Abstract
High grade serous ovarian cancer (HGSOC) is the most lethal gynecological malignancy in women worldwide and the fifth most common cause of cancer related deaths among U.S. women. New therapies are needed to treat HGSOC particularly since most patients develop resistance to current frontline therapies. Many natural product and fungal metabolites exhibit anti-cancer activity and represent an untapped reservoir of potential new agents with unique mechanism(s) of action. Verticillin A, an epipolythiodioxopiperazine (ETP) alkaloid, is one such compound and our recent advances in fermentation and isolation are now enabling evaluation of its anti-cancer activity. Verticillin A demonstrated cytotoxicity in HGSOC cell lines in a dose-dependent manner with a low nM IC50. Furthermore, treatment with verticillin A induced DNA damage and caused apoptosis in HGSOC cell lines OVCAR4 and OVCAR8. RNA-Seq analysis of verticillin A treated OVCAR8 cells revealed an enrichment of transcripts in the apoptosis signaling and the oxidative stress response pathways. Mass spectrometry histone profiling confirmed reports that verticillin A caused epigenetic modifications with global changes in histone methylation and acetylation marks. To facilitate in vivo delivery of verticillin A and to monitor its ability to reduce HGSOC tumor burden, verticillin A was encapsulated into an expansile nanoparticle (verticillin A-eNP) delivery system. In an in vivo human ovarian cancer xenograft model, verticillin A-eNPs decreased tumor growth and exhibited reduced liver toxicity compared to verticillin A administered alone. This study confirmed that verticillin A has therapeutic potential for treatment of HGSOC and that encapsulation into expansile nanoparticles reduced liver toxicity.
Keywords: Verticillin A, natural products, apoptosis, high grade serous ovarian cancer, cancer therapeutics, nanoparticles
Introduction
Ovarian cancer is the fifth leading cause of death among women and is the most lethal tumor type of the female reproductive tract (1). In 2019, an estimated 22,530 women in United States will be diagnosed with ovarian cancer and about 13,980 women will succumb to the disease (2). The most common and deadly histological subtype of ovarian cancer is high grade serous ovarian cancer (HGSOC). Current treatment for HGSOC entails cytoreductive surgery and chemotherapy. However, most patients develop chemoresistance and ultimately die from recurrent disease. Chemoresistance is multi-faceted and likely arises both from mechanisms of resistance to the current therapy, paclitaxel and carboplatin, or through immune cell evasion (3–6).
One avenue to identify new therapies that may avert the chemoresistance problem is through the investigation of natural product drug leads with both unique chemical structures and modes of action, such as the verticillins (7). The verticillins are epipolythiodioxopiperazine (ETP) alkaloids, and largely exist as dimers containing both diketopiperazine moieties and disulfide bridges. This group of secondary metabolites is usually isolated from terrestrial and marine filamentous fungi, such as Verticillium sp., Penicillium sp. and Gliocladium sp. that belong to Sordariomycetes and Eurotiomycetes (8,9). Verticillin A was the first discovered analogue in 1970, and to date, 27 verticillin analogues are described in the literature (8–10).
Enabled by recent advances in fungi fermentation and isolation, the role of verticillin A as an anti-cancer agent is actively being investigated (8,11). Verticillin A induces cytotoxicity in human breast carcinoma, large cell lung carcinoma, astrocytoma, colorectal adenocarcinoma and melanoma cell lines (8). Mechanistically, verticillin A selectively inhibits histone methyltransferases (HMTases) such as SUV39H1, SUV39H2, G9a, HTM, MLL1 and GLP (12,13). In colon carcinoma, verticillin A inhibited H3K9 methylation on the FAS promoter, restored Fas expression, and caused cell death by apoptosis (12). Furthermore, mice treated with a combination of verticillin A and 5-fluorouracil displayed significantly smaller tumors and sensitized metastatic colon carcinoma cells to 5-fluorouracil (12). Additionally, verticillin A suppressed metastatic colon cancer cell immune evasion and chemoresistance (12). In pancreatic cancer cells, verticillin A inhibited MLL1, a HMTase responsible for H3K4 methylation, leading to decreases in H3K4me3 levels and PD-L1 expression. Lower PD-L1 expression led to reduced binding to PD-1 and activation of an immune response along with an enhanced chemotherapeutic response (13). In a recent study with pancreatic ductal adenocarcinoma cells, verticillin A differentially altered H3K9me3 and H3K4me3 levels leading to expression of pro-apoptotic genes and inhibition of anti-apoptotic genes (14).
Since verticillin A acts through a variety of HMTases resulting in epigenome changes by histone marks, the genomic landscape of different tumor types that arise from different tissues will dictate gene expression; thus, defining the anti-cancer action. Therefore, in order to determine if ovarian cancer might respond to this class of compounds, this study focused on the mode of action of verticillin A in HGSOC cells. Verticillin A induced cell death in HGSOC cells and, when encapsulated into a nanoparticle, it reduced toxicity and tumor burden in vivo. Specifically, verticillin A altered histones, significantly changed gene expression, and induced DNA damage via oxidative stress. DNA damage leading to γH2.AX foci occurred rapidly and co-treatment with n-acetyl-cysteine blocked apoptosis caused by verticillin A. Animals treated with verticillin A loaded expansile nanoparticles (verticillin A-eNPs) reduced tumor burden in vivo and showed no morphological signs of liver toxicity compared to free verticillin A. Together, these findings validate verticillin A as a potential anti-cancer compound with a novel mechanism of action in HGSOC and highlight verticillin A-eNPs as a promising drug delivery strategy.
Materials and Methods
Verticillin A purification and synthesis of expansile nanoparticles
Verticillin A (Supplementary Figure S3A) was isolated and characterized from Clonostachys rogersoniana (strain MSX59553) as detailed previously following a recently published fermentation optimization strategy (8,11). The purity of the isolated verticillin A was assessed via UPLC and 1H-NMR (Supplementary Figure S3B–C). The synthesis of verticillin A-eNPs and eNPs was performed using a miniemulsion polymerization method (Supplementary Figure S2A) as previously described(15–17). Scanned electron microscope images were taken (Supplementary Figure S2B), and a calibration curve of verticillin A was built to calculate the encapsulation efficiency, which was found to be 76.9% (Supplementary Figure S2C). These data demonstrate the presence of 0.9 mg of verticillin A/ml of the expansile nanoparticles solution. Nanoparticles were stored at 4°C until the time of dosage.
Compounds/ Reagents
Verticillin A, UNC0638 (Sigma #U4885) and taxol/paclitaxel (Sigma #T7402) were resuspended in dimethyl sulfoxide (DMSO). N-Acetyl-L-cysteine (Sigma #A9165) was freshly prepared using sterile deionized water. Hydrogen peroxide (H2O2) (Thermo Fisher #H325) was freshly diluted with the media before addition to the cells. Final vehicle concentration < 0.1%(v/v).
Cell culture
OVCAR8 and OVCAR8-RFP (OVCAR8 cells expressing red fluorescent protein (RFP)) were a gift from Sharon Stack at the University of Notre Dame, immortalized human ovarian surface epithelial cells (IOSE80) were a gift from Nelly Auersperg at the University of Vancouver. Normal human fallopian tube cells FT33 were a gift from Ronny Drapkin at the University of Pennsylvania. OVSAHO cells (Japanese Cell Bank) were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (P/S-final concentration: 100 I.U./ml and 100 μg/ml, respectively). OVCAR4 cells (National Cancer Institute) were grown in RPMI 1640 supplemented with L-glutamine (2 mM), 10% FBS and P/S. OVCAR8, OVCAR8-RFP, HEPG2 and U87 cells were grown in DMEM with 10% FBS and P/S. IOSE80 cells were grown in v/v 50% Medium 199 and v/v 50% MCBD with 15% FBS, P/S, L-glutamine (2 mM), and 11 ng/ml epithelial growth factor. FT33 cells were grown in DMEM-Ham’s F12 supplemented with P/S and 2% Ultroser-G. All cultured cells were mycoplasma free and validated by short tandem repeat analysis in 2017. Cells were passaged a maximum of 20 times and maintained in a humidified incubator at 37°C in a 5% CO2 environment.
