Abstract
Eukaryotic transcription factor IIH (TFIIH) is a 500 kDa-multiprotein complex that harbors two SF2-family DNA-dependent ATPase/helicase subunits and the kinase activity of Cyclin-dependent kinase 7. TFIIH serves as a general transcription factor for transcription initiation by eukaryotic RNA polymerase II and plays an important role in nucleotide excision DNA repair. Aiming to understand the molecular mechanisms of its function and regulation in two key cellular pathways, the high-resolution structure of TFIIH has been pursued for decades. Recent breakthroughs, largely enabled by methodological advances in cryo-electron microscopy, have finally revealed the structure of TFIIH and its interactions in the context of the Pol II-pre-initiation complex, and provide a first glimpse of a TFIIH-containing assembly in DNA repair. Here, we review and discuss these recent structural insights and their functional implications.
Functions of TFIIH in transcription and DNA repair
Transcription initiation by eukaryotic RNA polymerase II (Pol II) requires the assembly of a transcription preinitiation complex (Pol II-PIC) on promoter DNA, a process that has been structurally characterized in detail using X-ray crystallography and cryo-electron microscopy (cryo-EM) [1,2]. The Pol II-PIC is formed by general transcription factors that recognize the transcription start site, recruit the multi-subunit Pol II complex, facilitate opening of the transcription bubble, and are involved in post-translational modification of Pol II [3,4]. Eukaryotic transcription factor IIH (TFIIH) is one of these general transcription factors for Pol II. Aided by TFIIE [5–7,8•], it joins the Pol II-PIC after recruitment of Pol II to the promoter (Figure 1a), and contributes to Pol II transcription initiation and promoter escape by facilitating transcription bubble opening and by phosphorylating the C-terminal heptapetide repeat domain of the Pol II subunit RPB1, respectively (reviewed in Ref. [9]).
Figure 1.
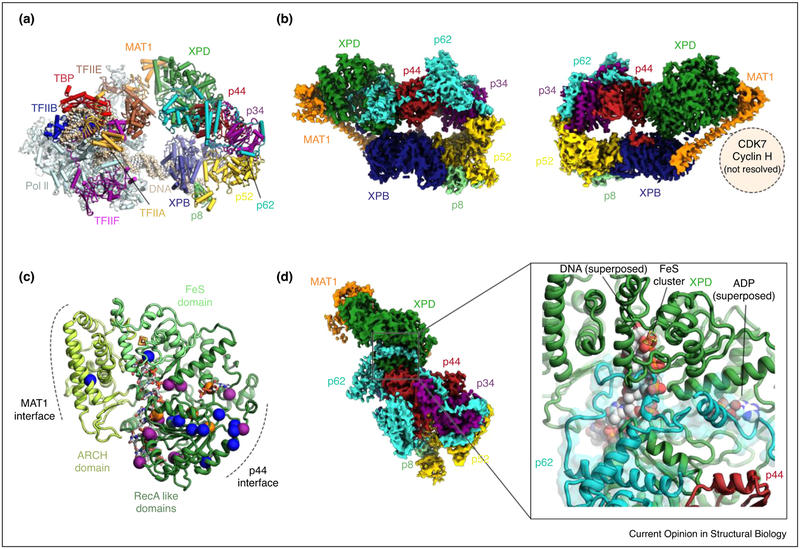
Structure and function of TFIIH. (a) Structure of the human Pol II-PIC with TFIIH, assembled from the models of free TFIIH [27••] (PDB ID 6NMI) and the core-PIC [12•] (PDB ID 5IY9) according to the cryo-EM maps of TFIIH-bound Pol II-PICs [12•,14••] (EMD-8133, EMD-3846). (b) Cryo-EM map of free TFIIH [27••] (EMD-0452), with subunits colored and labeled. The approximate position of the flexible CAK subcomplex is indicated in the right panel. (c) Mapping of human disease mutations onto XPD (blue: TTD; purple: XP; orange: XP-CS; PDB ID 6NMI). Note the prevalence of XP and XP-CS mutations near the enzymatic core (indicated by superposed DNA and nucleotide [42]; see (d) for details) and the clustering of TTD mutations near interaction sites with other TFIIH subunits. (d) Overview (left) and close-up view (right) of the interactions between XPD and p62 in human TFIIH [27••] (EMD-0452) with XPD substrates (DNA and ADP) superposed based on the structure of DinG [42] (PDB ID 6FWS). p62 partially overlaps with the DNA and occludes the access to the ATP-binding pocket.
