SUMMARY
Plasmacytoid dendritic cells (pDCs) are sensor cells with diverse immune functions, from type I interferon (IFN-I) production to antigen presentation, T cell activation, and tolerance. Regulation of these functions remains poorly understood but could be mediated by functionally specialized pDC subpopulations. We address pDC diversity using a high-dimensional single-cell approach: mass cytometry (CyTOF). Our analysis uncovers a murine pDC-like population that specializes in antigen presentation with limited capacity for IFN-I production. Using a multifaceted cross-species comparison, we show that this pDC-like population is the definitive murine equivalent of the recently described human AXL+ DCs, which we unify under the name transitional DCs (tDCs) given their continuum of pDC and cDC2 characteristics. tDCs share developmental traits with pDCs, as well as recruitment dynamics during viral infection. Altogether, we provide a framework for deciphering the function of pDCs and tDCs during diseases, which has the potential to open new avenues for therapeutic design.
Graphical Abstract

In Brief
Dendritic cells (DCs) are unique therapeutic targets given their capacity to modulate immune responses. Yet complete alignment of the DC network between species is lacking. Using a multidimensional approach, Leylek et al. identify the mouse homolog of human AXL+ DCs, named transitional DCs (tDCs), and reveal their similarities with pDCs.
INTRODUCTION
Dendritic cells (DCs) are specialized antigen-presenting cells that bridge innate and adaptive immune responses (Banchereau and Steinman, 1998). In both mouse and human, DCs are divided into functionally specialized subpopulations, i.e., plasmacytoid DCs (pDCs) and two subsets of classical DCs (cDCs), cDC1 and cDC2 (Guilliams et al., 2014; Merad et al., 2013). pDCs are quite distinct from their cDC counterparts. Ontologically, pDCs differentiate from both myeloid and lymphoid precursors but display lymphoid features regardless of their developmental path (Corcoran et al., 2003; Naik et al., 2007; Onai et al., 2007; Sathe et al., 2013; Shigematsu et al., 2004). Functionally, while cDC1s and cDC2s are specialized for interaction with T cells, pDCs are well known for their innate capacity to rapidly produce large quantities of type I interferon (IFNα and IFNβ, abbreviated IFN-I) upon viral encounter (Asselin-Paturel et al., 2001; Siegal et al., 1999). However, aside from their innate role, pDCs can carry out adaptive immune functions upon activation, i.e., antigen presentation, T cell activation, and tolerance (Alculumbre et al., 2019; Leylek and Idoyaga, 2019). This represents a rare phenomenon in the immune system, in which one cell type is capable of mediating diverse immune tasks. Yet there is a lack of understanding of the mechanisms that control pDC functions. Dissecting these mechanisms is imperative for the development of pDC-based therapeutics.
One possible mechanism to enable diverse pDC functions is a division of labor among functionally distinct pDC subpopulations. pDC subpopulations have been described in mouse and human (Björck et al., 2011; Hadeiba et al., 2008; Matsui et al., 2009; Wilhelm et al., 2016; Zhang et al., 2017). However, there has been a general lack of consensus on these populations. Previous descriptions of pDC subpopulations were limited by inherently biased flow cytometry analyses with few markers. Furthermore, these reports fail to resolve findings between species. Advances in single-cell omics technologies have yielded an unprecedented resolution of the human DC compartment. Using transcriptomic and proteomic approaches that leverage unbiased analyses, we and others have unveiled a human pDC-like population with select features of cDCs (Alcántara-Hernández et al., 2017; See et al., 2017; Villani et al., 2017). However, it remains unclear whether the murine DC compartment contains such a population. A thorough analysis of pDC diversity in mice is critical to provide functional insights in vivo.
Here, we used a high-dimensional mass cytometry approach (CyTOF) and unbiased analysis to interrogate the murine DC compartment at the protein level. We found a pDC-like population in mouse that displays cDC features and functions but ultimately demonstrates close developmental relationship to pDCs. We performed parallel cross-species comparisons with the goal of aligning this pDC subpopulation in mouse and human, which is imperative for the translation of basic biology to human immunology. We found that this murine pDC-like population is equivalent to the recently described human AXL+ DCs (also known as ASDCs) (Alcántara-Hernández et al., 2017; See et al., 2017; Villani et al., 2017). Although in mouse these cells did not express the protein Axl, they did share a continuum of phenotypes that span pDCs and cDC2s. We herein propose to name this intermediate population transitional DCs (tDCs) in both species. Our high-dimensional analysis represents an easily accessible data resource on the phenotype, transcriptome, and function of pDCs and tDCs at steady state and during influenza infection. This dataset reveals information that has not been recognized using low-dimensional flow cytometry analyses and can be exploited for the development of pDC- and tDC-based therapeutics.