Cell viability assay
3000 cells were seeded in triplicates in a clear flat-bottomed 96-well plate and allowed to attach overnight. Compounds were suspended in DMSO and added to the cells. Cells were incubated for 24, 48 and 72 hours in the cell culture incubator and fixed with 20% Trichloroacetic acid. Cell viability was determined using 0.04% Sulforhodamine B (SRB) via colorimetric detection at 505 nm (18). Normalization was performed using vehicle control (DMSO) and dose-response curves were generated using GraphPad Prism Software.
Spheroid assay
OVCAR8 cells were trypsinized and 5000 cells were resuspended in 80 μl of media per well in 96 well round bottom Ultra Low Attachment plate (Corning 07-201-680). After 4 days the spheroids were treated with 20 μl of compound for 72 hrs and the plates were incubated at 37°C for 7 days. The plate was equilibrated to room temperature for 30 mins and 100 μl of CellTiter-Glo® 3D reagent (Promega) was added. The plate was incubated on a shaker for 30 minutes to obtain complete lysis and luminescence was recorded using Synergy Mx plate reader (BioTek).
Histone profiling
Cells were seeded in 10 cm dishes and treated with verticillin A and vehicle control (DMSO) for 24 hrs. Cells were pelleted, washed with PBS and epiproteomic histone modification profiling was performed at Northwestern Proteomics Core Facility at Northwestern University as previously described (19). Relative abundance (%) of histone marks was calculated based on peptide peak areas extracted from the raw LC/MS data obtained.
RNA-sequencing
RNA was isolated from verticillin A and vehicle (DMSO) treated cells using Qiagen RNAeasy Mini kit (#74104) as per manufacturer’s protocol. RNA libraries (3 technical replicates/treatment) were created. RNA quality determination, mRNA enrichment, library construction, sequencing, and transcriptome statistical analysis were performed at the Genomics Core Facility at Northwestern University.
cDNA synthesis and qRT-PCR analysis
Total RNA (1 μg) was converted to cDNA using iScript™ cDNA synthesis kit (Bio-Rad). qRT-PCR measurements were performed using the CFX connect Real-Time PCR detection system (Bio-Rad) and SYBR green (Roche) according to manufacturer’s protocol. Samples were normalized to the housekeeping gene, GAPDH.
Alkaline comet assay
Agarose coated slides were prepared using 1% agarose (Thermo Fisher # BP160) and allowed to dry. Cells were trypsinized, washed in PBS and resuspended in 0.5% low melting agarose (Sigma # A9414). 300 μl of cell suspension was added to the coated slide to form a thin uniform layer and allowed to gel at 4°C for 30 mins. Cells were lysed by incubating the slides for 3 hrs with chilled lysis buffer (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris pH 10, 1% N-laurylsarcosine, 0.5% Triton X-100). Cells were subjected to alkaline electrophoresis (25V, 300mA, 40 mins) using electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13) at 4°C. Slides were coated with neutralization buffer (0.5 M Tris pH 7.5) and stained with 2 μg/ml ethidium bromide, washed and covered with a coverslip before visualization. Images were acquired using 20X objective on a Nikon Eclipse E600 microscope using DS-Ri1 digital camera and NIS Elements software (Nikon Instruments). Comet tail moments were analyzed by TriTek CometScore™ software.
Reactive oxygen species detection assay
Cells plated on glass coverslips were treated with verticillin A, H2O2 and vehicle control (DMSO). CellROX™ Green reagent (Invitrogen) was added (final concentration 5 μM) and incubated for 30 mins at 37°C. Cells were washed with PBS and fixed with 4% paraformaldehyde. Images were acquired using 40X objective on a Nikon Eclipse E600 microscope using a DS-Ri1 digital camera and NIS Elements software (Nikon Instruments).
2D foci assay
200 cells were seeded in a 60 mm plate and allowed to attach overnight. Cells were treated with verticillin A and vehicle control (DMSO) for 8 hrs. Following 15 days incubation, cells were fixed using 4% paraformaldehyde and stained with 0.05% crystal violet. Plates were washed using distilled water to minimize background. Images were acquired using FlourChem™ E system (ProteinSimple, Santa Clara, CA). Colonies were counted using ImageJ (imagej.nih.gov).
Immunoblot analysis
Cell lysates were prepared using RIPA lysis buffer (50 mM Tris pH 7.6, 150 mM NaCl, 1% Triton X-100, 0.1% SDS) supplemented with protease (Roche Applied Science #4693159001) and phosphatase (Sigma-Aldrich #P0044) inhibitors. Concentration of protein in lysates was determined by Bradford assay (Biorad #5000205) and proteins were resolved on SDS-PAGE gel. Proteins were transferred to nitrocellulose membrane and blocked in 5% non-fat milk. Primary antibody (Table 1) was incubated with the membrane overnight at 4°C. Membrane was washed and incubated in secondary antibody (Table 2) conjugated with Horseradish peroxidase (HRP). The membrane was washed, and protein bands were detected with SuperSignal™ West Femto substrate (Thermo Scientific #34095) and imaged on a FluorChem E system (ProteinSimple).
Table 1.
Primary antibodies:
| Antibody | Source | Dilution for WB | Dilution for Immunofluorescence (IF)/Immunohistochemistry (IHC) |
|---|---|---|---|
| Anti-rabbit PARP | CST #9542 | 1:1000 | - |
| Anti-rabbit cPARP | CST #9541 | - | 1:100 (IHC) |
| Anti-rabbit γH2A.X | CST #9718 | 1:500 | 1:100 (IHC, IF) |
| Anti-rabbit Bax | CST #5023 | 1:500 | - |
| Anti-rabbit actin | Sigma #A2066 | 1:5000 | - |
Table 2.
Secondary antibodies:
| Antibody | Source | Dilution for WB | Dilution for immunofluorescence |
|---|---|---|---|
| Anti-rabbit IgG-HRP | CST #7074 | 1:10000 | - |
| Anti-rabbit Alexa Fluor 488 | Invitrogen #A-11034 | - | 1:1000 |
Immunofluorescence analysis of γH2.AX foci
Cells grown on glass coverslips were fixed using 4% paraformaldehyde, permeabilized with 0.2% TritonX-100 and blocked with 1% BSA in PBS. Cells were then incubated with γH2.AX primary antibody (diluted in blocking solution) for 1 hr at room temperature. The cells were washed with washing buffer (PBS with 0.05% TritonX-100) and then incubated with fluorescent secondary antibody (diluted in blocking solution) for 1 hr at room temperature. Cells were washed thrice with washing buffer. Nuclei were stained with DAPI (0.1 μg/ml) (Thermo Fisher # EN62248) for 10 mins at room temperature. Cells were washed using washing buffer and coverslips were mounted on glass slides using mounting media (Vector laboratories #H-1000). Images were acquired using 40X objective on a Nikon Eclipse E600 microscope using DS-Ri1 digital camera and NIS Elements software (Nikon Instruments).
Annexin V/Propidium iodide staining
Cells were seeded in a 60 mm plate and allowed to attach overnight. Cells were treated with verticillin A, taxol and vehicle control (DMSO) for 24 hrs. Media was collected and cells were trypsinized and subjected to Annexin V-FITC/propidium iodide apoptosis assay (Nexcelom Biosciences) according to manufacturer’s instructions. K2 Cellometer was used to detect fluorescence using FCS express software. Gating channels were applied as per manufacturer’s protocol.