TFIIH is a ten-subunit protein complex whose two largest subunits, XPB and XPD, harbor ATP-dependent double-stranded DNA translocase and DNA helicase activities, respectively (reviewed in Ref. [9]). XPB and XPD are part of the TFIIH core complex, which includes five additional subunits (p62, p52, p44, p34, and p8) that form an intricate scaffold and recruit the two ATPases. Additionally, TFIIH harbors a kinase activity in its CDK-activating kinase (CAK) subcomplex, comprising Cyclin-dependent kinase 7 (CDK7), Cyclin H and MAT1. Transcription initiation relies on the DNA translocase activity of XPB [10,11], which has been shown to engage the downstream DNA in both human and yeast PIC cryo-EM structures [12•,13,14••,15] (Figure 1a), and on the presence of the CAK subcomplex [16]. In agreement with recent yeast Pol II-PIC structures showing spontaneous promoter melting [17], subsequent studies revealed that in yeast, some promoters melt more easily than others, which enables XPB-independent promoter opening [18]. Similar observations were made in biochemical studies of the human system, leading to the proposal that in some instances, XPB may act in a checkpoint-like manner to allow promoter melting only after successful assembly of the Pol II-PIC [19]. Nevertheless, the human Pol II-PIC, including TFIIE and TFIIH, was successfully reconstituted and characterized structurally with a closed promoter that did not open spontaneously during assembly [12•], suggesting that XPB DNA translo-case activity may be required for opening of some promoters.
In addition to its transcriptional role, the TFIIH core complex is essential in nucleotide excision repair (NER) [16]. NER can repair diverse UV-induced and chemically-induced DNA lesions that typically lead to distortions of the DNA backbone, such as 6–4 photoproducts or cyclobutane pyrimidine dimers. Initial DNA damage detection involves either specialized lesion sensors, such as XPC or DDB2-containing complexes in a subpathway termed global genome NER, or the stalled Pol II in transcription-coupled NER (reviewed in Ref. [20]). Once DNA damage has been detected, TFIIH is recruited to the DNA lesion. TFIIH engaged in NER employs the activities of both XPD and XPB to open duplex DNA to verify the presence of damaged DNA and enable its access to the downstream repair machinery [21–23]. Because the CAK subcomplex inhibits the activity of XPD [24–26], NER requires the release of the CAK subcomplex from the core [16,24], facilitated by the repair factor XPA ([24] and see below).
Defining the structure of human TFIIH
While structures of human and budding yeast TFIIH in the context of the Pol II-PIC were limited to medium resolution[12•,14••,15], the structure of human apo-TFIIH (Figure 1b), in the absence of substrate or components of the Pol II-PIC, enabled modeling of the seven core complex subunits and a part of MAT1, first at 4.4 Å and more recently at 3.7 Å resolution [27••,28•]. These structures revealed the intricate network of protein–protein interactions required for the assembly of the TFIIH core complex, among them interactions of p62 with several other subunits of the core complex, including XPD (Figure 1d), as well as the functionally important interaction between the N-terminal domain (NTD) of XPB and the so-called clutch domain of p52. Together with p8, the p52 clutch and the XPB NTD may form a cradle-like structure that pre-arranges the RecA-like domains of XPB for nucleotide-binding and hydrolysis, thereby increasing its activity [10,27••]. This idea is supported by the observation that mutations in these domains not only lead to TFIIH assembly defects [29], but also affect the ATPase activity of XPB [22], even though none of the mutated residues are in the immediate vicinity of the XPB active site [27••].
