SUMMARY
Pyruvate kinase is an important enzyme in glycolysis and a key metabolic control point. We recently observed a pyruvate kinase liver isoform (PKL) phosphorylation site at S113 that correlates with insulin resistance in rats on a 3 day high-fat diet (HFD) and suggests additional control points for PKL activity. However, in contrast to the classical model of PKL regulation, neither authentically phosphorylated PKL at S12 nor S113 alone is sufficient to alter enzyme kinetics or structure. Instead, we show that cyclin-dependent kinases (CDKs) are activated by the HFD and responsible for PKL phosphorylation at position S113 in addition to other targets. These CDKs control PKL nuclear retention, alter cytosolic PKL activity, and ultimately influence glucose production. These results change our view of PKL regulation and highlight a previously unrecognized pathway of hepatic CDK activity and metabolic control points that may be important in insulin resistance and type 2 diabetes.
Graphical Abstract

In Brief
Gassaway et al. identify a diet-induced, cyclin-dependent kinase-regulated phosphorylation site at S113 on pyruvate kinase. Although they determine that neither phosphorylation of this site nor the canonical PKA-regulated S12 site directly impacts enzyme kinetics, they demonstrate that S113 phosphorylation alters pyruvate kinase subcellular localization and influences glucose production.
INTRODUCTION
Gluconeogenesis is classically thought to be regulated by glucagon by protein kinase A (PKA) phosphorylation of pyruvate kinase liver isoform (PKL) (Feliú et al., 1976). PKL catalyzes the hydrolysis of phosphoenolpyruvate (PEP) into pyruvate, generating ATP in the process. PKL is a tightly regulated metabolic control point because PEP has the highest free energy of hydrolysis of any central carbon metabolite and unfettered PEP hydrolysis would short circuit gluconeogenesis. Glucagon is thought to inhibit PKL and prevent this futile cycle through cyclic AMP (cAMP)-activated PKA phosphorylation at position S12 (pS12) leading to an increase in the PKL Km (Michaelis constant) for PEP (Blair et al., 1976; Ekman et al., 1976; Feliú et al., 1976). This view of the PKL regulatory mechanism grew from activity measurements in murine liver lysates under conditions that promoted PKA phosphorylation of PKL (Berglund, 1978; Blair et al., 1976) or in vitro reaction of PKA with PKL (Ekman et al., 1976).
Recently, in a study investigating the role of PKCε in hepatic insulin resistance in high-fat-fed rats, we observed a PKL phosphorylation site at S113 (pS113) that increased in the high-fat fed, insulin-resistant state and returned to baseline with PKCε knockdown in vivo (Gassaway et al., 2018) (Figures 1A and S1). PKCε has previously been shown to drive lipid-induced hepatic insulin resistance through phosphorylation of the INSR and other substrates. Furthermore, knockdown of hepatic PKCε in rats protects against high-fat-diet (HFD)-induced insulin resistance (Gassaway et al., 2018; Petersen et al., 2016; Samuel et al., 2007). Thus, because PKL pS113 was correlated with the insulin-resistant state, we hypothesized that this site might play a role in PKL regulation and contribute to the underlying insulin resistance. Furthermore, PKL pS113 was not identified as a direct PKCε target, nor does it contain a PKA phosphorylation motif, which suggests an alternative pathway. Although PKL pS113 has appeared in phosphoproteomic datasets (Lundby et al., 2012; Zhou et al., 2013), its relevance and functional role in regulating PKL have not been previously investigated.
Figure 1. Identification and Characterization of S113, a Regulatory Phosphosite on PKL.
(A) Of the 90 phosphosites we observed on metabolic enzymes, PKL pS113 is the most prominent, with greater than 2-fold change in both the HFD-fed compared to chow-fed samples and the HFD-fed compared to the HFD-PKCε ASO. Data are plotted as in Figure S1B except only phosphorylation on metabolic enzymes is shown. Phosphosites <2-fold change in both comparisons are in green, phosphosites >2-fold change in 1 comparison are in blue, and PKL pS113 (>2-fold change in both comparisons) is in red.
(B) Western blot of chow, HFD-fed, and HFD-PKCε ASO rat liver lysates showing PKL pS113 (at 140 kDa), PKL pS12 (59 kDa), and total PKL (59 kDa), normalized to β actin.
(C) Kapp (allosteric sigmoidal Michaelis constant, see Eq. 1) for PEP for PKL WT (blue), pS12 (red), and pS113 (green) at various concentrations of FBP. n = 2 for each isoform and concentration of FBP. Error bars represent standard error.
(D) Vmax for PKL WT, pS12, and pS113. n = 2 for each isoform. Error bars represent standard error.
(E) Crystal structure of PKL pS113 (PDB: 6ECK). Tetrameric assembly is shown as ribbon cartoons with pS113 site shown in CPK representation.
(F) 2mFo-DFc electron density carved around the PKL pS113 site contoured at 1.0σ is shown in semitransparent surface and with grid lines with key residues shown in stick representation (magenta) showing the unambiguous density for the phosphorylation site.
(G) Superposition of pS113 loop residues (PDB: 6ECK, magenta) with that of PKL pS12 (PDB: 6ECH, blue) and PKL S12D (PDB: 4IP7, green).
(H) Crystal structure of PKL pS113 (magenta) aligned with PKL pS12 (blue) and PKL S12D (4IP7, green) showing limited movement in the FBP allosteric binding site. FBP is shown as sticks with key amino acid residues shown as thin lines, with dotted lines representing hydrogen-bond interactions.
RESULTS
Site-Specific Roles of PKL Phosphorylation
In order to characterize the contribution of PKL pS12 relative to the S113 site, two phospho-specific antibodies were made and validated as specific for their intended proteoform (Figure S2). In comparison to purified protein, which electrophoresed at the expected ~59 kDa, the PKL pS113 band was observed as a complex that ran at ~140 kDa despite stringent denaturing conditions (Figure 1B), whereas PKL pS12 was observed at the expected molecular weight (MW). Mass spectrometry (MS) of the immunoprecipitate from the high-molecular-weight band confirmed the identity as PKL pS113 (Figure S3). Given similar patterns of phosphorylation by western blot (Figure 1B), we sought to determine if PKL S113 phosphorylation may regulate enzyme activity given that S12 phosphorylation has been shown to inhibit PKL activity (Blair et al., 1976; Ekman et al., 1976; Feliú et al., 1976).
Because the upstream kinase for PKL pS113 is not known and in vitro PKA-driven phosphorylation of pS12 is suboptimal (being prone to artifacts and off target phosphorylation), in order to study the role of site phosphorylation in regulating PKL the individual site-specific proteins were directly synthesized (Park et al., 2011; Pirman et al., 2015). This method utilizes a orthogonal translation system, consisting of an aminoacyl tRNA synthetase (SepRS) that charges phosphoserine onto a tRNA (tRNASep) and an engineered EF-Tu (EF-Sep) to bring the tRNASep to the ribosome to generate site-specific phosphorylated proteins (Figure S4) (Park et al., 2011; Pirman et al., 2015). This system produces purified phosphorylated enzyme that avoids the artifacts, off-target phosphorylation, and scalability issues of in vitro phosphorylation with kinases. Using direct synthesis of recombinant phosphoproteins, we obtained highly pure PKL proteoforms (~6 mg/L of culture) that were 71.2% and 82.8% phosphorylated at pS12 and pS113, respectively (Figure S4B). Native gel electrophoresis and size-exclusion chromatography (SEC) confirmed the oligomeric state of the phosphoproteins as predominantly tetrameric and dimeric species of PKL and that phosphorylation did not significantly change the oligomeric state of the enzyme (Figures S4C and S4D).
To test whether PKL pS113 regulates enzyme activity in a similar manner to the pS12 site, enzymatic activity was assayed for the pure wild type (WT) (referring to non-phosphorylated PKL), pS113, and pS12 proteoforms. The WT enzyme demonstrated both substrate-dependent and fructose-1,6-bisphosphate (FBP)-dependent increases in activity, consistent with fully active PKL (Tanaka et al., 1967) (Figure S5). PKL pS12 is classically described as inhibiting PKL function by increasing the Km for PEP (Blair et al., 1976; Ekman et al., 1976; Feliú et al., 1976) by disrupting an activating interaction between the N-terminal unstructured domain and the main body of the protein (Fenton and Tang, 2009). Quite surprisingly, no significant change in Km (represented as Kapp using an allosteric sigmoidal equation, see Eq. 1) or Vmax was observed for either the pS113 or pS12 proteoforms (Figures 1C and 1D; Figure S5) with physiological concentrations of FBP (from ~4 μM in fasted rats to ~27 μM in high-carbohydrate-fed rats; Claus et al., 1979) (Figures 1C and 1D). These results are consistent with early observations by Ekman et al. (1976) and Kohl and Cottam (1977) using in-vitro-PKA-phosphorylated PKL or PKL precipitated from fed and fasted rat liver; as little as 5 μM FBP was sufficient to negate any differences in Km. By genetically encoding phosphoserine at the intended positions, we are able to demonstrate that single phosphorylation at either S12 or S113 in the presence of physiologic FBP is not sufficient to alter PKL kinetics, without the potential confounding effects from other PTM’s or co-immuno-precipitated proteins common to tissue-derived PKL.
It has been demonstrated previously that partial cysteine oxidation at position C436 to sulfenic acid (as opposed to full oxidation to cysteic acid) can increase the Km of PKL for PEP, especially when S12 is mutated to aspartic acid to mimic phosphorylation (Holyoak et al., 2013). If our PKL proteoforms were differentially oxidized (especially WT and pS113 relative to pS12), the Kms might not show a difference. To rule out this possibility, we specifically labeled oxidized cysteines in freshly prepared proteoforms with isotope-coded dimedone (ICDMD) (light form), followed by reduction and alkylation of non-modified cysteines with dithiothreitol (DTT) and iodoacetamide (IAA). Next, we measured the ratio of the carbamidomethyl (IAA)-labeled peptide to the ICDMD-labeled peptide by precursor reaction monitoring (PRM) to estimate the amount of oxidized cysteineon the proteoforms (there are two cysteines at C435/C436 that are indistinguishable in this analysis). We observed very little oxidation in PKL proteoforms by this method (Figures S6A and S6B) and, importantly, did not observe a significant difference in IAA:ICDMD ratio between proteoforms (Figure S6B). In parallel, we analyzed freshly prepared PKL proteoforms by intact liquid chromatography-mass spectrometry (LC-MS) and did not observe sulfenic acid modification but did observe some cysteic acid oxidation. Although this intact analysis shows more oxidation than the bottom-up PRM analysis, there was no significant difference in oxidation state between proteoforms (Figure S6C). When we “aged” our enzymes with H2O2 as previously described (Holyoak et al., 2013), we observed an increase in Km in all proteoforms compared to untreated PKL. The Km was increased in the “aged” PKL pS12 proteoform as previously reported (Holyoak et al., 2013) (Figure S6D). However, even though aging changes the Km, cystine oxidation was comparable in all proteoforms and the overall amount of oxidation (unless aged) was low. Therefore, differential cysteine oxidation does not explain the similar Km values for all proteoforms.