Mitochondrial toxicity assay
HEPG2 (hepatocellular carcinoma) and U87 (glioblastoma) cell lines were used to determine if verticillin A caused mitochondrial toxicity, as mitochondrial toxicity is prone to occur in the brain and liver tissue. Mitochondrial toxicity was evaluated using a Promega Mitochondrial ToxGlo™ kit according to the manufacturer’s protocol. Briefly, 10,000 cells/well were seeded in 96 well plates in either glucose (25 mM) or galactose (10 mM) supplemented media and allowed to adhere for 8 hrs. Cells were incubated with dilutions of oligomycin A (positive control) and verticillin A for 90 mins prior to assay. Cells were incubated for 30 mins with a cell impermeable, fluorogenic substrate, and fluorescence (Ex/Em 485/525nm) was measured. Lysis buffer was added, and net ATP levels were determined by luminescence measurement.
Animals and xenograft experiments
All animals were treated in accordance with NIH Guidelines for the Care and Use of Laboratory Animals and the established Institutional Animal Use and Care protocol at the University of Illinois, Chicago. Xenograft studies utilized NCr nu/nu athymic female mice 6-8 weeks in age (Taconic). Mice were housed in a temperature and light-controlled environment (12 hours light and 12 hours dark) and provided food and water ad libitum. For xenograft experiments, OVCAR8-RFP cells (5 x 106) were injected intraperitoneally (IP) per mouse and tumor growth was monitored using Xenogen IVIS® Spectrum In Vivo Imaging System (PerkinElmer) as previously described (20). Once all the mice formed tumors (~4 weeks), the mice were separated into 2 treatment groups and dosed once every two days with 0.5 mg/kg of verticillin A encapsulated nanoparticles (eNP-VA) and empty nanoparticles (eNP) for a total of 12 days. Mice were IVIS imaged twice weekly (535 nm excitation, and 620 nm emission, Exposure time: 2 seconds, F stop: 2). Living Image 4.0 software was used to quantify the average abdominal radiant efficiency and normalization was performed using Day 0 radiant efficiency. At week 7, all animals were sacrificed and tumors were collected for histological analysis.
Tumor immunohistochemistry
Tumors were fixed with 4% paraformaldehyde followed by dehydration and paraffin embedding. Paraffin blocks were sectioned using standard histologic procedures. Hematoxylin and eosin staining (H&E) and immunohistochemistry were performed as previously described (21). Sections were incubated with primary antibody (Table 1) followed by incubation with biotin conjugated secondary antibody (Table 2). Sections were developed using 3,3-diaminobenzidine (DAB) to enable chromogenic detection. Tissues with primary antibody treatment were used as a negative control. Images were acquired on a Nikon Eclipse E600 microscope using a DS-Ri1 digital camera and NIS Elements software (Nikon Instruments).
Statistical analysis
Data presented are mean ± standard error of the mean (SEM) and represent at least 3 independent biological replicates. Statistical analysis was carried out using GraphPad Prism software. Statistical significance was determined by Student’s unpaired t–test, one-way ANOVA, or two-way ANOVA with Dunnett’s multiple comparisons or Tukey’s posthoc test as mentioned in figure legends. p<0.05 was considered statistically significant (* p<0.05; ** p <0.01; *** p<0.001; **** p<0.0001).
Results
Verticillin A induces cytotoxicity in HGSOC cell lines in vitro
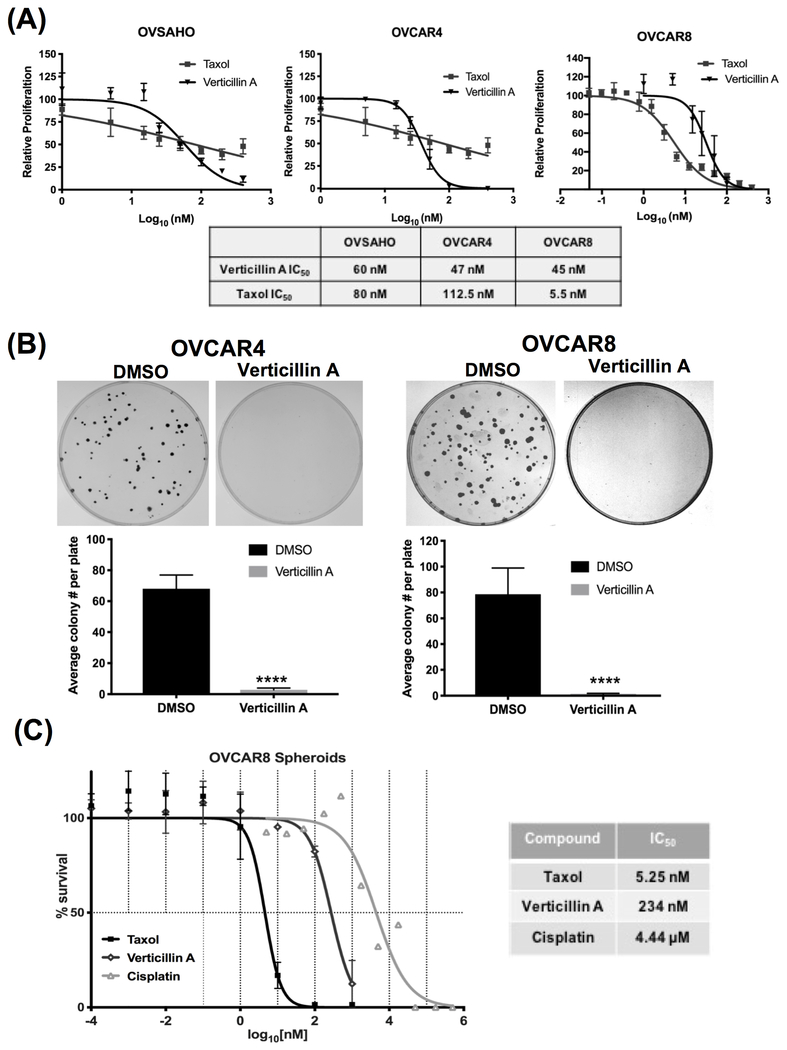
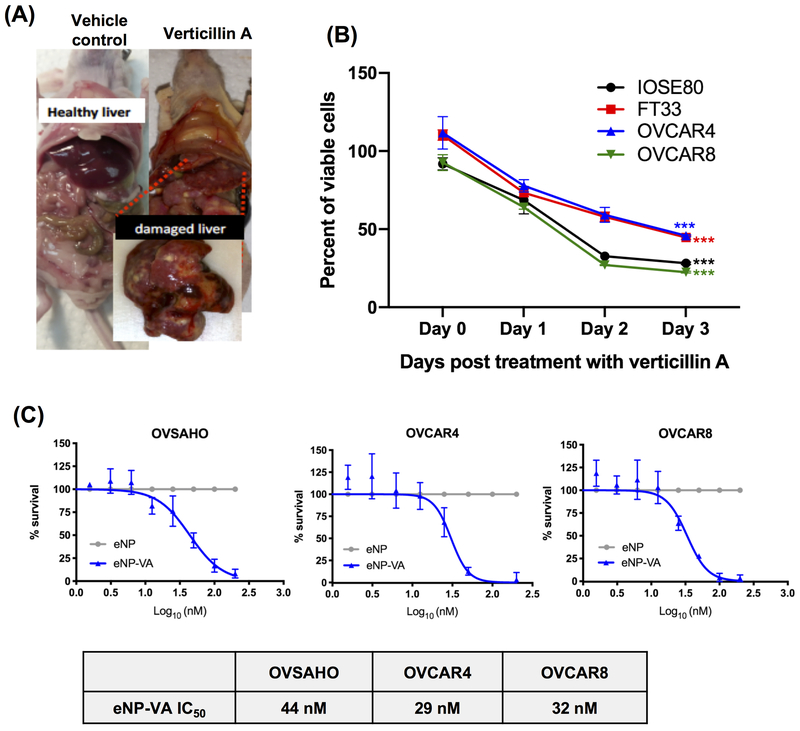
Verticillin A has been shown to inhibit the growth of colon cancer, pancreatic cancer, leiomyosarcoma and malignant peripheral nerve sheath tumor cells (12–14,22). In order to determine if verticillin A inhibited growth of ovarian cancer cells, three validated models of HGSOC, OVSAHO, OVCAR4 and OVCAR8 were used. Taxol was used as positive control. Verticillin A was found to inhibit the growth of all three cell lines with IC50 values of 60 nM, 47 nM and 45 nM in OVSAHO, OVCAR4 and OVCAR8, respectively (Figure 1A). In order to determine whether the growth inhibition was due to cytotoxic effects exerted by verticillin A, a 2D foci assay was performed. Cells were treated with verticillin A for 8 hrs and foci were allowed to form for 2 weeks. As shown in Figure 1B, treatment with verticillin A completely abrogated foci formation in OVCAR4 and OVCAR8 cells compared to vehicle control suggesting a cytotoxic effect and not a cytostatic effect. Furthermore, to determine if verticillin A affected cell viability of tumor spheroids, OVCAR8 spheroids were generated and treated with vehicle (DMSO), verticillin A, taxol and cisplatin. A spheroid viability assay was performed, and it was found that verticillin A treatment caused disintegration of OVCAR8 spheroids. As shown in Figure 1C, verticillin A treatment reduced spheroid viability with an IC50 value of 234 nM.
