psp2 and ATG1 mRNA. WT (SEY6210) and PSP2-PA cells (YZY051) were grown in YPD medium to mid-log phase, then shifted to SD-N medium. Cells were harvested in both conditions and subjected to RIP as described in Fig. 2a. Mean ratios ± SEM of n = 3 independent experiments of ATG1 mRNA relative enrichment in Psp2-PA SD-N RIP (normalized to Psp2-PA growing condition RIP) are shown. PGK1 mRNA served as a negative control. Student’s t-test, *P < 0.05, **P < 0.01. d Deleting HMT1 increased binding of ATG1 mRNA with Psp2. WT (non-tag) (SEY6210), PSP2-PA (YZY051), and PSP2-PA hmt1∆ (YZY177) cells were grown in YPD medium to mid-log phase and then subjected to RNA immunoprecipitation as described in Experimental Procedures. qRT-PCR experiments were performed and quantified to show the relative enrichment of different ATG1 mRNA fragments in Psp2-PA:non-tag RIP. Mean ratios ± SEM of n = 3 independent experiments of ATG1 mRNA relative enrichment in hmt1∆ Psp2-PA RIP (normalized to WT Psp2-PA RIP) are shown. PGK1 mRNA served as a negative control. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001; ns not significant. e Unmethylated arginines in RGG3 are important for the function of Psp2. The Atg1 protein level was measured by western blot in the indicated strains after 1 day of nitrogen starvation. A representative image and quantification are shown. Mean ± SEM of n ≥ 3 independent experiments are indicated. Student’s t-test; *P < 0.05, ****P < 0.0001; ns not significant. f The Pgi1-GFP processing assay was performed in the indicated strains (WT [YZY213], psp2∆ [YZY212], 5K [YZY252], 5F [YZY253]). Cells were grown in YPD medium until mid-log phase and then starved for nitrogen for 3 days. The processing of Pgi1-GFP was quantified as in Fig. 1f. Mean ± SEM of n ≥ 3 independent experiments are indicated. Student’s t-test; **P < 0.01, ***P < 0.001; ns not significant
Starvation conditions or rapamycin treatment can promote the inactive monomerization of Hmt1, thus abolishing its arginine methyltransferase activity.67 In addition, Hmt1 protein level also decreased after nitrogen starvation (Supplementary information, Fig. S5b). Therefore, we next proceeded to examine if the methylation status of Psp2 changes before and/or after starvation. Although the protein level of Psp2-MYC was stable even during prolonged nitrogen starvation as detected by anti-MYC antibody, the level of methylated Psp2 as indicated by MMA markedly dropped after nitrogen starvation; thus, the portion of unmethylated Psp2 largely increased (Fig. 5b). Similar results were observed with amino acid starvation (Supplementary information, Fig. S5c). Because there is no known non-histone arginine demethylation,68 we speculated that the decreased Psp2 methylation results from the turnover of methylated Psp2, whereas newly synthesized Psp2 is unmethylated during nitrogen starvation.
Unmethylated arginine in the RGG motif is required for normal Atg protein expression and autophagy activity during nitrogen starvation
Because Psp2 promotes ATG1 expression during starvation conditions but has no, or a minor, effect in growing conditions (Fig. 1a, b), which correlates with the methylation status of Psp2, we wondered if arginine methylation functions as a molecular switch that turns off Psp2 function. To this end, because starvation enhanced Psp2-ATG1 mRNA interaction (Fig. 5c), we first investigated if methylation of Psp2 alters its binding of ATG1 mRNA. Psp2-PA RIP and untagged-Psp2 RIP were performed in both WT and hmt1∆ cells. There was significantly higher enrichment of ATG1 mRNA when Psp2 was not methylated (Fig. 5d), suggesting that arginine methylation of Psp2 impairs its binding to ATG1 mRNA.
