Supplemental Digital Content is available in the text.
Keywords: heart rate, hypertension, machine learning, mitral valve prolapse, work flow
Background:
The ECG remains the most widely used diagnostic test for characterization of cardiac structure and electrical activity. We hypothesized that parallel advances in computing power, machine learning algorithms, and availability of large-scale data could substantially expand the clinical inferences derived from the ECG while at the same time preserving interpretability for medical decision-making.
Methods and Results:
We identified 36 186 ECGs from the University of California, San Francisco database that would enable training of models for estimation of cardiac structure or function or detection of disease. We segmented the ECG into standard component waveforms and intervals using a novel combination of convolutional neural networks and hidden Markov models and evaluated this segmentation by comparing resulting electrical intervals against 141 864 measurements produced during the clinical workflow. We then built a patient-level ECG profile, a 725-element feature vector and used this profile to train and interpret machine learning models for examples of cardiac structure (left ventricular mass, left atrial volume, and mitral annulus e-prime) and disease (pulmonary arterial hypertension, hypertrophic cardiomyopathy, cardiac amyloid, and mitral valve prolapse). ECG measurements derived from the convolutional neural network-hidden Markov model segmentation agreed with clinical estimates, with median absolute deviations as a fraction of observed value of 0.6% for heart rate and 4% for QT interval. Models trained using patient-level ECG profiles enabled surprising quantitative estimates of left ventricular mass and mitral annulus e′ velocity (median absolute deviation of 16% and 19%, respectively) with good discrimination for left ventricular hypertrophy and diastolic dysfunction as binary traits. Model performance using our approach for disease detection demonstrated areas under the receiver operating characteristic curve of 0.94 for pulmonary arterial hypertension, 0.91 for hypertrophic cardiomyopathy, 0.86 for cardiac amyloid, and 0.77 for mitral valve prolapse.
Conclusions:
Modern machine learning methods can extend the 12-lead ECG to quantitative applications well beyond its current uses while preserving the transparency that is so fundamental to clinical care.
WHAT IS KNOWN
Although computerized interpretation algorithms for ECGs have existed for decades, they have been constrained in that they aim to replicate the rules-based approach to ECG analysis used by human readers.
Conventional ECG reading approaches cannot readily account for high-level interactions between ECG signals from multiple leads, or small visually imperceptible yet informative changes which may exist in the signal, particularly in early disease stages.
WHAT THE STUDY ADDS
Using a combination of machine learning methods, including convolutional neural networks and hidden Markov models, we have developed an automated, scalable, interpretable method to perform detailed longitudinal tracking and comparison of ECGs.
As demonstration examples, we indicate how a personalized ECG vector profile can be used to estimate continuous measures of cardiac structure and function such as left ventricular mass and mitral annular e′ velocity.
We also use the ECG vector to train models to detect and track diseases such as cardiac amyloid, hypertrophic cardiomyopathy, and pulmonary arterial hypertension.
See Editorial by Kao
The ECG is the most commonly performed cardiovascular diagnostic procedure, with >100 million ECGs obtained annually in the United States,1 including use in 21% of annual health examinations2 and 17% of emergency department visits.3 The ECG tracing contains a large amount of information that directly reflects underlying cardiac physiology since its morphological and temporal features are produced from cardiac electrical and structural variations. However, the existing techniques physicians use to interpret ECGs4—using sets of rules that were initially established by empirical, manual review of ECGs from disease cohorts5—consider only a fraction of the total information available in the ECG.
Although computerized interpretation algorithms for ECGs have existed for decades,6 they have been constrained in that they aim to replicate the rules-based approach to ECG analysis used by human readers. Moreover, the prevailing policy, whether performed by humans or algorithms, aims to detect the presence or absence of disease (eg, left ventricular hypertrophy [LVH] or not), evaluating fairly simple criteria on only a small subset of the total information contained in the ECG, such as how the height of the R wave in lead aVL exceeding 11 mm suggests LVH.7 Such an approach cannot readily account for high-level interactions between ECG signals from multiple leads, or small visually imperceptible yet informative changes which may exist in the signal, particularly in early disease stages.
Given the physiological and structural correlates of ECG signals, we hypothesized that a modern algorithmic approach could be used to expand the clinical inferences derived from ECGs. Algorithmic analysis should aspire to estimate continuous attributes of cardiac disease and structure, as well as capture change in these attributes over time. Moreover, such an analysis should inform the clinician which components of the ECG signal are responsible for any given diagnosis,8 thus providing the algorithmic transparency needed to reassure physicians and patients about the basis and possible validity of resulting automated diagnosis. Although several recent efforts have applied machine learning techniques to ECG analysis,9,10 most of these innovative strategies suffer from being largely uninterpretable. In high-stakes fields such as medicine, this limits the ability to understand successes or troubleshoot failures, potentially dampening physician adoption of an unfamiliar technology. Presently, there does not exist an automated, scalable, interpretable method to perform detailed longitudinal tracking and comparison of ECGs.
In this work, we develop and test an algorithmic framework that facilitates scalable analysis of ECG data while preserving interpretable parallels to cardiac physiology. We demonstrate this framework on several examples of cardiac structure and disease, although we maintain that this approach can be used broadly across the spectrum of cardiac abnormalities. This approach aspires to expand the flexibility and scalability of algorithmic ECG analysis, laying the foundation to perform a wide range of novel ECG-based tasks, including improving accuracy, estimating quantitative cardiac traits, performing longitudinal tracking of serial ECGs, and monitoring disease progression and risk.
Materials and Methods
The source code for this project, including model weights, is available at https://bitbucket.org/rahuldeo/ecgai/.
Human Subjects Research
The University of California, San Francisco (UCSF) Institutional Review Board approval was obtained for this study.
Overview: Automated and Interpretable ECG Profiling for Disease Detection, Tracking, and Discovery
We sought to develop an automated, scalable, and interpretable method to characterize (1) cardiac structure, (2) diastolic function; and (3) detect and track disease using patient-specific ECG profiles. Figure 1 demonstrates the analysis pipeline, data inputs, and number of ECGs that were used in each step of algorithm development and validation. We termed the entire approach as ecgAI—referring to artificial intelligence.
Figure 1.