RESULTS
CyTOF Analysis Reveals a Transitional DC Subpopulation in Mouse Spleen
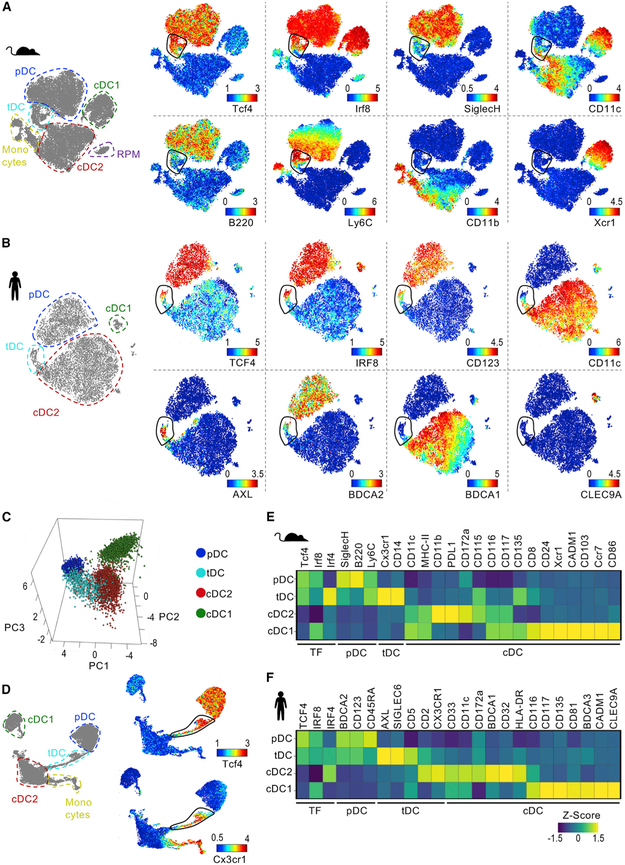
To dissect pDC diversity in mouse, we designed a myeloid-focused CyTOF panel (Table S1). In addition to surface markers, this panel incorporated transcription factors (TFs) differentially expressed in DC subsets: Tcf4 (pDCs), Irf8 (pDCs and cDC1s), and Irf4 (cDC2s) (Cisse et al., 2008; Schiavoni et al., 2002; Suzuki et al., 2004). To allow greater resolution of minor myeloid populations, we enriched mouse splenic DCs and plotted Lin− cells in a t-Distributed Stochastic Neighbor Embedding (tSNE) map (Figure 1A). We found a minimal number of Ccr2+ monocytes and CD64+ red pulp macrophages (RPMs) after enrichment (Figure S1). Based on their phenotype, we identified formally described DC subsets, i.e., pDCs, cDC1s, and cDC2s. As expected, we observed high Tcf4 expression in pDCs, but not in cDCs, and high Irf8 expression in cDC1s and pDCs, but not in cDC2s. Surprisingly, we observed a bridging population of cells localized between pDCs and cDC2s in the tSNE map. The phenotype of this bridging population ranged from pDC- to cDC2-like, i.e., high to low expression of Tcf4, Irf8, and the pDC marker SiglecH and low to high expression of CD11c.
Figure 1. Identification of a Transitional DC Population in Mouse and Human.
(A and B) tSNE plots of (A) Lin− events from BSA-enriched mouse splenocytes or (B) Lin− HLA-DR+ events from human PBMCs. One representative of two samples analyzed by CyTOF is shown. See Figures S1 and S2 for additional protein expression. tSNE with manual annotation (left) and colored by expression of indicated proteins (right).
(C) PCA of DC subsets in mouse spleen analyzed by CyTOF as in (A).
(D) UMAP of mouse splenocytes analyzed by CyTOF as in (A). UMAP with manual annotation (left) and colored by expression of indicated proteins (right).
(E and F) Heatmap of protein expression in mouse (E) and human (F) DC subsets (n = 2 for each species).
See also Table S1 for CyTOF panels.
The phenotype and tSNE map localization of this bridging population resembled human AXL+ DCs, a pDC-like population with cDC2 features described by us and others (Alcántara-Hernández et al., 2017; See et al., 2017; Villani et al., 2017). To evaluate DC populations’ homology between species, we performed parallel CyTOF analysis of human peripheral blood mononuclear cells (PBMCs) (Figure 1B). In line with our previous report, we observed that human AXL+ DCs bridge pDCs and cDC2s in a tSNE map, reflecting an intermediate phenotype, which was also true for human spleen (Figure S2). Similar to mouse, the human bridging population expressed high to low levels of TCF4, IRF8, and the pDC markers CD123 and BDCA2, while CD11c expression ranged from low to high. Thus, the presence of cells that display a transitional phenotype between pDCs and cDC2s is a conserved characteristic between mouse and human. We called these cells transitional DCs (tDCs) with the intent to emphasize not only their intermediate phenotype but also their continuum of pDC to cDC2 features.
Mouse tDCs consistently localized between pDCs and cDC2s when analyzed using other clustering methods, such as principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) (Becht et al., 2018) (Figures 1C and 1D). When we analyzed the expression of other markers present in our CyTOF panels, we again found that tDCs have intermediate expression of pDC and cDC markers (Figures 1E and 1F; see Figure S1 and S2 for tSNE plots). In contrast to pDCs, mouse and human tDCs expressed high levels of CX3CR1 and IRF4. Unlike cDC2s, tDCs were CD11b− in mouse and BDCA1− in human. Compared with cDC1s, tDCs were Xcr1− and Cadm1− in mouse and CLEC9A− in human. Altogether, our CyTOF analysis identified a distinct subpopulation of DCs in mouse that displays a transitional phenotype between pDCs and cDC2s and mirrors the phenotype of human blood and spleen AXL+ DCs.