Because ATG1 expression is regulated by many factors, and Hmt1 has broad substrate specificity, phenotypes of the hmt1∆ mutant may not be due only to defects in Psp2 methylation. To avoid off-target downstream effects and determine the significance of Psp2 methylation, we generated Psp2-PA constructs in which arginines within the RGG3 motif were mutated to lysines or phenylalanines. Positively charged lysine residues can functionally mimic unmethylated arginines and are not recognized by Hmt1 for methylation, whereas bulky hydrophobic phenylalanine residues can mimic constitutive methylated arginines.69–71 In line with the RIP experiments, cells expressing an arginine-methylation-mimetic variant, Psp2R551,553,557,559,563F-PA (RGG3-5F), showed a significantly lower level of Atg1 compared to cells with WT Psp2-PA (Fig. 5e). In contrast, cells expressing a non-methylatable arginine-mimic variant, Psp2R551,553,557,559,563K-PA (RGG3-5K), did not show any difference from WT, indicating that unmethylated arginines are required for Psp2 function. Consistent with changes in ATG1 expression level, cells expressing RGG3-5F, but not RGG3-5K, showed impaired autophagy activity based on the Pgi1-GFP processing assay (Fig. 5f). It is noteworthy that the phenotypes in RGG3-5F cells were similar to the ones observed for RGG3-5A and RGG3-deletion mutants. Taken together, these data indicate that arginine methylation in the RGG motifs (especially RGG3) functions as a switch that regulates Psp2 function, thus regulating ATG gene expression, in response to nutrient conditions.
Discussion
One of the major topics of focus in the field is how autophagy is kept at basal level in normal conditions and how it is quickly switched on upon certain types of stimulation. Cells respond to environmental stress, including nutrient deprivation, by promptly and precisely altering gene expression patterns. The control of mRNA translation in eukaryotes is an essential mechanism of gene regulation. The translational reprogramming under stress conditions often combines global translation shutdown and selective translation of stress-response genes.13 During starvation, the mechanism of how general protein synthesis is repressed has been described, whereas whether and how ATG genes are selectively translated remains unclear. In this study, we carried out a screen for translational regulators modulating ATG gene expression and identified Psp2 as a positive regulator of autophagy through promoting Atg protein synthesis.
During nitrogen starvation, the absence of Psp2 significantly reduced the expression of Atg1 and Atg13 proteins, whereas overexpression of Psp2 increased their expression, without changes in ATG1 and ATG13 mRNA levels, suggesting that Psp2 functions in translational control. The decreased autophagy activity and reduced cell survival in psp2∆ cells are likely primarily due to the decreased Atg1 and Atg13 protein levels. However, we cannot rule out the possibility that some other Atg or regulatory proteins might be upregulated by Psp2 because our screen was based on an RNA binding assay and only focused on Atg proteins that are in the core machinery or known to be upregulated during starvation. We showed that Psp2 directly bound ATG1 and ATG13 transcripts in an RGG motif-dependent manner. In line with this, the expression of Atg proteins, whose transcripts were not bound by Psp2, was not affected by PSP2 deletion. Further analysis of ATG1 mRNA by replacing its 3′ or 5′ UTR indicated that Psp2 targets the 5′ UTR of the ATG1 transcript. Atg1 and Atg13 physically interact with each other and are part of the Atg1 complex. Both of these proteins are at the convergence of multiple signaling pathways including the TOR, AMPK and PKA pathways,72 and are essential for initiating autophagosome formation. It is intriguing that the two corresponding genes are regulated by Psp2; perhaps the selective translation of Atg1 and Atg13 could cause a more robust induction and is important for the maintenance of autophagy activity during prolonged starvation.
Recently our lab determined that Dhh1, a DExD/H-box RNA helicase, is important for driving the translation of Atg1 and Atg13 by recognizing the structured regions of their mRNAs, which form proximal to the start codons;39 mutants that destabilize this structured region allow expression independent of Dhh1. To exclude the possibility that Psp2 and Dhh1 function in the same pathway, we introduced the structured region-mutated ATG1 into psp2∆ cells. We found that Psp2 was required for the efficient translation of both WT and mutated ATG1 (Supplementary information, Fig. S6), indicating that Psp2 and Dhh1 regulate the expression of Atg1 and Atg13 independently.
