Abstract
Introduction:
CMTM6 has been described as a PD-L1 regulator at the protein level by modulating stability via ubiquitination. In this study, we describe the patterns of CMTM6 expression and assess its association with response to PD-1 pathway blockade in non-small-cell lung cancer (NSCLC).
Methods:
We used multiplexed quantitative immunofluorescence to determine the expression of CMTM6 and PD-L1 in 438 NSCLCs represented in tissue microarrays, including two independent retrospective cohorts of immunotherapy treated (n = 69) and untreated (n = 258) patients, and a third collection of EGFR and KRAS genotyped tumors (n = 111).
Results:
Tumor and stromal CMTM6 expression was detected in approximately 70 % of NSCLCs. CMTM6 expression was not associated with clinical features or EGFR/KRAS mutational status and showed a modest correlation with T-cell infiltration (R2 < 0.40). We found a significant correlation between CMTM6 and PD-L1, higher in the stroma (R2 = 0.51) than tumor cells (R2 = 0.35). In our retrospective NSCLC cohort, neither CMTM6 nor PD-L1 expression alone significantly predicted immunotherapy outcomes. However, high CMTM6 and PD-L1 co-expression in the stroma and CD68 compartments (adjusted HR 0.38, p = 0.03), but not in tumor cells (p = 0.15), was significantly associated with longer OS in treated patients, but not observed in the absence of immunotherapy.
Conclusion:
This study supports the mechanistic role for CMTM6 in stabilization of PD-L1 in patient tumors and suggests that high co-expression of CMTM6 and PD-L1, particularly in stromal immune-cells (macrophages), might identify the greatest benefit from PD-1 axis blockade in NSCLC.
Keywords: CMTM6, PD-L1, macrophages, immune checkpoint blockade, NSCLC
Introduction
Monoclonal antibodies targeting the PD-1 pathway have transformed the treatment landscape of advanced-stage non-small-cell lung cancer (NSCLC). However, despite substantial improvements in overall survival (OS), only a minority of patients (≈ 20 %) truly benefit from these drugs when given as monotherapies(1). PD-L1 expression assessed by immunohistochemistry is the most widely used biomarker in the clinic, but it has only modest predictive performance. In NSCLC, although the benefit from PD-1 axis blockade is largely restricted to patients with PD-L1 expressing tumors, some PD-L1 negative NSCLCs respond to these drugs (≈ 8 %)(2–4), and the response rates in those patients most likely to benefit (PD-L1 tumor proportion score ≥ 50 %) are around 45 %(5). Given that the benefit from PD-1 pathway blockade in patients with NSCLC is higher in the presence of high PD-L1(6), we hypothesized that the study of regulatory pathways involved in PD-L1 expression, rather than PD-L1 alone, could be a way to improve the ability to predict outcomes to these agents.
Chemokine-like factor (CKLF)-like MARVEL transmembrane domain containing family member 6 (CMTM6) has been recently identified as one of the main PD-L1 regulators in two large-scale genetic screens(7,8). Mechanistically, both groups independently demonstrated that CMTM6 interacts with PD-L1 in the plasma membrane and recycling endosomes, preventing its lysosomal degradation(7,8). CMTM6 appears to function by inhibition of ubiquitination, and thus stabilization of PD-L1 in the membrane. CMTM6 depletion led to a robust decrease in constitutive and IFN-γ-induced PD-L1 protein levels in cancer cell-lines, dendritic cells and melanoma patient-derived xenografts, without affecting PD-L1 mRNA levels(7). CMTM6 was also shown to induce T-cell suppression by promoting a stable PD-L1 surface expression in the plasma membrane(7,8).
Despite these mechanistic associations, the distribution and patterns of CMTM6 protein expression, as well as the occurrence of CMTM6 and PD-L1 co-localization in human cancer tissue, have not been comprehensively assessed in lung cancer patients. In this study, we performed a simultaneous measurement of CMTM6 and PD-L1 protein levels in the tumor microenvironment in three independent cohorts of human NSCLC using multiplexed quantitative immunofluorescence (QIF). Our primary objective was to address CMTM6 expression, alone or co-expressed with PD-L1, as a potential predictive biomarker to PD-1 pathway blockade in NSCLC.
Methods
Patient cohorts and tissue microarray (TMA) construction
We analyzed retrospectively collected, formalin-fixed, paraffin-embedded (FFPE) tumor specimens represented in TMA format from three independent NSCLC cohorts from Yale. All tissue samples were collected and used with specific consent or waiver of consent under the approval from the Yale Human Investigation Committee protocol #9505008219. Cohort 1 (YTMA250) contained 288 tumors resected between 2004–2011 from patients that never received immune checkpoint inhibitors during their follow-up period(9); cohort 2 (YTMA310) included 138 tumors resected between 2011–2013 with known EGFR and KRAS genotypes, but without any further clinical annotation(10); and cohort 3 (YTMA404) contained 81 tumors resected between 2010–2016 from patients that received PD-1 pathway inhibitors for advanced disease (supplementary table S1). Thus, we used YTMA404 cohort to assess for biomarker predictive performance, whereas YTMA250 cohort was used to test for prognostic significance of biomarkers of interest. Table 1 summarizes the baseline characteristics of the patients included in YTMA404 and YTMA250 cohorts. The number of cases in which target proteins were quantified differs from the total number of cases included in each TMA due to loss of histospots during TMA construction or exclusion of cases after visual inspection for quality control.
Table 1.