To assess structural implications of PKL phosphorylation, homogeneous preparations of active, site-specific forms of phosphorylated PKL were used to generate crystal structures of PKL pS113 in complex with ATP, Mg+2, and FBP (PDB:6ECK), as well as PKL pS12 in complex with ATP, Mg+2, oxalate, and FBP (PDB: 6ECH); these structures were solved to 2.4 and 2.2 Å resolution, respectively, by alignment to the previously published S12D structure (PDB: 4IP7) (Holyoak et al., 2013). pS113 is located on an exposed loop on each subunit of the of the tetrameric enzyme (Figure 1E). The well-resolved electron density (Figure 1F) in this region suggests that the phosphoserine at position S113 is conformationally rigid and exposed to the solvent, with remarkable structural similarity to the same loop region in the non-phosphorylated enzyme (Figure 1G). Except for the B domain motions, we do not see any significant structural differences in the catalytic site in the A domain or FBP allosteric binding site in the C domain when compared with equivalent A and C domains of PKL S12D in both the pS113 and pS12 structures (Figure 1H). These results support our biochemical studies showing that single PKL phosphorylation sites do not alter the enzyme kinetics and suggest that the phosphorylation of this enzyme must regulate activity by a different mechanism than directly altering enzyme kinetics. This striking observation is contrary to the prevailing paradigm of direct modulation of enzyme activity by phosphorylation at pS12 (or possibly any position) and re-opens the question of the role of phosphorylation in general on this important class of enzymes.
CDKs Drive Changes in the HFD Phosphoproteome
Although it is known that PKL S12 is phosphorylated by PKA in response to glucagon, it is unclear whether PKL S113 is regulated by the same kinases and/or pathways. To determine whether PKL S113 is phosphorylated by PKA, rat primary hepatocytes were treated with IBMX, a phosphodiesterase inhibitor that increases cAMP, to activate PKA. Although PKL S12 phosphorylation was increased by this treatment, PKL S113 phosphorylation showed no response; however, when rat primary hepatocytes were nutrient challenged with pyruvate and lactate (mimicking a diabetic state), PKL S113 phosphorylation increased, whereas PKL S12 phosphorylation did not change (Figure 2A). These results suggest that PKL S113 is not regulated by PKA, unlike S12, and that PKL may be regulated by additional kinases and signaling pathways.
Figure 2. Identification of CDK as an Upstream Kinase for PKL pS113.
(A) Primary hepatocytes treated with glucose and the gluconeogenic precursors pyruvate and lactate showed increased PKL pS113 compared to just glucose or glucose with the PKA activator IBMX.
(B) Motif analysis was performed on phosphopeptides increased by the HFD by using the pLogo tool (O’Shea et al., 2013), revealing a CDK-like motif (+1P, +3–5 K/R). Residues shaded in gray are fixed, and the size of the residue correlates with the enrichment of that residue at a position. Residues taller than the red lines represent statistical significance (p < 0.05).
(C) Network analysis was performed using the Omicsintegrator software package (Tuncbag et al., 2016). The HFD-fed/chow-fed log2-normalized fold changes from of the 552 observed dynamically regulated phosphoproteins with the iRef protein interaction database were used to generate networks. Rectangular boxes are nodes observed in the proteomics data, and circular boxes are imputed nodes; background color indicates increasing betweenness centrality from red to green.
(D) An alignment showing the amino acid context of PKL S113 and the CDK consensus motif.
To determine which kinases and signaling pathways may be regulating PKL S113, we first examined the original phosphoproteomics data by using pLogo (O’Shea et al., 2013) to search for HFD responsive kinase motifs. Using this approach, we observed a striking cyclin-dependent kinase (CDK) motif (S*PxKKK; Figure 2B) in phosphosites upregulated by the HFD, and Motif-X analysis confirmed the statistical significance of the S*PxK motif (Figure S7) (Chou and Schwartz, 2011; Schwartz and Gygi, 2005). Next, the Omicsintegrator software package (Tuncbag et al., 2016) was used with the iRef protein interaction database (Razick et al., 2008) and the phosphoproteomic fold changes to identify central nodes within the data. This analysis revealed that CDK1 and CDKN1A were central nodes driving the changes resulting from the HFD (Figure 2C). This result supports the motif analysis and strongly suggests that a CDK is responsible for many of the changes in the phosphoproteome induced by the HFD.
Although we observed many HFD phosphosites that fit the consensus CDK motif, most of these predicted CDK sites have never been tested as direct CDK substrates. To validate many of these potential CDK substrates in parallel, we performed large-scale kinase assays by using a targeted library of rat-derived peptides (Figure S8). The library was prepared and analyzed in a similar manner to our recently published serine-oriented human library-kinase library reaction (SERIOHL-KILR) method (Barber et al., 2018). The library consisted of 72 potential CDK substrates that matched the CDK consensus motif along with 102 controls that either had no fold change in the phosphoproteomics data and no discernable motif or were substrates of a different kinase with a −3R preference. All 174 test substrates were expressed as glutathione S-transferase (GST) fusion proteins and assayed in vitro with different CDK isoforms with the resulting phosphorylation sites identified by LC-tandem MS (LC-MS/MS) (Table S2). Because CDKs 1, 2, 4, 5, and 6 have significant overlap in motif and substrate preference, all were tested with the peptide library. Consistent with the prediction of CDK activity in vivo, significantly higher kinase activity toward the 72 potential CDK substrates was observed compared to the 102 controls (Table S2). For example, CDK1 phosphorylated 62.5% of potential target peptides, while only phosphorylating 14.7% of the control peptides. Interestingly, most of the control library phosphorylation occurred on residues other than the CDK target site and many of these off-target sites included elements of the CDK consensus motif. These results support the network and motif analyses, suggesting that CDKs are indeed driving the in vivo phosphoproteome changes in response to the HFD.
Many known CDK substrates were validated in these experiments. For example, the CDK 1, 2, and 5 substrate Sirt2 was phosphorylated at S330 (homologous to S368 in mouse and human) by CDKs 1, 2, and 5 as well as CDK6, but not CDK4 (North and Verdin, 2007; Pandithage et al., 2008; Santamaría et al., 2007; Satyanarayana et al., 2008). In addition to Sirt2, the kinase-substrate relationship between CDK1 and many of its substrates were recapitulated, including CALD1, RSF1, LMNB1, MCM2, WHSC1, UFD1l, LIG1, MAP4, NDE1, DSN1, STMN1, NUCKS1, CENPC, TOP2A, NPM1, and TPX2 (Blethrow et al., 2008; Goss et al., 1994; Lee et al., 2015; Montagnoli et al., 2006; Okuwaki et al., 2002; Ookata et al., 1997; Petrone et al., 2016; Yamashiro et al., 1995). In general, far fewer CDK 4 substrates were observed because CDK4 lacks the basic motif preference at +3 and +4 and instead prefers flanking proline residues. Similarly, the reduced number of CDK6 substrates may be a result of its much weaker +3/+4 basic preference than CDKs 1, 2, and 5 (Miller et al., 2008; O’Shea et al., 2013). Among metabolic enzymes that were represented in this group, only LDHA T309 had an apparent CDK motif, although it was not validated as a CDK substrate in this experiment. Taken together, our results show that CDKs respond in vivo to the HFD to directly phosphorylate a large network of protein substrates (Table S2) and may play a more prominent role than previously recognized in the mechanisms of HFD-induced insulin resistance.
Regulation of PKL S113 by CDKs
The amino acid sequence surrounding PKL pS113, EATESFAT-pS-PLSYR, has features of a CDK substrate because of the +1P, +5R (CDK1, 2), and/or −4S (CDK 5) residues that are preferred by these kinases (Figure 2D) (Miller et al., 2008; O’Shea et al., 2013). Because PKL pS113 increased in vivo under HFD conditions where we have shown CDKs are active, we tested whether CDKs can directly phosphorylate PKL at position S113. Full-length, non-phosphorylated PKL was purified and incubated with our panel of active CDKs in vitro. CDKs 1, 2, and 5 and to a lesser degree 6 phosphorylated PKL at position S113 (Figure 2E), suggesting that PKL S113 is a direct CDK substrate.
To further establish the cellular basis of PKL pS113 as a CDK substrate, CDK activity was manipulated with two established cell cycle inhibitors (aphidicolin and nocodazole) in rat hepatoma cells. Aphidicolin prevents entry into S phase (Rosner et al., 2013) and maintains low CDK activity (Hochegger et al., 2008), whereas nocodazole prevents entry into M phase (Rosner et al., 2013) and maintains high CDK activity (Hochegger et al., 2008). We also used the established CDK small-molecule inhibitor Dinaciclib (CDKi) (Parry et al., 2010). PKL pS113 phosphorylation was low when CDK activity was reduced by aphidicolin or CDKi treatment; in contrast, PKL pS113 phosphorylation was high when CDK activity was stimulated by nocodazole (Figure 2F). To confirm CDK activation, a CDK substrate motif antibody (S*PxK/R) showed the same phosphorylation patterns in response to the small molecules as PKL pS113. We also noted that the CDK motif antibody also detected PKL pS113, as well as many other phosphoproteins. These results confirmed that CDK activity controls PKL pS113 levels in cells.
It is known that some metabolic enzymes can be regulated by localization. For example, glucokinase (GK) bound by glucokinase regulatory protein (GKBP) translocates to the nucleus where the enzyme is held in an inactive reserve (Shiota et al., 1999). To investigate whether PKL was regulated in a similar manner, we utilized our pS113-specific antibody to determine the intracellular localization of PKL pS113. Experiments in rat hepatoma cells and primary rat hepatocytes revealed a striking nuclear localization of PKL pS113 (Figures 3A and 3B). Consistent with localization studies with intact cells, nuclear and cytosolic fractions from both cell types showed significant nuclear enrichment of PKL pS113, whereas PKL pS12 and total PKL were mainly cytosolic (Figures 3C and 3D). Similar fractionation studies with livers from rats on normal chow, HFD, and PKCε antisense oligonucleotide (ASO) also displayed strong PKL pS113 nuclear localization and the cellular pool of PKL pS12 was cytosolic (Figure 3E). These observations support the conclusion that distinct signaling pathways and cellular localization separate different populations of phosphorylated PKL in vivo.
Figure 3. PKL S113 Is Phosphorylated in the Nucleus, Leading to Nuclear Retention and a Distinct Pool of PKL from the Cytosolic pS12 Population.
(A and B) Rat hepatoma cells (A) and primary hepatocytes (B) were immunostained for PKL pS113 and show strong nuclear localization (4‘,6-diamidino-2-phenylindole [DAPI]) of the pS113 signal. Scale bar represents 50 μm.
(C–E) Rat hepatoma cells (C), primary hepatocytes (D), and samples from control, HFD, and PKCε ASO rat liver lysates (E) were fractionated into nuclear and cytosolic fractions, and then blotted for pS113, pS12, total PKL, total PKM2, and Histone H3 (as a nuclear marker).
(F) Nuclear and cytosolic fractions from rat hepatoma cells treated overnight with DMSO or 40 μM importazole.
(G) Model of PKL translocation and phosphorylation.