Because only two mRNA targets of Psp2 have been found, we were not able to identify the RNA regulatory sequences or secondary structures that allow recognition and selective recruitment. RGG motifs have the biochemical properties to bind both RNA and proteins.21 Several yeast RGG motif proteins, including Scd6, Sbp1 and Npl3, bind eIF4G through their RGG motifs and repress translation in vitro.24 This is distinct from the case of Psp2 because this protein bound eIF4E in an RGG motif-independent manner. We also for the first time showed that an RGG motif protein in yeast could promote translation.
We found that PSP2 deletion leads to reduced expression of Atg1 and Atg13 in prolonged nitrogen starvation but has a minor or no effect in nutrient-rich conditions. Therefore, we speculated that there is a switch to activate the function of Psp2 upon starvation. Because the expression level of Psp2 is stable even after prolonged nitrogen starvation, we considered that posttranslational modifications of Psp2, such as phosphorylation and methylation, might control its activity and subsequently the expression of its target mRNAs. The phosphorylation status of RBPs regulates their RNA- and protein-binding activity as well as cellular localization.73,74 Previous phosphoproteome analyses have identified several phosphorylation sites in Psp2 including S236, S238, S340 and S522, among which phosphorylation by Cdk1 at S340 is significantly increased upon rapamycin treatment.75–77 Indeed, we observed that rapamycin treatment and nitrogen starvation can induce hyperphosphorylation of Psp2 (Supplementary information, Fig. S7a). However, neither non-phosphorylatable nor phosphomimetic mutations of those sites affected Atg1 synthesis during nitrogen starvation (Supplementary information, Fig. S7b), suggesting that the phosphorylation status of Psp2 may not be involved in its role in regulating translation.
Instead, we found that arginine methylation, in the context of the RGG motifs, by the PRMT Hmt1 functions as a molecular switch. Hmt1 is phosphorylated and active in the form of an oligomer in growing conditions. Starvation or rapamycin treatment leads to Hmt1 dephosphorylation by the PP2A phosphatase Pph22, disassembly and loss of methyltransferase activity.67 Thus, Hmt1 serves as a sensor for nutrient conditions. Although a subset of JmjC histone lysine demethylases were shown to be able to catalyze arginine demethylation,78 the existence of a specific arginine demethylase and whether arginine methylation is dynamic remains controversial.68 Therefore, we speculated that it is the newly synthesized unmethylated Psp2 that carries out the function of promoting ATG mRNA translation. Arginine methylation deregulation has been implicated in various types of cancers, and PRMTs have become a popular target for small molecule inhibition.68 Our finding suggests a possible mechanism of how PRMTs play a role in cancers through regulating autophagy, and provides potential therapeutic targets.
Psp2 does not have a conserved homolog in higher eukaryotes. However, the regulation mechanisms are likely to be conserved. First, RNA-binding proteins have modular structures, and they are often conserved or functionally conserved at the level of protein domain rather than the entire protein sequence. RGG/RG domains are the second most common RNA-binding domain in the human genome and numerous translation regulators in mammalian cells contain RGG domains. When we blasted amino acids 418–449 (including the RGG1 and RGG2 motifs) or amino acids 551–576 (including the RGG3 and RGG4 motifs), we found those regions are highly conserved in many other multicellular organisms including Xenopus. Furthermore, if we only looked for the conserved domain for RGG3 (amino acids 551–564) in the human proteome, more than 15 RBPs contain a domain that is highly conserved. These proteins could potentially be involved in similar types of regulation. Moreover, Psp2 contains many intrinsically disordered regions (IDRs). IDRs can rapidly evolve and often are not conserved at the amino acid sequence level. However, despite near-complete sequence divergence, orthologous IDRs can preserve regulatory functions.39 Therefore, there might be a functionally conserved homolog for Psp2, but it would be hard to identify using a protein blast, which is based on amino acid sequence similarity.
In summary, we identified the RGG motif protein Psp2 as a novel regulator of autophagy that promotes Atg1 and Atg13 synthesis during nitrogen starvation (Fig. 6). This information helps fill the void in our understanding of how Atg proteins are selectively translated. We also revealed a previously unidentified role for protein arginine methylation in autophagy regulation. In future studies, it will be interesting to explore the mechanism by which other ATG genes are translationally regulated.