Baseline characteristics of the patients in cohort 1 and cohort 3
| Characteristic | Immunotherapy treated cohort (YTMA404) | Immunotherapy untreated cohort (YTMA250) | |
|---|---|---|---|
| All patients | Monotherapy and pretreatment specimens | ||
| Total quantified tumors | 69 | 56 | 258 |
| Type of immunotherapy | |||
| Single-agent anti-PD1/PD-L1 | 58 (84) | 56 (100) | |
| Anti-PD-1/PD-L1 + anti-CTLA4 | 9 (13) | 0 | |
| Chemotherapy + anti-PD-1/PD-L1 | 1 (1) | 0 | |
| Other combinations | 1 (1) | 0 | |
| Specimen type for biomarker assessment | |||
| Pre-immunotherapy | 62 (90) | 56 (100) | |
| Post-immunotherapy | 7 (10) | 0 | |
| Gender | |||
| Male | 38 (55) | 30 (54) | 106 (41) |
| Female | 31 (45) | 26 (46) | 131 (51) |
| *Missing | 21 | ||
| Age | |||
| < 70 yo | 35 (51) | 25 (45) | 132 (51) |
| >= 70 yo | 34 (49) | 31 (55) | 104 (40) |
| *Missing | 22 | ||
| ECOG performance status | |||
| 0 | 6 (9) | 5 (9) | |
| 1 | 54 (79) | 43 (77) | |
| 2 | 8 (12) | 7 (12) | |
| 3 | 1 (1) | 1 (2) | |
| Smoking history | |||
| Never smoker | 13 (19) | 10 (18) | 38 (15) |
| Current smoker | 16 (23) | 12 (21 | 62 (24 |
| Former smoker | 39 (56) | 33 (59) | 121 (47) |
| *Missing | 1 | 1 | 37 |
| Histology | |||
| Adenocarcinoma | 50 (72) | 41 (73) | 135 (52) |
| Squamous-cell carcinoma | 15 (22) | 12 (21) | 63 (24) |
| Large-cell carcinoma | 3 (4) | 3 (5) | 12 (5) |
| Others | 1 (1) | 24 (9) | |
| *Missing | 24 | ||
| Stage | |||
| I | 147 (57) | ||
| II | 45 (17) | ||
| III | 2 (3) | 2 (4) | 30 (11) |
| IV (M1a) | 18 (26) | 15 (27) | 10 (4) |
| IV (M1b) | 10 (14) | 9 (16) | |
| IV (M1c) | 39 (57) | 30 (54) | |
| *Missing | 26 | ||
| EGFR mutation status | |||
| Wild type | 44 (64) | 37 (66) | |
| Mutant | 9 (13) | 6 (11) | |
| *Missing | 16 | 13 | |
| KRAS mutation status | |||
| Wild type | 32 (46) | 25 (45) | |
| Mutant | 18 (23) | 15 (27) | |
| *Missing | 19 | 16 | |
| CNS metastasis | |||
| No | 50 (73) | 42 (75) | |
| Yes | 18 (26) | 13 (23) | |
| *Missing | 1 | 1 | |
| Liver metastasis | |||
| No | 56 (81) | 45 (80) | |
| Yes | 12 (17) | 10 (18) | |
| *Missing | 1 | 1 | |
| LIPI score | |||
| Good | 28 (41) | 22 (39) | |
| Intermediate | 26 (38) | 20 (36) | |
| Poor | 4 (6) | 4 (7) | |
| *Missing | 11 | 10 | |
| Prior therapies | 15 (22) | 9 (16) | |
| 0 | 34 (49) | 30 (54) | |
| 1 | 19 (27) | 16 (28) | |
| > 1 | 1 | 1 | |
Multiplexed immunofluorescence staining protocol
Briefly, after TMA sections were deparaffinized, we subjected them to antigen retrieval with EDTA pH 8 buffer at 97°C for 20 min in a pressure boiling container (PT module, Lab Vision). Next, we incubated the slides with a solution of 0.3% hydrogen peroxide in methanol to inactivate endogenous peroxidase for 30 min, followed by another 30 min incubation with 0.3% bovine serum albumin with 0.05% tween-20 blocking solution. Subsequently, we performed a sequential multiplexed immunofluorescence staining (panel #1) with primary antibodies to detect epithelial tumor cells (pancytokeratin, polyclonal, Agilent), macrophages (CD68, clone PG-M1, Agilent), CMTM6 (clone RCT6, Absea) and PD-L1 (clone E1L3N, Cell Signaling) in the same tissue section. Isotype-specific HRP-conjugated secondary antibodies and tyramide-based amplification systems were used for signal detection. Residual horseradish peroxidase activity between sequential secondary antibody incubations was eliminated by exposing the slides twice for 7 min to a solution containing 100 mmol/L benzoic hydrazide and 50 mmol/L hydrogen peroxide. We used DAPI to highlight all nuclei. Control slides from a NSCLC titer array (YTMA295) were included in each staining experiment to ensure reproducibility.
To analyze the association between CMTM6 expression and tumor-infiltrating lymphocytes (TILs), we performed a previously standardized multiplexed immunofluorescence TIL staining protocol(11) (panel #2) in serial tissue sections of YTMA404 cohort. Briefly, after tissue sections were subjected to the same deparaffinization, antigen retrieval and blocking protocol mentioned above, we applied primary antibodies to detect epithelial tumor cells (cytokeratin, clone Z0622, Agilent), helper T-cells (CD4, clone SP35, Spring Bioscience), cytotoxic T-cells (CD8, clone C8/144B, Agilent) and B-cells (CD20, clone L26, Agilent), using a similar sequential protocol with isotype-specific HRP-conjugated secondary antibodies and tyramide-based amplifications systems as described above. Control slides from morphologically normal human tonsil were included in each staining batch as positive controls and to ensure reproducibility.