The detailed models of the XPB and XPD ATPases have enabled the mapping of human mutations that result in DNA repair defects (Figure 1c) and cause the human congenital diseases Xeroderma pigmentosum (XP) and XP combined with Cockayne syndrome [30], which are generally characterized by premature ageing or cancer predisposition [31]. These mutations map to the well conserved regions near the XPD active site and DNA-binding regions (Figure 1c) [27••,28•] where they impair DNA binding and translocation or nucleotide hydrolysis, thereby impairing NER. These observations are in excellent agreement with previous ideas derived from the analysis of structures of homologous enzymes from bacteria and archaea [32–35]. In contrast, in trichothio-dystrophy, a human disease with phenotypes ranging from brittle hair to mental retardation and premature death [30,31], mutations result in mild transcriptional defects in addition to impaired DNA repair [36] and are generally found in more peripheral regions of the ATPase subunits (Figure 1c), or in p8, the smallest subunit of TFIIH. These mutations impair TFIIH assembly and are associated with reduced stability and lower cellular levels of the full complex [36–39].
The cryo-EM structure of human apo-TFIIH is in excellent overall agreement with the cryo-EM structure of yeast TFIIH in the context of the Pol II-PIC [14••], indicating that, in spite of minor structural differences and well-defined conformational changes linked to presence of TFIIH within the Pol II-PIC (see below), the overall architecture and functional mechanisms of TFIIH are likely to be conserved from yeast to humans. The current most complete model of a human Pol II-PIC [40•] was computationally derived by re-interpretation of previously published cryo-EM maps [12•,14••,28•], revealing previously uncharacterized details about the TFIIH-Pol II-PIC interface, as well as conformational differences of the contact-forming PH-domain of p62 between the human and yeast systems [40•]. The overall topology of the TFIIH domains in this reinterpreted structure [40•] agrees with the models derived from the most recent higher-resolution cryo-EM maps of TFIIH [27••,41••]. However, the accuracy of the sequence register assignment in newly modeled regions of PIC-bound human TFIIH in this computational study [40•] may be limited by the use of lower resolution cryo-EM maps (previously published in [12•,14••,28•]).
Structure of the TFIIH-XPA-DNA complex
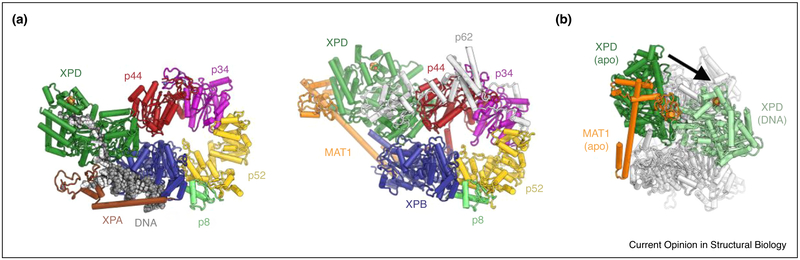
The structure of the TFIIH core complex — without any subunits of the CAK subcomplex — bound to a forked DNA substrate and to the NER factor XPA [41••] shows major conformational rearrangements between XPD and the remainder of the complex (Figure 2a,b). These conformational transitions may be important to activate XPD for DNA binding and translocation. The double-stranded portion of the DNA substrate is bound by XPB and locked in place by XPA (Figure 2a), which binds to multiple sites on the TFIIH core complex. Single-stranded DNA extends towards XPD, contacting the DNA-binding motifs of the RecA-like domains of XPD and then stretching across the XPD pore [41••], as predicted previously based on modeling according to DNA-bound helicase structures [33–35], and as observed in the DNA-bound bacterial XPD homolog DinG [42]. Interestingly, the 5’-end of the DNA strand engaging XPD binds at a site that has been proposed to serve as a nucleotide binding and DNA lesion verification site based on biochemical experiments (Figure 3) [23].
Figure 2.
Conformational changes in TFIIH upon binding to XPA and DNA. (a) Left: Overview of the structure of the TFIIH-XPA-DNA complex [41••] (PDB ID 6RO4). Even though partial density is present in the cryo-EM map, p62 was not modeled in this structure. Right: The structure of apo-TFIIH for comparison [27••] (PDB ID 6NMI; structures superposed on XPB). The conformational difference of the p44-XPD module between the structures is evident. (b) Visualization of the conformational change of XPD between apo-TFIIH (XPD dark green, MAT1 orange) and the TFIIH-XPA-DNA (XPD light green; structures superposed on XPB). MAT1 may not be able to reach far enough to contact both XPB and XPD in the DNA-bound complex and is known to be no longer present in the DNA-bound complex.