Figure 1. Verticillin A induces apoptosis in HGSOC cell lines in vitro.
(A) OVSAHO, OVCAR4 and OVCAR8 cells were treated with vehicle (DMSO), verticillin A (50 nM) and chemotherapeutic control taxol (10 nM) for 72 hrs. Dose-response curves were generated and normalized to vehicle control. IC50 values are denoted in the table. (B) OVCAR4 and OVCAR8 cells were treated with vehicle and verticillin A (50 nM) for 8 hrs. Following drug incubation, media was changed and cells were incubated for 2 weeks to form colonies. Representative images of 2D foci assay performed in OVCAR4 and OVCAR8 cells are shown. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Significance tested by Student’s t-test in comparison to vehicle control. (C) OVCAR8 spheroids were treated with vehicle, verticillin A, taxol and cisplatin for 72 hours. Spheroid viability was determined using CellTiter-Glo® Cell Viability assay. IC50 values are denoted in the table. Data represents mean ± SEM from two biological replicates.
RNA Seq analysis shows upregulation of apoptosis and oxidative stress in verticillin A treated OVCAR8 cells
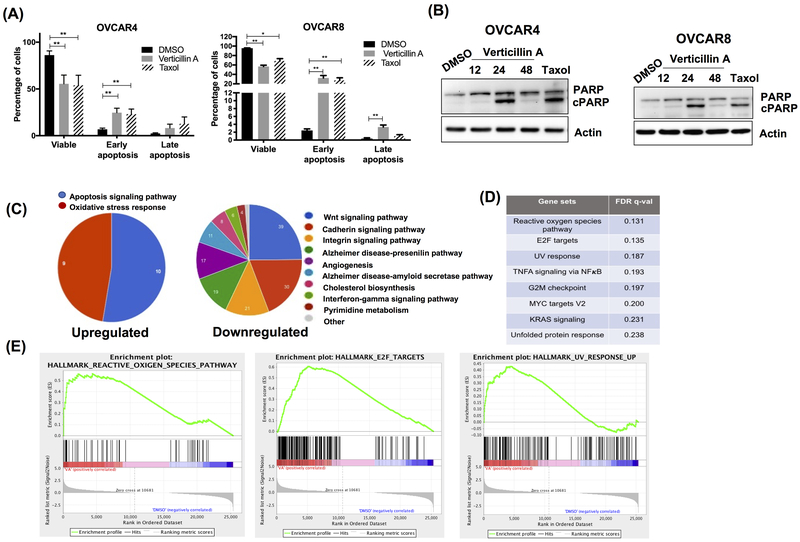
To further investigate whether verticillin A induced cytotoxicity through apoptotic cell death, annexin V-FITC (AV) and propidium iodide (PI) staining was performed. As shown in Figure 2A, verticillin A treatment significantly increased the rate of early apoptosis (AV+, PI−) in both OVCAR4 and OVCAR8 cells relative to vehicle control. Consistent to the AV-PI staining, treatment with verticillin A markedly increased level of cleaved poly ADP-ribose polymerase (cPARP), a marker for apoptosis (Figure 2B). Together, these results indicated that verticillin A inhibited growth of HGSOC cells by causing early apoptosis.
Figure 2. RNA Seq analysis shows upregulation of apoptosis and oxidative stress in Verticillin A treated OVCAR8 cells.
(A) OVCAR4 and OVCAR8 cells were treated with vehicle (DMSO), verticillin A (50 nM) and taxol (10 nM) for 24 hrs, stained with Annexin V-FITC (AV) and propidium iodide (PI) and analyzed by Nexcelom Cellometer. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Statistics were generated with one-way ANOVA with Dunnett’s multiple comparisons to vehicle control within each group. (B) Immunoblot analysis of whole cell lysates of OVCAR4 and OVCAR8 cells probed for apoptotic marker (cPARP) and actin was used as a loading control. (C) OVCAR8 cells were treated with vehicle and verticillin A for 24 hrs. Chart denotes pathway analysis of the upregulated genes by PANTHER analysis. (D) Table represent pathways identified by gene set enrichment analysis for OVCAR8 cells treated with vehicle control and verticillin A. (E) Gene set enrichment plots based upon GSEA of transcripts altered by verticillin A treatment in OVCAR8 cells.
Studies in pancreatic and colon cancer cells demonstrated that verticillin A alters histone modifications likely through inhibiting HMTases and reducing H3K9me3 levels to increase expression of Bax, a pro-apoptotic protein (14,23). Importantly, due to inhibition of HMTases and the genomic landscape of tumors, other targets such as Fas and PD-L1 were investigated in verticillin A treated HGSOC cell lines and were not significantly different. Since Bax was consistently changed in all cell lines studied in the literature, we investigated its expression and found that verticillin A treatment increased Bax expression (Supplementary Figure S1A). In order to confirm if histones were modified, a global histone modification profiling was performed using mass spectrometry. OVCAR8 cells were treated with verticillin A and vehicle control (DMSO) for 24 hrs and the relative abundance of 20 different post-translational histone modifications were quantified by LC-MS analysis. Verticillin A induced 13 histone modifications including a reduction in H3K9me2, which was consistent with findings reported in colon cancer cells (Supplementary Figure S1B). To further define the significance of histone modifications on global transcription, an RNA-sequencing analysis was performed with verticillin A treated OVCAR8 cells. 9196 genes were found to be significantly altered by verticillin A treatment demonstrating that this molecule causes major genome-wide alterations in gene expression. Compared to vehicle control, a significant upregulation of apoptosis and oxidative stress pathway genes were observed in verticillin A treated cells (Figure 2C). qRT-PCR analysis was performed to validate the top hits from apoptosis and oxidative stress pathway (Supplementary Figure S1C). Gene set enrichment analysis (GSEA) of the transcriptome identified the reactive oxygen species (ROS) pathway to be significantly upregulated in verticillin A treated cells (Figure 2D–E).