Mouse tDC Phenotypes Range from pDC-like to cDC-like
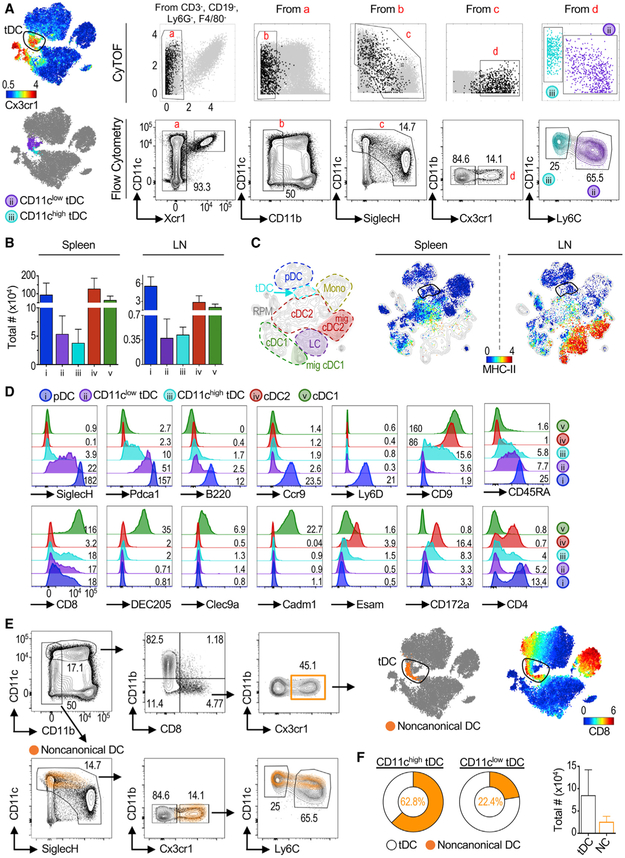
We then explored the diversity of mouse tDCs. We designed a flow cytometry gating strategy to enable tDC characterization, quantification, and purification (Figure S3). Because tDCs did not express the cDC1 marker Xcr1 or the cDC2/monocyte marker CD11b (Figure 1A), we used these parameters to eliminate these cells (Figure 2A). Biaxial gating of CyTOF data showed that tDCs (black dots) were among cells expressing high to low levels of SiglecH and low to high levels of CD11c. Furthermore, as shown by tSNE and UMAP analysis, tDCs, but not pDCs, were Cx3cr1high (Figures 1D and 2A, upper left). Finally, we observed that tDCs could be separated into two populations representing different parts of the spectrum: CD11clow Ly6Chigh tDCs (ii, purple) represent cells that are more similar to pDCs, while CD11chigh Ly6Clow tDCs (iii, cyan) correspond to cells that are more similarto cDC2s (Figure 2A, lower left). This partition mirrored our previous analysis of human AXL+ DCs, which can also be split based on expression of CD11c to facilitate characterization of their heterogeneity (Alcántara-Hernández et al., 2017). We validated our observations by flow cytometry and quantified tDCs in lymphoid organs (Figures 2A and 2B; see Figure S3 for the complete gating strategy). Compared with other DC subsets, tDCs were a minor population of cells in spleen and lymph nodes (LNs), representing ~0.02%–0.07% of all leukocytes. Notably, tSNE analysis revealed that LN tDCs mapped between pDCs and cDC2s and away from migratory major histocompatibility complex class II (MHC class II)high DCs (Figures 2C and S3C).
Figure 2. Mouse tDC Phenotype Transitions from pDC-like to cDC-like.
(A) tSNE plot of CyTOF data as in Figure 1, colored by Cx3cr1 expression (top left). Top row: biaxial plots of CyTOF data. Black dots represent gated Cx3cr1+ Tcf4+ cells. Cells were divided based on the expression of CD11c and Ly6C and mapped to the CyTOF tSNE plot (lower left). Bottom row: flow cytometry gating strategy (see Figure S3A for the full gating strategy). One representative of two independent experiments (exp).
(B) Total numbers of DC subsets in mouse spleen (n = 14) and skin-draining lymph nodes (LNs; n = 7). Shown is mean ± SD.
(C) tSNE map of spleen and LNs analyzed by CyTOF, with manual annotation (left) and colored by MHC class II expression (right). One representative of two exp.
(D) Surface markers analyzed in each DC subset by flow cytometry representative of n ≥ 3. Numbers indicate geometric Mean Fluorescence Intensity (gMFI) × 102.
(E) Noncanonical DCs were gated as described (Bar-On et al., 2010) and overlaid in the tSNE plot of mouse spleen CyTOF data (orange dots, upper panel). Noncanonical DCs were also overlaid in our flow cytometry gating strategy described in (A) (bottom panels).
(F) Frequency of noncanonical DCs within CD11chigh and CD11clow tDC gates (pie charts) and total number of tDCs and noncanonical DCs in the spleen (n = 14).
Flow cytometry analysis of CD11clow and CD11chigh tDCs corroborated their transitional phenotype (Figure 2D). CD11clow tDCs had a phenotype closer to that of pDCs, with higher levels of SiglecH, Pdca1, and B220 than CD11chigh tDCs. CD11clow and CD11chigh tDCs were negative for the pDC markers Ccr9 and Ly6D; however, CD11chigh tDCs had intermediate levels of CD9, a marker previously associated with pDC subpopulations (Björck et al., 2011). Although tDCs featured some levels of CD8, both CD11clow and CD11chigh tDCs lacked expression of more specific cDC1 markers such as DEC205, Cadml, and surface Clec9A. We did not detect significant levels of the cDC2 marker Esam, but CD11chigh tDCs were CD172alow.
The phenotype of mouse tDCs, particularly their expression of Cx3cr1 and CD8, resembled a previously described population of mouse DCs that has features of both pDCs and cDCs, termed noncanonical DCs (Bar-On et al., 2010). We hypothesized that noncanonical DCs could be represented within the tDC population. To test this, we mapped noncanonical DCs, gated as described by Bar-On et al. (2010), onto our mouse tSNE plot and confirmed the high expression of CD8 in this part of the map (orange on top of the upper-right tSNE map, Figure 2E). We also overlaid noncanonical DCs in our flow cytometry gating strategy (orange on top of the contour plots, Figure 2E). In both cases, we found that noncanonical DCs mapped within the tDC population but only captured a fraction of it: 62.8% of CD11chigh tDCs and 22.4% of CD11clow tDCs (Figure 2F). Noncanonical DCs represented ~34% of the entire tDC population (Figure 2F, right bar graph). Thus, murine tDCs include the previously described noncanonical DCs.
In summary, we present a flow cytometry gating strategy that captures tDCs and allows for enrichment of different parts of the phenotypic spectrum based on CD11c and Ly6C expression.
