Abstract
A nontargeted plasma metabolomic analysis was conducted to compare differentially expressed metabolites in women with and without fibromyalgia (FM) using data and samples collected from two parent studies in women with FM (n = 20) and comparative data collected from newly recruited age‐matched women (n = 20). Blood plasma samples were analyzed for metabolite content using liquid chromatography mass spectrometry. Consolidation of positive and negative ion mode metabolomics data with fold change (>2 or <0.5) and variable importance of projection scores ≥1 revealed statistically significant metabolites comparing samples from women with and without FM. Metabolite profiles in patients with FM differed from the comparison group in energy, lipid and amino acid metabolites reflecting heightened oxidative stress, inflammation, and tryptophan degradation in patients with FM. Study results may contribute to further identification of unique metabolomic profiles enhancing understanding of the pathophysiology of FM and for the development of effective therapeutic options.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Fibromyalgia (FM) is a chronic pain condition with no known etiology and no known cure. A variety of physiological factors are purported to be involved in the pathophysiology of FM; however, no one etiologic target or set of targets has been determined.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ Examination of metabolic differences in women with and without FM for purposes of identifying potential targets for personalized treatment strategies.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Study findings of altered energy and lipid and amino acid metabolism in women with FM support the premise that metabolomics is a useful method to better understand putative alterations in conditions such as FM.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The finding of altered levels of energy and lipid and amino acid metabolites in FM could lead to a better understanding of biological mechanisms, improve phenotypic specificity, and ultimately identify biomarkers for improved diagnosis and treatment options for this vulnerable patient population.
Fibromyalgia (FM) is a chronic pain condition on the extreme end of the centralized pain spectrum, which affects up to 8% of the US population and 3% of the global population.1 FM is characterized by a constellation of symptoms that include chronic pain, fatigue, distressed mood, and sleep dysfunction.2 Although there are ongoing efforts to accurately diagnose persons with FM,3 at present, the diagnosis of FM is assessed using the 2016 revised FM criteria (American College of Rheumatology‐2016),4 which is based on the Fibromyalgia Research Criteria.5, 6 Given the level of subjectivity in the diagnostic rubric for FM, the recognition of objective key causal factors/mechanisms leading to the development or confirmed diagnosis of FM and FM‐related symptoms, particularly pain, and objective measures to confirm FM diagnosis have not been identified.1, 2 The obscurity regarding the pathogenesis of FM underscores the pressing need to enhance understanding of the biological mechanisms that may distinguish patients with FM from individuals without FM.7, 8
A variety of physiological factors are purported to be involved in the pathophysiology of FM, including genetic vulnerability, altered levels of neurotransmitters, and immune system/inflammatory factors; however, no one etiologic target or set of targets has been determined.9, 10, 11 For example, researchers have suggested that chronic low‐grade systemic inflammation may contribute to both the initiation and maintenance of the centralized pain state, focusing their studies on peripheral blood markers in FM, including inflammatory cytokines.12, 13 However, there have been inconsistencies in these findings with results from previous studies (i.e., plasma levels of various catecholamines have been increased, decreased, or unchanged).14
Despite intensive research focusing on multiple biologically plausible markers to aid in the diagnosis and prognostication of disease activity in FM, further understanding of the biological perturbations is needed to better explain potential underlying mechanisms related to FM and to guide personalized health strategies for symptom management. Of the multi‐omics that are driving precision health, metabolomics is an approach that quantifies the final products of various biological processes and holds promise as a method to holistically and accurately identify biomarkers that reflect upstream biological events, such as genetic mutations and environmental changes. Recent innovations in metabolomics allow researchers to produce a “metabolic snapshot,” which provides a comprehensive approach to depict both the steady‐state physiological condition of a cell or organism and its dynamic responses to environmental modulation.15 Thus, the goal of this study was to conduct nontargeted metabolomic analysis in women with FM compared with age‐matched women without FM to continue the discovery phase of metabolic differences in FM. This study was designed to identify differential plasma metabolite levels from the case and comparison groups using state‐of‐the‐art high‐throughput methods.
Methods
Plasma samples (N = 20) collected as part of two larger (parent) studies in women diagnosed with FM16, 17 were compared with 20 age‐matched women comparators. Sample selection from the two parent studies was based on the availability of a sufficient amount of frozen plasma per sample, plasma that had been immediately isolated, processed, and frozen at the data collection time point and only thawed once for initial data analysis associated with parent studies. For the current study, all samples included were frozen and rethawed x1. As per protocol, all plasma from FM and comparison samples was immediately isolated at the time of sample collection; plasma was aliquoted into 5–10 microcentrifuge tubes at 500 μL per tube and immediately snap‐frozen at −80°C to avoid degradation until ready for batch analysis. Due to our uniform treatment of each sample, and the absence of multiple freeze thaws, we expect that any latent variation due to environmental differences would be randomly distributed between FM cases and comparison samples. A determination of the age range associated with available samples provided the selection of age‐matched women without FM as comparators. For both parent studies, researchers enrolled women with a confirmed FM diagnosis based on the 1990 American College of Rheumatology criteria18 who were recruited from healthcare clinics affiliated with a major academic health center in the mid‐Atlantic region of the United States between 2009 and 2011. Data collection was conducted and completed in 2017 for the age‐matched women comparators in the same manner and in the same location as those in the parent studies.
Inclusion criteria for parent study participants were age ≥18 years; female; with a current diagnosis of FM (as confirmed by a letter from participants’ primary care provider or rheumatologist); able to speak and read standard English; a minimum of a sixth grade education level; and an ability to understand and sign the consent form and complete study instruments. Exclusion criteria were other systemic rheumatologic conditions or immune disorders such as systemic lupus erythematosus, rheumatoid arthritis, or HIV/AIDS; systemic use of corticosteroids; history of epilepsy; currently pregnant; or presence of a psychiatric disorder, such as a history of psychosis, schizophrenia, or bipolar disorder. The age‐matched women control group, herein designated as the comparison group, met the same inclusion/exclusion criteria as the FM group except for a diagnosis of FM. All participants were recruited using (i) brochures and flyers placed in ambulatory care clinics and waiting rooms affiliated with the academic medical center and (ii) weekly email blasts via the employee bulletin. All participants involved in this research provided voluntary written informed consent. The institutional review board at the original study site approved the parent studies and the current pilot study protocol, and the institutional review board at the affiliate site approved the metabolomics analysis as nonhuman research. Frozen samples from the two parent case studies and comparison group were shipped on dry ice using standard procedures to the Southeast Center for Integrated Metabolomics (SECIM) in Gainesville, Florida, where they were stored in the Center for Translational Science Institute biorepository and analyzed in the SECIM laboratory.