Verticillin A causes oxidative stress and DNA damage in HGSOC cells without inducing mitochondrial toxicity
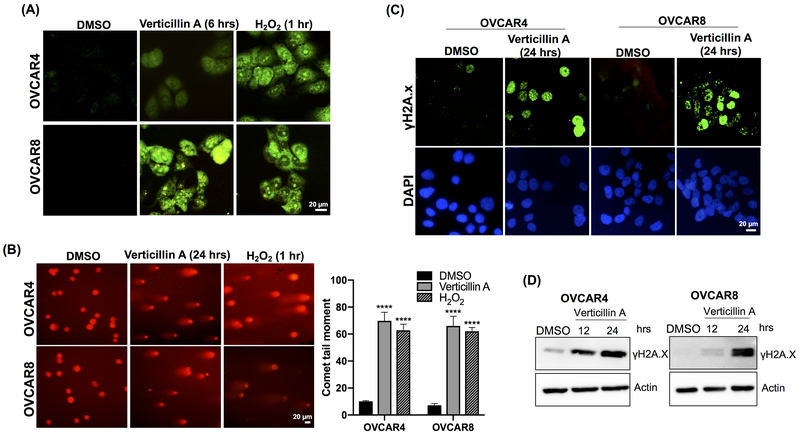
Verticillin A caused apoptosis in OVCAR4 and OVCAR8 cells and RNA-sequencing identified apoptosis and oxidative stress pathways to be significantly upregulated in OVCAR8 cells (Figure 2C–D). Since alterations in ROS had not been previously reported, the effect of verticillin A on ROS generation was examined by using the CellROX™ reagent, a cell-permeable fluorogenic probe. Upon oxidation, the probe becomes fluorescent, which was analyzed via fluorescence microscopy. H2O2 is known to induce DNA damage via ROS generation and was used as a positive control. As shown in Figure 3A, treatment with verticillin A and H2O2 led to ROS formation.
Figure 3. Verticillin A causes oxidative stress and DNA damage in HGSOC cells.
(A) Verticillin A induced oxidative stress in OVCAR4 and OVCAR8 cells was measured with CellROX™ Green reagent in cells treated with vehicle (DMSO), verticillin A (50 nM for 6 hrs) and H2O2 (50 μM for 1 hr). Images were acquired using 40X objective of a fluorescent microscope. Scale bar: 20 μm. (B) OVCAR4 and OVCAR8 cells were treated with vehicle, verticillin A (50 nM) and H2O2 (50 μM) and alkaline comet assay was performed to analyze DNA damage. Comet tail moment was used to quantify DNA damage by TriTek CometScore™ software. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Statistics were generated with one-way ANOVA with Dunnett’s multiple comparisons to vehicle control within each cell line. Scale bar: 20 μm. (C) Representative images of immunofluorescence staining of γH2A.X foci for OVCAR4 and OVCAR8 cells treated with vehicle and verticillin A (50 nM) for 24 hrs. Nuclei were stained by DAPI (0.1 μg/ml). Scale bar: 20 μm (D) Immunoblot analysis of whole cell lysates of OVCAR4 and OVCAR8 cells treated with vehicle and verticillin A (50 nM) for different time points. Lysates were probed for DNA damage marker γH2A.X and actin was used as a loading control.
Oxidative stress mediated apoptosis is frequently caused by damage to DNA (24). To determine if verticillin A treatment caused DNA damage in HGSOC cells, OVCAR4 and OVCAR8 cells were treated with verticillin A for 24 hrs and an alkaline comet assay was performed. As shown in Figure 3B, verticillin A caused DNA damage, which was observed by DNA migration smear (comet tail) under electrophoretic conditions. Cells treated with vehicle control did not form a comet tail whereas cells treated with H2O2 formed a comet tail. Analysis of comet tails showed a 5-fold increase in comet tail moment in verticillin A treated cells relative to vehicle control.
To confirm this observation, an immunofluorescence staining was performed for nuclear γH2A.X, which is a marker for DNA damage response pathways. As shown in Figure 3C, a significant increase in number of γH2A.X foci was observed for verticillin A treated cells relative to vehicle control. Consistent with the immunofluorescence analysis, expression as measured by western blots of γH2.AX was found to be increased in verticillin A treated OVCAR4 and OVCAR8 cells in a time-dependent manner (Figure 3D). A pathway leading to oxidative stress might be expected to induce mitochondrial toxicity, which is a common mode of failure for experimental agents in clinical trials. However, using a standard Mitochondrial ToxGlo™ kit, verticillin A did not demonstrate mitochondrial toxicity (Supplementary Figure S4).
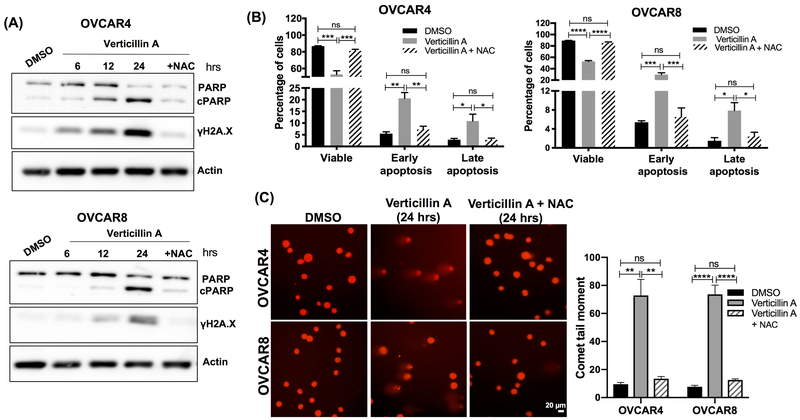
Verticillin A mediated apoptosis and DNA damage is reversed by free radical quencher N-acetyl-L-cysteine
In order to confirm if blocking oxidative stress would reduce DNA damage and apoptosis, cells were co-treated with verticillin A and antioxidant N-acetyl-L-cysteine (NAC). NAC is a ROS quencher and a combined treatment of verticillin A and NAC completely blocked apoptosis based on PARP cleavage (Figure 4A). Consistent with the western blot analysis, AV-PI staining showed that NAC blocked verticillin A stimulated apoptosis in OVCAR4 and OVCAR8 cells (Figure 4B). Similarly, induction of γH2A.X expression and comet tail was attenuated by NAC as seen in Figure 4A, 4C. This suggested that verticillin A induced DNA damage and apoptosis was mediated by oxidative stress.
Figure 4. Verticillin A mediated apoptosis and DNA damage is reversed by free radical quencher N-acetyl cysteine.
(A) Immunoblot analysis of whole cell lysates of OVCAR4 and OVCAR8 cells treated with vehicle (DMSO), verticillin A (50 nM) and combination of verticillin A (50 nM) and NAC (1 mM) for 24 hrs. Lysates were probed for apoptosis and DNA damage markers. Actin was used as a loading control. (B) OVCAR4 and OVCAR8 cells were treated with vehicle, verticillin A (50 nM) and combination of verticillin A (50 nM) and NAC (1 mM) for 24 hrs, stained with Annexin V-FITC (AV) and propidium iodide (PI) and analyzed by Nexcelom Cellometer. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Statistics were generated with one-way ANOVA with Tukey’s multiple comparisons within each group. (C) OVCAR4 and OVCAR8 cells were treated with vehicle, verticillin A (50 nM) and combination of verticillin A (50 nM) and NAC (1 mM) for 24 hrs. Alkaline comet assay was performed to analyze DNA damage. Comet tail moment was used to quantify DNA damage by TriTek CometScore™ software. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Statistics were generated with one-way ANOVA with Tukey’s multiple comparisons within each cell line. Scale bar: 20 μm.
Verticillin A encapsulated nanoparticles demonstrated reduced liver toxicity relative to free drug
To determine whether verticillin A affected tumor burden in vivo, OVCAR8-RFP cells were xenografted IP into female athymic nude mice. Tumors were allowed to form for 4 weeks and once all the mice displayed detectable tumors, the animals were treated with verticillin A and vehicle control (Cremophor EL/EtOH). However, animals treated with verticillin A showed significant gross morphological liver damage at the time of sacrifice (Figure 5A). These observations suggested that verticillin A exerted a non-specific cytotoxic effect in vivo. To confirm this, dose response curves were performed using two non-cancerous cell lines. Human ovarian surface epithelial cells IOSE80 and human fallopian tube epithelial secretory cells FT33 were treated with verticillin A and vehicle control (DMSO) for 3 days. As shown in Figure 5B, verticillin A inhibited growth of both cancerous and non-cancerous cells indicating a non-specific cytotoxic effect.