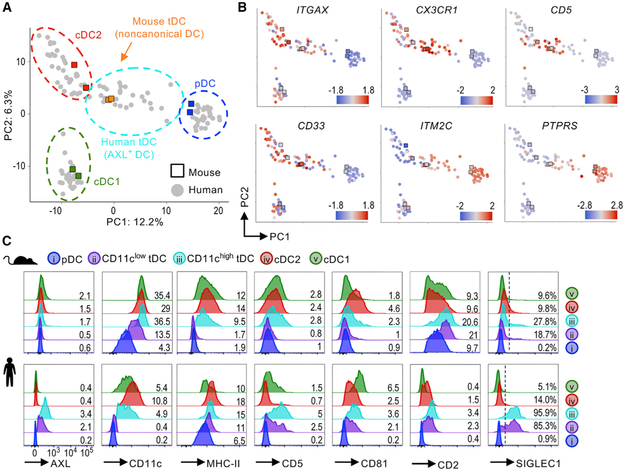
Transcriptome Analysis Aligns Mouse and Human tDCs
To further query the homology of mouse and human tDCs (also known as AXL+ DCs), we expanded our proteomic CyTOF approach to transcriptomics, which allows genome-wide comparison between species. We leveraged publicly available mouse bulk RNA sequencing (RNA-seq) data of individually purified DC subsets and human single-cell RNA-seq data of myeloid cells (Lau et al., 2016; Villani et al., 2017). The mouse dataset included noncanonical DCs, which we used to represent mouse tDCs for this analysis. PCA using nearly 1,000 differentially expressed genes overlapped mouse and human pDCs, cDC2s, and cDC1s, validating our analysis (Figure 3A; see Figure S4A for analysis details). Mouse tDCs localized between pDCs and cDC2s, overlapping with human tDCs. In both species, tDCs shared intermediate expression of several pDC genes (e.g., ITM2C and PTPRS), cDC genes (e.g., CD33 and ITGAX), CX3CR1, and CD5 (Figure 3B).
Figure 3. Mouse and Human tDCs Align Transcriptionally and Phenotypically.
(A) RNA-seq of mouse (Lau et al., 2016) and human (Villani et al., 2017) DC subsets analyzed by PCA and shown with manual annotation (see Figure S4A for analysis details).
(B) Expression Z-scores of select genes.
(C) Protein expression measured by flow cytometry in mouse (top) and human (bottom) DC populations. Histograms represent n ≥ 3. Numbers on histograms indicate gMFI × 102 or frequency (%) of positive cells.
Next, we inquired whether mouse tDCs share protein expression of surface markers described for human tDCs (Figure 3C). To mirror our mouse tDC analysis and characterize diversity within the population, we divided human tDCs based on their expression of CD11c, as previously described (Alcántara-Hernández et al., 2017). Human tDCs display high levels of the receptor tyrosine kinase AXL; however, Axl was undetectable in mouse tDCs using two antibody clones that efficiently labeled macrophages, as shown in Figure S4B. Human tDCs expressed CD5 and CD81 (Zhang et al., 2017), which was also true for mouse tDCs, especially CD11chigh tDCs. Lastly, both CD2 and SIGLEC1/CD169, two markers that have been used to define human pDC subpopulations (Matsui et al., 2009; Wilhelm et al., 2016), were enriched in tDCs compared with other DC subsets in both species. However, CD2 was not a unique marker for tDCs, and Siglec1 was only detected in a fraction (~20%–30%) of murine tDCs. We were not able to evaluate SIGLEC6, a marker of human tDCs, because it does not have a mouse homolog. Collectively, mouse and human tDCs overlap transcriptionally and phenotypically. Furthermore, many previous reports referring to pDC subpopulations can be explained by the heterogeneous phenotype of tDCs in both mouse and human.
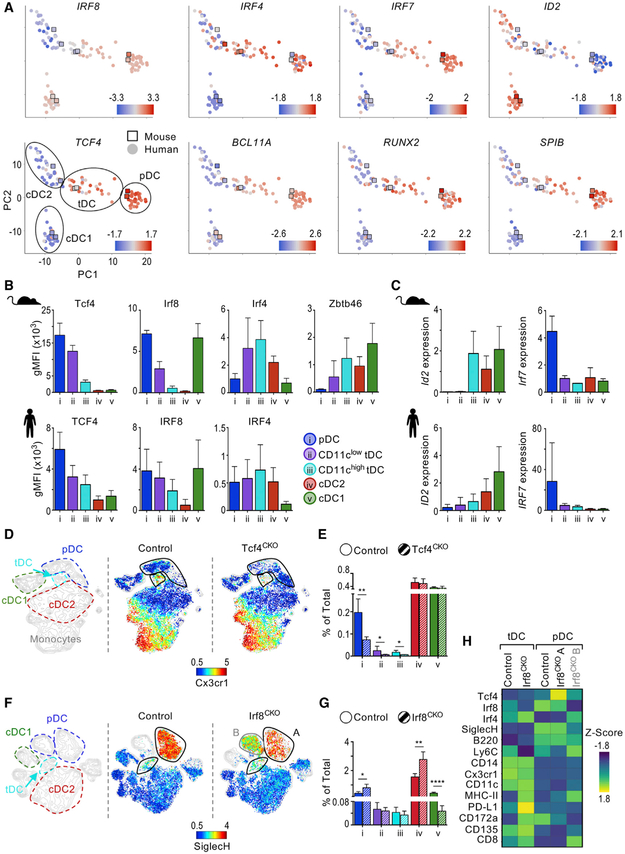
Mouse and Human tDCs Share TF Profiles
DC subsets are characterized by their expression of a combination of TFs, which are essential for each subset’s development, phenotype, and function. The TF TCF4 is required for pDC development and function (Cisse et al., 2008; Ghosh et al., 2010). IRF8 and IRF4 are required for cDC1 and cDC2 development, respectively (Schiavoni et al., 2002; Suzuki et al., 2004). Zbtb46 is uniquely expressed in cDCs and required for their function, but not their development (Meredith et al., 2012a, 2012b; Satpathy et al., 2012). Thus, we evaluated the TF signature of mouse and human tDCs in comparison to other DC subsets (Figures 4A–4C). At the RNA and protein level, both TCF4 and IRF8 expression ranged from intermediate to low in CD11clow and CD11chigh tDCs, respectively. We found high levels of IRF4 in tDCs, particularly CD11chigh tDCs; however, different from mouse, IRF4 was also present in human pDCs. Finally, Zbtb46 protein was detected in mouse tDCs, with intermediate to high expression in CD11clow and CD11chigh tDCs, respectively.