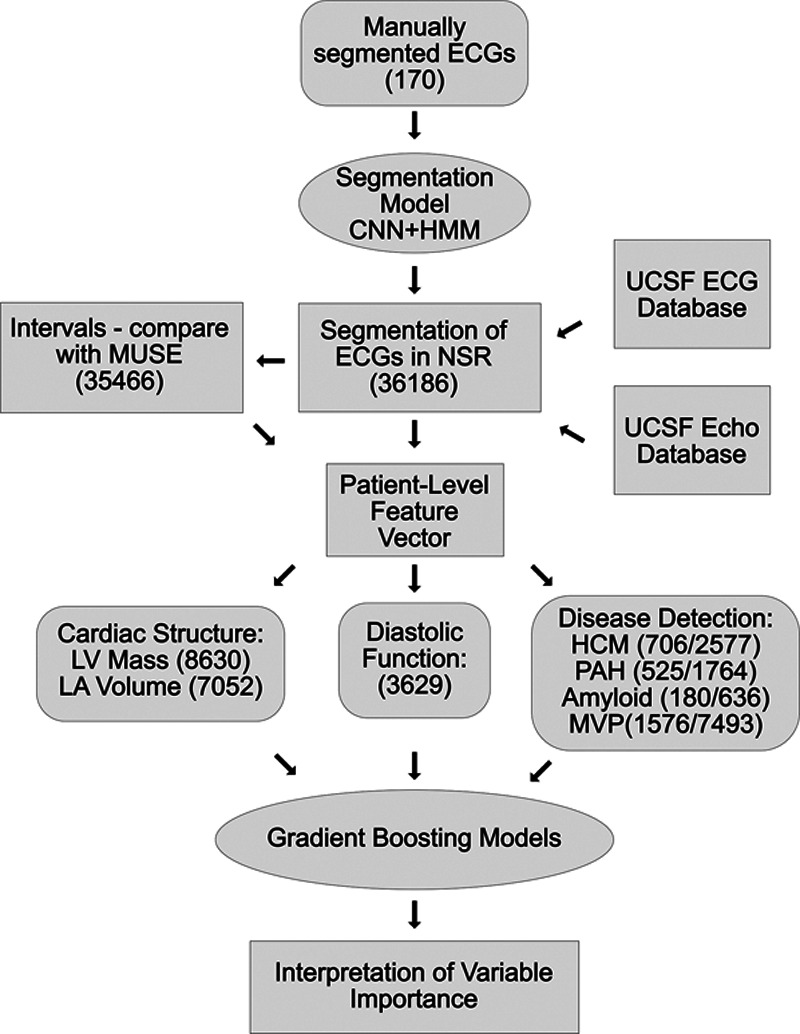
Workflow for ecgAI project. The workflow consisted of training an ECG segmentation model and using a selected group of ECGs to train interpretable models to estimate cardiac structure and function and detect and track disease. Concordance with measurements from the GE MUSE system was used for validation of segmentation as well as a filter for segmentation quality. The number of ECGs used for the various tasks is indicated in parentheses. For disease detection, a slash separates the number of cases and control ECGs used. Curved rectangles represent training data; ellipses represent algorithms; and standard rectangles represent other data types. CNN indicates convolutional neural network; HCM, hypertrophic cardiomyopathy; HMM, hidden Markov model; MVP, mitral valve prolapse; NSR, normal sinus rhythm; PAH, pulmonary arterial hypertension; LA, left atrium; and LV, left ventricle.
ECG Data
We selected a subset of ECGs from the UCSF ECG database to train models for estimation of cardiac structure and function and detection of disease. Standard 12-lead ECG data from 2010 to 2017 was obtained in XML format from the UCSF clinical MUSE ECG database (MUSE Version 9.0 SP4, GE Healthcare, Wauwatosa, WI). Based on the presence of concurrent clinical and echocardiographic information which was used for cardiac structure and disease correlations (described below), we selected 36 186 ECGs from the UCSF database, each of which included the standard 10 seconds of raw ECG voltage data across each of the 12 individual leads; 60% of data had been sampled at a frequency of 500 Hz, and 40% had been sampled at 250 Hz. As part of routine clinical care, each clinical ECG undergoes initial analysis by the GE software (MAC 5500 HD, Version 10, Revision F; Marquette 12SL; GE Healthcare, Wauwatosa, WI), and the interpretation is subsequently changed or confirmed by a UCSF cardiologist. We extracted standard ECG MUSE measurements, as well as final cardiologist-confirmed ECG diagnostic interpretations. Data from the UCSF electronic health record were obtained for relevant patients, including medical diagnoses, medications, specialty clinic referrals, and echocardiographic measurements.
Selection of Studies for Model Development
To facilitate model development, we restricted the analyses to those ECGs for which the GE/UCSF rhythm interpretation was normal sinus rhythm.
For cardiac structure models, we searched the UCSF echocardiographic database for all instances of patients with echocardiograms and ECGs collected within 30 days of one another and who had recorded measurements either of left ventricular mass or left atrial volume. We found 10 082 (Table I and Figure I in the Data Supplement) and 8289 (Table II and Figure II in the Data Supplement) studies, respectively, that met these criteria. For cardiac diastolic function, we performed a similar search and found 4205 instances of patients with an ECG and a recorded mitral annulus medial e′ value on echocardiographic within 30 days of each other (Table III and Figure III in the Data Supplement). There were fewer instances of lateral e′ values recorded within our database and we thus focused our efforts on the medial e′ metric.
We selected 4 diseases for which to perform a clinical demonstration of automated detection and tracking of disease using patient ECG profiles: pulmonary arterial hypertension (PAH), hypertrophic cardiomyopathy (HCM), cardiac amyloidosis (CA), and mitral valve prolapse (MVP). We previously identified the PAH, HCM, and CA patients as part of a parallel study on developing a computer vision pipeline for automated echocardiographic interpretation.11 Briefly, on chart review HCM patients met guideline-based criteria12 (Figure IV in the Data Supplement); CA patients had both echocardiographic evidence of hypertrophy and confirmation of amyloidosis by biopsy or imaging (Figure V in the Data Supplement); and PAH patients had an echocardiographic-indication of PAH and were on one of 4 PAH specific medications (Figure VI in the Data Supplement). Patients with MVP were identified by querying the UCSF echocardiographic database for patients with single or bileaflet MVP (Figure VII in the Data Supplement). Echocardiographic studies were subsequently over-read by a second board-certified cardiologist to confirm the diagnosis. We selected all ECGs corresponding to these patients that were available in XML format. To build classification models, we also matched each ECG to up to 5 ECGs matched by age (in 10 years bins), sex, year of study, and race (the patient demographic information for ECGs in our archive has been organized in a python dictionary to facilitate the control selection process). Patient and study characteristics are described in Tables IV through VII in the Data Supplement.