To distinguish whether PKL phosphorylation preceded translocation or whether PKL was phosphorylated in the nucleus and trapped, rat hepatoma cells were treated with and without importazole, a potent nuclear import inhibitor (Soderholm et al., 2011). PKL pS113 should accumulate in the cytosol when nuclear import is inhibited if phosphorylation of PKL pS113 was driving translocation. Instead, there was a general decrease in PKL pS113 with no change in total PKL (Figure 3F), suggesting PKL translocates to the nucleus and is then phosphorylated. Similarly, primary hepatocytes treated with importazole also had decreased S113 phosphorylation (Figure S9). These results support a model whereby PKL is translocated to the nucleus, phosphorylated by CDKs, and retained (Figure 3G).
CDK Inhibition Modulates Cytosolic PKL Activity and Glucose Production
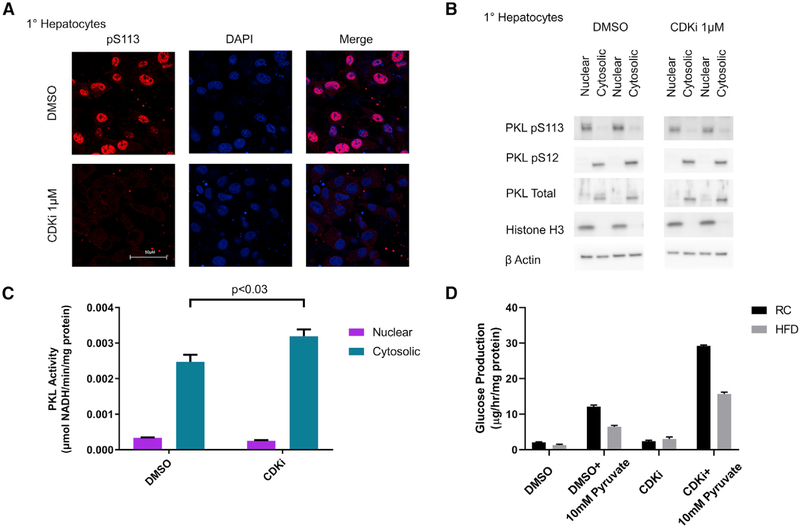
Given that direct phosphorylation of PKL S113 by CDKs led to retention of PKL in the nucleus, we investigated how PKL activity was distributed in the cell and whether CDK phosphorylation of PKL S113 would affect this distribution. In rat primary hepatocytes treated overnight with DMSO or CDKi, we observed significant reduction of nuclear PKL pS113 signal by both immunofluorescence and by western blot of nuclear and cytosolic fractions (Figures 4A and 4B) without an effect on PKL pS12 signal. When PKL activity was measured in nuclear and cytosolic fractions, we observed a significant increase in cytosolic PKL activity with CDK inhibition and a measurable nuclear PKL activity that did not change with CDK inhibition (Figure 4C). This result is consistent with the nuclear sequestration model (Figure 3G) in that reducing PKL S113 phosphorylation allows PKL to escape the nucleus and increase cytosolic PKL activity, which suggests that PKL S113 phosphorylation and nuclear retention could sequester PKL activity from the cytosol, and also shows that, potentially, there is an active pool of PKL in the nucleus.
Figure 4. Manipulation of CDK Activity Regulates PKL pS113 Phosphorylation and Alters Glucose Production in Rat Primary Hepatocytes.
(A) Rat primary hepatocytes were treated with and without 1 μM CDKi and then immunostained for PKL pS113. DAPI (blue) and PKL pS113 (red); scale bar represents 50 μm.
(B) Primary hepatocytes were treated with 1 μM CDKi, separated into nuclear and cytosolic fractions, and then blotted for pS113, pS12, total PKL, and Histone H3 (as a nuclear marker).
(C) Rat primary hepatocytes were treated overnight with DMSO or 1 μM CDKi and separated into nuclear and cytosolic fractions, which were assayed for PK activity. Nuclear and cytosolic PK rates represent mean ± SEM of n = 2 replicates.
(D) After overnight treatment with either DMSO or 1 μM CDKi, primary hepatocytes were starved for 2 h and then provided 10 mM pyruvate as a gluconeogenic substrate; the concentration of glucose in the media was measured after 3 h. Glucose production rates represent mean ± SEM of n = 6.
Because a reduction in PKL S113 phosphorylation results in increased cytosolic PKL activity and because this pathway was under the control of both CDKs and diet, we investigated whether gluconeogenesis was similarly altered in cultured hepatocytes. Primary hepatocytes from rats fed normal chow or HFD were treated with DMSO or CDKi. Glucose production with and without 10 mM pyruvate as a gluconeogenic substrate was measured. If activating CDKs by HFD increased PKL S113 phosphorylation to increase nuclear PKL, then the resulting reduced cytosolic PKL activity could increase glucose production. Conversely, if inhibiting CDKs reduced PKL S113 phosphorylation to reduce nuclear PKL, then the increased cytosolic PKL activity could decrease glucose production. Surprisingly, activation of CDKs by the HFD actually reduced glucose production, whereas inhibition of CDKs with CDKi increased glucose production (Figure 4D). These striking observations suggest that CDK phosphorylation of PKL may be a protective adaptation to suppress glucose production in the setting of a HFD. Although the mechanism of the CDK pathway to suppress glucose production includes PKL S113 phosphorylation, the mechanism is likely distributed across the CDK phosphoproteome. Collectively, our data show that CDKs are part of a hepatic signaling pathway controlling gluconeogenesis and are susceptible to dysregulation by HFD-induced insulin resistance. In addition, our data suggest that this CDK pathway may operate in parallel and possibly independent from known insulin and glucagon signaling pathways.
DISCUSSION
The widely accepted paradigm that PKL S12 phosphorylation by PKA in response to glucagon inhibits the enzyme by increasing the Km for its substrate PEP has persisted since the 1970s. Although it is clear that glucagon activation of PKA leads to increased PKL S12 phosphorylation, the hypothesis that S12 phosphorylation directly inhibits PKL is now challenged by the lack of a significant structural change or change in enzymatic activity under physiological FBP concentrations. Although the present work establishes that neither phosphorylation at pS12 nor S113 appears sufficient to inhibit PKL directly, it is important to note that combinatorial effects of additional phosphorylation sites, other posttranslational modifications, or protein-protein interactions were not investigated in this study. Although we did produce phosphospecific antibodies for both PKL pS12 and pS113, we did not perform absolute quantitation of these sites in vivo or in cultured cells by using isotopically labeled peptide standards and MS. These approaches would be required to determine the molarity of pS12- and pS113-modified PKL proteins in the cytoplasm and nucleus, respectively. We did observe, as have others (Holyoak et al., 2013), that C435/C436 oxidation may increase the Km of the PKL pS12 proteoform more than WT or pS113 proteoforms. Even though together these modifications may contribute to PKL inhibition, the physiological abundance and relevance of this oxidation site in vivo require additional study. Importantly, our reported examination of non-oxidized, phosphorylated enzymes indicate that single-site phosphorylation alone did not result in altered enzymatic activity. Thus, reopening the question of how PKA/glucagon signaling through PKL S12 phosphorylation may inhibit PKL. This important question will require further study to establish a physiologically relevant mechanism.
We speculate that these other mechanisms for inhibiting PKL may include manipulating the concentration of FBP, thereby inhibiting PKL activity, or using S12 phosphorylation to drive a protein-protein interaction, which sequesters PKL away from FBP. Previous work has shown that CDK6 is able to phosphorylate and inhibit phosphofructokinase to decrease the levels of FBP, as well as directly phosphorylate and inhibit PKM2 (at a different, non-homologous site to PKL) to regulate glycolytic flux (Wang et al., 2017); glucagon and PKA may work through a similar mechansim, however, this was not directly addressed in our study. Studies with yeast PK showed that FBP levels, rather than phosphorylation, are the primary regulators of PK activity (Xu et al., 2012). Alternatively, PKL pS12 may drive a protein-protein interaction that sequesters PKL away from FBP in a manner similar to hepatic GK regulation by GKBP (Shiota et al., 1999). In any case, the mechanism by which the glucagon-PKA signaling axis leads to PKL inhibition and the role that S12 phosphorylation plays in that process require further investigation.
Our results show that CDKs can directly regulate hepatic metabolism and control the localization, activity, and phosphorylation of PKL. The contributions of the individual CDK isoforms in different cellular signaling contexts remain to be elucidated. The role of CDKs in regulating metabolism is just beginning to be appreciated, as various CDKs have been implicated in glucose homeostasis (Lee et al., 2014), PPARγ signaling (Banks et al., 2015), and recently direct regulation of PKM2 activity in some cancers (Wang et al., 2017). In this study, we demonstrate that a short-term HFD activates CDKs in liver, and we have identified a network of proteins that CDKs may be able to phosphorylate in this setting, including PKL at position S113. PKL S113 phosphorylation results in PKL retention in the nucleus, whereas inhibition of CDKs leads to a reduction in nuclear PKL S113 and increased cytosolic PKL activity. Two separate approaches, CDK activation by HFD and CDK inhibition by a small-molecule inhibitor, both modulated glucose production in primary hepatocytes, recapitulating the signaling trends we observed in vivo. The extent to which this pathway may exert direct control over gluconeogenesis through PKL phosphorylation at S113 and/or the additional network of proteins that CDKs act upon in the HFD setting requires additional studies that can distinguish enzymatic and non-enzymatic compartmentalized roles. Given that CDK isoforms and drugs that target them could have overlapping or off-target contributions, mutations in the CDK pathway or hepatic PKL, either in vivo or transiently in primary hepatocytes, will be needed to establish the physiological relevance and role of PKL phosphorylation. In addition, more selective CDK inhibitors, perhaps in combination with mutagenesis, may narrow in on the major physiological driver of the pS113 PKL pathway.
Here we identify steady-state pools of nuclear PKL pS113 in vivo and in cultured cells, opening the possibility that PKL, like PKM2, may play additional physiologic roles in the nucleus. Nuclear PKL may play a role in transcriptional activation similar to PKM2 (Yang et al., 2012) and/or may provide a source of local ATP production in the nucleus. Recently, it has been suggested that nuclear ATP generation is required for the intensive chromatin rearrangement required of rapidly diving cells (Wright et al., 2016). Increased PKL S113 phosphorylation and retention in rapidly dividing cells is supported by our observed increase in nuclear PKL pS113 signal in rat hepatoma cells compared to rat primary hepatocytes (Figure 3C) and is consistent with this hypothesis, although it was not not directly tested in this study. However, these additional roles are speculative and require future investigation. Future studies will need to determine the concentration of metabolites and intermediates to more accurately assess the role of PKL and any other component of central carbon metabolism in the nucleus.
Here, we show that CDKs are part of a normal signaling pathway controlling gluconeogenesis and that this pathway is altered by insulin resistance in vivo. Furthermore, simple models of enzyme inhibition by phosphorylation are not always the correct explanation of how signaling changes across phosphoproteomes can be interpreted. Our work also opens up many new questions. How are CDKs activated by the HFD? How is PKL S113 phosphorylation rescued upon PKCε knockdown? How does CDK activity modulate hepatic glucose production, and how is PKL S113 phosphorylation and nuclear retention involved? Additional understanding of the mechanistic roles of hepatic CDKs in general as well as additional mechanistic insight into PKL regulation could have implications in our understanding and treatments of type 2 diabetes and cancer.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Jesse Rinehart (jesse.rinehart@yale.edu).