Fig. 6.
Model for Psp2-mediated translational control of Atg1 and Atg13 synthesis during nitrogen starvation. In nutrient-rich conditions, Psp2 is methylated by Hmt1 at its RGG motif, resulting in a lower binding affinity towards ATG mRNAs. Atg proteins are normally synthesized to maintain autophagy at a basal level. After nutrient deprivation, global translation is downregulated through both the TOR and general amino acid control (GAAC) pathways. TOR inhibition results in the loss of Hmt1 methyltransferase activity. Newly synthesized Psp2 remains unmethylated and binds ATG1 and ATG13 mRNA in an RGG motif-dependent manner. Psp2 also interacts with the eIF4F complex to promote the translation of targeted transcripts. As a result, Atg1 and Atg13 bypass the general translation inhibition and become highly expressed to support the increased demand for autophagy activity
Experimental procedures
Yeast strains, media, and growth conditions
Yeast strains used in this study are listed in Supplementary information, Table S1. Gene deletions and chromosomal tagging were performed using standard methods.79–81
Under growing conditions, yeast cells were grown in YPD (1% yeast extract, 2% peptone and 2% glucose). To induce autophagy, cells in mid-log phase were shifted from YPD to nitrogen starvation medium (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, containing 2% glucose) or amino acid starvation medium (0.69% yeast nitrogen base without amino acids, containing 0.02% uracil, 0.03% adenine, vitamins and 2% glucose) for the indicated times.
Plasmids
The pRS406-ATG1 and pRS406-ATG1-ATG7[3′UTR] plasmids were described previously.82 The pRS406-ATG7p-ATG1 plasmid was made via FastCloning as described previously.83 The native ATG1 promoter in pRS406-ATG1 was replaced with the ATG7 promoter (616 bps upstream of the ATG7 ORF) to make pRS406-ATG7p-ATG1.
Plasmid pRS406-PSP2-PA was constructed by amplifying the PSP2 promoter region (−550-0), PSP2 ORF and the sequence encoding two tandem repeats of PA by PCR from genomic DNA. The PCR product was digested with XhoI and SpeI and ligated into pRS406 digested with the same enzymes. Plasmids pRS406-PSP2R440,443,447A-PA (RGG2-3A), pRS406-PSP2R551,553A-PA (RGG3-2A), pRS406-PSP2R557,559,563A-PA (RRG3-3A), pRS406-PSP2R551,553,557,559,563A-PA (RGG3-5A), pRS406-PSP2(∆2-51)-PA, pRS406-PSP2(∆51-100)-PA, pRS406-PSP2(∆224–252)-PA, pRS406-PSP2(∆274–322)-PA, pRS406-PSP2(∆323–418)-PA, pRS406-PSP2R551,553,557,559,563F-PA (RGG3–5F) and pRS406-PSP2R551,553,557,559,563K-PA (RGG3-5K) were made by site-directed mutagenesis based on plasmid pRS406-PSP2-PA.
For overexpression in bacteria, PSP2-His6 and PSP2∆175-His6 were cloned into the plasmid pMCSG7 through FastCloning. To obtain RNA transcripts, ~500 bps upstream of the ATG1 and ATG7 start codon were cloned into pSOS354, a kind gift from Dr. Shu-ou Shan (Caltech). In this pUC19-based plasmid, transcription of these genes is regulated under the T7 promoter region. In each construct, the 3′ end is flanked by a HindIII restriction site.
mRNA in vitro transcription
pUC19-ATG1-5′UTR and pUC19-Atg7-5′UTR were linearized using HindIII (New England Biolabs) in 1 × Cut Smart buffer for at least 2 h at 37 °C prior to transcription. The digestion reaction was monitored using agarose gel electrophoresis to ensure that all of the circular plasmids had been linearized. mRNA transcription was carried out using the HiScribe T7 Quick High Yield RNA Synthesis Kit from New England Biolabs. The resulting RNA was purified using an RNeasy column from Qiagen.