A Novel Machine Learning-Based Approach to ECG Segmentation
To develop novel models with the goal of expanding ECG clinical utility, we needed an efficient way to derive patient-specific ECG profiles consisting of vectors of uniform length that capture the variation in ECG voltage over different leads. This first required a method to segment ECGs into their standard, interpretable components.
We first trained a convolutional neural network (CNN)-based model to delineate individual segments within the ECG. As training data, we downloaded raw ECG voltage data from 2 sources: 112 ECGs from the Physikalisch-Technische Bundesanstalt diagnostic database13 and 58 ECGs from the UCSF database. For each ECG, we extracted a 2-second strip and manually assigned to each 1 ms block 1 of 6 possible labels: P wave, PR segment (termination of P wave to start of QRS), QRS complex, ST segment, T wave, and TP segment. Before manual labeling, we performed linear interpolation so that, regardless of the initial ECG sampling frequency, 2 seconds corresponded to an input vector of length 2000.
We then trained a multilayered neural network to detect these segments within an ECG. The architecture of our network was based on the U-net network14 (Figure 2A). Our network accepted a 12×2000 input array and was composed of sequential contracting and expanding paths with a total of 32 convolutional layers, 5 max pool layers, and 3 deconvolutional layers. The output of this CNN is a 6×2000 array of ECG segment classes, identical in length to the input vector, and including a probability for each potential segment label at each position along the ECG trace (Note I in the Data Supplement).
Figure 2.
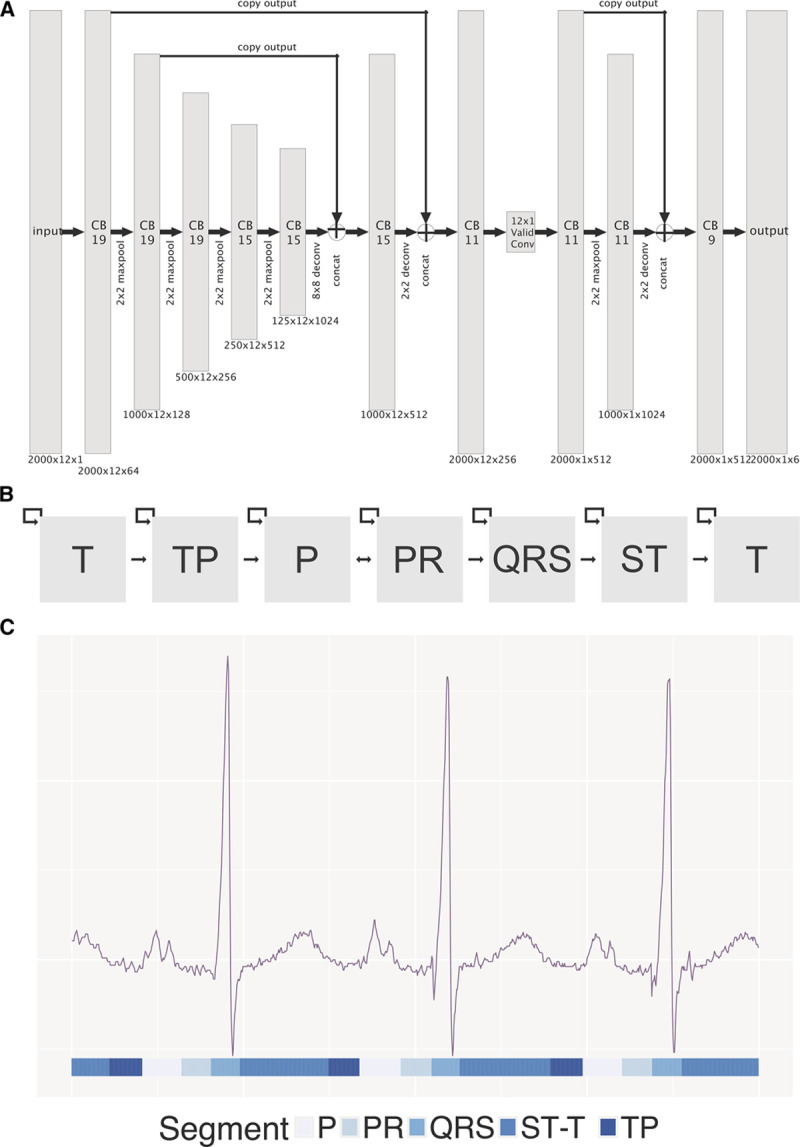
ecgAI method of ECG segmentation. A, Architecture of convolutional neural network used for ECG segmentation. Gray rectangles represent layers with dimensions listed below. The notation for each layer indicates the size of the input (eg, 2000 ms, initially) by the number of leads (eg, 12) by the number of filters. The size of the filter is specified in the body of the rectangle. B, Architecture of hidden Markov model (HMM) used after convolutional neural network (CNN) based segmentation. Gray boxes represent states that are traversed in order in the ECG. C, Example of CNN-HMM output for an ECG. CNN-HMM based classes are shown below the image. The ST and T wave segments have been combined.
Although U-Nets can provide accurate segmentation of objects, they fail to take advantage of the obligate ordering of elements in a typical ECG. We thus trained a hidden Markov model (HMM) to accept the output of the U-Net and provide improved segmentation.15 As a final step, we introduced a simple heuristic filter to eliminate implausibly short ECG complexes (ie, <10 ms), which was a consistent hallmark of poor HMM performance. The combined CNN-HMM–heuristic filter pipeline was run on all ECGs. ECG segmentation was validated by comparison to manual segmentation and by comparison against measurements a total of 141 864 measurements derived from the GE muse software for 35 466 ECGs (Note II in the Data Supplement). These ECGs were primarily selected for the various classification and estimation tasks related to cardiac structure, function, and disease detection described below and so thus may be biased towards more challenging cases.