Further details regarding incubation times, antibody clones and concentrations, and fluorescent reagents used can be found in supplementary tables S2 and S3.
Fluorescence signal quantification and cut-point selection
We used the AQUA (Automated Quantitative Analysis) method (NavigateBP), to quantify the fluorescence signal of CMTM6, PD-L1 and TILs as previously described(12). CMTM6 and PD-L1 were measured within three compartments: 1) tumor compartment, created by binarizing the cytokeratin signal; 2) stromal compartment, created by excluding the tumor mask (a dilated cytokeratin compartment) from a dilated DAPI mask representing the total tissue; and 3) CD68 positive macrophage compartment, created by binarizing the CD68 signal. A representative image showing these three tissue compartments generated with the AQUA software can be found in supplementary figure S1. TILs were measured in the DAPI mask, which was created by dilating and then binarizing the DAPI signal to generate a compartment that includes all cells in the histospot. QIF scores were calculated by dividing the target pixel intensity by the area of the compartment of interest (supplementary figure S1), and then normalized to the exposure time and bit depth at which the images were captured. Those cases with staining artifacts or presence of less than 2% compartment area were systematically excluded after visual inspection.
For each cohort, we performed staining and target measurement in two independent TMA blocks, each block containing one non-adjacent tumor core per patient, and the average target QIF scores were calculated for each case. Then, we split tumors into high and low CMTM6/PD-L1 expression using the median as the cut-point.
Immunotherapy efficacy assessment
We used response evaluation criteria in solid tumors (RECIST) v1.1 to retrospectively evaluate treatment response to immune checkpoint blockade. As previously described(13), we defined clinical benefit (CB) as having experienced partial response or stable disease lasting ≥ 6 months as best response, whereas non-clinical benefit (NCB) was defined as primary progressive disease or stable disease lasting < 6 months. Patients with stable disease who did not progress and were censored before 6 months of follow-up were non-evaluable. Overall survival (OS) and progression-free survival (PFS) were calculated from the treatment start date to the date of death or loss of follow-up, or the date of disease progression, death or loss of follow-up, respectively. For those patients who did not die or progress during the study period, the outcome was considered left-censored. For the purposes of this study, we focused the assessment for predictive significance to the subgroup of patients with pre-immunotherapy specimens receiving single-agent PD-1 axis blockade (n = 56) (Table 1). Out of these 56 patients, 47 received nivolumab (84 %), 4 received pembrolizumab (7 %), and 5 received atezolizumab (9 %).
Statistical analysis
We used the Pearson correlation coefficient to analyze the linear association between two continuous variables, and t-test or one-way ANOVA to compare the means between two or more groups respectively. Chi-squared test was used to compare proportions. Survival curves were estimated with the Kaplan-Meier product-limit method and compared using the log-rank test. Hazard ratios for OS were calculated using the Cox regression model. All hypothesis testing was performed at a two-sided significance level of α=0.05.
Results
Initially, we tittered three different anti-CMTM6 antibodies targeting non-overlapping epitopes in a NSCLC test array (YTMA295) containing 35 lung tumor cores with presumed variable CMTM6 expression (supplementary figure S2a–2c). We observed a specific CMTM6 staining pattern localized largely in the plasma membrane with the three antibodies, and selected clone RCT6 (Absea) targeting a c-terminal peptide as the reference for validation (supplementary figure S2d–2f). Then, we compared the CMTM6 QIF scores obtained with clone RCT6 with a polyclonal anti-CMTM6 antibody (ab198284, Abcam) targeting a c-terminal peptide, and a second anti-CMTM6 monoclonal antibody (clone KT174, Absea) targeting a n-terminal peptide, showing a high correlation coefficient with both the polyclonal antibody (R2 = 0.71) and clone KT174 (R2 = 0.81) (supplementary figure S2g–2h). In addition, CMTM6 measurement with RCT6 clone showed good reproducibility between two independent experiments (R2 = 0.82) (supplementary figure S2i). Thus, the RCT6 clone was considered validated and used for the remainder of the studies in this effort.
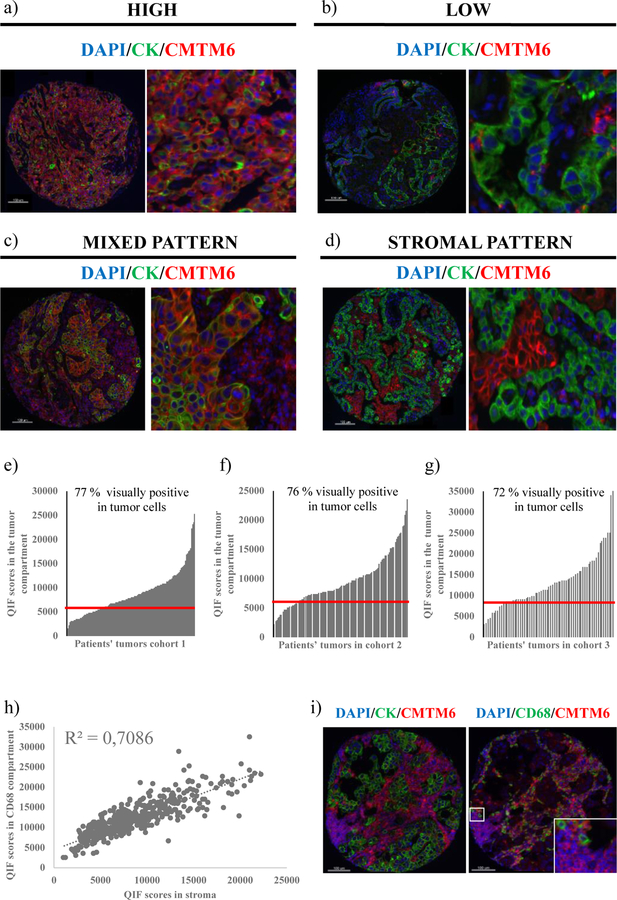
First, we evaluated the patterns of CMTM6 expression in human NSCLC. Predominantly membranous/cytoplasmic CMTM6 expression was detected in both tumor cells and stromal cells (figure 1a–1d). In the quantitative analysis, CMTM6 QIF scores showed a continuous distribution, both in the tumor compartment and the stromal compartment, the scores being comparable between the three cohorts (figure 1e–1g). Visually, tumor CMTM6 positivity was detected in about 70 % of NSCLCs. In the stroma, almost all tumors were visually positive for CMTM6 to some degree. There was a tight correlation between the CMTM6 QIF scores in the stromal compartment and in the CD68 compartment (figure 1h), suggesting that CD68 positive macrophages are a major immune cell type expressing CMTM6 in the stroma. However, some CD68 negative cells also stained positive for CMTM6 in the stroma (figure 1i).