Metabolic profiling
Global metabolomics profiling was performed on a Thermo Q‐Exactive Oribtrap mass spectrometer with Dionex UHPLC and autosampler. All samples were analyzed in positive and negative heated electrospray ionization. Extraction was performed by aliquoting 100 μL of plasma into a microcentrifuge tube, adding 20 μL of internal standard mixture, and then adding 800 μL of 8:1:1 acetonitrile:acetone:methanol, followed by vortex mixing and centrifugation (20,000 rcf) to pellet the proteins. The supernatant was transferred and dried under a gentle stream of nitrogen. The dried material was reconstituted in 100 μL of 0.1% formic acid in water. The mass spectrometry scan time is dependent on the resolution setting for this instrument, and we collected at a mass resolution of 35,000 (equates roughly to 6 Hz) at mass‐to‐charge ratio 200 as separate injections. Tandem mass spectrometry was collected as separate injections using a data‐dependent mode choosing the top five peaks. Collision energy was scanned from 2 to 60 V. Separation was achieved with gradient elution on an ACE C18‐PFP 100 × 2.1 mm, 2 μm column, with mobile phase A as 0.1% formic acid in water and mobile phase B as acetonitrile with full details described by Liu et al.19 This is a polar embedded stationary phase that provides comprehensive coverage but does have some limitation in the coverage of very polar species. The flow rate was 350 μL/minute with a column temperature of 25°C; 4 μL was injected for negative ions and 2 μL for positive ions. The metabolites met level one identification guidelines as stipulated by the metabolomics standards initiative.20 Metabolites were identified by reference to an in‐house retention time library developed by running authentic standards. Verification included accuracy of <5 ppm (positive ion) or 10 ppm (negative ion) and retention time tolerance of ±0.15 minutes. Data from positive and negative ion modes were separately subjected to statistical analyses. There were a total of 3,792 features detected from the positive mode and 3,771 features in the negative mode (Figure 1). Adducts and complexes were identified based on retention time and accuracy of expected mass difference. Identified adducts and complexes were removed from the data set before processing. Normalization was performed after MZmine peak picking and identification. MZmine (freeware)21 was used to identify features, deisotope, align features, and perform gap filling to fill in any features that may have been missed in the first alignment algorithm.
Figure 1.
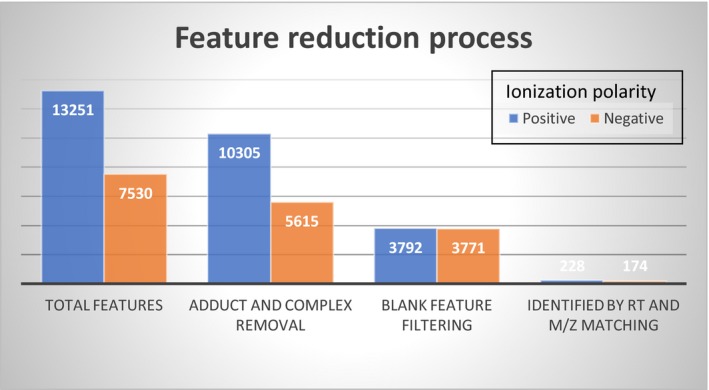
Feature reduction process in total sample (fibromyalgia n = 20 and comparison group n = 20). Ionization polarity. M/Z, mass‐to‐charge ratio; RT, retention time.
Statistical analyses
Statistical analysis was performed using statistical analysis software R.22 Descriptive statistics (mean, SD, frequency, and percentage) were generated for demographic variables including age, race, ethnicity, relationship status, income, and employment. Categorical demographic variables between the two groups were compared using Fisher's exact test, whereas continuous demographic variables were compared using a two‐sample t‐test. For comparison of demographic variables, a P value <0.05 was considered to be statistically significant.
The metabolomic data were preprocessed using half‐minimum imputation, log transformation, normalization by median, and autoscaling. In negative ionization, only 6.2% of the data had missing values. Ten features were removed that were not present in at least half of the samples. Those remaining were imputed with half‐minimum value. In positive ionization, only 4% of the data had missing values and 11 features were removed that were not present in at least half of the samples. The remaining missing values were imputed with half‐minimum value. To compare metabolite concentration between women with FM and the comparison group, independent t‐tests were performed. The resulting P values were adjusted using the Benjamini–Hochberg procedure for multiple testing,23 with P <0.01 considered to be statistically significant. Principal component analysis was performed for known metabolites detected under the positive and the negative ion modes separately. In addition, the Variable Importance in Projection was calculated for each metabolite to provide a quantitative estimation of the importance of each individual metabolite in both the positive and negative ion modes in the prediction models based on projections to latent structures discriminant analysis (PLS‐DA), a supervised pattern recognition approach.24 Our PLS‐DA analysis was conducted separately for positive and negative ion modes using known metabolites as predictors. Finally, a heat map of the identified metabolites of interest was created where hierarchical clustering (Euclidean distance, complete linkage) was applied to both samples and metabolites. Data and methods associated with this report are available upon request.
Results
Demographic characteristics of sample population and comparison group
The mean age of participants was 49.1 (6.9) years in each group (n = 20; n = 20), with the majority of the N = 40 study participants reported as white, non‐Hispanic, and married (Table 1). The FM group was significantly less educated (P = 0.009) and had higher rates of disability and unemployment (P = 0.027) and lower income (P = 0.005) compared with the comparison group. Other demographic variables were not significantly different between the two groups.
Table 1.