Deriving Patient-Level ECG Profiles
Because ECG waveforms and intervals have corollaries to electrical and structural cardiac physiology, a crucial principle to our approach aimed to create a representation of the raw ECG data which preserves these features while still decreasing the feature space, making it tractable for analysis by interpretable machine learning algorithms. To achieve this, we developed a 725-component ECG vector representation derived from segments of the CNN/HMM-segmented ECG. For 3 ECG segments—the PR interval, the QRS complex, and the ST-T-wave complex (including both the ST segment and the T wave)—the vector of raw-voltage amplitude from each of the 12 leads was resized to 20 samples by linear interpolation, averaged across all cardiac cycles, and included as ECG vector components (totaling 720 components). The PR interval, P wave duration, QRS interval, heart rate, and QT intervals were calculated based on the segmentation boundaries and averaged across all cardiac cycles and across 12 leads, and the 5 averaged values were included as 5 additional components in the ECG vector—resulting in a total of 725 components.
ECG-Derived Estimates of Cardiac Structure and Function
The 725-component ECG patient vector can be used as an input to train models to estimate a variety of cardiac structure and functional estimates. In this article, we examined the ability to estimate continuous measurements of left ventricular mass (indexed for body surface area), left atrial volume (indexed), and mitral annular medial e′ (medial e′) as demonstration examples. Anticipating complex interactions among input features, as well as heterogeneity among patients,16 we used a machine learning algorithm known as a Gradient Boosted Machine (GBM),17 which is an ensemble regression-tree based technique. Individual GBM models were trained to estimate the 3 continuous structure and function metrics. We also generated dichotomous measures for each of these, treating controls as individuals with values below (for left ventricular mass indexed and left atrial volume indexed) or above (medial e′) the median value, and cases as individuals above or below the tenth percentile (Tables VIII through X in the Data Supplement). Given that we noted occasional inaccuracy in both our CNN-HMM segmentation model as well as in the MUSE values, we limited our models to ECGs with substantial agreement (mean difference <10%) across the RR, PR, QRS, and QT intervals. This filter merely served as a quick check on the quality of our segmentation, which is essential to building an accurate phased patient profile vector. There was no appreciable difference in patient characteristics for this subset (Tables I through III in the Data Supplement). Models were fit using the GBM function in the R caret package. Tuning parameters were selected in an automated manner using 3-fold cross validation.
Accuracy was assessed using 5-fold cross validation, with area under the receiver operating characteristic curve (AUROC) curves used to evaluate classification tasks and absolute differences (50th, 75th, and 95th percentiles) and Bland-Altman18 plots used for continuous measures. Variable importance was extracted for each of the 725 features and averaged over cross-validation runs. To facilitate interpretation of variable importance rankings, we binned the variable importance scores for the lead-specific voltage values so that each segment (eg, QRS) was represented by 5 rather than 20 bins. We note that there is still redundancy between voltages in highly similar leads (eg, leads I and aVL), but we elected not to bin across different leads. Overall, the redundancy in voltages within and across leads may reduce the variable importance of these features compared with minimally redundant measures such as the ECG intervals.
Disease Detection and Tracking
In addition to quantifying cardiac structure, we also trained GBM models to detect PAH, HCM, CA, and MVP. Separate GBM models were trained to output a probability for each disease based on an input ECG vector using a similar approach as outlined above for structure and function. We also assessed the discriminative ability of conventional features, such as maximum voltage in aVL (for HCM) and maximum voltage in lead V1 (for PAH). To demonstrate the use of this approach to track longitudinal changes in disease over time, we selected all patients who had ECGs in 2 or more years and took the median score per patient for each year. Scores were plotted as a function of year.
Statistical Methods
All analyses were performed using R 3.3.2 or Python 2.7. Differences between case and control characteristics for the disease detection models were performed using 2-tailed Wilcoxon-Mann-Whitney, t, or χ2 tests. Only a single value was taken per patient in these pairwise comparisons. The AUROC for disease detection models were computed using held-out values from 5-fold cross validation with the help of the pROC and hmeasure packages in R. CIs for the AUROC were generated by the nonparametric method of Delong,19 as implemented in the pROC package. The only predictor for these models was the patient-level disease score, as output by the GBM model.
CNNs were developed using the TensorFlow Python package.20 Signal manipulation (such as linear interpolation for resizing) was performed using scikit-image.21
Results
Validation of ecgAI Machine Learning-Based ECG Segmentation
Our ecgAI segmentation pipeline (Figure 2A and 2B) was trained on 170 manually segmented ECGs and deployed on 36 186 sinus rhythm ECGs (Figure 1). ECG segmentation forms the basis for subsequent steps in the ecgAI pipeline and example output from the ecgAI segmentation model is shown in Figure 2C, with every time-step along the ECG tracing being classified as belonging to one of the 6 segments (illustrated in the Figure by separate colors). The ecgAI segmentation performed reasonably well versus manual annotations as demonstrated by the IoU metrics of 91±3 (P wave), 85±2 (PR segment), 94±4 (QRS complex), 88±3 (ST segment), 91±3 (T wave), and 92±5 (TP segment). As a second indirect validation of segmentation performance, standard ECG interval measurements were calculated based on ecgAI segmentation on 35 466 ECGs not included in the training set and compared against the reference MUSE values (Tables XI and XII in the Data Supplement), overall demonstrating good agreement with MUSE calculated intervals. Intervals from ecgAI-measurements demonstrated a strong correlation with those from MUSE (ρ=0.77–0.98, Figure 3; Figure VIII in the Data Supplement). The HMM and, to a lesser extent, the heuristic filter, contributed substantially to the accuracy of interval estimation (Table XIII in the Data Supplement).
Figure 3.

Comparison of ecgAI (hidden Markov model [HMM]+convolutional neural network [CNN]) derived measurements and MUSE/University of California, San Francisco (UCSF) values for 4 commonly reported ECG measurements. The scatterplot depicts 35 466 comparisons. The line y=x is drawn to help identify any bias. The unit for heart rate is beats per minute while that of the other 3 metrics is milliseconds.
ecgAI Performance to Quantify Cardiac Structure and Function
In contrast to the binary detection of cardiac structural diagnosis on ECG using existing methods, the ecgAI approach enables estimation of the severity of structural abnormalities using continuous metrics. For the 3 demonstration examples of cardiac structural and function, we used echocardiographic measurements for training and validation. Median absolute deviation of ecgAI predictions against reference echocardiographic measurements varied by structure: the lowest deviation was for left ventricular mass indexed (16.5%), intermediate deviation was for mitral annulus medial e′ (19.1%), and the greatest deviation was for left atrial volume indexed (22.9%; Table XI in the Data Supplement). For all 3 structural measurements, there was a tendency to overestimate low values and underestimate high values (Figure 4A and 4B; Figure IX in the Data Supplement), suggesting a more limited dynamic range for ECG compared with echocardiography. When the continuous measurements for the cardiac structures were dichotomized, the model demonstrated strong discrimination for both LVH and diastolic dysfunction with AUROCs of 0.87 (95% CI, 0.86–0.89) and 0.84 (95% CI, 0.82–0.86), respectively (Figure 4C and 4D). Left atrial enlargement had a much lower AUROC of 0.62 (95% CI, 0.60–0.64), most likely reflecting a failure of the ECG to correctly estimate large atrial volumes (Figure IX in the Data Supplement).