Figure 1. Patterns of CMTM6 expression in human NSCLC tissue.
(a)-(b) Representative cases with high and low CMTM6 expression; (c)-(d) Representative cases with mixed stromal and tumor CMTM6 expression (c) and stroma-predominant expression (d); (e)-(g) Dynamic range of CMTM6 expression in the tumor compartment in the three tested cohorts. Red line represents the visual CMTM6 detection threshold in tumor cells; (h) Correlation between CMTM6 QIF scores in the stromal compartment vs. the CD68 compartment (three cohorts combined, n = 438); (i) Representative image of CMTM6 expression in CD68 positive and CD68 negative cells in the stroma
In order to study the association between CMTM6 expression in the tumor microenvironment with lymphocyte infiltration, we stained near tissue sections of YTMA404 with a multiplexed TIL immunofluorescence panel. There was a moderate/weak correlation between tumor or stromal CMTM6 expression and the presence of CD8 T-cells (R2 = 0.30 for tumor and R2 = 0.31 for stromal CMTM6) and CD4 T-cells (R2 = 0.34 for tumor and R2 = 0.39 for stromal CMTM6). CMTM6 expression showed no correlation with the presence of B lymphocytes (R2 = 0.09 for tumor and R2 = 0.008 for stromal CMTM6).
We found no significant or consistent association between tumor or stromal CMTM6 expression and clinical-pathological factors (supplementary tables S4 and S5). Tumor CMTM6 expression was higher in squamous-cell carcinomas in YTMA250 cohort (p = 0.02), but these differences were not significant in YTMA404 cohort (supplementary table S4). Also, tumor or stromal CMTM6 expression levels were not significantly different across EGFR- and KRAS-mutant NSCLC subgroups (supplementary figure S3a–3b). Similarly, PD-L1 expression levels were not significantly different between these NSCLC genomic subgroups (supplementary figure S3c–3d).
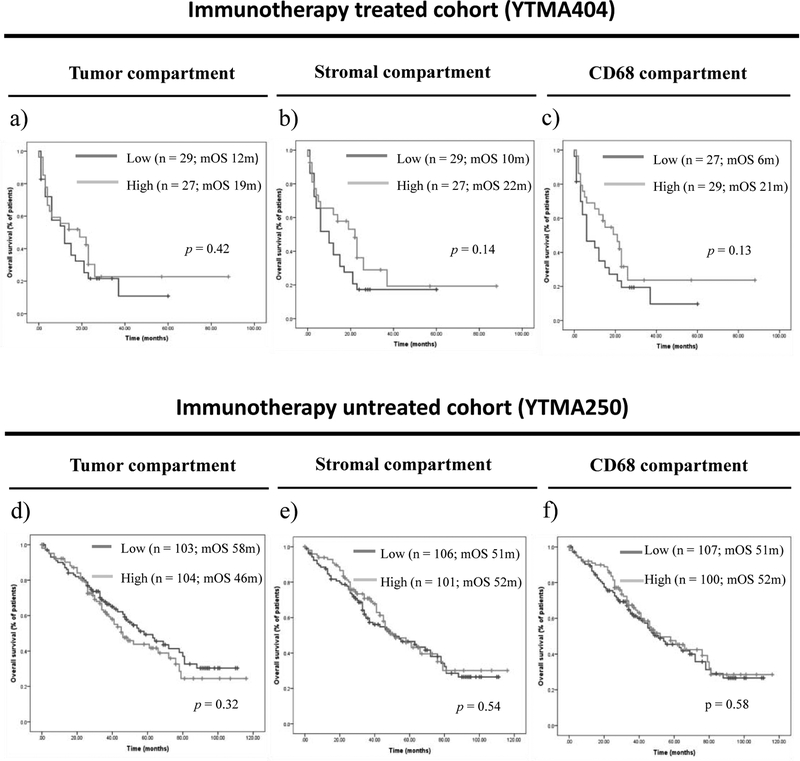
CMTM6 expression levels were not significantly different between patients with clinical benefit and those with no clinical benefit to single-agent PD-1 axis blockade in YTMA404 cohort (supplementary figure S4a–4c). Objective response rates were numerically higher in patients with high CMTM6 expression in the stroma (24 %) or the CD68 compartments (21.4 %) as compared to those with low CMTM6 expression (6.9 % in the stroma and 7.7 % in the CD68 compartment), but these differences did not reach statistical significance (p = 0.07 and p = 0.15, respectively) (supplementary table S6). Similarly, CMTM6 expression did not significantly predict PFS (supplementary figure S4d–4f) or OS under single-agent PD-1 axis blockade, although those patients with high CMTM6 expression in the stromal and CD68 compartments had a trend towards longer median OS as compared to those with low CMTM6 expression (p = 0.14 and p = 0.13, respectively) (Figure 2a–2c). In historical control patients that were not treated with immunotherapy, median OS was comparable and not statistically significant between patients with high and low CMTM6 expressing tumors in the YTMA250 cohort (Figure 2d–2f).