Figure 5. Verticillin A encapsulated nanoparticles demonstrate reduced liver toxicity relative to free drug.
(A) OVCAR8-RFP cells were xenografted IP to form tumors. Mice were dosed with verticillin A and vehicle (Cremophor EL/EtOH) once in 7 days (Dosage: 0.5 mg/kg). Representative images show liver damage in verticillin A treated animals. (B) Four ovarian cell lines (IOSE80, FT33, OVCAR4 and OVCAR8) were treated with vehicle (DMSO) and verticillin A (50 nM) for 72 hrs. Dose-response curves were generated and normalized to vehicle control. Each experiment was performed in three biological replicates, and data represent mean ± SEM. Statistics were generated with Student’s t-test for Day 0 and Day 3 within each cell line. (C) OVSAHO, OVCAR4 and OVCAR8 cells were treated with verticillin A encapsulated nanoparticles (eNP-VA) and empty nanoparticles (eNP) for 72 hrs. Dose-response curves were generated and normalized to eNP. Data represents mean ± SEM from three biological replicates. IC50 values are denoted in the table.
To improve the drug specificity towards tumor cells, verticillin A was encapsulated in expansile nanoparticles (verticillin A-eNPs) that have been shown to, following IP administration, localize to IP tumors of ovarian, mesothelial and pancreatic origin (16,17,25–27). Unloaded-eNPs were used as a negative control. These eNPs localize to tumors via materials-based targeting and, following internalization via macropinocytosis, release the encapsulated verticillin A upon encountering the acidic late endosome, thus reducing non-specific toxicity and increasing drug delivery to the tumor (16,17). To determine whether verticillin A loaded nanoparticles were still active, verticillin A-eNPs and eNPs were evaluated in an in vitro cytotoxicity assay using 3 HGSOC cell lines (OVSAHO, OVCAR4 and OVCAR8). As shown in Figure 5C, verticillin A-eNPs showed potent cytotoxicity with IC50 values of 44 nM, 29 nM and 32 nM in OVSAHO, OVCAR4 and OVCAR8 cells respectively. Interestingly, IC50 values of verticillin A-eNPs were similar to free compound in all three HGSOC cell lines tested, confirming that encapsulation in nanoparticles did not reduce the activity and potency of verticillin A (Figure 1A, 5C). Unloaded-eNPs did not exert a cytotoxic effect on any of the cell lines (Figure 5C).
Verticillin A reduces tumor burden in vivo
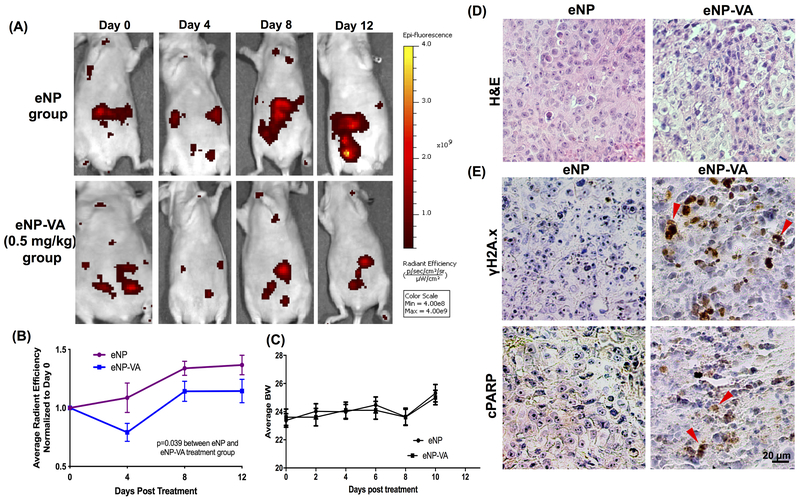
To evaluate the ability of verticillin A-eNPs to reduce in vivo tumor burden, OVCAR8-RFP cells were xenografted IP in athymic nude mice and tumors were allowed to form for 4 weeks. The mice were treated with unloaded-eNPs and verticillin A-eNPs every other day for 12 days at a dose of 0.5 mg/kg. As shown in Figure 6A–C, animals treated with verticillin A-eNPs had significantly less tumor burden based on average radiant efficiency measured with IVIS and had no measurable change in body weight. Interestingly, eNPs and verticillin A-eNPs did not cause the same gross morphological liver damage that was seen in mice treated with free verticillin A suggesting that encapsulation into the eNPs allowed for drug efficacy while reducing liver toxicity. Immunohistochemical analysis showed an increased staining for γH2A.X and cleaved PARP for tumors from verticillin A-eNP treated animals (Figure 6E). This indicates that verticillin A reduced tumor burden by causing DNA damage and apoptosis, which was consistent with the in vitro findings.
Figure 6. Verticillin A reduces tumor burden in vivo.
(A) OVCAR8-RFP cells were xenografted IP to form tumors. Mice were dosed with eNP-VA and eNP once every 2 days for total 12 days (Dosage: 0.5 mg/kg). Representative images show IVIS images of tumors in mice on Day 0, 4, 8 and 12 of drug treatment. (B) Quantification of tumor burden (average radiant efficiency as measured with IVIS imaging) normalized to Day 0. Statistics were performed using two-way ANOVA relative to vehicle control. (C) Quantification of average bodyweight of the mice during the course of treatment with eNP and eNP-VA. (D) H&E staining showing the histomorphology of paraffin embedded tumor tissue sections of eNP and eNP-VA treated animals. (E) Immunohistochemistry of γH2A.X and cPARP staining as markers of DNA damage and apoptosis is shown from tumors of animals treated with eNP and eNP-VA. Scale bar: 20 μm.
Discussion
New therapies are needed to treat HGSOC and metastasis related lethality from the disease. Nearly 60% of all FDA approved anti-cancer drugs are derived from nature, and it remains a promising source for new drug leads, particularly due to the chemical diversity (28,29). This study found that verticillin A, a fungal metabolite, caused cytotoxicity in HGSOC cells and when encapsulated into a nanoparticle, it reduced tumor burden in vivo. Mechanistically, treatment with verticillin A caused DNA damage and induced apoptotic cell death in HGSOC cells. A complete reversal of DNA damage and apoptosis occurred by combining verticillin A with the anti-oxidant, NAC. This indicated that verticillin A-mediated cytotoxicity occurred in part due to induction of DNA damage from oxidative stress. RNA-sequencing analysis of verticillin A treated OVCAR8 cells supported this finding as oxidative stress and apoptosis pathways were significantly upregulated relative to vehicle treated cells. Encapsulation of verticillin A in eNPs improved drug specificity, reduced toxicity and, more importantly, caused a significant reduction of tumor burden in vivo.
Epigenetic alterations such as histone modifications are reversible and regulate gene expression by transcriptional activation and/or repression (30,31). A number of studies have shown that alterations in histone methylation and acetylation levels play a role in ovarian cancer metastasis, re-sensitization towards cisplatin, and overall patient survival (30,32–35). Mechanistically, verticillin A targets HMTases, causing apoptotic cell death and improved chemotherapeutic response in colon and pancreatic tumor cell models (12,13,36). Consistent with previous studies in colon and pancreatic cancer, verticillin A altered histone modifications in OVCAR8, a HGSOC cell line. Global histone modification mapping using mass spectrometry revealed several histone modifications after treatment of OVCAR8 cells with verticillin A, including a reduction in H3K9me2, which was reported in colon cancer, along with changes in H3K14 acetylation marks (12). Interestingly, a reduction in H3K36me2 was found after verticillin A treatment. H3K36me2 is reported to play a role in double strand break repair, specifically by stabilizing DNA repair factors (37). In addition to our findings that verticillin A induced DNA damage, reduction in H3K36me2 also suggests that verticillin A may impact DNA double strand break repair.