Figure 4. TF Profiles Are Shared between Mouse and Human tDCs.
(A) PCA denoting expression Z-scores of TFs in mouse and human DC subsets. Manual annotation of PCA is shown in the bottom left panel.
(B) gMFI of TF expression measured by flow cytometry in mouse (top, n = 2–3) and human (bottom, n = 4–5).
(C) Expression of ID2 and IRF7 in sorted mouse splenic and human blood DC subsets measured by qPCR. Expression represents ΔΔCq relative to the internal control gene RPL13A/Rpl13a and cDC2s (n = 2–4).
(D) CyTOF analysis of BSA-enriched splenocytes from CD11cCRE Tcf4fl/fl (Tcf4CKO) and control (Tcf4fl/fl and B6) mice manually annotated (left) and colored by protein expression (right). One representative of two exp.
(E) Frequency of DC subsets in spleen of Tcf4CKO and control mice (n = 3 in 2 exp).
(F) CyTOF analysis of BSA-enriched splenocytes from CD11cCRE Irf8fl/fl (Irf8CKO) and control (Irf8fl/fl) mice manually annotated (left) and colored by protein expression (right). One representative of two exp.
(G) Frequency of DC subsets in spleen of Irf8CKO and control mice (n = 3 in 2 exp).
(H) Heatmap of protein expression in pDCs and tDCs from Irf8CKO and control mice (n = 2). pDCs from Irf8CKO were divided in two based on Irf8 expression; i.e., population A is present in control mice, whereas population B is present in Irf8CKO mice.
Bar graphs indicate mean ± SD. Statistics determined by t-test. *p < 0.05, **p < 0.01, ****p < 0.0001.
Next, we analyzed the TF ID2, which promotes cDC development by antagonizing TCF4 (Grajkowska et al., 2017). We found that ID2 expression inversely correlated with TCF4 expression, as expected (Figure 4C; see Table S2 for primers). In mouse, Id2 was not detected in pDCs and CD11clow tDCs but was noted in CD11chigh tDCs at levels comparable to cDCs. Similarly, low ID2 expression was detected in human CD11chigh tDCs. Finally, we evaluated IRF7, a TF associated with the capacity of pDCs to produce IFN-I (Honda et al., 2005; Izaguirre et al., 2003). We found that unlike pDCs, all tDCs in mouse and human expressed negligible levels of IRF7, suggesting functional differences between these cells. In summary, tDCs demonstrate a mixed pattern of TFs, correlating with their transitional phenotype between pDCs and cDCs.
To test whether tDC development depends on Tcf4 expression, we analyzed CD11cCRE Tcf4fl/fl (Tcf4CKO) mice (Figures 4D and 4E). Conditional deletion of Tcf4 in this model results in pDC loss, as expected (Cisse et al., 2008; Ghosh et al., 2010). The frequencies of both CD11chigh and CD11clow tDCs were also significantly decreased in Tcf4CKO mice, while cDC1s and cDC2s were unaffected. This indicates that similar to pDCs, tDC development depends on Tcf4.
IRF8 expression is shared between pDCs and cDC1s. However, while cDC1s require IRF8 for development, pDCs require it for their function (Schiavoni et al., 2002; Sichien et al., 2016). Depletion of Irf8 from pDCs generates a population with some features of cDCs, i.e., lower levels of SiglecH and higher levels of CD11c, Irf4, and MHC class II, which resembles the tDC population described here. Therefore, we analyzed pDCs and tDCs in CD11cCRE Irf8fl/fl (Irf8CKO) mice by CyTOF. As expected, cDC1s failed to develop in these mice (Figures 4F and 4G). On the contrary, pDC and tDC numbers were unaffected in Irf8CKO mice. Most pDCs from Irf8CKO mice clustered separately from control mice but did not overlap with tDCs (population B in Irf8CKO versus population A in control mice). Indeed, pDCs from Irf8CKO mice had decreased expression of several markers, including SiglecH, and increased expression of MHC class II, as previously shown (Figure 4H; Sichien et al., 2016). Similarly, tDC phenotype was slightly altered in Irf8CKO mice, although to a lower extent, corresponding to their lower expression of Irf8. We conclude that similar to pDCs, Irf8 is not required for tDC development. Furthermore, our results show that Irf8 depletion does not promote pDC differentiation into tDC-like cells.
tDCs Display Lymphoid Characteristics, Similar to pDCs
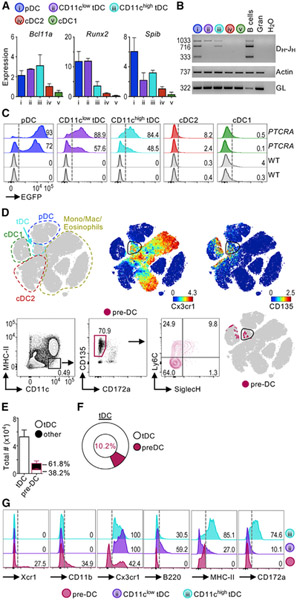
Our transcriptome analysis showed that mouse and human tDCs express Bcl11a, Runx2, and SpiB, i.e., TFs associated with pDC development and function (Figure 4A; Chopin et al., 2016; Ippolito et al., 2014; Sawai et al., 2013; Schotte et al., 2004; Wu et al., 2013). We confirmed this analysis by evaluating the expression of these TFs within mouse CD11clow and CD11chigh tDCs (Figure 5A). We detected high levels of Bcl11a, Runx2, and SpiB in pDCs and tDCs but little to none of these TFs in cDC1s or cDC2s. Bcl11a was expressed similarly by all tDCs and pDCs, whereas Runx2 expression varied from high to low in tDC fractions. SpiB expression was higher in tDCs than in cDCs, but lower than in pDCs. These results show that tDCs share multiple transcriptional regulators with pDCs.