Figure 2. Indicative and prognostic performance of CMTM6 expression.
(a)-(c) OS according to CMTM6 expression in the tumor compartment (a), the stromal compartment (b), and the CD68 compartment (c) in patients treated with single-agent PD-1 axis blockade; (d)-(f) OS according to CMTM6 expression in the tumor compartment (d), the stromal compartment (e), and the CD68 compartment in patients non-treated with immune therapies
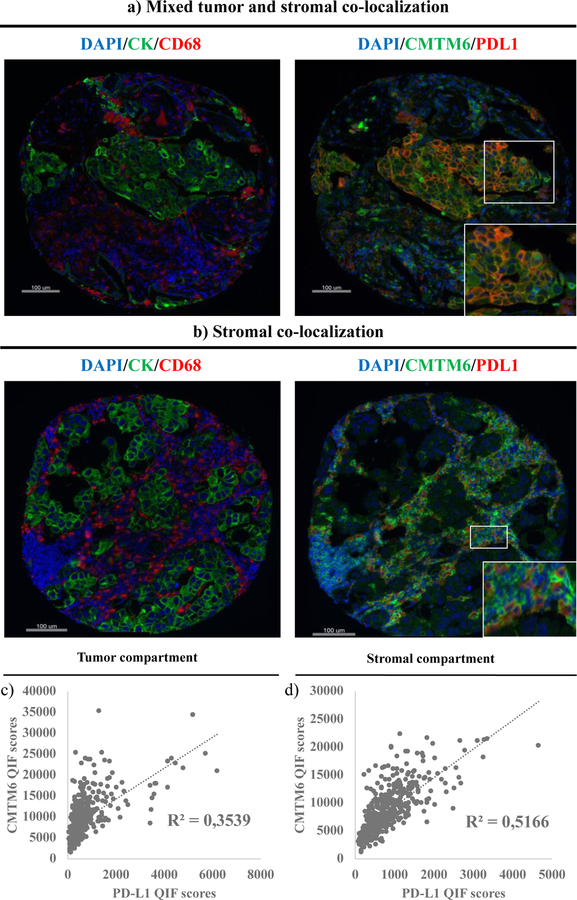
Next, based on the mechanistic evidence for interaction, we evaluated the association between CMTM6 and PD-L1 expression and their co-localization patterns in human lung cancer tissue. Visually, CMTM6 and PD-L1 co-localized both in tumor cells and immune cells in the stroma (figure 3a–3b). In the stroma, CMTM6 and PD-L1 co-localization mostly occurred in CD68 positive macrophages (figure 3b). Combining all tumors from the three cohorts together (n = 438), we found a statistically significant (p < 0.0001) but modest correlation between CMTM6 and PD-L1 levels, higher in the stromal compartment (R2 = 0.51), and lower in the tumor compartment (R2 = 0.35) (figure 3c–3d). Notably, all cases with elevated PD-L1 showed moderate to high levels of CMTM6, but some cases with elevated CMTM6 displayed low or negative PD-L1 expression. This association was similarly observed when the three cohorts were independently analyzed (supplementary figure S5).
Figure 3. CMTM6 and PD-L1 expression in NSCLC.
(a)-(b) CMTM6 and PD-L1 co-localization in tumor and stroma (a) and stroma only (predominantly in CD68+ macrophages) (b); (c)-(d) Correlation between CMTM6 expression levels and PD-L1 expression levels in the tumor compartment (c) and the stromal compartment (d) (n = 438)
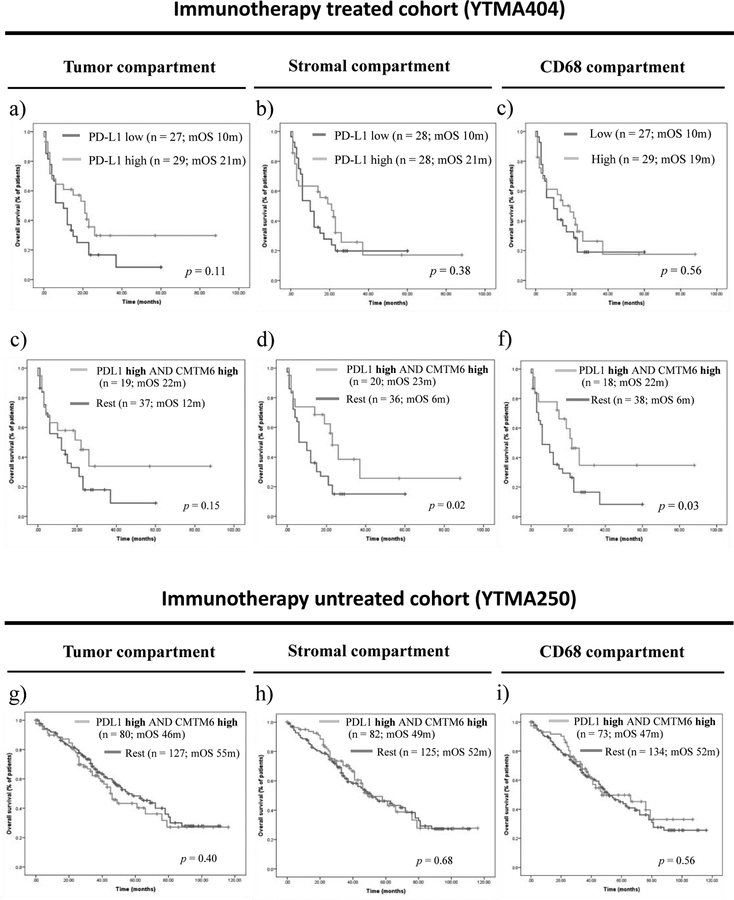
Mechanistically, CMTM6 has been shown to stabilize PD-L1 in the cell, which would thus predict their interaction could indicate PD-L1 function. To test the hypothesis that high CMTM6 and PD-L1 co-expression would exceed the value of either biomarker alone to predict survival benefit from single-agent PD-1 pathway blockade, we first evaluated the predictive performance of PD-L1 expression alone. In this cohort, PD-L1 expression levels in the tumor microenvironment were comparable in patients with clinical benefit and those with no clinical benefit (supplementary figure S6a–6c). Objective response rates were significantly higher in patients with high versus those with low PD-L1 expression in the stromal compartment (26.9 % vs. 3.9 %; p = 0.016), but not in the tumor compartment (p = 0.44) (supplementary table S7). PFS was not significantly different for patients with high versus low PD-L1 expression in the tumor microenvironment (supplementary figure S6d–6f). With regard to OS, PD-L1 expression, measured in either the tumor, stromal, or CD68 compartments, and using either the median cut-point (figure 4a–4c) or the top 30th percentile (supplementary figure S7), trended toward, but did not significantly predict longer OS.