- Plasmids generated in this study have been deposited to Addgene:
PCRT7 tetR pLtetO MBP-rPKL S12TAG Amp Addgene Cat#107155 PCRT7 tetR pLtetO MBP-rPKL WT Amp Addgene Cat#107156 PCRT7 tetR pLtetO MBP-rPKL S113TAG Amp Addgene Cat#107157 PCRT7 tetR pLtetO MBP-rPKM2 Amp Addgene Cat#107158 Targeted CDK peptide library in pCRT7 NT Topo arabinose Addgene Cat#73446 PKL pS113 and PKL pS12 antibodies generated in this study will be made available on request but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal Experiments
All experimental protocols involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine prior to study initiation. Male Sprague-Dawley rats weighing 160 g (~4–5 wks old*) were obtained from Charles River Laboratories. Rats received free access to food and water and were housed with 12 h light/dark cycles at 23°C. Rats received 4 weeks of intraperitoneal ASO injection (either scrambled or PKCε-targeted) as described in Method Details. Seven days before sacrifice, jugular venous and carotid artery catheters were placed. After 4 days of recovery, control rats were maintained on regular chow (Harlan TD2018: 18% fat, 58% carbohydrate, 24% protein) while HFD rats were switched to a high-fat safflower oil-based diet (Research Diets D12492: 59% fat, 26% carbohydrate, 15% protein) supplemented with 6% w/v sucrose water for 3 days (Kraegen et al., 1991; Samuel et al., 2004, 2007).
Primary Hepatocytes
Male Sprague-Dawley rats were obtained from Charles River Laboratories at 200–230 g (~6–7 wks old*). Hepatocytes were isolated by the Yale University Liver Center from overnight fasted rats (maintained on regular chow unless HFD is specified, in which case rats were treated with HFD for 3 days [see above]). Isolated hepatocytes were suspended and washed two times in recovery medium containing DMEM high glucose (20 mM) (Sigma, D5648) with 10% FBS, 1 nM insulin, 1 nM dexamethasone, and antibiotics (10,000 units/ml penicillin and 10 mg/ml streptomycin, Invitrogen). Cell count and viability were estimated by trypan blue exclusion. Primary hepatocytes were cultured under 5% CO2 and 95% O2 in air at 37°C. For glucose production assays, the cells were plated at 5 × 105 cells/cm2 in collagen-I-coated (BD Biosciences) 6-well plates in recovery medium. For all other experiments, cells were plated at 2.5 × 106 cells/cm2 in collagen-I-coated (BD Biosciences) 10cm dishes. After 4 h, cells were washed with PBS, and the medium was changed to DMEM low glucose (5 mM) (Sigma, D5648) with 10% FBS, 1 nM insulin, 1 nM dexamethasone, and antibiotics (10,000 units/ml penicillin and 10 mg/ml streptomycin, Invitrogen).
Rat Hepatoma Cell Line
ATCC Cell line McA-RH7777 (ATCC CRL-1601) was obtained from ATCC (Sex: Female, Strain: Buffalo, Disease: Morris Hepatoma 7777). Cells were cultured in DMEM + 10% FBS at 37°C and 5% CO2, passaged 1:10 approximately every 4 days.
*Rats are ordered based on weight rather than age; this allows better comparison between diets, blood glucose, and insulin levels. As all rats were purchased from the same vendor, rats of a similar weight will also be of a similar age.
METHOD DETAILS
Animal Experiments
All experimental protocols involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Yale University School of Medicine prior to study initiation. Male Sprague-Dawley rats weighing 160 g were obtained from Charles River Laboratories. Rats received free access to food and water and were housed with 12 h light/dark cycles at 23°C.
Antisense Oligonucleotide (ASO) Knockdown and Diets
2′-O-methoxyethyl chimeric antisense oligonucleotides were synthesized and screened as described (Geary et al., 2001). Rats received either control nonspecific ASO (5′-CCTTCCCTGAAGGTTCCTCC-3′) or ASO targeting PKCε (5′-GCCAGCTCGATC TTGCGCCC-3′) by intraperitoneal injection at a dose of 75 mg kg−1 wk−1 for 4 weeks as described previously (Samuel et al., 2007). Seven days before sacrifice, jugular venous and carotid artery catheters were placed. After 4 days of recovery, control rats were maintained on regular chow (Harlan TD2018: 18% fat, 58% carbohydrate, 24% protein) while study rats were switched to a high-fat safflower oil-based diet (Research Diets D12492: 59% fat, 26% carbohydrate, 15% protein) supplemented with 6% w/v sucrose water for 3 days (Kraegen et al., 1991; Samuel et al., 2004, 2007).
Animal Phenotyping
After 3 days of high-fat feeding, rats were fasted 6 h (0700–1300h). Jugular venous blood samples (2 ml) were collected, and used to determine basal plasma glucose (YSI Biochemistry Analyzer, Yellow Springs Instruments), insulin (radioimmunoassay, Millipore), and NEFA (NEFA-HR, Wako). Rats were anesthetized with pentobarbital sodium injection (150 mg kg−1), and livers were taken within 1 min, snap-frozen with liquid N2–cooled aluminum tongs, and stored at −80°C for subsequent analysis.
The DAG extraction and analysis were performed as previously described (Cantley et al., 2013). DAG was extracted from cytosolic/lipid droplet and membrane-associated subcellular fractions and measured by LC/MS/MS. Total DAG content is expressed as the sum of individual species. Liver triglyceride was extracted by the method of Bligh and Dyer (Bligh and Dyer, 1959) and measured with Triglyceride-SL reagent (Sekisui).
Phosphoproteomics
Our workflow is similar to those reported previously (Kettenbach et al., 2015) with the exception that we label prior to TiO2 enrichment and employ ERLIC fractionation to separate the enriched phosphopeptides from each other rather than using ERLIC to enrich for phosphopeptides (Alpert, 2008).
Lysis and protein extraction
50 mg of ground livers from each animal selected was lysed using 1mL Cytobuster (Millipore) supplemented with protease inhibitor (cOmplete, Roche), phosphatase inhibitor (Phosphatase Inhibitor Cocktail 1, Sigma), 14.3 mM β-mercaptoethanol, 50mM NaF, and 1mM NaVO4, incubated for 45 min on ice, and sonicated (30 s on 30 s off for 4 cycles at 55 A) on a pedestal sonicator. Protein was precipitated using methanol and chloroform as described (Wessel and Flügge, 1984), washed 1x with methanol, and dried under vacuum.
Digestion
5mg of dry protein was weighed out, and reconstituted with a 2.5% solution of ALS-110 (Protea) in 50mM Tris-HCL pH 8.5 with 5mM EDTA, and 50 mM DTT. Following sonication (30 s on 30 s off for 4 cycles at 55 A), samples were diluted with Tris-HCL to 0.83% ALS-100. Cystines were reduced by incubating for 35 min at 55°C, alkylated with iodoacetamide for 30 min in the dark, and quenched with the addition of excess DTT. Samples were digested with LysC (Wako) at 1:100 enzyme to protein ratio for 4 hr at 37°C, diluted under 0.5% ALS-100 with Tris-HCL pH 8.5 and CaCl2, and digested at 1:50 enzyme to protein ratio with trypsin (Promega) for 16 hr at 37°C. ALS-110 was cleaved with the addition of 20% TFA to under pH 3.0 and incubated at room temperature for 15 min. Liver digests were pooled, desalted on SepPak Classic SPE cartridges (Waters), and lyophilized.
Dimethyl Labeling
Lyophilized peptides were reconstituted in 50% ACN 0.1% TFA and concentration was estimated by A280 using a Nanodrop 2000 (Thermo). 16mg of peptides were labeled using reductive dimethylation as described (Boersema et al., 2009) using NaCH3CN and CH2O (light-8mg) or CD2O (medium-8mg) in 100mM MES buffer pH 5.5 (Wilson-Grady et al., 2013), mixed 1:1, and desalted on Sep-Pak Classic SPE cartridges. Samples were labeled in technical duplicate, but with switched heavy/light labels.
TiO2 Enrichment
Phosphopeptide enrichment was performed by resuspending 8mg peptides in 50% ACN 2M lactic acid and iteratively incubating with Titansphere TiO2 beads (GL Sciences) with a 10:1 TiO2 to peptide ratio 3 times, eluting peptides from the beads 1x with 0.2M Sodium Phosphate pH 7.8, 2x with 5% NH4OH, and 1x with 5% pyrrolidine and combining all elutions into 70% formic acid (Kettenbach and Gerber, 2011). Enriched phosphopeptides were desalted with C18 Microspin Columns (The Nest Group). Phosphopeptides were reconstituted in 85% ACN 0.1% formic acid, quantified, and separated on an PolyWAX LP column (150 × 1.0 mm, 5 μm particle size, 300 Å pore size, PolyLC) using an 70min non-linear gradient from 85% ACN 0.1% formic acid to 30% ACN 0.1% formic acid (adapted from Hao et al., 2013) and 32 fractions were collected (2 min fractions for 30 min, 2–5 min fractions from 60–70min). Fractions were dried using vacuum centrifugation, and reconstituted for ERLIC.
Offline ERLIC Chromatography
TiO2 enriched samples were reconstituted in 50% ACN 0.1% formic acid, and concentration was measured by A280 (Nanodrop, Thermo). Samples were further diluted to 85% ACN 0.1% formic acid. 50 μg of peptides were injected onto a PolyWAX LP 150×1.0mm, 5 μm, 300Å resin column (PolyLC) and separated on a 70 min non-linear gradient (adapted from Hao et al., 2013) from 85% ACN 0.1% formic acid to 30% ACN 0.1% formic acid at a flow rate of 50 μl/min. 30 2-min fractions were collected across the entire gradient, with the addition of 2 5-min fractions at the end. Each sample was separated in triplicate on the ERLIC column for a total of 150ug. Fractions were dried in a rotary evaporator (Thermo) and reconstituted for LC-MS.
LC-MS/MS
Samples were separated using a NanoAquity (waters) HPLC with a vented split on a 30mm 150 μM ID trap column with a Kasil frit packed with Magic C18AQ 3 mM 200Å resin (Bruker) and a 20cm 75 μM ID self-packed Pico-frit column (New Objective) packed with 1.9 μM 120Å reprosil-Pur C18-AQ resin (Dr. Maisch) on a non-linear 120min gradient from 5% ACN 0.1% formic acid to 95% ACN 0.1% formic acid and analyzed with a LTQ Orbitrap Velos (Thermo) using a Top 10 method. ERLIC samples were run in duplicate, with the first ERLIC fraction of each set of 3 run separately, while the 2nd and 3rd fraction of each set were pooled for a single run. In addition, for each sample 2 μg of the pre-enriched peptides and 2 μg of the total (unfractionated) enriched phosphopeptides were run using a non-linear 200min gradient from 5% ACN 0.1% formic acid to 95% ACN 0.1% formic acid.