Characteristics of the study participants
| Characteristic | Fibromyalgia n = 20 | Female control n = 20 | P value |
|---|---|---|---|
| Age (mean/SD) | 49.1 (6.9) | 49.1 (6.9) | 1.00 |
| Race | |||
| Black | 35% | 20% | 0.144 |
| Indian | 5% | 0% | |
| Unknown | 10% | 0% | |
| White | 50% | 80% | |
| Ethnicity | |||
| Hispanic or Latino | 5% | 0% | 1.00 |
| Non‐Hispanic or Latino | 95% | 95% | |
| Unknown | 0% | 5% | |
| Marital status | |||
| Divorced/separated | 30% | 20% | 0.215 |
| Married/partner | 70% | 65% | |
| Single/never married | 0% | 15% | |
| Education | |||
| High school | 20% | 0% | 0.009 |
| Secondary school | 45% | 20% | |
| Bachelors+ | 35% | 80% | |
| Income | |||
| < $30,000 | 20% | 5% | 0.005 |
| $30,000–$59,999 | 55% | 20% | |
| ≥ $60,000 | 25% | 75% | |
| Work status | |||
| Disabled | 15% | 0% | 0.027 |
| Full‐time | 60% | 85% | |
| Other/not reported | 5% | 0% | |
| Part‐time | 5% | 5% | |
| Unemployed | 15% | 0% | |
| Comorbidities | |||
| Anxiety | 60% | 25% | 0.025 |
| Depression | 75% | 30% | 0.004 |
| Diabetes | 15% | 10% | 1.000 |
| Heart disease | 5% | 0% | 1.000 |
| High blood pressure | 40% | 5% | 0.019 |
| Overweight | 55% | 35% | 0.203 |
| Obesity | 20% | 25% | 1.000 |
Bachelors+ = Individuals who have degrees in higher education, ranging from Bachelors to PhD.
Differential metabolites between the FM and comparison groups
There were 1,462 statistically significant (P < 0.05) known metabolites and unknown spectral features (n = 739 positive ion mode; n = 723 negative ion mode). In subsequent analysis, we focused on known metabolites. Principal component analysis plots for the positive and negative ion mode data including only known metabolites were created to assess distance and relatedness of metabolites between patients with FM and comparators. There was a clear separation between the two groups, indicating that patients with FM and comparators had different metabolomic profiles (Figures 2 and 3). PLS‐DA models using positive mode data and negative mode data both provide excellent prediction performance (Q 2 = 0.82 for positive mode data and Q 2 = 0.91 for negative mode data; P < 0.001 for both). Consolidating the positive and negative mode data and focusing on known metabolites with adjusted P value < 0.01 and Variable Importance in Projection scores ≥1, the generally accepted minimum threshold of significance,25 and fold change (FC) >2 or <0.5, we found n = 48 statistically significant metabolites with FC >2 (Table 2) and n = 23 with FC <0.5 (Table 3). The metabolites that differed between groups included metabolites from multiple classes, including energy metabolism, lipid metabolism, and amino acid metabolism. Two metabolites with FC >2 and four metabolites with FC <0.5 were unclassified. A heat map of differential metabolites for classifying FMs vs. comparators is shown in Figure 4, demonstrating a clear separation of the patients with FM and the comparison group.
Figure 2.
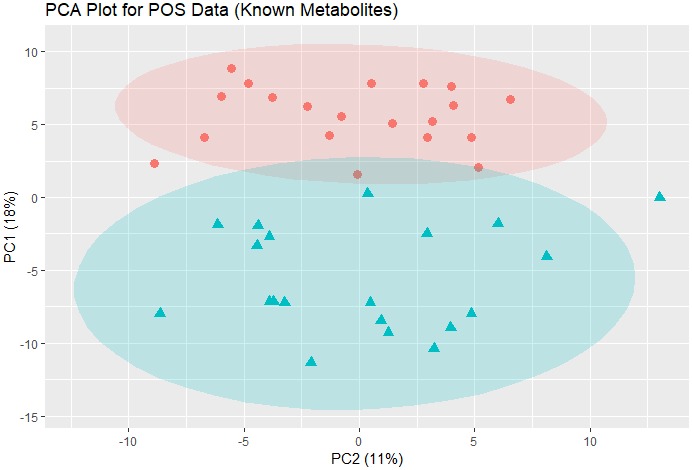
PCA plot for POS ion mode data (219 known metabolites). The data were autoscaled before PCA was performed. Pink dots: comparison group; blue triangles: fibromyalgia group. The shaded ovals are the 95% data ellipses. PCA, principal component analysis; POS, positive.
Figure 3.
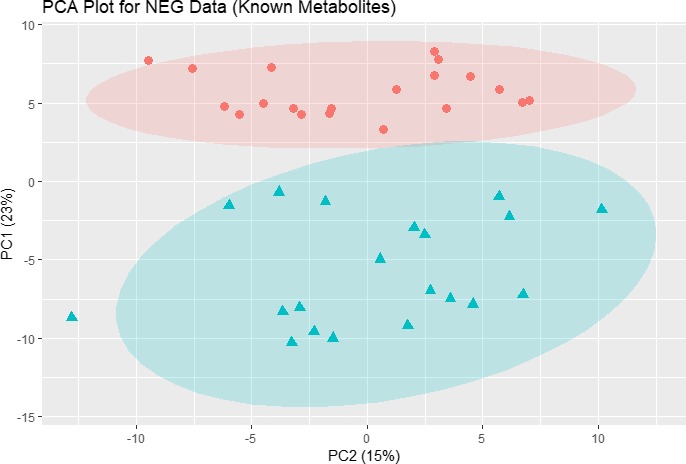
PCA plot for NEG ion mode data (174 known metabolites). The data were autoscaled before PCA was performed. Pink dots: comparison group; blue triangles: fibromyalgia group. The shaded ovals are the 95% data ellipses. NEG, negative; PCA, principal component analysis.
Table 2.