Figure 5. tDCs Display Lymphoid Characteristics Associated with pDCs.
(A) TF expression in sorted mouse splenic DC subsets measured by qPCR (n = 3 in 3 exp). Expression represents ΔΔCq relative to the internal control gene Rpl13a and cDC2s.
(B) Sorted mouse splenic DC subsets analyzed by PCR assay for IgH D-J rearrangement. Actin and IgH germline (GL) are also shown (1 representative of 6 exp).
(C) EGFP expression in splenic DC subsets from PTCRA-EGFP mice compared with wild-type (WT) mice. Two mice for each condition shown. Numbers indicate frequency of EGFP+ cells.
(D) tSNE map of Lin− events from negatively enriched (anti-CD3 and anti-CD19) mouse splenocytes analyzed by CyTOF. Left: manual annotation; right: colored by protein expression. Bottom panels show splenic pre-DCs gated as CD3− CD19− CD335− B220− and CD11c+ MHC class II− CD135+ CD172a− as described (Liu et al., 2009). Pre-DCs were overlaid in the tSNE map (pink dots). See Figure S5 for a comparison between spleen tDCs and BM pre-DCs.
(E) Total numbers of tDCs and pre-DCs (n = 5 in 3 exp). The frequency of pre-DCs that overlap tDCs (white) or other cells (black) is shown.
(F) Frequency of tDCs that correspond to pre-DCs (average of n = 5 in 3 exp).
(G) Surface marker expression in spleen pre-DCs and tDCs representative of 3 exp. Numbers indicate frequency of positive cells.
Bar graphs indicate mean ± SD.
Distinct from cDCs, pDCs exhibit lymphoid characteristics independent of their myeloid or lymphoid origin, i.e., rearranged immunoglobulin heavy chain (IgH) D-J genes and expression of the pre-T cell receptor α (PTCRA) (Sathe et al., 2013; Shigematsu et al., 2004). Thus, we asked whether tDCs also bear these lymphoid characteristics. We detected IgH D-J rearrangement in pDCs, CD11clow tDCs, and CD11chigh tDCs, but not in cDC2s or granulocytes (Figure 5B). As previously described, cDC1s have a very low frequency of rearrangement, which we detected sporadically (data not shown; Corcoran et al., 2003). It is possible that some fraction of cells have rearrangements that were not detected, as the assay used can only detect arrangement with particular DH segments.
We next took advantage of transgenic mice that allow evaluation of PTCRA-EGFP at the single-cell level (Figure 5C; Shigematsu et al., 2004). As expected, we detected strong PTCRA-EGFP labeling in pDCs, but not cDC1s or cDC2s. tDCs were also labeled with PTCRA-EGFP, and levels varied across the population, correlating with their phenotypic similarity to pDCs. Moreover, PTCRA-EGFP was detected in ~50%–90% of tDCs despite their high expression of Zbtb46 (Figure 4B), a TF usually associated with cDCs. Thus, comparable to bona fide pDCs, tDCs display several lymphoid characteristics such as a unique TF profile, IgH D-J rearrangements, and PTCRA-EGFP labeling.
Human tDCs have been suggested to be cDC precursors or pre-DCs (See et al., 2017). However, different from cDCs, tDCs harbor lymphoid characteristics and require Tcf4 for their development, which is inconsistent with these cells being pre-DCs. To further analyze the relationship between tDCs and pre-DCs, we performed CyTOF profiling and unbiased analysis. We were unable to identify tDCs in the bone marrow (BM); thus, we first compared splenic tDCs with BM pre-DCs. We found that splenic tDCs did not overlap with BM pre-DCs in a tSNE map, indicating that they are phenotypically distinct (Figure S5). Next, we analyzed the relationship between splenic tDCs and splenic pre-DCs, gated as Lin− CD11chigh MHC class II− CD172a− CD135+ (Figure 5D; Liu et al., 2009). Unbiased analysis revealed that splenic pre-DCs were heterogeneous and localized to various CD135high areas of the tSNE map. Most pre-DCs mapped near cDC1s, and only a fraction of pre-DCs mapped with Cx3cr1+ tDCs in the tSNE map (Figure 5D). Furthermore, splenic pre-DCs were considerably less numerous than tDCs, and only 10% were encompassed within the total tDC population (Figures 5E and 5F). Closer phenotypic analysis revealed that ~60% of pre-DCs expressed either Xcr1 or CD11b, markers that tDCs lack (Figures 1A and 5G). In addition, only ~40% of pre-DCs expressed Cx3cr1, which is expressed by all tDCs. Conversely, tDCs expressed variable levels of B220, MHC class II, and CD172a, which are excluded from the pre-DC definition. Finally, splenic tDCs express SiglecH, while splenic pre-DCs are mostly SiglecH−, as previously described (Figures 1A and 5D; Schlitzer et al., 2015). Thus, our unbiased analysis revealed that there is minimal overlap between splenic tDCs and pre-DCs.
Altogether, our findings show that tDCs follow a developmental program similar to pDCs and suggest that tDCs represent a distinct population of cells that are different from traditional pre-DCs.