Figure 3.
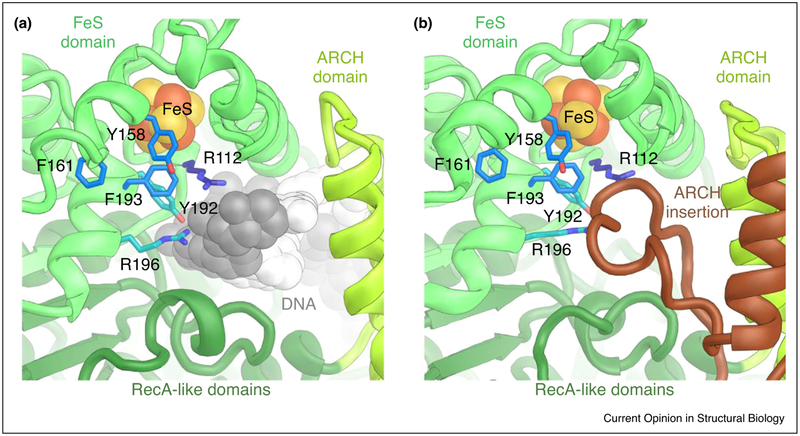
Insights into XPD inhibition. (a) In DNA-bound XPD (from the TFIIH-XPA-DNA NER complex structure; PDB ID 6RO4) [41••], DNA reaches across the pore between the ARCH and FeS domains and approaches residues R196 and Y192, which are implicated in DNA lesion verification [23]. Neighboring residues Y158, F161, and F193 are known to be important for helicase activity [43]. Residue R112 is affected by a TTD mutation in human patients [30]. (b) In apo-TFIIH [27••] (PDB ID 6NMI), the pore between the XPD ARCH and FeS domains is occupied by an insertion within the ARCH domain.
Insights into TFIIH regulation
Biochemical studies have shown that the enzymatic activity of XPD is not required in transcription initiation [43], and that the presence of the CAK subcomplex — which is important in transcription initiation — inhibits XPD [24–26]. Conversely, both XPD and XPB are positively regulated by interactions with p44 and p52, the protein subunits that recruit the ATPases into the complex [22,29,36,44], hinting at a complex regulatory interplay between TFIIH subunits. This hypothesis is further reinforced by the latest structures of apo-TFIIH, which revealed that functionally important regions of XPD, including its DNA-binding cavity, the active site cleft, and the mobile ARCH domain, are contacted by other TFIIH components (Figure 1b,d), suggesting that these contacts play a role in XPD regulation [27••,28•]. Specifically, the interactions of MAT1 with XPD and XPB may inhibit global conformational changes associated with TFIIH activation in the context of DNA repair (Figure 2a,b) [41••], as well as reduce the flexibility of the ARCH domain of XPD [27••]. Motions of the XPD ARCH domain have been shown to correlate with the ATPase cycle of the bacterial XPD homolog DinG [42], and ARCH domain flexibility is required to allow access of substrate DNA to a pore-like structure in XPD [45]. Indeed, the structure of DNA-bound TFIIH in the absence of MAT1 [41••] showed that a eukaryotic-specific insertion in the XPD ARCH domain is disordered (Figure 3a), while the same insertion is ordered and occupies the entrance to the XPD DNA-translocating pore in the presence of MAT1 (Figure 3b) [27••,28•]. These observation provide a possible explanation for how the presence of the CAK subcomplex may directly inhibit XPD [27••,41••].
Further regulatory interactions of XPB and p62, which contact DNA-binding sites in XPD [27••], were initially proposed based on comparisons with substrate-bound structures of related helicases (Figure 1d) [42,46]. Additionally, a short segment of p62 occludes the entrance to the ATP-binding cavity of XPD in apo-TFIIH (Figure 1d) [27••]. Given that these structural elements are not visualized in the activated, DNA-bound and XPA- bound form of TFIIH [41••], they most likely cooperate with MAT1 in inhibiting XPD and are released from XPD to de-repress the enzyme during NER. Indeed, recent biochemical analysis implicated p62 in XPD regulation and DNA damage sensing [47•], in agreement with a role for p62 that goes beyond structural stabilization of TFIIH.