Statistically significant metabolites with VIP >1 and FC >2 in women with FM relative to plasma samples in women in a comparison group
| Metabolite | Positive ion mode | Negative ion mode | Metabolic pathway | ||||
|---|---|---|---|---|---|---|---|
| VIP | FC | P value | VIP | FC | P value | ||
| Energy metabolism | |||||||
| Adp | – | – | – | 1.80 | 5.87 | < 0.001 | Energy metabolism |
| c12h22o11‐Disaccharide‐(6c/6c; glc‐glc/glc‐frc/gal‐glc) | – | – | – | 1.22 | 3.38 | 0.002 | — |
| Lactose + k | 2.22 | 17.84 | < 0.001 | – | – | – | Galactose metabolism |
| Nicotinamide | 2.20 | 4.55 | < 0.001 | – | – | – | Nicotinate metabolism |
| Lipid metabolism | |||||||
| Arachidonic acid (20:4) | – | – | – | 1.56 | 6.95 | < 0.001 | Fatty acid synthesis |
| Docosahexaenoic acid (22:6) | – | – | – | 1.44 | 4.47 | < 0.001 | Fatty acid synthesis |
| Eicosapentaenoic acid | – | – | – | 1.52 | 9.40 | < 0.001 | Fatty acid synthesis |
| LysoPC (16:0) | 1.60 | 4.39 | < 0.001 | – | – | – | Glycerophospholipid metabolism |
| LysoPC (16:1) | 1.24 | 2.29 | 0.007 | – | – | – | Glycerophospholipid metabolism |
| LysoPC (18:0) | 1.68 | 4.17 | < 0.001 | – | – | – | Glycerophospholipid metabolism |
| LysoPC (18:1) | 1.43 | 2.89 | 0.003 | 1.21 | 2.88 | 0.005 | Glycerophospholipid metabolism |
| LysoPC (20:4) | 1.50 | 3.27 | 0.001 | – | – | – | Glycerophospholipid metabolism |
| LysoPC (22:6) | 1.51 | 2.79 | 0.001 | – | – | – | Glycerophospholipid metabolism |
| LysoPE (16:0) | – | – | – | 1.38 | 4.03 | < 0.001 | Glycerophospholipid metabolism |
| LysoPE (18:0) | 1.70 | 4.62 | < 0.001 | 1.48 | 4.86 | < 0.001 | Glycerophospholipid metabolism |
| LysoPE (20:4) | 1.47 | 2.43 | < 0.001 | 1.44 | 2.80 | < 0.001 | Glycerophospholipid metabolism |
| LysoPE (22:6) | 1.34 | 2.15 | 0.007 | 1.19 | 2.06 | 0.003 | Glycerophospholipid metabolism |
| Phosphocholine | 1.77 | 2.39 | < 0.001 | – | – | – | Glycerophospholipid metabolism |
| Spermidine | 1.82 | 3.23 | < 0.001 | – | – | – | Arginine and proline metabolism |
| Sphingosine‐1‐phosphate | – | – | – | 1.46 | 4.06 | < 0.001 | Sphingolipid metabolism |
| Sulfoacetaldehyde | – | – | – | 1.56 | 2.15 | < 0.001 | Taurine metabolism |
| Amino acid metabolism | |||||||
| 3‐Sulfino‐l‐alanine | – | – | – | 1.66 | 2.87 | 0.000 | Cysteine and methionine metabolism |
| 4‐Guanidinobutanoate | 1.88 | 3.23 | < 0.001 | 1.54 | 3.07 | < 0.001 | Arginine and proline metabolism |
| 4‐Oxoproline | – | – | – | 1.71 | 3.48 | < 0.001 | Arginine and proline metabolism |
| 5‐Oxo‐l‐proline | 2.09 | 4.05 | < 0.001 | – | – | – | Glutathione metabolism |
| Allantoin | 1.91 | 2.10 | < 0.001 | 1.68 | 2.14 | < 0.001 | Purine metabolism |
| Allopurinol | 1.56 | 5.12 | < 0.001 | – | – | – | Purine metabolism |
| Anthranilate | 1.95 | 2.54 | < 0.001 | – | – | – | Tryptophan metabolism |
| Aspartate | 2.26 | 3.11 | < 0.001 | 1.27 | 1.42 | < 0.001 | Arginine biosynthesis |
| Carnosine | 1.52 | 6.02 | < 0.001 | – | – | – | Histadine metabolism |
| Gluconic acid | 2.03 | 2.70 | < 0.001 | 1.86 | 1.80 | < 0.001 | Pentose phosphate synthesis |
| Glyceric acid | – | – | – | 1.84 | 3.24 | < 0.001 | Pentose phosphate synthesis |
| Glycerophosphocholine | 2.20 | 7.92 | < 0.001 | – | – | – | Glycerophospholipid metabolism |
| Glycyl‐l‐leucine | 1.89 | 2.86 | < 0.001 | – | – | – | – |
| Homovanillate | – | – | – | 1.88 | 7.41 | < 0.001 | Tyrosine metabolism |
| Indole‐3‐acetate | – | – | – | 1.39 | 2.36 | < 0.001 | Tryptophan metabolism |
| L‐glutamic acid | – | – | – | 2.03 | 11.18 | < 0.001 | Arginine biosynthesis |
| L‐lysine | 2.11 | 2.19 | < 0.001 | – | – | – | Lysine biosynthesis |
| Leu pro | 1.43 | 2.42 | 0.001 | – | – | – | – |
| Ll‐2,6‐Diaminoheptanedioate | 1.88 | 2.36 | < 0.001 | 0.73 | 1.20 | 0.122 | Lysine biosynthesis |
| Methionine sulfoxide | 1.97 | 4.09 | < 0.001 | – | – | – | Cysteine and methionine metabolism |
| N‐acetylneuraminate | 2.23 | 4.20 | < 0.001 | 1.98 | 2.95 | < 0.001 | Amino/nucleotide sugar metabolism |
| N‐formylglycine | 1.88 | 1.61 | < 0.001 | 1.88 | 2.12 | < 0.001 | – |
| N‐methyl‐d‐aspartic acid | 2.32 | 10.19 | < 0.001 | – | – | – | Ion channel regulator |
| O‐phosphoserine | 2.29 | 8.18 | < 0.001 | – | – | – | Gly/Ser/Thr metabolism |
| Taurine | 1.99 | 2.33 | < 0.001 | 1.74 | 2.18 | < 0.001 | Bile acid metabolism |
| No classification | |||||||
| Citramalate | – | – | – | 1.44 | 2.13 | < 0.001 | – |
| Mono‐(2‐ethylhexyl) | |||||||
| Phthalate dimer | – | – | – | 1.35 | 3.02 | 0.001 | – |
FC, fold change; FM, fibromyalgia; Gly, glycine; Ser, serine; Thr, threonine; VIP, Variable Importance in Projection.
References used to pinpoint the pathway for each metabolite were the Human Metabolome Database30 (http://www.hmdb.ca/) and the Kyoto Encyclopedia of Genes and Genomes KEGG33 (https://www.genome.jp/kegg/).