Verticillin A showed concentration-dependent cytotoxicity in HGSOC cell lines OVSAHO, OVCAR4, and OVCAR8 by causing apoptosis. Moreover, verticillin A induced oxidative stress in HGSOC cells leading to DNA damage, as confirmed by comet assay and immunostaining for γH2.AX foci. This was supported by RNA-sequencing analysis, which showed significant upregulation of oxidative stress and apoptosis pathways after verticillin A treatment. Induction of apoptosis by verticillin A is reported in colon and pancreatic cancer, however, its role in oxidative stress was unknown (12,13,38). In colon and pancreatic cancer cells, verticillin A treatment reduced H3K9me3 levels which increased expression of pro-apoptotic protein Bax (12,36,38). This is consistent with our findings in OVCAR4 and OVCAR8 cells suggesting that histone modifications caused by verticillin A lead to apoptotic cell death by increasing Bax expression. Verticillin A is a selective inhibitor of HMTase G9a and expression of G9a correlates with tumor aggressiveness and reduced survival in ovarian cancer patients (12,39). Studies using small molecules targeting G9a indicated that blocking G9a caused oxidative stress in breast cancer cells (40). In another study using colorectal cancer cells, downregulation of G9a caused DNA damage and inhibited cell proliferation (41). Verticillin A reduced H3K9me2 levels as demonstrated by histone profiling of OVCAR8 cells and G9a is a repressive HMTase for H3K9me2 (12). Taken together, these finding suggest that verticillin A treatment induced oxidative stress and apoptosis possibly by G9a inhibition in HGSOC.
A challenge with chemotherapeutic agents, particularly hydrophobic ones derived from nature, is to identify a delivery method that enables drug to accumulate therapeutic concentrations within the target tissue while minimizing, or averting altogether, adverse systemic toxicities. For example, taxol, which was first reported in 1971, was not considered a promising drug in terms of solubility, toxicity and large-scale production (42–44). However, tremendous progress has been made in the past four decades making taxol one of the most effective chemotherapeutic drugs, particularly in frontline therapy for ovarian cancer (45–47). IP administration of free verticillin A, solubilized in Cremophor EL/EtOH as a means to address its low solubility, resulted in liver toxicity. Shorter studies using verticillin A IP did not report significant liver toxicity, however, studies performed using peripheral nerve sheath tumor cells report toxicity and weight loss in mice treated with verticillin A (12,22). The specific mechanism(s) of verticillin A induced hepatotoxicity is unknown and further toxicology studies are required. To overcome the toxic effects induced by verticillin A, and to enhance solubility, a well-characterized nanoparticle-based drug delivery system, the eNP was used (16,17,48). Previously, eNPs have been used to deliver other cytotoxic natural products (e.g., taxol) with high specificity to peritoneal tumors (16,17,25–27,49). As a result of their unique materials-based targeting mechanism, eNPs afford 10 to 100-fold higher intratumoral drug concentrations than are achieved with free drug and show remarkable efficacy (e.g., doubling of survival compared to free drug controls) (16). In terms of ovarian cancer, paclitaxel encapsulated eNPs demonstrated better uptake by tumor cells as well as reduced tumor reoccurrence in vivo compared to paclitaxel alone (27). In this study, eNPs delivered verticillin A to ovarian tumors and consistent with previous studies, the eNP formulation demonstrated improved drug efficacy with reduced hepatotoxicity. Verticillin A-eNP treated animals showed significant reduction in tumor burden in comparison to the eNP treated animals. Furthermore, histological studies confirmed induction of DNA damage and apoptosis in verticillin A-eNP treated animals consistent with our in vitro findings.
Another challenge with natural products, particularly with verticillin A, despite its enormous promise as an anti-cancer agent, is the inability to produce verticillin A in large quantities. However, sufficient quantities of verticillin A for pre-clinical studies are now possible by cultivation of a unique fungal strain and optimization of its fermentation conditions, such that 50-150 mg of verticillin A can be generated monthly on the laboratory scale (11). An adequate supply of verticillin A now exists for pre-clinical studies, which is being used to fully explore the pharmacological potential of this drug lead.
In conclusion, our findings validate verticillin A as a potent cytotoxic agent in HGSOC cells by inducing DNA damage. Encapsulation of verticillin A in an eNP improves drug efficacy, reduces toxicity and highlights the advantages of marrying natural products/anti-cancer research with nanoparticle drug delivery systems to address a clinical challenge. Continued investigation of verticillin A, verticillin analogs, and the eNP delivery system will provide key data for preparing an optimized efficacious formulation worthy of large animal pharmacokinetic studies and bring us one step closer to a promising therapy for ovarian cancer patients.
Supplementary Material
Table 3.
qRT-PCR primers:
| Target gene | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| TXN | TGAAGCAGATCGAGAGCAAGAC | TTCATTAATGGRGGCRRCAAGC |
| DUSP6 | CCTGAGGCCATTTCTTTCATAGA | GTCACAGTGACTGAGCGGCTAAT |
| NFKB2 | GAACAGCCTTGCATCTAGCC | TCCCAGTCGCTATCAGAGG |
| RELB | TCCCAACCAGGATGTCTAGC | AGCCATGTCCCTTTTCCTCT |
| TXNRD1 | GAAGATCTTCCCAAGTCCTATGAC | ATTTGTTGCCTTAATCCTGTGAGG |
Acknowledgements
This work was supported in part by grants P01CA125066 awarded to Dr Alan Kinghorn and R01CA227433 to Dr Joanna E. Burdette from the National Cancer Institute of the NIH (Bethesda, MD, USA). Grant # T32 AT007533 was awarded to Dr Guido Pauli for which Julia R. Austin is a recipient by the Office Of The Director, National Institutes Of Health (OD) and the National Center For Complementary & Integrative Health (NCCIH). IVIS imaging was performed using Xenogen IVIS® Spectrum Imager at University of Illinois, Chicago Research Resource Center’s Center for Cardiovascular Research and Physiology Core. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to Dr Neil L Kelleher at the Robert H Lurie Comprehensive Cancer Center and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569 awarded to Dr Neil L Kelleher. RNA-sequencing analysis was performed at the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, Feinberg School of Medicine, and Shared and Core Facilities of the University’s Office for Research.
Financial information: The authors gratefully acknowledge the support of grants P01CA125066 from the NCI of the NIH (Bethesda, MD).