These structural and biochemical findings suggest that free TFIIH has a partially auto-inhibited conformation that transitions to an activated form through a series of conformational changes upon CAK release and binding to DNA and/or XPA. Tight regulation of XPD is likely to be important to allow TFIIH to faithfully perform its dual function and do so at the right time. Such a regulation would ensure that during DNA repair, XPD activity is coordinated with the concerted assembly of the remaining DNA repair proteins at the repair bubble for accurate repair.
Conformational transitions of TFIIH upon Pol II-PIC binding
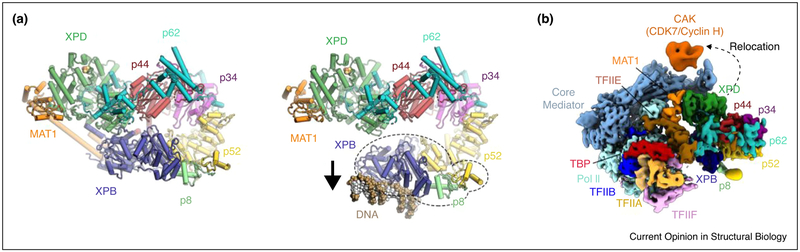
Comparisons of the structure of free TFIIH [27••,28•] with the structures of the human and yeast Pol II-PICs [12•,14••,15] have revealed the conformational rearrangement undergone by TFIIH upon incorporation into the Pol II-PIC (Figure 4a). A module consisting of XPB, p8, and the C-terminal half of p52 moves towards the DNA within the PIC [27••,28•], increasing the distance between the open ends of the horseshoe-shaped TFIIH core complex and breaking the interactions between the RecA-like folds of XPD and XPB that exist in the apo-complex (Figure 4a). This conformational change must dislodge or rearrange the long helix of MAT1 that stretches from the XPD ARCH domain to the XPB DRD-like domain in the free TFIIH structure [27••,28•] (only very weak density is observed in the PIC-bound TFIIH [12•,14••]). These conformational rearrangements of TFIIH are likely coordinated with the formation of additional interactions between TFIIH and other general transcription factors, and may be important for the regulation of XPB activity. These ideas are consistent with biochemical data [6] and with the observation of direct TFIIE-XPB interactions [14••] (Figure 4b).
Figure 4.
Conformational changes of TFIIH upon incorporation into the Pol II-PIC. (a) Left: Structure of TFIIH in the free form of the complex [27••] (PDB 6NMI), shown in the same orientation as the map in Figure 1b. Right: Model of DNA-bound TFIIH within the Pol II-PIC generated by docking the structure of free TFIIH [27••] into the cryo-EM maps of PIC-bound TFIIH [12•,14••] (EMD-8133, EMD-3846). (b) Cryo-EM map of the Mediator-bound yeast Pol II-PIC [14••] (EMD-3850), in which the CDK7-Cyclin H module of TFIIH is located near Mediator (orange, indicated).
In addition to this well-defined structural transition in the TFIIH core complex upon Pol II-PIC entry, the presence of the Mediator complex in the Pol II-PIC may induce repositioning of the flexibly tethered CDK7-Cyclin H module of the TFIIH CAK subcomplex. Mediator is a large co-activator complex that serves to integrate the signals from transcriptional activators or repressors and regulates various aspects of transcription initiation by Pol II [48]. In the absence of Mediator, weak density, possibly corresponding to the highly flexible CDK7-Cyclin H module of the CAK subcomplex [28•], is observed near XPD in human and yeast Pol II-PICs [12•,40•,49]. In the structure of the yeast Pol II-PIC bound to the Mediator complex, the CDK7-Cyclin H portion of the CAK becomes repositioned onto the surface of the Mediator middle module (Figure 4b), bringing it into proximity of its Pol II CTD substrate [14••,50]. The latter location is probably representative of the positioning of the CAK during transcription initiation in vivo, when Mediator is present. CDK7-Cyclin H in this position may contribute to the strong enhancement of Pol II CTD phosphorylation in the presence of Mediator [51,52].