Table 3.
Statistically significant metabolites with VIP >1 and FC <0.5 in women with FM relative to plasma samples of women in a comparison group
| Metabolite | Positive ion mode | Negative ion mode | Metabolic pathway | ||||
|---|---|---|---|---|---|---|---|
| VIP | FC | P value | VIP | FC | P value | ||
| Energy metabolism | |||||||
| 2‐Hydroxyglutarate | 1.65 | 0.30 | < 0.001 | 0.37 | 1.10 | 0.505 | Butanoate metabolism |
| Alpha‐ketoglutaric acid + 2h | – | – | – | 1.99 | 0.21 | < 0.001 | Citrate cycle |
| Pyruvate | – | – | – | 2.03 | 0.16 | < 0.001 | Glycolysis |
| Lipid metabolism | |||||||
| 3‐Hydroxykynurenine | 1.57 | 0.37 | < 0.001 | 1.47 | 0.40 | < 0.001 | – |
| 3‐Hydroxy‐3‐methylglutarate | – | – | – | 1.71 | 0.28 | <0.001 | Lipid metabolism |
| Azelaic acid | – | – | – | 1.53 | 0.42 | < 0.001 | – |
| Decanoate | – | – | – | 1.11 | 0.45 | 0.004 | Fatty acid biosynthesis |
| Amino acids | |||||||
| 4‐Coumarate | – | – | – | 1.51 | 0.27 | < 0.001 | Tyrosine metabolism |
| 3‐Methyl‐2‐oxovaleric acid | – | – | – | 1.56 | 0.24 | < 0.001 | Val/Leu/Ile biosynthesis |
| 4‐Methyl‐2‐oxo‐pentanoic acid | – | – | – | 1.56 | 0.20 | < 0.001 | Val/Leu/Ile biosynthesis |
| 4‐Hydroxy‐l‐phenylglycine | 1.29 | 0.31 | 0.005 | – | – | – | Cyanoamino acid metabolism |
| Cys‐gly | 2.03 | 0.04 | < 0.001 | – | – | – | Glutathione metabolism |
| Decanoylcarnitine | 1.23 | 0.49 | 0.007 | – | – | – | – |
| Homocysteine | 1.61 | 0.45 | < 0.001 | 1.48 | 0.42 | < 0.001 | Cysteine and methionine metabolism |
| L‐cysteine | 0.23 | 0.92 | 0.870 | 1.91 | 0.13 | < 0.001 | Cysteine and methionine metabolism |
| L‐cystine | 1.52 | 0.50 | < 0.001 | 1.86 | 0.20 | < 0.001 | Gly/Ser/Thr metabolism |
| L‐glutamine | 1.73 | 0.34 | < 0.001 | 1.54 | 0.29 | < 0.001 | Arginine biosynthesis |
| L‐methionine | 1.46 | 0.49 | 0.002 | 1.34 | 0.50 | 0.001 | Cysteine and methionine metabolism |
| N‐acetylputrescine | 1.57 | 0.40 | 0.001 | – | – | – | Arginine and proline metabolism |
| Unclassified | |||||||
| Ascorbic acid | – | – | – | 1.77 | 0.08 | < 0.001 | Ascorbate and aldarate metabolism |
| D‐saccharic acid | 2.22 | 0.16 | < 0.001 | 1.97 | 0.13 | < 0.001 | Ascorbate and aldarate metabolism |
| L‐methionine sulfoximine | – | – | – | 1.84 | 0.16 | < 0.001 | – |
| S‐carboxymethyl‐l‐cysteine | 1.89 | 0.28 | < 0.001 | 1.78 | 0.23 | < 0.001 | – |
FC, fold change; FM, fibromyalgia; Gly, glycine; Ile, isoleucine; Leu, leucine; Ser, serine; Thr, threonine; Val, valine; VIP, Variable Importance in Projection.
References used to pinpoint the pathway for each metabolite were the Human Metabolome Database30 (http://www.hmdb.ca/) and the Kyoto Encyclopedia of Genes and Genomes KEGG33 (https://www.genome.jp/kegg/).
Figure 4.
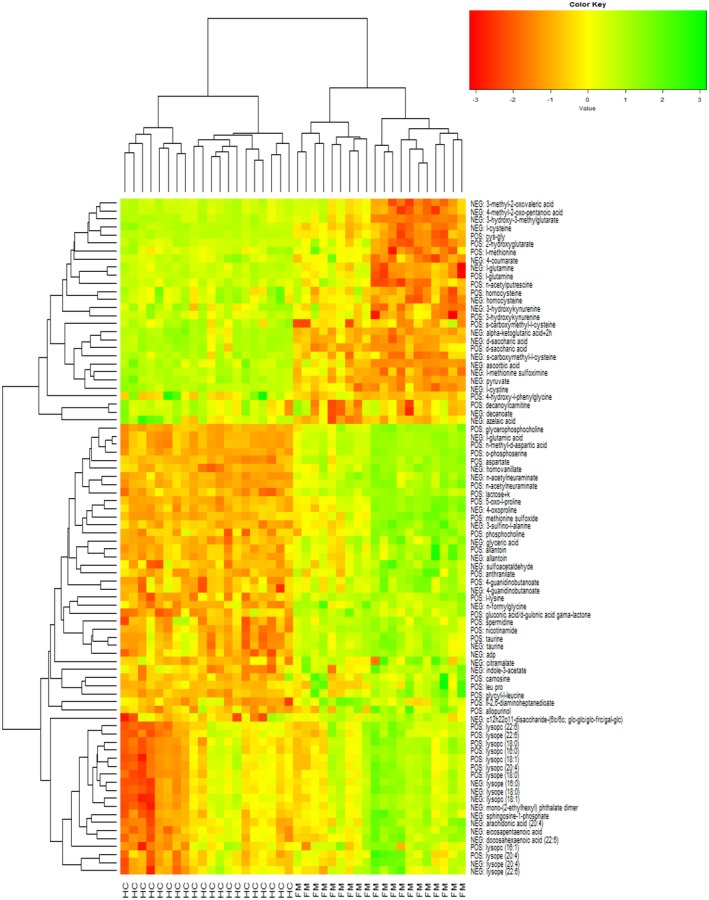
Statistically significant differential metabolites with fold change (FC) >2.0 or FC <0.5 in women with FM relative to comparison samples. FM, fibromyalgia; HC, comparison group; NEG, negative ion mode; POS, positive ion mode.