Bioinformatics
Data was searched using Maxquant version 1.5.1.2 (Cox and Mann, 2008) with Acetyl (N-Term), Deamidation (NQ), Oxidation (M), and Phospho(STY) as variable modifications and Carbamidomethyl (C) as a fixed modification with up to 3 missed cleavages, 5 AA minimum length, and 1% FDR against the Uniprot Rat database (Downloaded Dec 11, 2013). The pre-enriched, total enriched, and ERLIC fractionated peptides were combined into 1 search for each sample. Searches were analyzed with Perseus version 1.5.0.31. This analysis included removed of contaminant and reverse protein hits, normalization of ratios (correction for H/L channels in label switched samples), log2 transformation, and averaging across technical (label-switch) replicates.
Phosphoprotein Synthesis and Purification
Cloning and Phosphoprotein Synthesis
G-blocks containing the full-length C-term 6-his tagged WT Rat PKL sequence, S12TAG, or S113TAG mutations, and WT PKM2 were assembled into addgene #53225 (removing the MEK gene, but maintaining the MBP and Factor Xa cleavage sites) (Addgene 107155–107158). Plasmids were transformed into rEColi (Lajoie et al., 2013) (Addgene 68306) along with SepOTSλ (Pirman et al., 2015) (Addgene 68292). Strains were streaked from glycerol stocks stored at −80°C and grown on plates containing 100 A 25 kan. 5–6 colonies were inoculated into 5–6mL media, grown 8–10hr, then transferred to 50mL of culture for overnight growth. Cultures were back diluted to OD600 0.15 in 500 mL of LB with antibiotics and 2mM O-phospho-L-serine (Sep), grown to OD600 0.7–0.8, then induced with arabinose (0.2%) and IPTG (1mM). Cultures were grown for 20–24hrs at 20°C. Cultures were harvested by centrifugation at 4000x g, removal of media, and frozen at −80°C for future analysis.
Protein Purification
Cells were lysed in buffer containing 10% glycerol, 1 mg/mL lysozyme, 50mM Tris-HCL pH 7.8, 500mM NaCl, 0.5mM EDTA, 0.5mM EGTA, 1mM DTT, 50mM NaF, 1mM NaVO4, and protease inhibitor (cOmplete, Roche). Proteoforms were dual-tag purified on a NiNTA column (QIAGEN), followed by Amylose resin (NEB) according to manufacturer’s instructions. MBP was cleaved by addition of Factor Xa (NEB) at a 1:50 ratio protease:protein and overnight incubation at 4°C, followed by a second NiNTA column purification to remove the cleaved MBP. Protein was buffer exchanged into storage containing 50mM Tris/HCl pH 7.4, 150mM NaCl, and 20% glycerol using Amicon 30K MWCO spin columns. “Freshly purified” protein prior to aging with H2O2 was exchanged into the same buffer with the addition of 1mM DTT.
Protein Characterization
Western Blotting
Ground livers were lysed in buffer containing 10mM Tris-HCL pH 8.0, 1mM EDTA, 0.5mM EGTA, 1% Triton X-100, 0.1% sodium de-oxycholate, 0.1% SDS, 140mM NaCl, protease inhibitor cocktail (cOmplete, Roche), and phosphatase inhibitor (Phosphatase Inhibitor Cocktail 1, Sigma), sonicated, normalized using a Bradford Assay (Bio-Rad), and resolved by SDS-PAGE electrophoresis. Proteins were transferred to PVDF membranes using semi-dry transfer cells (Bio-Rad), and blocked in 5% milk.
Cell cultures were washed 1x with PBS, lysed in buffer containing 2% Triton x-100, 100mM Tris-HCl pH 7.4, 300mM NaCl, 10% Glycerol, 5mM EDTA, 5mM EGTA, 2mM DTT, protease and phosphatase inhibitors (cOmplete, Roche; Phosphatase Inhibitor Cocktail 1, Sigma), spun at 21000 g for 10 min at 4°C, mixed with SDS-PAGE loading buffer and resolved by electrophoresis.
Phostag SDS-PAGE
Samples were prepared described as in Western Blotting. Phostag SDS-PAGE gels were made with 12% acrylamide and 100 mM Phos-tag acrylamide (Waco), and transferred to PVDF membranes as described.
Native PAGE
Proteoforms were analyzed using the NativePAGE Novex Bis-Tris system (Life Technologies) according to manufacturer’s specifications on 4%–16% Gradient gels, with and without Coomassie G-250 in the running buffer. Gels ran without G-250 in the running buffer were stained for 20 min using Coomassie R-250, and destained with buffer containing 10% ethanol and 10% acetic acid.
Coupled Enzymatic Activity Assay
Pyruvate Kinase enzymatic activity was analyzed using a coupled enzymatic assay adapted from Bergmeyer (1974). Briefly, pyruvate kinase activity (from purified construct or liver lysate) was measured under the following conditions: 50 mM Tris pH 7.5, 100 mM KCl, 15 mM MgCl2, 0.18 mM NADH, 0.005 U/μL LDH, 1 mM DTT, 0.03% BSA, 2 mM ADP (final concentrations) with varying concentrations of PEP and FBP. Reactions were assembled without ADP and incubated for 10–15 min. The reactions were then initiated by adding ADP and the disappearance of NADH was followed at 340 nm.
Initial rates were measured for each progress curve and were fit to a modified version of an allosteric sigmoidal equation (Equation 1) to account for the biphasic shapes of the velocity versus PEP curves, as previously described (Fenton and Alontaga, 2009): for each PKL enzyme, Vmax values were globally fit across FBP concentrations to obtain a single Vmax value for each enzyme form, while other parameters were allowed to float.
| Eq. 1 |
Data were normalized to protein concentration measured at an absorbance of 280nm with a reaction coefficient of 41,000.
Aging
Freshly purified enzymes prepared immediately prior to the PK activity assay were diluted with 0.25% BSA to prevent unspecific binding then incubated for 8 min. at 30°C either with 5mM H2O2 final concentration to age the enzyme or in the absence of H2O2 for the fresh enzyme control. The samples were then further diluted similarly in 0.25% BSA to optimize for a measurable activity range and a fraction of these enzyme samples (20% of the final reaction volume) was added into each reaction well of the PK activity reaction plate, and assayed as stated above.
PRM Analysis of Cysteine Oxidation
This method was modified from Martínez-Acedo et al. (2014). Briefly, 12 μg of purified PKL was diluted to a final volume 18uL by adding 6uL 8M urea/50mM HEPES pH7.0. Dimedone (Sigma) was added to 1mM, and incubated at 25°C for 30 min with constant mixing at 1200rpm in thermomixer. Reaction was quenched with addition of DTT to 2mM, and incubated at 1200rpm in thermomixer. A second reaction was performed at 500X excess (5mM) of IAA (Sigma), and incubated at 1200rpm in thermomixer. The second reaction was quenched by putting samples on ice and precipitating protein with addition of 100% TCA to 20% v/v, followed by incubation on ice for 30 mi and freezing at −80°C. After thawing, samples were spun at max speed in cooled centrifuge for 30 min, and supernatant removed. The pellet was washed with ice cold 10% TCA, then ice cold acetone. The pellets were dried by vacuum centrifugation, and resuspended in 50mM ammonium bicarbonate pH 8.0/10% ACN. Samples were digested with 100ng trypsin at 37°C for 4 h, desalted using a Hypersep Retain Pep 30mg cartridge (Thermo Scientific), and analyzed by LC-MS/MS on a Thermo Fusion Tribrid Mass Spectrometer using a Precursor Reaction Monitoring (PRM) method that triggered an HCD MS2 on 610.3305 (CC), 650.8538 (IC or CI), and 691.3771 (II) m/z. Data were analyzed using Skyline v 4.2.0.19072.
Intact LC-MS Analysis
In each analysis, 5ul of sample was injected onto a self-packed reversed phase column (1/32” O.D. × 500 um I.D., 5 cm of POROS 50R2 resin). After desalting for four min, protein was eluted with an HPLC gradient (0%–100% B in 1 min, A = 0.2M acetic acid in water, B = 0.2 M acetic acid in acetonitrile, flow rate = 30 μL/min) into an LTQ ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA) that acquired profile MS spectra (m/z 300–2000). Mass spectra were deconvoluted using MagTran software v 1.03b2 (Zhang and Marshall, 1998).
Crystallization, data collection, and structure determination
PKL proteoforms (pS12, pS113) were further purified by size exclusion chromatography (SEC) column using a Superdex S200 column (GE Healthcare Life Sciences). SEC eluted protein in the buffer 10 mM MES, 150 mM KCl, 5 mM MgCl, 1 mM DTT, pH 6.8 was pooled, concentrated to 9 mg/mL. The concentrated protein was incubated with 2mM FBP and 10mM ATP at 4°C for 30 min prior to setting up crystallization. Co-crystals of the complex were grown by hanging drop vapor diffusion by equilibrating the above mixture with a reservoir solution containing 0.1 M HEPES pH 7.5, 10% ethylene glycol, 8~12% PEG8000 in a 2:1 ratio in 18°C. Plate shaped crystals appeared in 3 days and grew to full size within 7 days. Crystals were equilibrated in a cryo-protectant buffer containing reservoir solution containing 20% (v/v) ethylene glycol as a cryo-protectant and were flash-frozen in liquid nitrogen.
X-ray characterization and data collection for the PKL pS113·FBP co-complex crystals were performed at the Shanghai Synchrotron Radiation Facility beamline 19U1 with whereas the PKL pS12·FBP were collected at Canadian Light Source beamline ID08–1, both equipped with Pilatus 6M detector. Diffraction data were processed using HKL2000 (Otwinowski and Minor, 1997). Crystals were characterized to belong to orthorhombic form with space group of P212121 for PKL pS12 and P21212 for PKL pS113 respectively. Statistics of data collection, processing, and refinement are summarized in Table S1. The structure was determined by molecular replacement with Phaser (McCoy et al., 2007) using the structure of human PKL S12D variant structure (PDB: 4IP7) as a search model. Iterative model building and refinement were initially carried out using COOT (Emsley et al., 2010), Refmac5 (Murshudov et al., 2011) from the CCP4 software suite (Winn et al., 2011). Final stages of refinement were carried out using Phenix (Adams et al., 2010). Examination of difference Fourier map (mFo–DFc) calculations showed unambiguous density for the presence of phospho-Serine 113 in PKL pS113 dataset. However, phosphoserine 12 was not observed in the electron density map calculations in the PKL pS12 dataset most likely due to the dynamic disorder of the N-terminal region.
Kinase Substrate Validation
Candidate Kinase Substrate Selection
Of the 406 phosphosites observed upregulated in the HFD, we selected the 72 peptides that had +1P. We also selected 52 peptides that had a −3R (PKC) motif and 50 peptides that had 0 fold change in the HFD and that had no elements of either the PKC motif (−3R) or the CDK motif (+1P) surrounding the central phosphorylated residue to serve as controls.