Purification of His6-tagged proteins
Both C-terminally His6-tagged Psp2 constructs were overexpressed in BL21-CodonPlus (DE3)-RIL E. coli cells (Agilent Technologies) using 0.5 mM IPTG for 4 h at 37 °C. Cells were lysed by sonication in Buffer A (50 mM Tris, pH 7.5, 500 mM NaCl, 4 mM 2-mercaptoethanol, 2 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 × protease inhibitor cocktail [Roche]). After removal of cell debris (12,000 g, 30 min), the supernatant fraction was purified using Ni-NTA Agarose (Qiagen; 1 ml of resin per 1 liter of cells). Protein was loaded and washed with Buffer A supplemented with 20 mM imidazole, and eluted with Buffer A containing 200 mM imidazole. Elution fractions were dialyzed against Buffer A to remove imidazole prior to further purification by size exclusion chromatography using a Superdex 200 Increase 10/300 column (GE Healthcare Life Sciences) in Buffer A. Purified proteins were exchanged into assay buffer (1 × PBS, 5% glycerol, 2 mM MgCl2, 2 mM DTT) using Bio-Gel P-6 desalting columns (Bio-Rad). All purification steps were carried out at 4 °C. The concentrations of full-length Psp2 and Psp2∆175 (Psp2∆RGG) were determined using absorbance at 280 nm and an extinction coefficient of 41,830 M−1 cm−1 and 26,930 M−1 cm−1, respectively.
Yeast viability assay
Yeast cells were grown in YPD to mid-log phase and then shifted to SD-N medium. The attenuance of each culture was adjusted to 0.8 and the cells were starved for the indicated times. At each time point, the same volume of culture was collected and subjected to serial dilution. An aliquot (2 µl) of each dilution was spotted on YPD plates; the cells were grown at 30 °C for 2 days before being imaged.
Autophagic flux assays
The Pgi1-GFP, Fba1-GFP and Pgk1-GFP processing assays are based on the vacuolar delivery of the chimera through non-selective autophagy; the GFP moiety is relatively resistant to vacuolar hydrolases, such that the generation of free GFP is a measure of autophagy. Yeast cells were cultured in YPD to mid-log phase and then shifted to SD-N medium for the indicated times. Cells were harvested and subjected to western blot. Monoclonal anti-YFP antibody was used to recognize GFP. The Pho8∆60 assay was performed as previously described.41
RNA immunoprecipitation
The RNA immunoprecipitation assay was adapted from a previously published protocol.43 Psp2-PA, untagged Psp2 and the indicated mutant strains were cultured in YPD to mid-log phase and then starved for 2 h in SD-N medium. The cells were subjected to cross-linking by incubation with 0.8% formaldehyde for 10 min at room temperature with slow shaking. To stop cross-linking, glycine was added to a final concentration of 0.2 M and the cells were incubated for another 5 min. The samples were collected, washed with ice-cold PBS twice and resuspended in ice-cold FA lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) containing 5 mM PMSF, 1 tablet of cOmplete™ protease inhibitor cocktail (Roche) and RNasin® PLUS RNase inhibitor (Promega, 40 units/µl). Samples were subjected to vortex with glass beads at 4 °C to lyse the cells. The lysates were collected and sonicated at 4 °C with three rounds of 15 s pulses of 45% amplitude with a 1-min break in between. After centrifuging the sonicated sample, the supernatant was collected and divided into input and immunoprecipitate (IP) fractions. Input fractions were frozen in liquid nitrogen and kept at −80 °C for later use. IP fractions were incubated with IgG Sepharose™ beads (GE healthcare Life Sciences) overnight at 4 °C. After incubation, IgG beads were washed with FA lysis buffer 3 times and 1 time with TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). The proteins and RNAs were eluted from beads in RIP elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% SDS) with RNase inhibitor at 70 °C for 10 min. To remove cross-linked peptides and reverse cross-linking, both input and IP samples were incubated with proteinase K for 1 h at 42 °C, followed by 1 h at 65 °C. Next, the RNA in the samples was recovered with acid-phenol:chloroform; then 25 ml 3 M sodium acetate, 20 mg glycogen, and 625 ml ice-cold 100% ethanol were added to precipitate RNA for 1–2 h at −80 °C. Samples were centrifuged and the pellets were washed with 70% ethanol and dried for 15 min. The precipitated RNA was then treated using a TURBO DNA-free kit (Thermo Fisher Scientific) to eliminate residual DNA; RNasin® PLUS RNase inhibitor was also added in this reaction. After inactivating DNase activity, samples were subjected to RT-qPCR.