Discussion
In this exploratory metabolomics study, untargeted (global) metabolomics analyses were performed to characterize plasma profiles of women with FM compared with women without FM with the goal of identifying differential metabolites as potential biomarkers involved with FM pathogenesis. Because of the small sample size and the cross‐sectional design of our study, we did not conduct pathway analysis but report on select metabolites that suggest a potential for clinically meaningful health consequences in persons diagnosed with FM.26 Study findings are intended to inform the direction for conducting future targeted metabolomic research. A number of metabolites were higher (Table 2), and several lower, in the FM group (Table 3), relative to the comparison group. Although some metabolites showed a discrepancy between positive and negative mode data (e.g., 2‐hydroxyglutarate, positive mode P < 0.001 and negative mode P = 0.505), we note that this is due to the fact that some metabolites, such as 2‐hydroxyglutarate, have a stronger signal in positive mode than in negative mode, which is relative to the signal‐to‐noise ratio between modes. The metabolites that differed between groups included metabolites from multiple classes, including energy metabolism, lipid metabolism, and amino acid metabolism. We report differential levels of LysoPCs (16:0), (16:1), (18:0), (18:1), (20:4), and (22:6), taurine, methionine sulfoxide, anthranilate, and alpha‐ketoglutaric acid in our FM participants. A product of lipid peroxidation fragmentation, lysoPCs, act through a lipid mediator, the platelet‐activating factor receptor.27 It has been suggested that lysoPCs function as proinflammatory agents whose mechanisms seem also to be mediated via platelet‐activating factor receptor.7, 27 Our findings of elevated levels of lysoPC 16:0 corroborate those of Caboni et al.,7 who found an over‐representation of lysoPCs (14:0; 16:0) in the metabolomic profiles of individuals with FM, and Fais et al.,28 who found altered levels of plasma phospholipases, which are involved in lysoPC production. Caboni et al.7 suggested that a higher degree of oxidative stress with lipid peroxidation induced by reaction oxygen species may be relevant to the pathogenesis of FM. Similarly, we found elevated levels of taurine, a sulfur‐containing amino acid whose pathway is understood to be associated with alterations in lipid metabolism in women.29 Elevated levels of taurine have been previously reported in serum30, 31 and urine32 in women diagnosed with FM. Similarly, elevated levels of methionine sulfoxide and anthranilate have been reported.8, 30 Methionine sulfoxide is a primary oxidation product of methionine (Met) via its nucleophilic oxidation and is sometimes considered a biomarker of oxidative stress,33 whereas anthranilate is a tryptophan metabolite that has been implicated in brain/immune communication.34 In addition to higher levels of the metabolites described above, we found lower levels of alpha‐ketoglutaric acid and metabolites associated with cellular metabolism in individuals with FM. Alpha‐ketoglutarate is a crucial intermediate of the Krebs cycle and plays a critical role in multiple metabolic processes and acts as an antioxidant agent.35
Among the metabolites that suggest potential clinical significance to the pathogenesis of FM, the differential values of Met and anthranilate are particularly noteworthy. Met is one of nine essential amino acids (provided by food) and is required for growth and tissue repair. Met protects cells from pollutants, slows cell aging, and provides antioxidant defense. The finding of increased methionine sulfoxide in women with FM vs. comparators may be linked to the lower levels of l‐Met, as oxidation of Met by reaction oxygen species produces methionine sulfoxide (both the R and S diastereomers).36 Similar to the present study, Bazzichi et al.37 reported that individuals with FM (n = 34) had significantly lower plasma concentration of Met (P < 0.0001) as compared with the comparison group (n = 18), again suggesting that lower levels of Met may reflect reduced protection against oxidative stress. Other studies have linked high levels of plasma methionine sulfoxide with aging and memory impairment.38 The novel finding of significantly higher plasma concentration of methionine sulfoxide suggests a mechanism of involvement in accelerated aging as well as the development of cognitive complaints39 in persons with FM that warrants further investigation.
The presence of differential levels of anthranilate between our two study groups suggests potential dysregulation of kynurenine (KYN) metabolism. KYN, a central metabolite of the kynurenine pathway (KP), is catabolized to generate the metabolite anthranilic acid. The KP is understood as the primary route in the metabolism of tryptophan (Trp), an essential amino acid known to be a precursor to both serotonin and melatonin. Altered metabolism along the KP has been associated with psychiatric disorders such as depression40 and neurological disorders such as mild cognitive impairment and dementia.26, 41 For example, in a sample of 2,067 dementia‐free Framingham Offspring Cohort participants, Chouraki et al.26 reported an association between higher plasma anthranilic acid and a greater risk of dementia. Other studies have reported elevated levels of KYN8 or lower levels of Trp37 in persons with FM. Together, these studies suggest that disruption of KP metabolic homeostasis may have potential clinical consequences in persons with FM, the symptomatology of which has been reported to include cognitive impairment described as “fibrofog”39 and distressed mood such as depression and anxiety.1 Deciphering the complexity of differences in metabolites and their expression in different cells and tissues under different circumstances and relating them back to biological function and clinical outcomes is a current challenge for the field of metabolomics;40 however, with preliminary data such as we have reported here, there is potential for future studies designed to target Met, oxidative stress, and FM symptoms as well as the metabolites associated with the KP and Trp metabolism and clinical outcomes in individuals with FM.