Substrate Peptide Design
Predicted kinase substrate phosphosites were encoded as peptides at least 21 amino acids in length. The central phosphorylatable S/T (position 0) was flanked by at least 10 amino acids on either side, or further extended to the next basic residue within the parent protein to maintain native trypsin cleavage sites. Ser at position 0 was encoded by an amber (TAG) codon, allowing incorporation of either Ser or phosphoserine, as described below. Gene sequences were codon optimized for expression in K12 E. coli using IDT’s codon optimization tool (http://www.idtdna.com/site/account/login?returnurl=%2FCodonOpt) and contained a 5′ SfoI site and a 3′ TAA stop codon followed by a HindIII site. These genes were synthesized as 400–750 base pair concatemers by Genewiz.
Cloning of Library Members
Gene libraries were inserted into a modified pCRT7/NT-TOPO vector (Heinemann et al., 2012). A cassette under a pBad inducible promoter was encoding GST, a HRV3C protease cleavage site, and SfoI and HindIII restriction sites. This plasmid and sequence information is available from Addgene as plasmid #73446. The plasmid and synthetic DNA were digested with SfoI and HindIII, and ligation was performed with mixed insert populations. Ligation reactions were transformed into ElectroMAX™ DH10B™ E. coli cells (Invitrogen), obtaining > 100,000 cfu for each population. Plasmid libraries harvested from these cells were then introduced into genomically recoded C321.ΔA.ΔserB E. coli including either a plasmid containing the serine tRNA amber suppressor supD or SepOTSλ, allowing Ser or phosphoserine incorporation at amber codons, respectively.(Lajoie et al., 2013; Pirman et al., 2015)
Gene Expression and Purification
2 L of LB supplemented with 50 μg/mL ampicillin and 25 mg/mL kanamycin were inoculated with cells containing the plasmid library from stationary overnight culture to OD600 = 0.15. Cells were grown at 30°C ad 230rpm until OD600 = 0.6–0.8, and protein expression was induced with 1 mL of 20% arabinose. Cells were grown for an additional three h and pelleted by centrifugation. Lysis and sonication were performed as in Pirman et al. (2015).
2 mL bed volume equilibrated HiCap GST resin (QIAGEN) was incubated with lysates for 90 min on a rotisserie shaker at 4°C. The resin was then washed with 5 mL wash buffer (50 mM Tris/HCl pH 7.5, 500 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 10% glycerol, 1 mM DTT, 50 mM NaF, 1 mM NaVO4), and transferred to a gravity column, and washed with an additional 5 mL wash buffer. Proteins were eluted with 3×1 mL elution buffer (wash buffer supplemented with 50 mM reduced glutathione, pH 8).
Eluates were buffer exchanged using a 10 kDa cutoff spin column (Amicon) according to manufacturer’s recommendations into buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, and 20% glycerol. GST was cleaved by addition of 10 μl PreScission protease (GE Healthcare) in a 500 μL volume and incubated overnight at 4°C on rotisserie shaker. Peptides were separated from GST using a 30 kDa spin column (Amicon) and concentrated using a 3 kDa spin column (Amicon) according to manufacturer’s protocol. Protein concentration was determined by BCA assay (Pierce).
in vitro Kinase Reactions
Purified CDKs 1, 2, 4, 5, and 6 were purchased from Signal Chem, CDKs were normalized by specific activity and diluted in Kinase Dilution Buffer III (Signal Chem) to the following final concentrations: CDK1, 1.6 ng/μl; CDK2, 1 ng/μl; CDK4, 4 ng/μl; CDK5, 2 ng/μl; CDK6, 4 ng/μl.
Kinase reactions were performed in 50 μL containing 5 μL Kinase Assay Buffer I (Signal Chem), 10 μg of purified substrate peptide library, 5 μL of 250 uM ATP mix, diluted kinase (see above), and ddH2O to 50 μl. Reactions were incubated for 30 min at 30°C.
Sample Workup
Samples were desalted using C18 Ultramicrospin columns (The Nest Group) according to manufacturer’s protocol, and dried on a rotary evaporator.
Samples were digested with trypsin and desalted as described previously (Amiram et al., 2015). Peptides were reconstituted in 3:8 (volumetric) 70% formic acid: 0.1% TFA; 1/5th of the sample was placed in an HPLC vial and diluted to 7 μL with 3:8:1 70% formic acid: 0.1% TFA:50 mM sodium phosphate pH 7.4 for mass spectrometry. The remaining sample was dried, and phosphopeptides were enriched using TiO2 (Rappsilber et al., 2003, 2007). Eluates were dried on a rotary evaporator, and reconstituted in 3:8:1 70% formic acid:0.1% TFA:50 mM sodium phosphate pH 7.4 for mass spectrometry.
LC-MS/MS
Samples were separated using a NanoAquity (waters) HPLC with a vented split on a 30mm 150 μM ID trap column with a Kasil frit packed with 3.0 μM 120Å reprosil-Pur C18-AQ resin (Dr. Maisch) and a 20cm 75 μM ID self-packed Pico-frit column (New Objective) packed with 1.9 μM 120Å reprosil-Pur C18-AQ resin (Dr. Maisch) on a non-linear 90min gradient from 5% ACN 0.1% formic acid to 95% ACN 0.1% formic acid and analyzed with a LTQ Orbitrap Velos (Thermo) using a Top 10 method.
Bioinformatics
Data were searched using Maxquant version 1.5.1.2 (Cox and Mann, 2008) with Acetyl (N-Term), Deamidation (NQ), Oxidation (M), and Phospho (STY) as variable modifications and Carbamidomethyl (C) as a fixed modification with up to 3 missed cleavages, 5 AA minimum length, and 1% FDR against the EcoCyc E. coli database (v17) and a custom database containing all 204 GST-tagged kinase substrate peptides.
Cell Culture
Cell Lines and Maintenance
McA-RH7777 cells (rat hepatoma, ATCC CRL-1601) were grown in DMEM +10% FBS in 150 cm2 flasks at 37°C, 5% CO2, and 100% humidity.
Cell Culture Experiments
For starvation, media was removed, cells were washed with PBS, and then given the indicated media. For cell cycle and CDK inhibition experiments, the indicated small molecules were added at the indicated concentrations: Nocodazole (400ng/mL, Sigma), Aphidicolin (5μg/mL, Sigma), Dinaciclib (1μM, Selleckchem). Treatment was overnight (16–18hr).
Fractionation
Rat hepatoma cells grown to 70% confluency or primary hepatocytes (obtained from the Yale Liver Center) plated in 10cm dishes were washed 2x with PBS, 1x with iso-osmotic buffer (65mM Sucrose, 215mM D-Mannitol, 5mM HEPES, 3mM MgCl2, 5mM KH2PO4, 5mM KHCO3, pH 7.4), and scraped from the plate with 0.5mL iso-osmotic buffer containing protease (cOmplete protease inhibitor, Roche) and phosphatase (PHOS-Stop, Roche) inhibitors; for intact rat liver 0.5mL of iso-osmotic buffer with inhibitors was added to ~30mg of pulverized liver powder. Lysate was dounced 50x in a pre-chilled 2mL β-pestle dounce after which the whole cell fraction was set aside. 250 μL was placed in an Eppendorf tube, and centrifuged at 1800 g for 3.5min at 4°C, and supernatant (Cytosolic fraction) was transferred to a new tube. Pellet was re-suspended in 250 μL iso-osmotic buffer (Nuclear fraction).
Immunoprecipitation and In-gel Digestion
20 μL PKL total (ProteinTech pAb, or Santa Cruz mAb) and PKL pS113 antibodies were added to 10 μL bed volume protein A/G PLUS-Agarose beads (Santa Cruz), incubated for 1hr at RT and washed 2x with PBS. Beads were added to 5mg rat liver lysate in 0.1% triton with protease (cOmplete, Roche) and phosphatase (Phosphatase Inhibitor Cocktail 1, Sigma) inhibitors and incubated overnight at 4°C in a lab rotator. Beads were pelleted by centrifugation and washed 3x with PBS. Protein was eluted by adding 10ul laemmli sample buffer, incubating at 95°C for 5 min, and separated by SDS-PAGE. Gel bands were cut as indicated, and digested according to previous published methods (Sawyer et al., 2014). Briefly, Gel pieces were sequentially washed with fresh solutions containing 50% (v/v) acetonitrile (ACN), 25 mM NH4HCO3 and 50% S7(v/v) ACN, 5 mM NH4HCO3. Proteins were then digested overnight at 37°C in a solution containing 45 mM NH4CO3, 5% ACN, and 13.33 ng/μl trypsin (Promega). Peptides were extracted from gel pieces with 1.67% FA in 66.6%ACN, dried in a roto-evaporator, reconstituted for LC/MS/MS analysis, and analyzed as described for the Kinase Substrate Validation. Peptides were searched using MASCOT v2.5.1 (www.matrixscience.com) against the Uniprot Rat Database.
Immunocytochemistry and Confocal Imaging
Cells were cultured in a glass coverslip mounted multi-well chamber (LiveAssay) coated with 0.1% gelatin. Post treatment, samples were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 20 min at room temperature. Thereafter, cells were permeabilized using 0.1% Triton X-100 (Sigma-Aldrich) for 15 min, washed twice with ice cold PBS, and blocked with a reconstituted blocking buffer (10% goat serum constituted in 0.1% Triton X-100) for 4 h. Samples were incubated with the primary antibody diluted in the blocking buffer (1:100) overnight, washed three times with PBS, and incubated with Alexa Fluor-conjugated secondary antibody (Life Technologies) diluted in blocking buffer (1:200) for 1 h. After washing with PBS thrice, samples were directly imaged using a Leica Laser Scanning SP8 Microscope using 40x oil objective (NA 1.3), illuminated using EL6000 White light illumination. Images were analyzed using ImageJ, and fluorescence intensity per cell determined using manual masking.
Glucose Production Assay
Primary hepatocytes from overnight fasted rats were obtained from the Yale Liver Center, plated on collagen coated plates, and treated overnight with either DMSO or 1μM CDKi. Hepatocytes were washed with PBS and incubated for 2 h in DMEM with 1% HEPES, 2% BSA, 44mM NaHCO3. Hepatocytes were then provided with 10mM Pyruvate as a substrate, and glucose concentration was measured by colorimetric assay using the Sekisui Glucose-SL assay according to manufacturer’s directions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass spectrometry data were quantified using Maxquant version 1.5.1.2 (Cox and Mann, 2008), and analyzed using Perseus 1.5.0.31, including averaging (mean) of H/L ratios of 3 biological replicates and 3 technical replicates for each biological replicate at 1% target-decoy based FDR as described in Method Details.
Purified PKL proteoform activity data for Figures 1C and 1D was analyzed using Graphpad Prism version 7.01. Kapp1 and Vmax values were calculated by fitting to Equation 1 as described in Method Details. Values represent the fit ± standard error (where each value of the V versus [PEP] curve at various concentrations of FBP was the average (mean) of two technical replicates; see Figures S5A–S5C).
Crystallography diffraction data were processed with HKL2000 (Otwinowski and Minor, 1997), and structure was determined by molecular replacement with Phaser (McCoy et al., 2007). Iterative model building and refinement were carried out using COOT (Emsley et al., 2010), Refmac5 (Murshudov et al., 2011) from the CCP4 software suite (Winn et al., 2011), and Phenix (Adams et al., 2010) as described in Method Details. Statistics of data collection, processing, and refinement are summarized in Table S1.