Figure 4. Indicative and prognostic performance of high CMTM6 and PD-L1 co-expression.
(a)-(c) OS according to PD-L1 expression alone in the tumor compartment (a), the stromal compartment (b), and the CD68 compartment (c) in patients treated with single-agent PD-1 axis blockade; (d)-(d) OS in patients with high CMTM6 and PD-L1 co-expression in the tumor compartment (d), the stromal compartment (e), and the CD68 compartment (f) in patients treated with single-agent PD-1 axis blockade; (g)-(i) OS in patients with high CMTM6 and PD-L1 co-expression in the tumor compartment (g), the stromal compartment (h), and the CD68 compartment (i) in patients non-treated with immunotherapy
Next, we evaluated immunotherapy outcomes in four NSCLC subgroups based on high or low CMTM6 and PD-L1 expression levels (supplementary figure S8). Objective response rates were significantly higher in patients whose tumors showed high CMTM6 and PD-L1 co-expression as compared to the other three phenotypes combined, but only when both were high in the stroma (33.3 % vs. 3.6 %; p = 0.007) or the CD68 compartment (29.4 % vs. 8.1 %; p = 0.041), and not when co-expressed in the tumor compartment (22.1 % vs. 11.1 %; p = 0.27) (supplementary table S8). Median PFS was comparable between the four CMTM6/PD-L1 phenotypes (supplementary figure S9). However, OS was significantly longer in patients whose tumors had high CMTM6 and PD-L1 co-expression as compared the other three expression phenotypes combined, but only when both markers where high in the stromal compartment (23 months vs. 6 months, p = 0.02) or CD68 compartments (22 months vs. 6 months, p = 0.03), and not in the tumor compartment (22 months vs. 12 months, p = 0.15) (figure 4c–4f). In the multivariable Cox proportional hazard analysis, high CMTM6 and PD-L1 co-expression in the CD68 compartment remained as an independent predictor of OS after adjusting by age, performance status, smoking history, histology, LIPI score and baseline liver metastasis (HR = 0.38, CI 95 % 0.16–0.92; p = 0.03). As an exploratory analysis, we expanded the OS analysis to the full immunotherapy treated cohort (n = 69). Similar to the monotherapy subgroup, high CMTM6 and PD-L1 co-expression significantly predicted for longer OS in the stromal compartment (p = 0.005) and the CD68 compartment (p = 0.004) but not in the tumor compartment (p = 0.09), and neither PD-L1 expression alone, nor CMTM6 expression alone, reached significance for OS prediction (supplementary figure S10).
To be certain that tumors with highest levels of expression of both CMTM6 and PD-L1 were truly co-localized, we created a formula for AQUA analysis in the YTMA404 cohort to calculate the percentage of pixels per unit area where the pixels were above the threshold for both CMTM6 and PD-L1, then divided that by the number of pixels within the compartment of interest (tumor, stroma, or CD68). The colocalization was significantly higher in the CD68 compartment than either the tumor or stromal compartments (supplementary figure S11). Using this same algorithm, we found that the tumors with high expression of both CMTM6 and PD-L1 were also the tumors that showed high level of pixel by pixel co-localization, independent of compartment (supplementary figure S12).
Finally, we analyzed whether CMTM6 and PD-L1 co-expression had prognostic significance, where we observed that patients with high CMTM6 and PD-L1 co-expression in the tumor microenvironment showed no survival benefit in the absence of immunotherapy (Figure 4g–4i).
Discussion
CMTM6, recently described as one of the main positive regulators of PD-L1 expression at the protein level(7,8), was found to be upregulated in most tumor types at the mRNA level(7), but little was known regarding protein expression patterns in human cancer tissue. Here, we developed a multiplexed immunofluorescence panel with primary antibodies to detect epithelial tumor cells, CD68 positive macrophages, PD-L1, and CMTM6 in the same tissue section (panel #1). We decided to target CD68 positive macrophages because a parallel work from our lab showed that CD68 positive macrophages are the predominant immune cell type expressing PD-L1 (Liu et al., under review). We found that CMTM6 was broadly expressed in NSCLC, with most tumors showing both tumor and stromal expression. We showed that CD68 positive macrophages are a major immune cell type expressing CMTM6 in the stroma, but we could not estimate the precise percentage of CD68/CMTM6 double positive cells in the tumor microenvironment becausethe AQUA software does not perform cell segmentation and counting.