Mouse and Human tDCs Display Similar Functional Capabilities
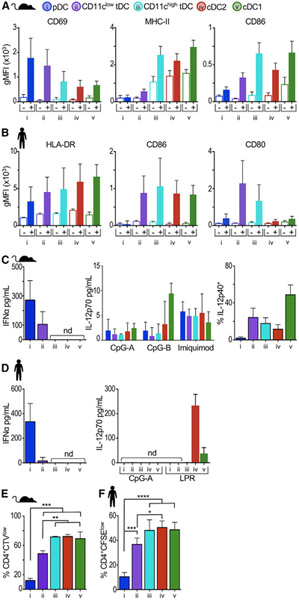
Next, we compared the functional capacities of mouse and human tDCs, i.e., their capacity to respond to Toll-like receptor (TLR) stimulation, secrete cytokines, and activate T cell proliferation. Following in vivo stimulation with the TLR9 agonist CpG-A, mouse pDCs displayed high levels of CD69, an early marker of lymphocyte activation, while MHC class II and CD86 were only slightly upregulated (Figures 6A and S6). In contrast, both cDC2s and cDC1s strongly upregulated MHC class II and CD86, but not CD69. In accordance with their transitional phenotype, the CD11clow tDC fraction upregulated CD69 at comparable levels to pDCs, whereas the CD11chigh fraction upregulated MHC class II and CD86 similar to cDCs. Human DC subsets behaved similarly when stimulated ex vivo with CpG-A; i.e., HLA-DR and CD86 were less upregulated in pDCs and scaled up from tDCs to cDCs (Figure 6B). Interestingly, we found that human tDCs were particularly efficient at upregulating the costimulatory marker CD80 upon CpG-A stimulation.
Figure 6. Mouse and Human tDCs Display Similar Functional Capabilities.
(A) Splenic DCs were analyzed 6 h after intravenous (i.v.) inoculation of CpG-A (filled bars) or PBS control (empty bars). gMFI of activation markers is shown as the mean ± SD (n = 2–4 in 2–4 exp). See Figure S6 for histograms of activation markers.
(B) Human DCs were sorted from PBMCs and analyzed at time 0 (empty bars) or after 24 h of culture with 5 μg/mL of CpG-A (filled bars). gMFI of activation markers is shown as the mean ± SD (n = 2–4 in 2–4 exp).
(C) IFNα and IL-12p70 measured by ELISA in supernatants from sorted mouse DCs stimulated with CpG-A for 16–18 h (n = 4 in 4 exp). Frequency of IL-12p40-positive cells was measured by intracellular cytokine staining after 4 h of stimulation with CpG-A in the presence of brefeldin A (BFA).
(D) IFNα measured by ELISA and IL-12p70 measured by cytometric bead array in the supernatants from sorted human DCs stimulated as in (B) or with an adjuvant cocktail (lipopolysaccharide-poly(I:C)-R848 [LPR]).
(E) Frequency of CellTrace violet (CTV)low mouse CD4+ T cells in mixed leukocyte reactions. DCs were sorted from B6 mice and cocultured with CTV-labeled CD4+ T cells from BALB/c mice for 5 days (n = 3 in 3 exp).
(F) Frequency of carboxyfluorescein succinimidyl ester (CFSE)low human CD4+ T cells in mixed leukocyte reactions. Human DC populations were sorted and cocultured with allogeneic CFSE-labeled T cells for 5–6 days (n = 3–4 in 3 exp).
Bar graphs indicate mean ± SD; nd, not detected. Statistics determined by one-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In accordance with their lower expression of IRF7 (Figure 4C), tDCs had lower capacity to produce IFNα than pDCs. Mouse CD11clow tDCs, but not CD11chigh tDCs, were able to produce IFNα, although to a lower extent than pDCs (Figure 6C). Similarly, human CD11clow tDCs, but not CD11chigh tDCs, secreted very low levels of IFNα (Figure 6D). We also evaluated tDC capacity to produce interleukin-12 (IL-12), a TH1-activating cytokine suggested to be secreted by human tDCs (See et al., 2017; Villani et al., 2017). We found that mouse tDCs were unable to secrete the active form IL-12p70 following stimulation with CpG-A or CpG-B, whereas all mouse DC subsets secreted low levels of this cytokine in response to imiquimod. However, we did detect the precursor subunit IL-12p40 in tDCs and cDCs following CpG-A stimulation (Figure 6C). We observed similar results in human; i.e., in our hands, pDCs and tDCs did not secrete IL-12p70 with either stimulation tested (Figure 6D). These results show that mouse and human tDCs have a limited capacity to produce IFNα and IL-12p70.
Finally, we compared the capacity of mouse and human tDCs to promote allogeneic T cell proliferation in a mixed leukocyte reaction (MLR). Both mouse and human freshly isolated pDCs expressed low levels of MHC class II/HLA-DR (Figure 3C) and consequently failed to promote the proliferation of allogeneic CD4+ T cells at steady state (Figures 6E and 6F). The antigen presentation capabilities of freshly isolated tDCs transitioned across the population, correlating with their phenotypic similarity to pDCs and cDCs. Altogether, these results indicate that tDCs’ functional capabilities vary across the population, with CD11clow cells behaving more like pDCs and CD11chigh cells behaving more like cDCs. These transitional functions of tDCs were similar in mouse and human, cementing their homology.