Conclusions
Transcription initiation is a crucial process in the control of gene expression, and its deregulation is linked to human disease, including cancers. The recent high-resolution structures of human TFIIH [27••,28•,41••], together with those of the human and yeast Pol II-PICs [12•,14••,17], human TFIID [53,54], and fission yeast Mediator [55], represent a breakthrough in the structural and mechanistic understanding of the Pol II transcription initiation machinery. Future studies will be needed to address possible conformational changes of PIC-bound TFIIH during nucleotide binding and hydrolysis, and to elucidate how TFIIH is affected by Pol II-PIC remodeling during transition of Pol II to transcription elongation [8•]. In its DNA repair role, TFIIH acts as the centerpiece organizing the assembly of the NER bubble. This is a multi-step process, mechanistic understanding of which will greatly benefit from structure determination of the intermediates along the way.
Acknowledgements
This work was supported by National Institute of General Medical Sciences (NIGMS) grants R01-GM63072 and P01-GM063210 to E.N.; B.J.G. was supported by fellowships from the Swiss National Science Foundation (projects P300PA_160983, P300PA_174355). E.N. is a Howard Hughes Medical Institute Investigator.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nogales E, Louder RK, He Y: Structural insights into the eukaryotic transcription initiation machinery. Annu Rev Biophys 2017, 46:59–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sainsbury S, Bernecky C, Cramer P: Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 2015, 16:129–143. [DOI] [PubMed] [Google Scholar]
- 3.Roeder RG: The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci 1996, 21:327–335. [PubMed] [Google Scholar]
- 4.Goodrich JA, Cutler G, Tjian R: Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell 1996, 84:825–830. [DOI] [PubMed] [Google Scholar]
- 5.Maxon ME, Goodrich JA, Tjian R: Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev 1994, 8:515–524. [DOI] [PubMed] [Google Scholar]
- 6.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, Sancar A, Reinberg D: Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 1994, 368:769–772. [DOI] [PubMed] [Google Scholar]
- 7.Di Lello P, Miller Jenkins LM, Mas C, Langlois C, Malitskaya E, Fradet-Turcotte A, Archambault J, Legault P, Omichinski JG: p53 and TFIIEalpha share a common binding site on the Tfb1/p62 subunit of TFIIH. Proc Natl Acad Sci U S A 2008, 105:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.•.Compe E, Genes CM, Braun C, Coin F, Egly J-M: TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription. Nat Commun 2019, 10:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biochemical characterization of Pol II-PIC remodeling events during and after transcription initiation. This study proposes that the TFIIH CAK subcomplex is released not only during NER, but also at a late stage of Pol II transcription initiation.
- 9.Compe E, Egly J-M: nucleotide excision repair and transcriptional regulation: TFIIH and beyond. Annu Rev Biochem 2016, 85:265–290. [DOI] [PubMed] [Google Scholar]
- 10.Grünberg S, Warfield L, Hahn S: Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 2012, 19:788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishburn J, Tomko E, Galburt E, Hahn S: Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc Natl Acad Sci U S A 2015, 112:3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.•.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E: Near-atomic resolution visualization of human transcription promoter opening. Nature 2016, 533:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM study defining the structures of closed, open, and initially transcribing human Pol II core-PICs at better than 4Å resolution, thus visualizing the conformational changes occurring at the transitions between these states. It provided initial insight into the overall architecture of TFIIH.
- 13.He Y, Fang J, Taatjes DJ, Nogales E: Structural visualization of key steps in human transcription initiation. Nature 2013, 495:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.••.Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, Cramer P: Structures of transcription pre-initiation complex with TFIIH and mediator. Nature 2017, 551:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM reconstruction of the yeast Pol II-PIC, including TFIIH and Mediator, elucidating the overall architecture of the Mediator-containing Pol II-PIC and providing insights into the structure of TFIIH.