Gel-shift assay
Complex formations between Psp2 and variants with the 5′ UTR of the ATG1 and ATG7 mRNAs were monitored using a gel-shift assay. In brief, a fixed amount of messenger RNA was titrated with various amounts of protein in assay buffer at room temperature for 1 h before running the resulting complex on a 4% (29:1 acrylamide:bisacrylamide) native polyacrylamide gel. Each reaction contained 40 units of RNasin® PLUS RNase inhibitor. The unbound and bound mRNAs were separated in 0.8 × TAE buffer (32 mM Tris-acetate, 16 mM sodium acetate, 0.8 mM EDTA, pH 8) under 100 V at 4 °C. The RNA bands were visualized by incubating the native gel in a 1 × solution of SYBR Green stain (Invitrogen) for 30 min on an orbital shaker. Prior to imaging, the gel was briefly washed in water three times. Visualization of the stained RNA bands was carried out using 300-nm UV transillumination. The intensities of RNA bands were quantified using the ImageLab software from Bio-Rad. That data were fit to an allosteric sigmoidal curve with a Hill coefficient of 2 to obtain the apparent binding affinities.
RNA and RT-qPCR
Yeast cells were grown in YPD to mid-log phase and then shifted to SD-N medium for the indicated time. Total RNA was extracted using the NucleoSpin RNA kit (Takara). An additional DNase treatment was performed to eliminate genomic DNA contamination. One microgram of total RNA was reverse-transcribed in a 20-μl reaction system using the High-capacity cDNA Reverse Transcription kit (Applied Biosystems). The cDNA levels were then analyzed by real-time PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems). The transcript abundance in samples was determined using the CFX Manager Software regression method as previously described.9 The primers used for the RT-qPCR analysis are listed in Supplementary information, Table S2.
Polysome profiling and qRT-PCR
Yeast cells were grown to mid-log phase in YPD before shifting them for 6 h to SD-N for nitrogen starvation. Cells were harvested and prepared for sucrose-gradient fractionation as previously described.84 Clarified lysate from 7,500 OD units of cells (~100 μl) were loaded onto a freshly prepared 25–50% sucrose gradient and centrifuged for 3 h at 40,000 rpm in an SW-41Ti rotor (Beckman). Gradients were fractionated, and the following samples were collected: unbound RNAs, free subunits, 80S monosomes, and one each for mRNAs bound to two, three, four, etc. ribosomes. Peaks were resolved for up to 7 bound ribosomes.
qRT-PCR was carried out essentially as described previously.85 After the addition of 1 ng of Fluc mRNA (TriLink, L-7202) to each sample, which was used for normalization, RNA was isolated by phenol-chloroform extraction from a fixed percentage of each sample’s total volume, and reverse transcription was carried out per the manufacturer’s instructions using the Protoscript II Kit (New England Biolabs). qPCR was performed with Excella 2 × SYBR master mix (Worldwide Life Sciences) per the manufacturer’s instructions on a BioRad IQ2, using the primers listed in Supplementary information, Table S2. Each gene was normalized first to Fluc mRNA to account for differences in capture and precipitation of each sample. Next, the abundance of each mRNA in each fraction was normalized to the total amount of that mRNA on the gradient. The translation units (TU) were calculated from these data by multiplying the percentage of mRNA in each fraction with the number of ribosomes bound in that sample, and summing these over all gradient fractions. The ΔTU value was obtained by subtracting the TU of each gene in WT cells from the TU in psp2∆ cells.
Native protein immunoprecipitation
Protein A and MYC-epitope affinity isolations were performed essentially as previously described.86 IgG Sepharose™ 6 fast flow beads (GE Healthcare Life Sciences) and anti-MYC magnetic beads (Thermo Scientific) were used, respectively.