Figure 4.
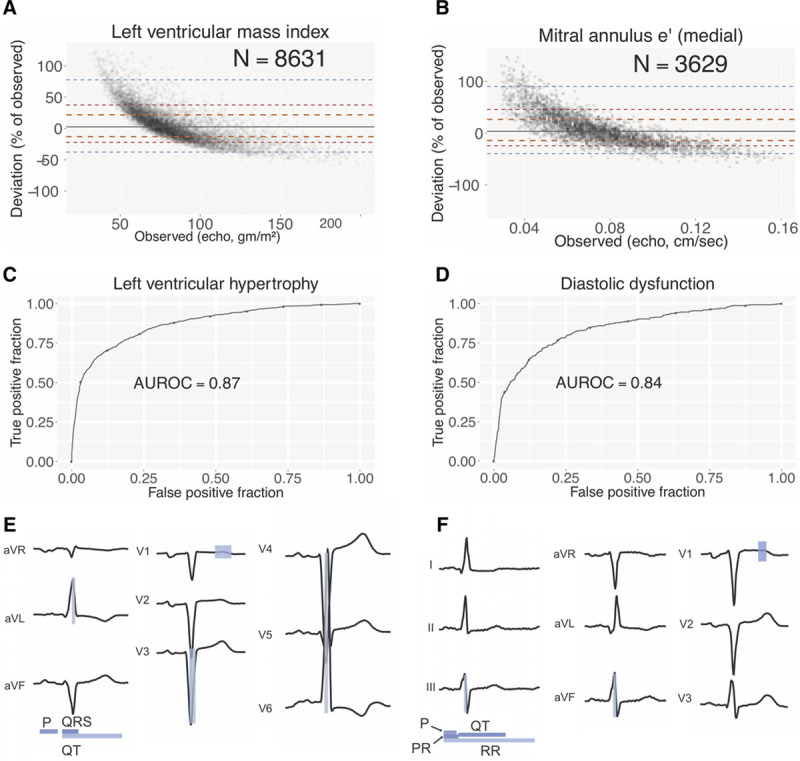
Estimating cardiac structure and function using patient-level ECG profiles. Bland-Altman plots comparing estimation of left ventricular mass index (LVMi; A) and mitral annulus medial e′ (B) values using ECG alone compared with echo-derived values. Number of studies depicted in comparison is shown. Orange, red, and blue dashed lines delineate the central 50%, 75%, and 95% of patients, as judged by difference between automated and manual measurements. The solid gray line indicates the median. Receiver operating characteristic (ROC) curves for classification models for left ventricular hypertrophy (C) and diastolic dysfunction (D). The area under the ROC curve (AUROC) is indicated. Variable importance for LVMi (E) and mitral annulus e′ (F) estimation models. The predictors most important for each model are highlighted with the relative importance indicated by the shading (white to blue). Informative intervals are depicted below the plot while lead-specific segments of the ECG are highlighted on the voltage trace.
The ecgAI approach also enables the identification of which ECG components (waveform voltages and intervals from the 725-component patient-level ECG profile) most strongly contributed to classification for each cardiac structural abnormality (Figure 4E and F; Table XIV in the Data Supplement). For left ventricular mass index, QRS duration was the strongest predictor with a variable score of 4.0, followed by P wave duration (3.3), QT duration (1.7), the middle portion of the QRS from lead V3 (1.5, segments 8–12 out of a total of 20), and the middle portion of the ST-T complex from lead V1 (1.3, segments 12–16; Figure 4E; Table XIV in the Data Supplement). Collectively, these reflect many of the classic criteria for LVH,7 while also highlighting potential new ECG-based predictors of LVH.
For medial e′ the strongest ECG predictors were PR duration (3.1), QT duration (2.9), P wave duration (2.4), the middle portion of the ST-T complex from lead V1 (1.8, segments 8–12), and heart rate (1.2). For left atrial volume indexed, top predictors were QT duration (4.6), P wave duration (4.5), QRS duration (1.4), PR duration (1.3), and the middle portion of the QRS from lead V6 (0.97).
ecgAI Performance for Cardiac Disease Detection
In addition to quantifying cardiac structure, we applied ecgAI toward classification of 4 example diseases, accompanied by the discovery of ECG predictors for each disease (Figure 5; Figure XI in the Data Supplement for precision-recall curves). The strongest discrimination was observed for a model for PAH, which had an AUROC of 0.94 (95% CI, 0.93–0.95). Key predictors for PAH included the middle portion of QRS from lead V1 (variable score =4.5, segments 8–12), reflecting a tall R′ (Figure 5G, P<2×10−16), followed by the latter and middle portions of the QRS from lead V1 (1.6, segments 12–16; 1.4 segments 12–16), reflecting a deep S wave; and the early portion of the P-PR complex from lead V3 (0.9, segments 4–8) and aVR (0.9, segments 4–8), presumably reflecting right atrial enlargement (Figure 5A and 5B; Table XV in the Data Supplement). The GBM model for PAH was better than one constructed solely using the maximum height of the QRS complex in V1 (as a proxy for right ventricular hypertrophy), which yielded an AUROC of 0.77 (95% CI, 0.73–0.78).
Figure 5.

Detecting disease using patient-level ECG profiles. Receiver operating characteristic (ROC) curves (with area under the ROC curve [AUROC] indicated) for disease detection models for pulmonary arterial hypertension (PAH; A), hypertrophic cardiomyopathy (HCM; C), and cardiac amyloid (CA; E). Corresponding variable importance plots (B, D, F, and H) with coloring as in Figure 4. Violin plots indicating distribution of the top predictive feature in cases and controls for PAH (G), HCM (H), and CA (I). Precision-recall curves are depicted in Figure II in the Data Supplement.