- 15.Murakami K, Tsai K-L, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD: Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci U S A 2015, 112:13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svejstrup JQ, Wang Z, Feaver WJ, Wu X, Bushnell DA, Donahue TF, Friedberg EC, Kornberg RD: Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 1995, 80:21–28. [DOI] [PubMed] [Google Scholar]
- 17.Plaschka C, Hantsche M, Dienemann C, Burzinski C, Plitzko J, Cramer P: Transcription initiation complex structures elucidate DNA opening. Nature 2016, 533:353–358. [DOI] [PubMed] [Google Scholar]
- 18.Dienemann C, Schwalb B, Schilbach S, Cramer P: Promoter distortion and opening in the RNA polymerase II cleft. Mol Cell 2018:1–24. [DOI] [PubMed] [Google Scholar]
- 19.Alekseev S, Nagy Z, Sandoz J, Weiss A, Egly J-M, Le May N, Coin F: Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell 2017, 65:504–513.e505. [DOI] [PubMed] [Google Scholar]
- 20.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ: Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol 2014, 15:465–481. [DOI] [PubMed] [Google Scholar]
- 21.Li C-L, Golebiowski FM, Onishi Y, Samara NL, Sugasawa K, Yang W: Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol Cell 2015, 59:1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coin F, Oksenych V, Egly J-M: Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 2007, 26:245–256. [DOI] [PubMed] [Google Scholar]
- 23.Mathieu N, Kaczmarek N, Rüthemann P, Luch A, Naegeli H: DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Curr Biol 2013, 23:204–212. [DOI] [PubMed] [Google Scholar]
- 24.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly J-M: Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell 2008, 31:9–20. [DOI] [PubMed] [Google Scholar]
- 25.Sandrock B, Egly JM: A yeast four-hybrid system identifies Cdk-activating kinase as a regulator of the XPD helicase, a subunit of transcription factor IIH. J Biol Chem 2001, 276:35328–35333. [DOI] [PubMed] [Google Scholar]
- 26.Abdulrahman W, Iltis I, Radu L, Braun C, Maglott-Roth A, Giraudon C, Egly J-M, Poterszman A: ARCH domain of XPD, an anchoring platform for CAK that conditions TFIIH DNA repair and transcription activities. Proc Natl Acad Sci U S A 2013, 110: E633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.••.Greber BJ, Toso D, Fang J, Nogales E: The complete structure of the human TFIIH core complex. eLife 2019, 8:e44771. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM structure of free human TFIIH at 3.7Å leading to a complete model of the human TFIIH core complex, along with most of the CAK-complex subunit MAT1. The structure describes a number of inhibitory interactions that block ATP and DNA binding sites in XPD.
- 28.•.Greber BJ, Nguyen THD, Fang J, Afonine PV, Adams PD, Nogales E: The cryo-electron microscopy structure of human transcription factor IIH. Nature 2017, 549:414–417 [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM structure of free human TFIIH providing detailed atomic models for some of subunits, and allowing the mapping of disease mutations onto human TFIIH that are associated with mild transcriptional defects and impaired DNA repair.