Footnotes
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer Facts & Figures 2019. Atlanta, Ga: American Cancer Society; 2019. [Google Scholar]
- 3.Bowtell DD, Böhm S, Ahmed AA, Aspuria P-J, Bast RC, Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korkmaz T, Seber S, Basaran G. Review of the current role of targeted therapies as maintenance therapies in first and second line treatment of epithelial ovarian cancer; In the light of completed trials. Crit Rev Oncol Hematol. 2016;98:180–8. [DOI] [PubMed] [Google Scholar]
- 5.Bitler BG, Watson ZL, Wheeler LJ, Behbakht K. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol Oncol. 2017;147:695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herzog TJ, Monk BJ. Bringing new medicines to women with epithelial ovarian cancer: what is the unmet medical need? Gynecol Oncol Res Pract. 2017;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinghorn AD, DE Blanco EJC, Lucas DM, Rakotondraibe HL, Orjala J, Soejarto DD, et al. Discovery of Anticancer Agents of Diverse Natural Origin. Anticancer Res. 2016;36:5623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueroa M, Graf TN, Ayers S, Adcock AF, Kroll DJ, Yang J, et al. Cytotoxic epipolythiodioxopiperazine alkaloids from filamentous fungi of the Bionectriaceae. J Antibiot. 2012;65:559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katagiri K, Sato K, Hayakawa S, Matsushima T, Minato H. Verticillin A, a new antibiotic from Verticillium sp. J Antibiot. 1970;23:420–2. [DOI] [PubMed] [Google Scholar]
- 10.Minato H, Matsumoto M, Katayama T. Studies on the metabolites of Verticillium sp. structures of Verticillins A, B, and C. J Chem Soc Perkin Trans I. 1973;17:1819–25. [DOI] [PubMed] [Google Scholar]
- 11.Amrine CSM, Raja HA, Darveaux BA, Pearce CJ, Oberlies NH. Media studies to enhance the production of verticillins facilitated by in situ chemical analysis. J Ind Microbiol Biotechnol. 2018;45:1053–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paschall AV, Yang D, Lu C, Choi J-H, Li X, Liu F, et al. H3K9 Trimethylation Silences Fas Expression To Confer Colon Carcinoma Immune Escape and 5-Fluorouracil Chemoresistance. J Immunol. 2015;195:1868–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, et al. The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion. J Natl Cancer Inst. 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C, Yang D, Sabbatini ME, Colby AH, Grinstaff MW, Oberlies NH, et al. Contrasting roles of H3K4me3 and H3K9me3 in regulation of apoptosis and gemcitabine resistance in human pancreatic cancer cells. BMC Cancer. 2018;18:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubris KAV, Liu R, Colby A, Schulz MD, Colson YL, Grinstaff MW. In Vitro Activity of Paclitaxel-Loaded Polymeric Expansile Nanoparticles in Breast Cancer Cells. Biomacromolecules. 2013;14:2074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colby AH, Oberlies NH, Pearce CJ, Herrera VLM, Colson YL, Grinstaff MW. Nanoparticle drug-delivery systems for peritoneal cancers: a case study of the design, characterization and development of the expansile nanoparticle: Nanoparticle drug-delivery systems for peritoneal cancers. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2017;9:e1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R, Colby AH, Gilmore D, Schulz M, Zeng J, Padera RF, et al. Nanoparticle tumor localization, disruption of autophagosomal trafficking, and prolonged drug delivery improve survival in peritoneal mesothelioma. Biomaterials. 2016;102:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nature protocols. 2006;1:1112–6. [DOI] [PubMed] [Google Scholar]
- 19.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewellen KA, Metzinger MN, Liu Y, Stack MS. Quantitation of Intra-peritoneal Ovarian Cancer Metastasis. J Vis Exp. 2016; [DOI] [PubMed] [Google Scholar]
- 21.King SM, Hilliard TS, Wu LY, Jaffe RC, Fazleabas AT, Burdette JE. The impact of ovulation on fallopian tube epithelial cells: evaluating three hypotheses connecting ovulation and serous ovarian cancer. Endocrine-related cancer. 2011;18:627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zewdu A, Lopez G, Braggio D, Kenny C, Constantino D, Bid HK, et al. Verticillin A Inhibits Leiomyosarcoma and Malignant Peripheral Nerve Sheath Tumor Growth via Induction of Apoptosis. Clin Exp Pharmacol. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu F, Liu Q, Yang D, Bollag WB, Robertson K, Wu P, et al. Verticillin A overcomes apoptosis resistance in human colon carcinoma through DNA methylation-dependent upregulation of BNIP3. Cancer Res. 2011;71:6807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochemical Journal. 1996;313:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colby AH, Berry SM, Moran AM, Pasion KA, Liu R, Colson YL, et al. Highly Specific and Sensitive Fluorescent Nanoprobes for Image-Guided Resection of Sub-Millimeter Peritoneal Tumors. ACS Nano. 2017;11:1466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera VL, Colby AH, Tan GA, Moran AM, O’Brien MJ, Colson YL, et al. Evaluation of expansile nanoparticle tumor localization and efficacy in a cancer stem cell-derived model of pancreatic peritoneal carcinomatosis. Nanomedicine. 2016;11:1001–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmore D, Schulz M, Liu R, Zubris KAV, Padera RF, Catalano PJ, et al. Cytoreductive surgery and intraoperative administration of paclitaxel-loaded expansile nanoparticles delay tumor recurrence in ovarian carcinoma. Ann Surg Oncol. 2013;20:1684–93. [DOI] [PubMed] [Google Scholar]
- 28.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J Nat Prod. 2016;79:629–61. [DOI] [PubMed] [Google Scholar]
- 29.El-Elimat T, Zhang X, Jarjoura D, Moy FJ, Orjala J, Kinghorn AD, et al. Chemical Diversity of Metabolites from Fungi, Cyanobacteria, and Plants Relative to FDA-Approved Anticancer Agents. ACS Med Chem Lett. 2012;3:645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh DJ, Shah JS, Cole AJ. Histones and their modifications in ovarian cancer - drivers of disease and therapeutic targets. Front Oncol. 2014;4:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawson MA, Kouzarides T, Huntly BJP. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–57. [DOI] [PubMed] [Google Scholar]
- 32.Abbosh PH, Montgomery JS, Starkey JA, Novotny M, Zuhowski EG, Egorin MJ, et al. Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res. 2006;66:5582–91. [DOI] [PubMed] [Google Scholar]
- 33.Wang R-H, Zheng Y, Kim H-S, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, et al. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer. 2010;127:1332–46. [DOI] [PubMed] [Google Scholar]
- 35.Balch C, Fang F, Matei DE, Huang TH-M, Nephew KP. Minireview: epigenetic changes in ovarian cancer. Endocrinology. 2009;150:4003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Liu Q, Yang D, Bollag WB, Robertson K, Wu P, et al. Verticillin A overcomes apoptosis resistance in human colon carcinoma through DNA methylation-dependent upregulation of BNIP3. Cancer Res. 2011;71:6807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA. 2011;108:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, Yang D, Sabbatini ME, Colby AH, Grinstaff MW, Oberlies NH, et al. Contrasting roles of H3K4me3 and H3K9me3 in regulation of apoptosis and gemcitabine resistance in human pancreatic cancer cells. 2018;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua K-T, Wang M-Y, Chen M-W, Wei L-H, Chen C-K, Ko C-H, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer. 2014;13:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Kim Y-S, Kim DE, Lee JS, Song JH, Kim H-G, et al. BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy. 2013;9:2126–39. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, He P, Xi Y, Geng M, Chen Y, Ding J. Down-regulation of G9a triggers DNA damage response and inhibits colorectal cancer cells proliferation. Oncotarget. 2015;6:2917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J Am Chem Soc. 1971;93:2325–7. [DOI] [PubMed] [Google Scholar]
- 43.Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell. 2014;25:2677–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberlies NH, Kroll DJ. Camptothecin and taxol: historic achievements in natural products research. J Nat Prod. 2004;67:129–35. [DOI] [PubMed] [Google Scholar]
- 45.Crown J, O’Leary M. The taxanes: an update. Lancet. 2000;355:1176–8. [DOI] [PubMed] [Google Scholar]
- 46.Camptothecin and taxol: historic achievements in natural products research. [DOI] [PubMed]
- 47.Tuma RS. Taxol’s Journey from Discovery to Use: Lessons & Updates. Oncology Times. 2003;25:52. [Google Scholar]
- 48.Griset AP, Walpole J, Liu R, Gaffey A, Colson YL, Grinstaff MW. Expansile Nanoparticles: Synthesis, Characterization, and in Vivo Efficacy of an Acid-Responsive Polymeric Drug Delivery System. J Am Chem Soc. 2009;131:2469–71. [DOI] [PubMed] [Google Scholar]
- 49.Colson YL, Liu R, Southard EB, Schulz MD, Wade JE, Griset AP, et al. The performance of expansile nanoparticles in a murine model of peritoneal carcinomatosis. Biomaterials. 2011;32:832–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.