HCM had the next strongest discrimination with an AUROC of 0.91 (95% CI, 0.90–0.92). The strongest predictors of HCM were the latter portion of the ST-T complex from lead V1 (3.8, segments 12–16), which can be markedly deeper in some patients with HCM (Figure 5H, P<2×10−16), the P wave duration (3.5), QT duration (2.7), PR duration (2.4), and the middle portion of the QRS from lead aVR (1.3, segments 12–16; Figure 5C and 5D; Table XV in the Data Supplement). The GBM model for HCM was superior to one constructed solely using the maximum height of the QRS complex in lead aVL (as a proxy for LVH), which yielded an AUROC of 0.69 (95% CI, 0.67–0.71).
CA had an AUROC of 0.86 (95% CI, 0.82–0.89), and the strongest predictors in this model were the early portion of the QRS from lead aVR (3.0, segments 4–8), which is blunted in voltage in CA patients (Figure 5I, P=3×10−7), QRS duration (1.3), the middle and early portions of the QRS from lead I (1.2, segments 8–12; 1.1, segments 4–8), and the earliest portion of the QRS from lead V1 (1.1, segments 0–4; Figure 5E and 5F; Table XV in the Data Supplement).
The MVP showed the weakest discrimination, with an AUROC of 0.77 (95% CI, 0.76–0.78; Figure III in the Data Supplement), a disease not known to strongly impact ECG morphology. The top predictors for MVP included PR duration (3.3), the early portion of the QRS from lead V2 (1.2, segments 4–8), the earliest portion of the QRS from lead V3 (1.2, segments 0–4), P wave duration (1.1), and QT duration (0.97; Figure X and Table XV in the Data Supplement).
Serial ECGs Analysis With ecgAI to Perform Within-Patient Disease Tracking
By applying ecgAI to serial ECGs of patients with PAH, we obtained a progression of scores over time corresponding to the degree to which the model estimated likelihood of PAH based on ECG features (Figure 6A). The dashed blue line represents the PAH score at which PAH is identified with 80% sensitivity and 90% specificity. Patients typically have scores that remain with a narrow range, but there are some exceptions—and we highlight the 3 most prominent ones. Figure 6B shows a time course of ECG tracings for the individual depicted by the purple trajectory in Figure 6A). In 2010 and 2011, ECG tracings do not have any marked abnormalities. In 2015 and 2017, ECG tracings appear increasingly abnormal, with a prominent R wave and T wave inversion in lead V1, and QRS changes and a tall prominent P wave in lead I. These progressive ECG changes over time correspond with the increasing PAH scores from 2010 to 2017.
Figure 6.

ECG-profile based models can be used to track changes in patient ECGs in pulmonary arterial hypertension (PAH). A, Scores for PAH detection for individual patients with measurements for 2 or more years. A median of all scores for each year is computed, and lines are drawn connecting scores for different years for each patient. The blue dashed line indicates a score threshold with 90% specificity and 80% sensitivity for a diagnosis of PAH. Purple, red, and yellow lines highlight score trajectories for 3 patients with dramatic variation in scores, crossing this threshold. B, Variation in ECG patterns for leads I and V1 from 2010 to 2017 for patient highlighted in purple in A. Over time, as the PAH score increases, the QRS axis swings progressively rightward (lead I), the P wave height grows, and the R′ wave in lead V1 increases in size.
Two other patients (trajectories colored in red and yellow) had precipitous decreases followed by subsequent increases in score (Figure XII in the Data Supplement). In both cases, the ECG tracings from the high PAH score year appear abnormal, featuring prominent R waves in V1 and a more negatively directed QRS vector in lead I. In contrast (and for unclear reasons), the subsequent low PAH score ECG tracings for both individuals appear substantially different and more normal, with a decrease in R wave prominence in V1 and normalization of the QRS in lead I (Figure XIIB and XIIC in the Data Supplement). The GBM PAH score thus tracks well with visible morphological change in the ECG.
Discussion
There has been a dramatic increase in publications applying machine learning methods to perform routine diagnostic tasks in medicine. Most of these have emphasized matching or even outperforming practicing physicians, whether it be for interpreting retinograms,22 skin disorders,23 bone x-rays,24 or heart rhythm abnormalities.24 Here, we outline a machine learning approach to ECG interpretation that differs crucially from these prior works in emphasizing 3 facets which are crucial when applying machine learning toward medical applications: (1) the use of machine learning to extend the utility of a diagnostic tool to applications beyond what would be possible by human readers; (2) the focus on eliciting interpretable features which can be used to both justify an automated diagnosis within clinical care and inspire new research on physiological correlates of disease; and (3) the demonstration of a flexible framework that permits estimation or classification for a broad range of cardiac metrics and diseases. Our machine learning approach not only outperforms existing rule-based binary diagnostic criteria for ECG diagnosis against which it was compared but also it more importantly expands the utility of the ECG as a clinical tool beyond present human capability. We demonstrate the ability to estimate continuous metrics of cardiac structure and function while also performing disease detection and longitudinal tracking of predicted disease. We think this constitutes a new paradigm in ECG analysis by expanding the clinical inferences that can be drawn from an ECG, increasing its potential utility to novel applications.
We think that the enormous potential of applying machine learning to medicine must lie in its ability to illuminate patterns across large quantities of data in a way that preserves clinical interpretability, both to maintain physician and patient agency in decision-making and to enable knowledge discovery. In the case of ECG-disease correlates, there is considerable evidence that previously recognized ECG predictors represent only a fraction of informative features of any disease,25,26 making the case for data-driven discovery of novel ECG correlates, which our approach uniquely enables. Most prior disease-focused studies have highlighted the association of various ECG features with disease status, rather than describing the global discrimination performance24,27–30—limiting our ability to directly compare our performance. In work bearing the most similarity to ours,24 the investigators used various ECG features to identify HCM patients, however, their focus remained the optimization of predictive performance rather than enabling clinical interpretability. In our study, the ECG-based features identified as most strongly contributing to prediction for each disease have clear physiological parallels—such as ECG correlates of right ventricular hypertrophy in PAH and myocardial infiltration in CA—which conforms to our expectations based on pathophysiology, and increases confidence in and acceptance of model performance. Furthermore, the novel predictors identified by our models may provide inroads into future investigation.