- 29.Jawhari A, Lainé J-P, Dubaele S, Lamour V, Poterszman A, Coin F, Moras D, Egly J-M: p52 Mediates XPB function within the transcription/repair factor TFIIH. J Biol Chem 2002, 277:31761–31767. [DOI] [PubMed] [Google Scholar]
- 30.Cleaver JE, Thompson LH, Richardson AS, States JC: A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum Mutat 1999, 14:9–22. [DOI] [PubMed] [Google Scholar]
- 31.Rapin I: Disorders of nucleotide excision repair. Handb Clin Neurol 2013, 113:1637–1650. [DOI] [PubMed] [Google Scholar]
- 32.Bienstock RJ, Skorvaga M, Mandavilli BS, Van Houten B: Structural and functional characterization of the human DNA repair helicase XPD by comparative molecular modeling and site-directed mutagenesis of the bacterial repair protein UvrB. J Biol Chem 2003, 278:5309–5316. [DOI] [PubMed] [Google Scholar]
- 33.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA: XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell 2008, 133:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolski SC, Kuper J, Hänzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C: Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 2008, 6:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie A-M, Brown SE, Naismith JH, White MF: Structure of the DNA repair helicase XPD. Cell 2008, 133:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubaele S, Proietti De Santis L, Bienstock RJ, Keriel A, Stefanini M, Van Houten B, Egly J-M: Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell 2003, 11:1635–1646. [DOI] [PubMed] [Google Scholar]
- 37.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NGJ, Raams A, Argentini M, van der Spek PJ et al. : A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet 2004, 36:714–719. [DOI] [PubMed] [Google Scholar]
- 38.Botta E, Nardo T, Lehmann AR, Egly J-M, Pedrini AM, Stefanini M: Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum Mol Genet 2002, 11:2919–2928. [DOI] [PubMed] [Google Scholar]
- 39.Kainov DE, Vitorino M, Cavarelli J, Poterszman A, Egly J-M: Structural basis for group A trichothiodystrophy. Nat Struct Mol Biol 2008, 15:980–984. [DOI] [PubMed] [Google Scholar]
- 40.•.Yan C, Dodd T, He Y, Tainer JA, Tsutakawa SE, Ivanov I: Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat Struct Mol Biol 2019, 26:397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]; The complete computational model of the human Pol II-PIC based on previously published cryo-EM maps defines the contacts between the human Pol II-PIC core and TFIIH.
- 41.••.Kokic G, Chernev A, Tegunov D, Dienemann C, Urlaub H, Cramer P: Structural basis of TFIIH activation for nucleotide excision repair. Nat Commun 2019, 10:2885. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM structure of a TFIIH-XPA-DNA complex provides a second high-resolution model of human TFIIH and insight into the activation of TFIIH for nucleotide excision repair, defining the large conformational transitions accompanying this process.
- 42.Cheng K, Wigley DB: DNA translocation mechanism of an XPD family helicase. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuper J, Braun C, Elias A, Michels G, Sauer F, Schmitt DR, Poterszman A, Egly J-M, Kisker C: In TFIIH, XPD helicase is exclusively devoted to DNA repair. PLoS Biol 2014, 12: e1001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JS, Saint-André C, Lim HS, Hwang C-S, Egly J-M, Cho Y: Crystal structure of the Rad3/XPD regulatory domain of Ssl1/ p44. J Biol Chem 2015, 290:8321–8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Constantinescu-Aruxandei D, Petrovic-Stojanovska B,Penedo JC, White MF, Naismith JH: Mechanism of DNA loading by the DNA repair helicase XPD. Nucleic Acids Res 2016, 44:2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Büttner K, Nehring S, Hopfner K-P: Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 2007, 14:647–652. [DOI] [PubMed] [Google Scholar]
- 47.•.Barnett J, Kuper J, Koelmel W, Kisker C, Kad N: The TFIIH components p44/p62 act as a damage sensor during nucleotide excision repair. bioRxiv 2019:43874 10.1101/643874 [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports that p44 and p62 are involved in DNA damage sensing in NER.
- 48.Soutourina J: Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 2018, 19:262–274. [DOI] [PubMed] [Google Scholar]
- 49.Tsai K-L, Yu X, Gopalan S, Chao T-C, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW et al. : Mediator structure and rearrangements required for holoenzyme formation. Nature 2017, 544:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei P-J, Burlingame AL, Kornberg RD: Structure of a complete Mediator-RNA polymerase II pre-initiation complex. Cell 2016, 166:1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F et al. : Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 2015, 518:376–380. [DOI] [PubMed] [Google Scholar]
- 52.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD: A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 1994, 77:599–608. [DOI] [PubMed] [Google Scholar]
- 53.Patel AB, Louder RK, Greber BJ, Grünberg S, Luo J, Fang J, Liu Y, Ranish J, Hahn S, Nogales E: Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 2018: eaau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E: Structure of promoter-bound TFIID and model of human preinitiation complex assembly. Nature 2016, 531:604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nozawa K, Schneider TR, Cramer P: Core Mediator structure at3.4Å extends model of transcription initiation complex. Nature 2017, 545:248–251. [DOI] [PubMed] [Google Scholar]