Western blot
Antisera were from the following sources: Atg1,87 Pgk1 (a generous gift from Dr. Jeremy Thorner, University of California, Berkeley), Atg9,88 monoclonal YFP (Clontech, 632381), antibody to PA (Jackson ImmunoResearch, 323-005-024), anti-MYC antibody (Sigma, M4439), and mono-methyl arginine (MMA)-specific antibody (Cell Signaling Technology, 8711). The blot was imaged using either a ChemiDoc Touch imaging system (Bio-Rad) or photographic film; images were quantified using either Bio-Rad Image Lab software or ImageJ software, respectively.
Structure prediction
The Psp2 structure prediction was conducted using the webserver at http://bioinf.cs.ucl.ac.uk/psipred/.
Statistical analyses
The two-tailed Student’s t-test was used to determine statistical significance. For all figures, P value < 0.05 were considered significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns not statistically significant.
Supplementary information
Acknowledgements
This work was supported by NIGMS grants GM131919 (DJK) and GM117093 (KK), and by HHMI Faculty Scholar grant 55108536 (KK).
Author contributions
Z.Y. performed most of the experiments. X.L. and M.J. designed and conducted the screen and autophagy activity assay, X.L. also developed the RIP assay, A.A. carried out protein purification and the gel-shift assay, H.H. performed the polysome profiling assay. Z.Y. drafted the manuscript with inputs from all authors and K.K. and D.J.K. revised the manuscript. Z.Y., and D.J.K. designed the project, and K.K. and D.J.K. supervised the project.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0246-4.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb. Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 8.Jin M, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr. Biol. 2014;24:1314–1322. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu G, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 2015;17:930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin S, et al. m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018;28:955–957. doi: 10.1038/s41422-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubas M, et al. eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 2018;19:e46072. doi: 10.15252/embr.201846072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25:354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 14.Hershey John W.B., Sonenberg Nahum, Mathews Michael B. Principles of Translational Control. Cold Spring Harbor Perspectives in Biology. 2018;11(9):a032607. doi: 10.1101/cshperspect.a032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova JB, et al. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol. Cell. Biol. 2005;25:9340–9349. doi: 10.1128/MCB.25.21.9340-9349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou K, Ouyang Q, Li H, Zheng J. A global characterization of the translational and transcriptional programs induced by methionine restriction through ribosome profiling and RNA-seq. BMC Genom. 2017;18:189. doi: 10.1186/s12864-017-3483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji, E. et al. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol. Cell. Biol. 39. 10.1128/MCB.00508-18 (2019). [DOI] [PMC free article] [PubMed]
- 18.Gebauer F, Preiss T, Hentze MW. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb. Perspect. Biol. 2012;4:a012245. doi: 10.1101/cshperspect.a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corley SM, Gready JE. Identification of the RGG box motif in Shadoo: RNA-binding and signaling roles? Bioinform. Biol. Insights. 2008;2:383–400. doi: 10.4137/bbi.s1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol. Cell. 2013;50:613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 24.Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thandapani, P. et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife4. 10.7554/eLife.06234 (2015). [DOI] [PMC free article] [PubMed]
- 26.Brandariz-Nunez A, Zeng F, Lam QN, Jin H. Sbp1 modulates the translation of Pab1 mRNA in a poly(A)- and RGG-dependent manner. RNA. 2018;24:43–55. doi: 10.1261/rna.062547.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang R, et al. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3’-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J. Biol. Chem. 2010;285:8887–8893. doi: 10.1074/jbc.M109.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formosa T, Nittis T. Suppressors of the temperature sensitivity of DNA polymerase alpha mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1998;257:461–468. doi: 10.1007/s004380050670. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao BS, Parker R. Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2017;114:E9569–E9578. doi: 10.1073/pnas.1712396114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 32.Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015;11:2114–2122. doi: 10.1080/15548627.2015.1099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pike LR, et al. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 34.Alemu EA, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft C, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakatogawa H, et al. The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation. J. Biol. Chem. 2012;287:28503–28507. doi: 10.1074/jbc.C112.387514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maitra PK, Lobo Z. Genetic studies with a phosphoglucose isomerase mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 1977;156:55–60. doi: 10.1007/BF00272252. [DOI] [PubMed] [Google Scholar]