Although we used a machine learning approach to segment ECGs in this work, other methods, including any of the existing heuristic-based segmentation algorithms, could also be used to derive patient-level ECG profiles.31 A limitation of our machine learning ECG segmentation pipeline is that they are currently only optimized to analyze ECGs in normal sinus rhythm. Similarly, we limited our cardiac disease and structure models to segmented ECGs with substantial agreement with clinical measurements. While these limitations of our existing pipeline would need to be optimized before clinical deployment, we made these decisions to demonstrate the performance of our approach of using ECG profiles for this proof of concept. An additional limitation is that our data, although large in scale, is derived from a single medical center.
Our primary intended applications of this work all relate to augmenting clinical practice, rather than replacing what is already performed by skilled practitioners. Patients with uncommon diseases, such as PAH, HCM, and cardiac amyloidosis, all of which have approved or emerging therapies, would benefit from early detection and referral to a specialty center. Combining low-cost testing—potentially even with mobile ECG devices—with an automated detection algorithm can help recognize and triage these individuals. Of course, criteria such as precision (ie, positive predictive value) and more broadly, a decision analysis regarding the costs of a false positive or negative result, should come into play when evaluating the viability of any such approach.31 The ECG is also currently not used as a quantitative detection of disease progression in an individual, although several studies suggest this is feasible.32–34 Our approach provides an additional method by which to achieve quantitative tracking of disease progression which benefits from being automatic and not limited to predefined disease criteria. ECG features that co-occur with hypertension35 and even obesity,36 diabetes mellitus,37 coronary artery disease,38 and aging39 may occur at variable rates in different individuals and artificial intelligence-assisted monitoring of ECG features using our approach may provide a low-cost noninvasive window into systemic processes that can be slowed with existing or emerging therapies.40 Such rates of minute change in ECG tracings would otherwise be challenging to assess with the human eye and will need automated systems for the development and the validation of quantitative models.
Such quantitative patient tracking using the output of multidimensional models is not performed routinely for ECGs or even echocardiographic, in part, because of long-standing fears that it might obscure the diagnostic process.41,42 With the current widespread availability of digital data, we strongly believe such concerns should be revisited, both for the benefit of the physician and patient. To this end, a primary motivation of this work is to demonstrate how we can extract much more knowledge from our current low-cost input data, all in an automated manner, and yet remain transparent to physicians, patients, and researchers about the provenance of these insights.
Acknowledgments
We acknowledge the assistance of Edward Mahoney and John Cooper in accessing records from the MUSE server. We thank Dr Pulkit Agrawal for advice on the ECG segmentation model.
Sources of Funding
The funding for this work was through National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) DP2 HL123228 to Dr Deo and NIH/NHLBI K23HL135274 to Dr Tison.
Disclosures
None.
Supplementary Material
Footnotes
Dr Tison and J. Zhang contributed equally to this work.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCOUTCOMES.118.005289.
References
- 1.Drazen E, Mann N, Borun R, Laks M, Bersen A. Survey of computer-assisted electrocardiography in the United States. J Electrocardiol. 1988;21(suppl):S98–104. doi: 10.1016/0022-0736(88)90068-4. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia RS, Bouck Z, Ivers NM, Mecredy G, Singh J, Pendrith C, Ko DT, Martin D, Wijeysundera HC, Tu JV, Wilson L, Wintemute K, Dorian P, Tepper J, Austin PC, Glazier RH, Levinson W. Electrocardiograms in low-risk patients undergoing an annual health examination. JAMA Intern Med. 2017;177:1326–1333. doi: 10.1001/jamainternmed.2017.2649. doi: 10.1001/jamainternmed.2017.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitts SR, Niska RW, Xu J, Burt CW. National hospital ambulatory medical care survey: 2006 emergency department summary. Natl Health Stat Report. 2008;7:1–38. [PubMed] [Google Scholar]
- 4.Schläpfer J, Wellens HJ. Computer-interpreted electrocardiograms: benefits and limitations. J Am Coll Cardiol. 2017;70:1183–1192. doi: 10.1016/j.jacc.2017.07.723. doi: 10.1016/j.jacc.2017.07.723. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn H, Keys A, Simonson E, Rautaharju P, Punsar S. The electrocardiogram in population studies. A classification system. Circulation. 1960;21:1160–1175. doi: 10.1161/01.cir.21.6.1160. doi: 10.1161/01.cir.21.6.1160. [DOI] [PubMed] [Google Scholar]
- 6.Pipberger HV, Stallman FW, Berson AS. Automatic analysis of the P-QRS-T complex of the electrocardiogram by digital computer. Ann Intern Med. 1962;57:776–787. doi: 10.7326/0003-4819-57-5-776. doi: 10.7326/0003-4819-57-5-776. [DOI] [PubMed] [Google Scholar]
- 7.Casale PN, Devereux RB, Kligfield P, Eisenberg RR, Miller DH, Chaudhary BS, Phillips MC. Electrocardiographic detection of left ventricular hypertrophy: development and prospective validation of improved criteria. J Am Coll Cardiol. 1985;6:572–580. doi: 10.1016/s0735-1097(85)80115-7. doi: 10.1016/s0735-1097(85)80115-7. [DOI] [PubMed] [Google Scholar]
- 8.Lipton ZC. The mythos of model interpretability. Communications of the ACM. 2018;61:36–43. [Google Scholar]
- 9.Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Friedman PA. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med. 2019;25:70–74. doi: 10.1038/s41591-018-0240-2. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 10.Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, Lassen MH, Fan E, Aras MA, Jordan C, Fleischmann KE, Melisko M, Qasim A, Shah SJ, Bajcsy R, Deo RC. Fully automated echocardiogram interpretation in clinical practice. Circulation. 2018;138:1623–1635. doi: 10.1161/CIRCULATIONAHA.118.034338. doi: 10.1161/CIRCULATIONAHA.118.034338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;58:e212–e260. doi: 10.1016/j.jacc.2011.06.011. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. Physio bank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. doi: 10.1161/01.cir.101.23.e215. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 14.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks For Biomedical Image Segmentation. Accessed April 1, 2017. https://arxiv.org/abs/1505.04597.