Study limitations
There are several limitations to this exploratory study. In this pilot study, we did not control for medications in either the FM or the comparison group and thus acknowledge this as a study limitation that can be addressed in future studies. We further address the issue of unknown spectrometry features in our study findings. Although mass spectrometry has become a valuable tool for accurately identifying vast numbers of molecules, da Silva et al.42 noted that <2% of spectra in untargeted metabolomics can be annotated, leaving many of the “chemical signatures” of novel metabolites unmatched to molecular structures that have yet to be characterized. The existence of “unknown spectra” is a limitation of not only this study but also all metabolomic studies in that reference spectra do not exist for molecules that have yet to be named or described. Metabolomics scientists are currently seeking to remedy this problem through developing targeted bioinformatics approaches and data sharing of newly characterized metabolite structures.42
Another potential limitation of this study is the difference in plasma sample storage times prior to metabolite analysis between the FM cases and the comparison samples. One recent study by Haid et al.43 found changes in metabolite concentrations ranging from ~8 to 15% depending on the length of storage time. However, their analyses were based on two freeze–thaw cycles because the initial pooled plasma had been frozen at −80°C prior to aliquoting the test volumes of 100 μL used for analysis. In contrast, other recent studies44, 45, 46 did not find any appreciable differences in storage times in plasma, serum, or sputum and, similar to our study, were analyzed after one freeze–thaw cycle. Nevertheless, our future studies will ensure equivalence of storage times between groups to avoid potential bias. In addition, study personnel at SECIM who processed the samples were not blinded to group status, which could have introduced bias in the analysis. Further, we did not try to match comparison study participants based on conditions and treatments that may have an impact on an individual's metabolomics, such as diabetes, hypertension, or hypercholesteremia.
To enhance study validity and reliability, future study considerations could include overnight fasting prior to study participation, data collection on dietary intake, and attention to exclusion criteria related to psychiatric disorders, medications, and other comorbidities. Controlling for medications would include “matching” individuals on pain medications prescribed for chronic pain as well as for sprains or other temporary musculoskeletal conditions. With a careful inclusion/exclusion criteria screening process combined with collaborative arrangements with referring healthcare providers, and a well‐powered sample size that compares individuals with and without FM, the potential for contributing to enhanced phenotyping and development of diagnostic signatures may be deemed strongly possible.
In summary, our findings of altered energy and lipid and amino acid metabolism, particularly in the Met and KYN metabolites in women with FM, support the premise that metabolomics may be a useful comparative technique to better understand putative biological alterations in conditions such as FM and other rheumatologic conditions.47 A deeper exploration of metabolites and their putative relationship to physical and mental well‐being in individuals diagnosed with chronic conditions such as FM is anticipated to shed light on FM phenotypes and ultimately identify biomarkers for improved diagnosis and treatment options for this vulnerable patient population.
Funding
This study was funded by Virginia Commonwealth University CTSA (UL1TR002649 from the National Institutes of Health's National Center for Advancing Translational Science) and the CCTR Endowment Fund of the Virginia Commonwealth University.
Conflicts of Interest
All authors declared no competing interests for this work.
Author Contributions
V.M., A.S., D.L.K., Y.Y., T.S.‐S., P.P., and D.L. wrote the manuscript. V.M., A.S., L.T., and D.L. designed the research. V.M. performed the research. L.T., Y.Y., and T.G. analyzed the data. T.G. contributed new reagents/analytical tools.
References
- 1. Chinn, S. , Caldwell, W. & Gritsenko, K. Fibromyalgia pathogenesis and treatment options update. Curr. Sci. Inc. 20, 25 (2016). [DOI] [PubMed] [Google Scholar]
- 2. Sluka, K. A. & Clauw, D. J. Neurobiology of fibromyalgia and chronic widespread pain. Neuroscience 338, 114–129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold, L.M. et al AAPT diagnostic criteria for fibromyalgia. J. Pain 20, 611–628 (2018). [DOI] [PubMed] [Google Scholar]
- 4. Wolfe, F. et al 2016 Revisions to the 2010/2011 fibromyalgia diagnostic criteria. Semin. Arthritis Rheum. 46, 319–329 (2016). [DOI] [PubMed] [Google Scholar]
- 5. Wolfe, F. et al Fibromyalgia criteria and severity scales for clinical and epidemiological studies: a modification of the ACR Preliminary Diagnostic Criteria for Fibromyalgia. J. Rheumatol. 38, 1113–1122 (2011). [DOI] [PubMed] [Google Scholar]
- 6. Hoskin, T.L. , Whipple, M.O. , Nanda, S. & Vincent, A. Longitudinal stability of fibromyalgia symptom clusters. Arthritis Res. Ther. 20, 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caboni, P. et al Metabolomics analysis and modeling suggest a lysophosphocholines‐PAF receptor interaction in fibromyalgia. PLoS One 9, e107626 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hackshaw, K.V. , Rodriguez‐Saona, L. , Plans, M. , Bell, L.N. & Buffington, C.A. A bloodspot‐based diagnostic test for fibromyalgia syndrome and related disorders. Analyst 138, 4453–4462 (2013). [DOI] [PubMed] [Google Scholar]
- 9. Ablin, J.N. & Buskila, D. Update on the genetics of the fibromyalgia syndrome. Best Pract. Res. Clin. Rheumatol. 29, 20–28 (2015). [DOI] [PubMed] [Google Scholar]
- 10. Knezevic, N.N. , Tverdohleb, T. , Knezevic, I. & Candido, K.D. The role of genetic polymorphisms in chronic pain patients. Int. J. Mol. Sci. 19, (2018). < 10.3390/ijms19061707>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang, Z. et al SNPs in inflammatory genes CCL11, CCL4 and MEFV in a fibromyalgia family study. PLoS One 13, e0198625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ernberg, M. et al Plasma cytokine levels in fibromyalgia and their response to 15 weeks of progressive resistance exercise or relaxation therapy. Mediators Inflamm. 2018, 3985154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sturgill, J. , McGee, E. & Menzies, V. Unique cytokine signature in the plasma of patients with fibromyalgia. J. Immunol. Res. 2014, 938576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rus, A. , Molina, F. , Del Moral, M.L. , Ramirez‐Exposito, M.J. & Martinez‐Martos, J.M. Catecholamine and indolamine pathway: a case‐control study in fibromyalgia. Biol. Res. Nurs. 20, 577–586 (2018). [DOI] [PubMed] [Google Scholar]