- 38.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, et al. Dhh1 promotes autophagy-related protein translation during nitrogen starvation. PLoS Biol. 2019;17:e3000219. doi: 10.1371/journal.pbio.3000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6:794–797. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 41.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 42.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 43.Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009;2009:pdb.prot5234. doi: 10.1101/pdb.prot5234. [DOI] [PubMed] [Google Scholar]
- 44.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 45.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yassour M, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. USA. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 48.Kenan DJ, Query CC, Keene JD. RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 49.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blackwell E, Zhang X, Ceman S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010;19:1314–1323. doi: 10.1093/hmg/ddq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, et al. Protein arginine methyltransferase 1 regulates herpes simplex virus replication through ICP27 RGG-box methylation. Biochem. Biophys. Res. Commun. 2010;391:322–328. doi: 10.1016/j.bbrc.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 52.Wall ML, Lewis SM. Methylarginines within the RGG-motif region of hnRNP A1 affect Its IRES trans-acting factor activity and are required for hnRNP A1 stress granule localization and formation. J. Mol. Biol. 2017;429:295–307. doi: 10.1016/j.jmb.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 53.Hinnebusch A. G., Lorsch J. R. The Mechanism of Eukaryotic Translation Initiation: New Insights and Challenges. Cold Spring Harbor Perspectives in Biology. 2012;4(10):a011544–a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 55.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 56.Schlecht U, Miranda M, Suresh S, Davis RW, St Onge RP. Multiplex assay for condition-dependent changes in protein-protein interactions. Proc. Natl. Acad. Sci. USA. 2012;109:9213–9218. doi: 10.1073/pnas.1204952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 58.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calnan BJ, et al. recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 61.Hyun S, Jeong S, Yu J. Effects of asymmetric arginine dimethylation on RNA-binding peptides. Chembiochem. 2008;9:2790–2792. doi: 10.1002/cbic.200800544. [DOI] [PubMed] [Google Scholar]
- 62.Rajyaguru P, Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11:2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hubers L, et al. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 64.Plank M, et al. Expanding the yeast protein arginine methylome. Proteomics. 2015;15:3232–3243. doi: 10.1002/pmic.201500032. [DOI] [PubMed] [Google Scholar]
- 65.Poornima G, Shah S, Vignesh V, Parker R, Rajyaguru PI. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 2016;44:9358–9368. doi: 10.1093/nar/gkw762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 67.Messier V, Zenklusen D, Michnick SW. A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell. 2013;153:1080–1093. doi: 10.1016/j.cell.2013.04.035. [DOI] [PubMed] [Google Scholar]
- 68.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Mostaqul Huq MD, et al. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006;25:5094–5104. doi: 10.1038/sj.emboj.7601389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stetler A, et al. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 71.Campbell M, et al. Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Biol. Chem. 2012;287:5806–5818. doi: 10.1074/jbc.M111.289496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 2016;39:61–68. doi: 10.1016/j.ceb.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paquin N, et al. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 74.Abdelmohsen K, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soulard A, et al. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swaney DL, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walport LJ, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 2016;7:11974. doi: 10.1038/ncomms11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 80.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gardner JM, Jaspersen SL. Manipulating the yeast genome: deletion, mutation, and tagging by PCR. Methods Mol. Biol. 2014;1205:45–78. doi: 10.1007/978-1-4939-1363-3_5. [DOI] [PubMed] [Google Scholar]
- 82.Gatica D, et al. The Pat1-Lsm Complex Stabilizes ATG mRNA during Nitrogen Starvation-Induced Autophagy. Mol. Cell. 2019;73:314–324 e314. doi: 10.1016/j.molcel.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li C, et al. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferretti MB, Ghalei H, Ward EA, Potts EL, Karbstein K. Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol. 2017;24:700–707. doi: 10.1038/nsmb.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abeliovich H, Zhang C, Dunn WA, Jr., Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noda T, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.