In this work, we did not attempt to characterize in detail what type of other immune cells express CMTM6, but we did observe that some CD68 negative immune cells were also visually positive for CMTM6. Based on morphological inspection, most of these cells were probably tumor-associated monocytes and CD68 negative macrophages. We found a modest/weak correlation between CMTM6 expression and the presence of CD4 and CD8 T-cells, but as this multiplexed TIL panel did not include a primary antibody against CMTM6 and was performed in near but not the same tissue section, we could not determine if T-lymphocytes also express CMTM6.
Supporting the findings from previous experimental studies(7,8), we found that CMTM6 and PD-L1 co-localize both in tumor cells and CD68 positive macrophages. In the quantitative analysis, the association between both markers was modest but statistically significant, and interestingly, it was higher in the stroma. Notably, cases with elevated PD-L1 co-expressed CMTM6, but a fraction of cases with high CMTM6 displayed low or negative PD-L1, suggesting that CMTM6 upregulation is not sufficient to mediate PD-L1 protein expression in NSCLC. In fact, CMTM6 does not regulate PD-L1 transcriptionally, either in the presence or absence of IFNγ(7,8). Moreover, CMTM6 expression has been shown to be independent of IFNγ pathway activation(8,14). As shown by Burr et al., CMTM6 levels were not influenced by IFNγ exposure in vitro in several cell line models(8), which further supports the existence of tumors with high levels of CMTM6 but low or undetectable PD-L1. Actually, CMTM6 has likely alternative functions other than PD-L1 binding, which could also explain the imperfect correlation that we observed in tumor tissue. For instance, CMTM6 has been recently found to be involved in lipid and LDL-cholesterol uptake by macrophages during atherogenesis(15,16). On the other hand, CMTM6 is not the only PD-L1 regulator at the posttranslational level. For example, CMTM4 has been shown to act as back-up PD-L1 protein stabilizer in CMTM6 deficient cells (7). Another proposed PD-L1 stabilizer is COP9 signalosome 5 (CSN5) via NF-κB signaling, which inhibits PD-L1 degradation by removing ubiquitin chains (17). In addition, somatic mutations within the intracytoplasmic domain of PD-L1 in motifs that are target for ubiquitination and posttranslational modifications have also shown to affect PD-L1 stability and function(18), again highlighting the importance of posttranslational modifications in modulating PD-L1 inhibitory functions.
The relationship between CMTM6 expression and outcome is unclear. We found no clear association between CMTM6 expression in the tumor microenvironment with clinical, genomic features or outcome in NSCLC. In contrast, previous reports have shown that high CMTM6 expression at the mRNA level was associated with poor prognostic clinical and molecular features in patients with gliomas (11,12). In other tumor types, high CMTM6 expression at the RNA level was associated with poor prognosis in pancreatic cancers(19), but with good prognosis in patients with triple-negative breast cancer(19). These differences between distinct tumor types might potentially reflect a tissue-specific CMTM6 upregulation or function. To our knowledge, this study is the first report addressing the predictive performance of CMTM6 expression in NSCLC patients treated with immune checkpoint inhibitors. While neither CMTM6 nor PD-L1 expression alone were significantly associated with better outcomes in patients treated with immunotherapy in this limited sized retrospective cohort, we found that high CMTM6 and PD-L1 co-expression was significantly associated with higher response rates and longer OS under single agent PD-1 pathway blockade, but, interestingly, only when both markers where co-expressed in the stromal and CD68 compartments and not in the tumor compartment. In the absence of prognostic significance in traditionally managed patients, high CMTM6 and PD-L1 co-expression in stromal immune cells (macrophages) might be a novel indicative biomarker to predict benefit from PD-1 axis blockade.
The observation that high CMTM6 and PD-L1 co-expression significantly predicts outcomes upon PD-1 pathway blockade only in the stroma suggests that the regulatory interaction between CMTM6 and PD-L1 might be biologically more relevant in immune cells compared to tumor cells. This finding is consistent with findings in many of other tumor types where the predictive value of PD-L1 seems to be largely restricted to immune-cell expression, such as urothelial (20), cervical(21), head and neck(22), and triple-negative breast cancers(23). In NSCLC, both tumor cell and immune cell PD-L1 expression have been associated with benefit from PD-1 axis blockade(4,24). However, detailed localization studies have not yet been done. While some studies suggest that PD-L1 in tumor cells are not concordant with PD-L1 expression in immune cells(24), other studies including one quantitative study finds a tight relationship between tumor cell and immune cell expression of PD-L1(25). If there is a tight correlation between tumor cell and immune cell PD-L1, then the predictive value of response to immune checkpoint inhibition could be uniform for all tumor types. This also concordant with our recent quantitative study of PD-L1 expression in macrophages (Liu et al, submitted).
This study should be considered in the context of a number of limitations. First, the immunotherapy treated cohort is a retrospective collection with mixed therapies, not a clinical trial. As such, it is not possible to calculate an interaction score since there is no patient subset that did not received immunotherapy. We are able to show absence of prognostic value in a historical cohort, and thus have used the term “indicative” rather than “predictive”. Furthermore, this study might be underpowered to demonstrate a significant association of PD-L1 expression with survival, as has been the case in some NSCLC clinical trials (26,27). Similarly, the small cohort size prevents us from determination of the extent to which the combination of CMTM6 and PD-L1 expression outperforms the established predictive power of PD-L1 expression alone in NSCLC(3–5,28). It is also a limitation that we used the median cut-point to split the population in high and low expression of CMTM6 and PD-L1. While that may be near the biologically correct cut-point, a validation set would be required to optimize the cut-points. Future studies addressing the predictive value of CMTM6 with or without PD-L1 co-expression will likely require the validation of an optimal and reproducible cut-point in larger and well-powered cohorts that can be split into training and validation sets. Finally, we used TMAs instead of whole tissue sections for biomarker assessment. As in all good TMA studies, we tried to partially avoid the issue of tumor heterogeneity by staining tumors in a two-fold redundancy (two histospots derived from two separate regions of the tumor).