- 15. Beckonert, O. et al Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Prot. 2, 2692–2703 (2007). [DOI] [PubMed] [Google Scholar]
- 16. Menzies, V. , Lyon, D.E. , Elswick, R.K. , Montpetit, A.J. & McCain, N.L. Psychoneuroimmunological relationships in women with fibromyalgia. Biol. Res. Nurs. 15, 219–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menzies, V. , Lyon, D.E. , Elswick, R.K. Jr , McCain, N.L. & Gray, D.P. Effects of guided imagery on biobehavioral factors in women with fibromyalgia. J. Behav. Med. 37, 70–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolfe, F. et al The American College of Rheumatology 1990 criteria for the classification of fibromyalgia. Report of the multicenter criteria committee. Arthritis Rheum. 33, 160–172 (1990). [DOI] [PubMed] [Google Scholar]
- 19. Liu, H. , Garrett, T.J. , Su, Z. , Khoo, C. & Gu, L. UHPLC‐Q‐Orbitrap‐HRMS‐based global metabolomics reveal metabolome modifications in plasma of young women after cranberry juice consumption. J. Nutr. Biochem. 45, 67–76 (2017). [DOI] [PubMed] [Google Scholar]
- 20. Sumner, L.W. et al Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pluskal, T. , Castillo, S. , Villar‐Briones, A. & Oresic, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry‐based molecular profile data. BMC Bioinform. 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fassa, A.A. , Himbert, D. & Vahanian, A. Transcatheter aortic valve replacement: current application and future directions. Curr. Cardiol. Rep. 15, 353 (2013). [DOI] [PubMed] [Google Scholar]
- 23. Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 24. Wold, S. , Sjöström, M. & Eriksson, L. PLS‐regression: a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 58, 109–130 (2001). [Google Scholar]
- 25. Mehmood, T. , Liland, K. , Snipen, L. & Saebo, S. A review of variable selection methods in partial least squares regression. Chemometr. Intell. Lab. Syst. 118, 62–69 (2012). [Google Scholar]
- 26. Chouraki, V. et al Association of amine biomarkers with incident dementia and Alzheimer's disease in the Framingham Study. Alzheimers Dement. 12, 1327–1336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oestvang, J. , Anthonsen, M.W. & Johansen, B. LysoPC and PAF trigger arachidonic acid release by divergent signaling mechanisms in monocytes. J. Lipids 2011, 532145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fais, A. , Cacace, E. , Atzori, L. , Era, B. & Ruggiero, V. Plasma phospholipase, gamma‐CEHC and antioxidant capacity in fibromyalgia. Int. J. Rheum. Dis. 20, 550–554 (2017). [DOI] [PubMed] [Google Scholar]
- 29. Sirdah, M.M. Protective and therapeutic effectiveness of taurine in diabetes mellitus: a rationale for antioxidant supplementation. Diabetes Metab. Syndr. 9, 55–64 (2015). [DOI] [PubMed] [Google Scholar]
- 30. Ruggiero, V. et al Free amino acids in fibromyalgia syndrome: relationship with clinical picture. Scand. J. Clin. Lab. Invest. 77, 93–97 (2017). [DOI] [PubMed] [Google Scholar]
- 31. Larson, A.A. , Giovengo, S.L. , Russell, I.J. & Michalek, J.E. Changes in the concentrations of amino acids in the cerebrospinal fluid that correlate with pain in patients with fibromyalgia: implications for nitric oxide pathways. Pain 87, 201–211 (2000). [DOI] [PubMed] [Google Scholar]
- 32. Malatji, B.G. et al A diagnostic biomarker profile for fibromyalgia syndrome based on an NMR metabolomics study of selected patients and controls. BMC Neurol. 17, 88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Umehara, H. et al Altered KYN/TRP, Gln/Glu, and Met/methionine sulfoxide ratios in the blood plasma of medication‐free patients with major depressive disorder. Sci. Rep. 7, 4855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris, G. , Carvalho, A.F. , Anderson, G. , Galecki, P. & Maes, M. The many neuroprogressive actions of tryptophan catabolites (TRYCATs) that may be associated with the pathophysiology of neuro‐immune disorders. Curr. Pharm. Des. 22, 963–977 (2016). [DOI] [PubMed] [Google Scholar]
- 35. Liu, S. , He, L. & Yao, K. The antioxidative function of alpha‐ketoglutarate and its applications. Biomed. Res. Int. 2018, 3408467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee, B.C. & Gladyshev, V.N. The biological significance of methionine sulfoxide stereochemistry. Free Radic. Biol. Med. 50, 221–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bazzichi, L. et al Altered amino acid homeostasis in subjects affected by fibromyalgia. Clin. Biochem. 42, 1064–1070 (2009). [DOI] [PubMed] [Google Scholar]
- 38. Tapia‐Rojas, C. et al Is L‐methionine a trigger factor for Alzheimer's‐like neurodegeneration?: changes in Abeta oligomers, tau phosphorylation, synaptic proteins, Wnt signaling and behavioral impairment in wild‐type mice. Mol. Neurodegener. 10, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walitt, B. et al Characterizing “fibrofog”: subjective appraisal, objective performance, and task‐related brain activity during a working memory task. Neuroimage Clin. 11, 173–180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lovelace, M.D. et al Recent evidence for an expanded role of the kynurenine pathway of tryptophan metabolism in neurological diseases. Neuropharmacology 112, 373–388 (2017). [DOI] [PubMed] [Google Scholar]
- 41. Clarke, G. , Stone, T.W. & Schwarcz, R. The kynurenine pathway: towards metabolic equilibrium. Neuropharmacology 112, 235–236 (2017). [DOI] [PubMed] [Google Scholar]
- 42. da Silva, R.R. , Dorrestein, P.C. & Quinn, R.A. Illuminating the dark matter in metabolomics. Proc. Natl. Acad. Sci. USA 112, 12549–12550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haid, M. et al Long‐term stability of human plasma metabolites during storage at ‐80 degrees C. J. Proteome Res. 17, 203–211 (2018). [DOI] [PubMed] [Google Scholar]
- 44. Breier, M. et al Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One 9, e89728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez‐Bascon, M.A. , Priego‐Capote, F. , Peralbo‐Molina, A. , Calderon‐Santiago, M. & Luque de Castro, M.D. Influence of the collection tube on metabolomic changes in serum and plasma. Talanta 150, 681–689 (2016). [DOI] [PubMed] [Google Scholar]
- 46. Wandro, S. , Carmody, L. , Gallagher, T. , LiPuma, J. J. & Whiteson, K. Making it last: storage time and temperature have differential impacts on metabolite profiles of airway samples from cystic fibrosis patients. mSystems 2, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta, L. , Ahmed, S. , Jain, A. & Misra, R. Emerging role of metabolomics in rheumatology. Int. J. Rheum. Dis. 21, 1468–1477 (2018). [DOI] [PubMed] [Google Scholar]