- 15.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–286. [Google Scholar]
- 16.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–1232. [Google Scholar]
- 18.Bland JM, Altman DJ. Regression analysis. Lancet. 1986;1:908–909. doi: 10.1016/s0140-6736(86)91008-1. doi: 10.1016/s0140-6736(86)91008-1. [DOI] [PubMed] [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 20.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yuan Y, Zheng X. Tensorflow: a system for large-scale machine learning. OSDI. 2016;16:265–283. [Google Scholar]
- 21.van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T scikit-image contributors. Scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 23.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90. [Google Scholar]
- 24.Rahman QA, Tereshchenko LG, Kongkatong M, Abraham T, Abraham MR, Shatkay H. Utilizing ECG-based heartbeat classification for hypertrophic cardiomyopathy identification. IEEE Trans Nanobiosci. 2015;14:505–512. doi: 10.1109/TNB.2015.2426213. doi: 10.1109/TNB.2015.2426213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Z, Wu M, Fu Y, Huang T, Wang T, Sun Y, Feng Z, Li C. Palmitic acid curcumin ester facilitates protection of neuroblastoma against oligomeric Aβ40 insult. Cell Physiol Biochem. 2017;44:618–633. doi: 10.1159/000485117. doi: 10.1159/000485117. [DOI] [PubMed] [Google Scholar]
- 26.Nahar J, Imam T, Tickle KS, Chen YP. Computational intelligence for heart disease diagnosis: a medical knowledge driven approach. Expert Syst Appl. 2013;40:96–104. [Google Scholar]
- 27.Cheng Z, Zhu K, Tian Z, Zhao D, Cui Q, Fang Q. The findings of electrocardiography in patients with cardiac amyloidosis. Ann Noninvasive Electrocardiol. 2013;18:157–162. doi: 10.1111/anec.12018. doi: 10.1111/anec.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumont CA, Monserrat L, Soler R, Rodríguez E, Fernandez X, Peteiro J, Bouzas A, Bouzas B, Castro-Beiras A. Interpretation of electrocardiographic abnormalities in hypertrophic cardiomyopathy with cardiac magnetic resonance. Eur Heart J. 2006;27:1725–1731. doi: 10.1093/eurheartj/ehl101. doi: 10.1093/eurheartj/ehl101. [DOI] [PubMed] [Google Scholar]
- 29.Lyon A, Mincholé A, Martínez J, Laguna P, Rodriguez B. Computational techniques for ECG analysis and interpretation in light of their contribution to medical advances. J Roy Soc Interface. 2018;15:20170821. doi: 10.1098/rsif.2017.0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fikrle M, Paleček T, Kuchynka P, Němeček E, Bauerová L, Straub J, Ryšavá R. Cardiac amyloidosis: a comprehensive review. Cor Vasa. 2013;55:e60–e75. [Google Scholar]
- 31.Greenland S. The need for reorientation toward cost-effective prediction: comments on ‘Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond’ by M. J. Pencina et al., Statistics in Medicine (DOI: 10.1002/sim.2929). Stat Med. 2008;27:199–206. doi: 10.1002/sim.2995. doi: 10.1002/sim.2995. [DOI] [PubMed] [Google Scholar]
- 32.Tonelli AR, Baumgartner M, Alkukhun L, Minai OA, Dweik RA. Electrocardiography at diagnosis and close to the time of death in pulmonary arterial hypertension. Ann Noninvasive Electrocardiol. 2014;19:258–265. doi: 10.1111/anec.12125. doi: 10.1111/anec.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henkens IR, Gan CT, van Wolferen SA, Hew M, Boonstra A, Twisk JWR, Kamp O, van der Wall EE, Schalij MJ, Vonk Noordegraaf A, Vliegen HW. ECG monitoring of treatment response in pulmonary arterial hypertension patients. Chest. 2008;134:1250–1257. doi: 10.1378/chest.08-0461. doi: 10.1378/chest.08-0461. [DOI] [PubMed] [Google Scholar]
- 34.Waligóra M, Tyrka A, Podolec P, Kopeć G. Corrigendum to “ECG markers of hemodynamic improvement in patients with pulmonary hypertension”. Biomed Res Int. 2018;2018:1541709. doi: 10.1155/2018/1541709. doi: 10.1155/2018/1541709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okin PM, Devereux RB, Jern S, Kjeldsen SE, Julius S, Nieminen MS, Snapinn S, Harris KE, Aurup P, Edelman JM, Wedel H, Lindholm LH, Dahlöf B LIFE Study Investigators. Regression of electrocardiographic left ventricular hypertrophy during antihypertensive treatment and the prediction of major cardiovascular events. JAMA. 2004;292:2343–2349. doi: 10.1001/jama.292.19.2343. doi: 10.1001/jama.292.19.2343. [DOI] [PubMed] [Google Scholar]
- 36.Simonson E, Keys A. The effect of age and body weight on the electrocardiogram of healthy men. Circulation. 1952;6:749–761. doi: 10.1161/01.cir.6.5.749. doi: 10.1161/01.cir.6.5.749. [DOI] [PubMed] [Google Scholar]
- 37.Kalliomaki JL, Mollerstrom J, Sollberger A. Observations on the standard electrocardiogram in diabetics with clinically normal hearts. Acta Med Scand. 1956;156:211–220. doi: 10.1111/j.0954-6820.1956.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 38.Simonson E. The effect of age on the electrocardiogram. Am J Cardiol. 1972;29:64–73. doi: 10.1016/0002-9149(72)90417-1. doi: 10.1016/0002-9149(72)90417-1. [DOI] [PubMed] [Google Scholar]
- 39.Bachman S, Sparrow D, Smith LK. Effect of aging on the electrocardiogram. Am J Cardiol. 1981;48:513–516. doi: 10.1016/0002-9149(81)90081-3. doi: 10.1016/0002-9149(81)90081-3. [DOI] [PubMed] [Google Scholar]
- 40.Levy D, Salomon M, D’Agostino RB, Belanger AJ, Kannel WB. Prognostic implications of baseline electrocardiographic features and their serial changes in subjects with left ventricular hypertrophy. Circulation. 1994;90:1786–1793. doi: 10.1161/01.cir.90.4.1786. doi: 10.1161/01.cir.90.4.1786. [DOI] [PubMed] [Google Scholar]
- 41.Brohet CR, Robert A, Derwael C, Fesler R, Stijns M, Vliers A, Braasseur LA. Computer interpretation of pediatric orthogonal electrocardiograms: statistical and deterministic classification methods. Circulation. 1984;70:255–262. doi: 10.1161/01.cir.70.2.255. doi: 10.1161/01.cir.70.2.255. [DOI] [PubMed] [Google Scholar]
- 42.Kors JA, van Bemmel JH. Classification methods for computerized interpretation of the electrocardiogram. Methods Inf Med. 1990;29:330–336. [PubMed] [Google Scholar]