In conclusion, this work supports the mechanistically hypothesized role for CMTM6 in stabilization of PD-L1 in patient tumors in that high co-expression levels of both CMTM6 and PD-L1 are associated with better outcome in the presence of immune checkpoint inhibitor therapy. Furthermore, co-expression in immune-cells, or more specifically macrophages, is consistent with current trends in other tumor types, where immune cells, not tumor cells are identified as the companion diagnostic test. We believe this works suggest that with extensive further validation of this biomarker may result in a more specific companion diagnostic test for immunotherapy.
Supplementary Material
Acknowledgements
The authors thank Lori A. Charette and the staff of Yale Pathology tissue services for expert histology services. J. Zugazagoitia was supported by a Rio Hortega contract from the Carlos III Research Institute (CM15/00196) and a fellowship from the Spanish Society of Medical Oncology (SEOM).
Funding
J. Zugazagoitia was supported by a Rio Hortega contract from the Carlos III Research Institute (CM15/00196) and a fellowship from the Spanish Society of Medical Oncology (SEOM). This work was supported by funds from Navigate BioPharma (Novartis) and Yale SPORE in lung cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M, et al. Five-Year Follow-Up of Nivolumab in Previously Treated Advanced Non-Small-Cell Lung Cancer: Results From the CA209–003 Study. J Clin Oncol. 2018; 36(17):1675–1684. [DOI] [PubMed] [Google Scholar]
- 2.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(17):1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. The Lancet. 2017;389(10066):255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–1833. [DOI] [PubMed] [Google Scholar]
- 6.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28. [DOI] [PubMed] [Google Scholar]
- 7.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burr ML, Sparbier CE, Chan Y-C, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altan M, Pelekanou V, Schalper KA, Toki M, Gaule P, Syrigos K, et al. B7–H3 Expression in NSCLC and Its Association with B7–H4, PD-L1 and Tumor-Infiltrating Lymphocytes. Clin Cancer Res. 2017;23(17):5202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toki MI, Mani N, Smithy JW, Liu Y, Altan M, Wasserman B, et al. Immune Marker Profiling and Programmed Death Ligand 1 Expression Across NSCLC Mutations. J Thorac Oncol. 2018;13(12):1884–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed quantitative analysis of CD3, CD8, and CD20 predicts response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2014;20(23):5995–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–7. [DOI] [PubMed] [Google Scholar]
- 13.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand (PD-L)-Ligand 1 Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol. 2018; 36(7):633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockmann M, Blomen VA, Nieuwenhuis J, Stickel E, Raaben M, Bleijerveld OB, et al. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature. 2017;546(7657):307–11. [DOI] [PubMed] [Google Scholar]
- 15.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domschke G, Linden F, Pawig L, Hafner A, Akhavanpoor M, Reymann J, et al. Systematic RNA-interference in primary human monocyte-derived macrophages: A high-throughput platform to study foam cell formation. Sci Rep. 2018;8(1):10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim S-O, Li C-W, Xia W, Cha J-H, Chan L-C, Wu Y, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30(6):925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gato-Cañas M, Zuazo M, Arasanz H, Ibañez-Vea M, Lorenzo L, Fernandez-Hinojal G, et al. PDL1 Signals through Conserved Sequence Motifs to Overcome Interferon-Mediated Cytotoxicity. Cell Rep. 2017;20(8):1818–29. [DOI] [PubMed] [Google Scholar]
- 19.Mamessier E, Birnbaum DJ, Finetti P, Birnbaum D, Bertucci F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Transl Med. 2018;6(3):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. The Lancet. 2016;387(10031):1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung HC, Schellens JHM, Delord J-P, Perets R, Italiano A, Shapira-Frommer R, et al. Pembrolizumab treatment of advanced cervical cancer: Updated results from the phase 2 KEYNOTE-158 study. J Clin Oncol. 2018;36(15_suppl):5522–2. [Google Scholar]
- 22.Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, De Castro G Jr, et al. LBA8_PRKEYNOTE-048: Phase III study of first-line pembrolizumab (P) for recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC). Ann Oncol. 2018;29(suppl_8). [Google Scholar]
- 23.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379(22):2108–2121. [DOI] [PubMed] [Google Scholar]
- 24.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. The Lancet. 2016;387(10030):1837–46. [DOI] [PubMed] [Google Scholar]
- 25.Rehman JA, Han G, Carvajal-Hausdorf DE, Wasserman BE, Pelekanou V, Mani NL, et al. Quantitative and pathologist-read comparison of the heterogeneity of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Mod Pathol. 2017;30(3):340–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376(25):2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barlesi F, Vansteenkiste J, Spigel D, Ishii H, Garassino M, De Marinis F, et al. Avelumab versus docetaxel in patients with platinum-treated advanced non-small-cell lung cancer (JAVELIN Lung 200): an open-label, randomised, phase 3 study. Lancet Oncol. 2018;19(11):1468–79. [DOI] [PubMed] [Google Scholar]
- 28.Herbst RS, Baas P, Kim D-W, Felip E, Perez-Gracia JL, Han J-Y, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. The Lancet. 2016; 387(10027):1540–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.