Motif analysis was performed using pLogo tool v 1.2.0 (O’Shea et al., 2013), with the central phosphorylated residue ± 15 amino acids on either side, and using the Uniprot Rat proteome as background. Grey shading represents fixed residues; red line represents p < 0.05.
Network analysis by Omicsintegrator (Tuncbag et al., 2016) was used with the following parameters: w10, b1, D6, mu0.01.
Nuclear and cytosolic PK rates in Figure 4C represent mean ± SEM of n = 2 biological replicates (1 replicate represents 1 10cm plate of primary hepatocytes). Rates were determined by n = 3 technical replicates for each biological replicate. Significance was determined using GraphPad Prism v 7.01 by 2-way ANOVA followed by Dunnett’s multiple comparisons test.
Glucose production rates in Figure 4D represent the mean ± SEM of n = 6 biological replicates (1 replicate represents 1 well of a 6-well plate of primary hepatocytes); p values are calculated in GraphPad Prism v 7.01 by ordinary 2-way ANOVA followed by Sidak’s multiple comparisons test.
Protein Identification in Figures S3C and s3D was performed using MASCOT v2.5.1 with target-decoy based FDR of 1% against the Uniprot Rat Database.
V versus [PEP] curves in Figure S5A–C were generated by a 12-point, 2-fold dilution curve of PEP starting at 4mM and ending at 0mM for each concentration of FBP. Values represent average (mean) +/− standard deviation of n = 2 technical replicates at each concentration of PEP and FBP.
Peak areas in Figures S6A and S6B were determined using Skyline v 4.2.0.19072; error bars represent mean ± SEM of 2 replicates of in vitro-treated PKL constructs.
Km’s in Figure S6D were calculated using Graphpad Prism v 7.01 by a Michaelis-Menten non-linear least-squares fit of V versus [PEP] rates using an 8-point 2-fold PEP dilution curve starting at 5mM PEP and ending at 0 mM PEP. n = 4 untreated and n = 5 in vitro treated replicates for each proteoform. Error bars represent 95% CI
Motif enrichment analysis in Figure S7 was performed using Motif-X using a width of 31, 20 occurrences, a significance of 0.000001, and the IPI Rat Proteome as background.
Peptides in Table S2 were analyzed as described in Method Details, and considered significant if replicated in at least 2 of 3 in vitro phosphorylation technical replicates.
DATA AND CODE AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Project Name: Rat Liver HFD versus Chow, HFD versus PKCepsilon ASO; Project accession: PXD010209. PKL pS113 is deposited in the wwPDB as PDB: 6ECK under deposition ID D_1000236082 and PKL pS12 as PDB: 6ECH under deposition ID D_1000236056.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Actin | Abcam | Cat#ab6276; RRID: AB_2223210 |
| Histone H3 | Cell Signaling Technologies | Cat#4499; RRID: AB_10544537 |
| Phospho-MAPK/CDK Substrate | Cell Signaling Technologies | Cat#2325; RRID: AB_331820 |
| PKL | ProteinTech | Cat#22456-1-AP; RRID: AB_10918271 |
| PKL | Santa Cruz | Cat#SC-D10; RRID: AB_2163668 |
| PKL pS113 (A/G purified) | Covance-Custom Antibody | N/A |
| PKL pS12 (A/G purified) | Covance-Custom Antibody | N/A |
| PKM2 | ProteinTech | Cat#15822-1-AP; RRID: AB_1851537 |
| Bacterial and Virus Strains | ||
| ReColi (Strain: C321.ΔA) | Lajoie et al., 2013 | Addgene Cat#68306 |
| ElectroMAX™ DH10B™ E. coli | Invitrogen | Cat#18290015 |
| Biological Samples | ||
| Primary Rat Hepatocytes | Yale Liver Center Cellular and Molecular Physiology Core Facility | https://medicine.yale.edu/intmed/livercenter/core-facilities/cellular-molecular-physiologycore-facility/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CDK1/CyclinA2 | SignalChem | Cat#C22-18G-05 |
| CDK2/CyclinA2 | SignalChem | Cat#C29-10G-05 |
| CDK4/CyclinD1 | SignalChem | Cat#C31-10G-05 |
| CDK5/p35 | SignalChem | Cat#C33-10BG-05 |
| CDK6/CyclinD3 | SignalChem | Cat#C35-10H-05 |
| Aphidicolin from Nigrospora sphaerica | Sigma | Cat#A0781 |
| Nocodazole | Sigma | Cat#M1404 |
| Dinaciclib | Selleckchem | Cat#S2768 |
| Dimedone (light) | Sigma | Cat#D153303 |
| Iodoacetamide | Sigma | Cat#I1149 |
| IBMX | Sigma | Cat#I7018 |
| Critical Commercial Assays | ||
| Insulin radioimmunoassay | Millipore | Cat#RI-13K |
| NEFA-HR(2) | Wako | Cat#434-91795 |
| Triglyceride-SL | Sekisui | Cat#236-99 |
| Glucose-SL | Sekisui | Cat#235-60 |
| Deposited Data | ||
| PKL pS113 | wwPDB | 6ECK |
| PKL pS12 | wwPDB | 6ECH |
| Mass Spectrometry-based Phosphoproteomics | ProteomeXchange | PXD010209 |
| Experimental Models: Cell Lines | ||
| McA-RH7777 | ATCC | Cat# CRL-1601; RRID: CVCL_0444 |
| Experimental Models: Organisms/Strains | ||
| Male Sprague-Dawley Rats | Charles River Laboratories | RRID: RGD_734476 |
| Oligonucleotides | ||
| Control ASO (5′- CCTTCCCTGAAGGTTCCTCC-3′) | Samuel et al., 2007 | Ionis Pharmaceuticals |
| PKCε ASO (5′-GCCAGCTCGATCTTGCGCCC-3′) | Samuel et al., 2007 | Ionis Pharmaceuticals |
| Recombinant DNA | ||
| PCRT7 tetR pLtetO MBP-rPKL S12TAG Amp | This Paper | Addgene Cat#107155 |
| PCRT7 tetR pLtetO MBP-rPKL WT Amp | This Paper | Addgene Cat#107156 |
| PCRT7 tetR pLtetO MBP-rPKL S113TAG Amp | This Paper | Addgene Cat#107157 |
| PCRT7 tetR pLtetO MBP-rPKM2 Amp | This Paper | Addgene Cat#107158 |
| Targeted CDK peptide library in pCRT7 NT Topo arabinose | This Paper | Addgene Cat#73446 |
| SepOTSλ | Pirman et al., 2015 | Addgene Cat#68292 |
| Software and Algorithms | ||
| Maxquant v 1.5.1.2 | Cox and Mann, 2008 | http://www.coxdocs.org/doku.php?id=maxquant:common:download_and_installation |
| Perseus v 1.5.0.31 | Tyanova et al., 2016 | http://www.coxdocs.org/doku.php?id=perseus:start |
| MASCOT | Perkins et al., 1999 | http://www.matrixscience.com |
| pLogo v 1.2.0 | O’Shea et al., 2013 | https://plogo.uconn.edu/ |
| MotifX v 1.2 | Schwartz and Gygi, 2005 | https://motif-x.med.harvard.edu/motif-x.html |
| Omicsintegrator | Tuncbag et al., 2016 | http://fraenkel-nsf.csbi.mit.edu/omicsintegrator/ |
| MagTran v 1.03 b2 | Zhang and Marshall, 1998 | Available from the developer, Dr. Zhongqi Zang |
| Skyline v 4.2.0.19072 | MacLean et al., 2010 | https://skyline.ms/project/home/software/Skyline/begin.view |
| Graphpad Prism v 7.01 | Graphpad Software | https://www.graphpad.com/ |
| Codon Optimization Tool | IDT | http://www.idtdna.com/site/account/login?returnurl=%2FCodonOpt |
| HKL2000 | HKL research, Inc. | http://www.hkl-xray.com/hkl-2000 |
| Phaser v 2.7.0 | McCoy et al., 2007 | https://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| COOT v 0.8.7 | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Refmac5.8.0230 | Murshudov et al., 2011 | http://www.ccp4.ac.uk/html/refmac5.html |
| CCP4 software suite | Winn et al., 2011 | http://www.ccp4.ac.uk/ |
| Phenix v 1.13-2998 | Adams et al., 2010 | https://www.phenix-online.org |
| Other | ||
| Regular Chow | Harlan | Cat#TD2018 |
| High-fat safflower oil-based diet | Research Diets | Cat#D12492 |
| Kinase Assay Buffer I | SignalChem | Cat#K01-09-01 |
| Kinase Dilution Buffer III | SignalChem | Cat#K23-09-01 |
| Lysyl Endopeptidase | Wako | Cat#125-02543 |
| Sequencing Grade Modified Trypsin | Promega | Cat#V511x |
| Titansphere TiO2 | GL Science | Cat#5020-75000 |
| Acid Labile Surfactant | Protea | Cat#ALS-110 |
| CytoBuster | Millipore | Cat#71009 |
| Superdex S200 Column | GE Healthcare Lifesciences | Cat# 17517501 |
| Hypersep Retain PEP 30mg Cartridge | Thermo Scientific | Cat# 60107-201 |
| Ultra-4 Centrifugal Filter Unit | Amicon | Cat#UFC8030 |
Highlights.
High-fat diet induces, whereas PKCε-knockdown rescues hepatic PKL S113 phosphorylation
PKL phosphorylation at S113 by CDKs or S12 by PKA increases gluconeogenesis
Neither S12 or S113 phosphorylation alters PKL kinetics or structure
CDK1-mediated phosphorylation of PKL S113 traps PKL in the nucleus
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants R01GM117230, P01DK017433, and P30DK45735 to J.R.; R01 DK110181 and R01 DK108283 to R.G.K.; and R01 DK-11674, R01 DK-11968, R01 DK-113984, and P30 DK-045735 to G.I.S., as well as generous funding from Agios Pharmaceuticals. B.M.G. and K.W.B. are supported by National Science Foundation Graduate Research Fellowship grant DGE1122492. M.C.P. is supported by NIH Medical Scientist Training Program grants T32GM007205 and F30DK104596. We thank Terence Wu of the West Campus Analytical Core and Vladimir Polejaev of the West Campus Imaging Core for training on the use and maintenance of equipment, James Lee at the Dana-Farber Cancer Institute Molecular Biology Core Facility (DFCI MBCF) for the intact MS analysis, the Yale Liver Center (supported by P30 DK034989) for hepatocyte isolation, and Amanda Kedaigle and Jennifer Wilson for their help in using Omicsintegrator.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.009.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert AJ (2008). Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal. Chem 80, 62–76. [DOI] [PubMed] [Google Scholar]
- Amiram M, Haimovich AD, Fan C, Wang YS, Aerni HR, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, et al. (2015). Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol 33, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, McAllister FE, Camporez JP, Zushin PJ, Jurczak MJ, Laz-nik-Bogoslavski D, Shulman GI, Gygi SP, and Spiegelman BM (2015). An ERK/Cdk5 axis controls the diabetogenic actions of PPARgamma. Nature 517, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber KW, Miller CJ, Jun JW, Lou HJ, Turk BE, and Rinehart J (2018). Kinase Substrate Profiling Using a Proteome-wide Serine-Oriented Human Peptide Library. Biochemistry 57, 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund L (1978). Phosphorylation of rat kidney pyruvate kinase type L by cyclic 3’,5’-AMP-dependent protein kinase. Biochim. Biophys. Acta 524, 68–77. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU (1974). Methods of Enzymatic Analysis (Academic Press; ). [Google Scholar]
- Blair JB, Cimbala MA, Foster JL, and Morgan RA (1976). Hepatic pyruvate kinase. Regulation by glucagon, cyclic adenosine 3’−5’-monophosphate, and insulin in the perfused rat liver. J. Biol. Chem 251, 3756–3762. [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, and Shokat KM (2008). Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. USA 105, 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, and Heck AJ (2009). Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc 4, 484–494. [DOI] [PubMed] [Google Scholar]
- Cantley JL, Yoshimura T, Camporez JP, Zhang D, Jornayvaz FR, Ku-mashiro N, Guebre-Egziabher F, Jurczak MJ, Kahn M, Guigni BA, et al. (2013). CGI-58 knockdown sequesters diacylglycerols in lipid droplets/ER-preventing diacylglycerol-mediated hepatic insulin resistance. Proc. Natl. Acad. Sci. USA 110, 1869–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MF, and Schwartz D (2011). Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13 (Unit 13), 15–24. [DOI] [PubMed] [Google Scholar]
- Claus TH, El-Maghrabi MR, and Pilkis SJ (1979). Modulation of the phosphorylation state of rat liver pyruvate kinase by allosteric effectors and insulin. J. Biol. Chem 254, 7855–7864. [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Ekman P, Dahlqvist U, Humble E, and Engström L (1976). Comparative kinetic studies on the L-type pyruvate kinase from rat liver and the enzyme phosphorylated by cyclic 3’, 5’-AMP-stimulated protein kinase. Biochim. Biophys. Acta 429, 374–382. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliú JE, Hue L, and Hers HG (1976). Hormonal control of pyruvate kinase activity and of gluconeogenesis in isolated hepatocytes. Proc. Natl. Acad. Sci. USA 73, 2762–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AW, and Alontaga AY (2009). The impact of ions on allosteric functions in human liver pyruvate kinase. Methods Enzymol 466, 83–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AW, and Tang Q (2009). An activating interaction between the unphosphorylated n-terminus of human liver pyruvate kinase and the main body of the protein is interrupted by phosphorylation. Biochemistry 48, 3816–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassaway BM, Petersen MC, Surovtseva YV, Barber KW, Sheetz JB, Aerni HR, Merkel JS, Samuel VT, Shulman GI, and Rinehart J (2018). PKCε contributes to lipid-induced insulin resistance through cross talk with p70S6K and through previously unknown regulators of insulin signaling. Proc. Natl. Acad. Sci. USA 115, E8996–E9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Sasmor H, Manoharan M, and Levin AA (2001). Pharmacokinetic properties of 2’-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther 296, 890–897. [PubMed] [Google Scholar]
- Goss VL, Hocevar BA, Thompson LJ, Stratton CA, Burns DJ, and Fields AP (1994). Identification of nuclear beta II protein kinase C as a mitotic lamin kinase. J. Biol. Chem 269, 19074–19080. [PubMed] [Google Scholar]
- Hao P, Ren Y, Dutta B, and Sze SK (2013). Comparative evaluation of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and high-pH reversed phase (Hp-RP) chromatography in profiling of rat kidney proteome. J. Proteomics 82, 254–262. [DOI] [PubMed] [Google Scholar]
- Heinemann IU, Rovner AJ, Aerni HR, Rogulina S, Cheng L, Olds W, Fischer JT, Söll D, Isaacs FJ, and Rinehart J (2012). Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 586, 3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Takeda S, and Hunt T (2008). Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat. Rev. Mol. Cell Biol 9, 910–916. [DOI] [PubMed] [Google Scholar]
- Holyoak T, Zhang B, Deng J, Tang Q, Prasannan CB, and Fenton AW (2013). Energetic coupling between an oxidizable cysteine and the phosphorylatable N-terminus of human liver pyruvate kinase. Biochemistry 52, 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, and Gerber SA (2011). Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal. Chem 83, 7635–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Sano H, Keller SR, Lienhard GE, and Gerber SA (2015). SPECHT—single-stage phosphopeptide enrichment and stable-isotope chemical tagging: quantitative phosphoproteomics of insulin action in muscle. J. Proteomics 114, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl EA, and Cottam GL (1977). Kinetic studies with L-type pyruvate kinase from rats either fed a high carbohydrate, low protein diet or starved. Biochim. Biophys. Acta 484, 49–58. [DOI] [PubMed] [Google Scholar]
- Kraegen EW, Clark PW, Jenkins AB, Daley EA, Chisholm DJ, and Storlien LH (1991). Development of muscle insulin resistance after liver insulin resistance in high-fat-fed rats. Diabetes 40, 1397–1403. [DOI] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni H-R, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, et al. (2013). Genomically recoded organisms expand biological functions. Science 342, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, Chim H, Lim JH, Ruan HB, Yang X, et al. (2014). Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park YY, Cho MY, Chae S, Yoo YS, Kwon MH, Lee CW, and Cho H (2015). The chromatin remodeller RSF1 is essential for PLK1 deposition and function at mitotic kinetochores. Nat. Commun 6, 7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundby A, Secher A, Lage K, Nordsborg NB, Dmytriyev A, Lundby C, and Olsen JV (2012). Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun 3, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Acedo P, Gupta V, and Carroll KS (2014). Proteomic analysis of peptides tagged with dimedone and related probes. J. Mass Spectrom 49, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ML, Jensen LJ, Diella F, Jørgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. (2008). Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal 1, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, and Santocanale C (2006). Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem 281, 10281–10290. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, and Vagin AA (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, and Verdin E (2007). Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J. Biol. Chem 282, 19546–19555. [DOI] [PubMed] [Google Scholar]
- O’Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, and Schwartz D (2013). pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Tsujimoto M, and Nagata K (2002). The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 13, 2016–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookata K, Hisanaga S, Sugita M, Okuyama A, Murofushi H, Kitazawa H, Chari S, Bulinski JC, and Kishimoto T (1997). MAP4 is the in vivo substrate for CDC2 kinase in HeLa cells: identification of an M-phase specific and a cell cycle-independent phosphorylation site in MAP4. Biochemistry 36, 15873–15883. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, Lüscher-Fir-zlaff J, Vervoorts J, Lasonder E, Kremmer E, Knöll B, and Lüscher B (2008). The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol 180, 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, and Söll D (2011). Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, Seghezzi W, Paruch K, Dwyer MP, Doll R, et al. (2010). Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther 9, 2344–2353. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, and Cottrell JS (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Petersen MC, Madiraju AK, Gassaway BM, Marcel M, Nasiri AR, Butrico G, Marcucci MJ, Zhang D, Abulizi A, Zhang X-M, et al. (2016). Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J. Clin. Invest 126, 4361–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone A, Adamo ME, Cheg C, and Kettenbach AN (2016). Identification of candidate CDK1 substrates in mitosis by quantitative phosphoproteomics. Mol. Cell. Proteomics 15, 2448–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirman NL, Barber KW, Aerni HR, Ma NJ, Haimovich AD, Rogulina S, Isaacs FJ, and Rinehart J (2015). A flexible codon in genomically recoded Escherichia coli permits programmable protein phosphorylation. Nat. Commun 6, 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, and Mann M (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663–670. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896–1906. [DOI] [PubMed] [Google Scholar]
- Razick S, Magklaras G, and Donaldson IM (2008). iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M, Schipany K, and Hengstschläger M (2013). Merging high-quality biochemical fractionation with a refined flow cytometry approach to monitor nucleocytoplasmic protein expression throughout the unperturbed mammalian cell cycle. Nat. Protoc 8, 602–626. [DOI] [PubMed] [Google Scholar]
- Samuel VT, Liu ZX, Qu X, Elder BD, Bilz S, Befroy D, Romanelli AJ, and Shulman GI (2004). Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J. Biol. Chem 279, 32345–32353. [DOI] [PubMed] [Google Scholar]
- Samuel VT, Liu ZX, Wang A, Beddow SA, Geisler JG, Kahn M, Zhang XM, Monia BP, Bhanot S, and Shulman GI (2007). Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J. Clin. Invest 117, 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, and Barbacid M (2007). Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Hilton MB, and Kaldis P (2008). p21 Inhibits Cdk1 in the absence of Cdk2 to maintain the G1/S phase DNA damage checkpoint. Mol. Biol. Cell 19, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer N, Gassaway BM, Haimovich AD, Isaacs FJ, Rinehart J, and Regan L (2014). Designed phosphoprotein recognition in Escherichia coli. ACS Chem. Biol 9, 2502–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, and Gygi SP (2005). An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol 23, 1391–1398. [DOI] [PubMed] [Google Scholar]
- Shiota C, Coffey J, Grimsby J, Grippo JF, and Magnuson MA (1999). Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J. Biol. Chem 274, 37125–37130. [DOI] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Ue-hara-Bingen M, Weis K, and Heald R (2011). Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol 6, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Harano Y, Sue F, and Morimura H (1967). Crystallization, characterization and metabolic regulation of two types of pyruvate kinase isolated from rat tissues. J. Biochem 62, 71–91. [DOI] [PubMed] [Google Scholar]
- Tuncbag N, Gosline SJ, Kedaigle A, Soltis AR, Gitter A, and Fraenkel E (2016). Network-Based Interpretation of Diverse High-Throughput Datasets through the Omics Integrator Software Package. PLoS Comput. Biol 12, e1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Hein M, Geiger T, Mann M, and Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Wang H, Nicolay BN, Chick JM, Gao X, Geng Y, Ren H, Gao H, Yang G, Williams JA, Suski JM, et al. (2017). The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, and Flügge UI (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Wilson-Grady JT, Haas W, and Gygi SP (2013). Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods 61, 277–286. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, Nacht AS, Samino S, Font-Mateu J, Vicent GP, et al. (2016). ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 352, 1221–1225. [DOI] [PubMed] [Google Scholar]
- Xu YF, Zhao X, Glass DS, Absalan F, Perlman DH, Broach JR, and Rabinowitz JD (2012). Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation. Mol. Cell 48, 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Yamakita Y, Yoshida K, Takiguchi K, and Matsumura F (1995). Characterization of the COOH terminus of non-muscle caldesmon mutants lacking mitosis-specific phosphorylation sites. J. Biol. Chem 270, 4023–4030. [DOI] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, and Lu Z (2012). ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol 14, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, and Marshall AG (1998). A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom 9, 225–233. [DOI] [PubMed] [Google Scholar]
- Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, and Mohammed S (2013). Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res 12, 260–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Project Name: Rat Liver HFD versus Chow, HFD versus PKCepsilon ASO; Project accession: PXD010209. PKL pS113 is deposited in the wwPDB as PDB: 6ECK under deposition ID D_1000236082 and PKL pS12 as PDB: 6ECH under deposition ID D_1000236056.