tDCs Are Recruited to the Site of Influenza Infection
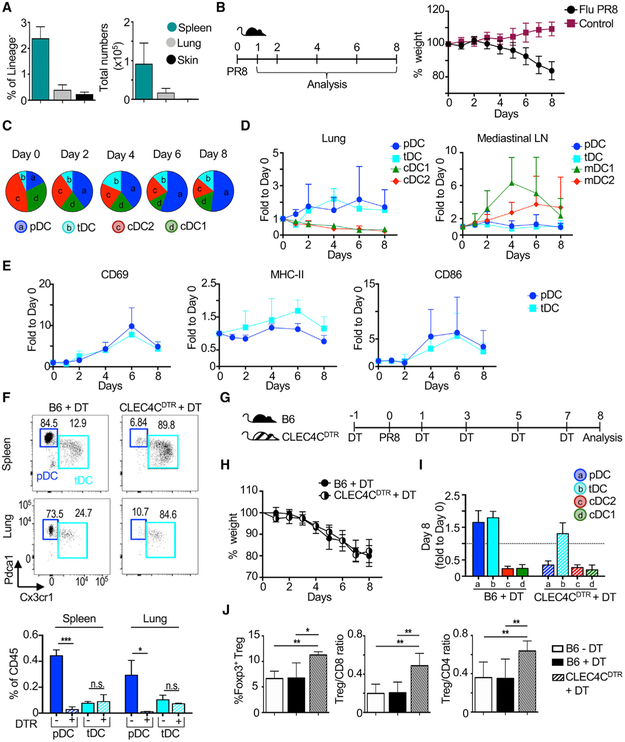
We found that tDCs are hardly present in tissues such as lung or skin at steady state, i.e., in the absence of inflammation (Figure 7A). The few lung and skin tDCs found by CyTOF expressed Tcf4, Pdcal, and Cx3cr1 but showed decreased expression of SiglecH compared with splenic tDCs (Figures S7A and S7B). This observation is in accordance with a lack of tDCs in healthy human skin, as described (Alcántara-Hernández et al., 2017).
Figure 7. tDCs Are Recruited to the Lung during Influenza Infection.
(A) Frequency and number of tDCs in spleen (n = 6), lung (n = 6), and skin (n = 3) quantified by flow cytometry and CyTOF. See Figure S7A and S7B for CyTOF of lung and skin.
(B) Intranasal influenza (PR8) infection of B6 mice was followed by daily weight loss (n = 6 mice per time point, combined from 3 exp).
(C) Composition of the lung DC compartment by subset over time post-infection. Pie charts were drawn from total numbers. See Figure S7C for the lung DC gating strategy.
(D) Fold change of each DC subset to day 0 in the lung and mediastinal lymph nodes (LNs). In LNs, migratory cDC subsets (mDC1s and mDC2s) were analyzed.
(E) Activation marker expression on lung pDCs and tDCs over time. Fold change of gMFI relative to day 0 is shown.
(F) Depletion of pDCs, but not tDCs, after a single dose of diphtheria toxin (DT) in CLEC4CDTR or control B6 mice in the spleen and lung. Statistics determined by t-test.
(G) Timeline of influenza infection and DT administration in CLEC4CDTR mice.
(H) Weight of DT-inoculated mice was evaluated daily (n = 5–6 mice, combined from 3 exp).
(I) Fold change of each DC subset to day 0 in the lung at day 8 post-infection is shown.
(J) Lung T cells analyzed at day 8 (n = 5–6 mice, combined from 3 exp). Statistics determined by one-way ANOVA with Tukey’s multiple comparison test.
Bar graphs indicate mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
We then evaluated tDC accumulation in the lung during respiratory viral infection with influenza virus. Different from cDCs that migrate from the lung to local draining mediastinal LNs, pDCs are known to accumulate in the infected lung (Grayson et al., 2007; Helft et al., 2012). Given their developmental similarities with pDCs, we hypothesized that tDCs will demonstrate similar behavior and accumulate in the lung during infection. B6 mice were infected intranasally with influenza (Flu PR8), and the course of infection was tracked by animal weight loss (Figure 7B). Before infection, cDC2s and cDC1s were represented in higher numbers than pDCs and tDCs in the lung. However, shortly after infection, lung cDC2 and cDC1 numbers decreased dramatically, whereas pDC and tDC numbers increased 2- to 3-fold (Figures 7C and 7D; see Figure S7C for the gating strategy). By days 4–6, pDCs and tDCs occupied more than 50% of the DC compartment. The loss of cDC1s and cDC2s in the lung corresponded to an increase in migratory cDCs in the lung-draining mediastinal LNs, with a peak at day 4 (Figure 7D). We did not observe accumulation of tDCs in mediastinal LNs, suggesting that these cells were not following the same migratory pattern as cDCs. Finally, we observed that both lung tDCs and pDCs upregulated the activation markers CD69, MHC class II, and CD86 (Figure 7E). We conclude that similar to pDCs, tDCs are recruited to the site of influenza infection and display an activated phenotype.
Several pDC depletion models have been developed and show contradictory results regarding the role of pDCs during viral infection (Cervantes-Barragan et al., 2007, 2012; Swiecki et al., 2013; Wolf et al., 2009). However, it is unknown whether tDCs are also depleted in these models, which may account for inconsistent conclusions regarding the necessity of pDCs for anti-viral responses. Therefore, we evaluated tDC depletion in one prominent model, a transgenic mouse that expresses diphtheria toxin receptor (DTR) under the promoter of the human pDC marker CLEC4C/BDCA2 (Swiecki et al., 2010). We found that diphtheria toxin (DT) administration in CLEC4CDTR mice eliminated pDCs, but not tDCs, in the spleen and lung (Figure 7F). Furthermore, we did not find depletion of any other DC subset in this model, as previously described (data not shown; Swiecki et al., 2010). We conclude that CLEC4CDTR mice are a good model for the specific depletion of pDCs without affecting tDC numbers.
We then used CLEC4CDTR mice to investigate the dynamics of tDC accumulation in the absence of pDCs during influenza infection (Figure 7G). Interestingly, pDC depletion did not alter the overall course of influenza infection or the accumulation of tDCs in the lung (Figures 7H and 7I). Similarly, cDC decrease from the lung was not impaired in the absence of pDCs (Figure 7I). However, we found that the absence of pDCs promoted a significant increase in lung Foxp3+ regulatory T (Treg) cells (Figure 7J). Consequently, the ratio of Treg cells to CD8+ and CD4+ T cells was significantly higher in pDC-depleted mice, suggesting that lung-recruited pDCs and tDCs may have different roles during influenza infection.
DISCUSSION
In this study, we provide formal proof of the existence of a population of mouse DCs closely related to pDCs and equivalent to the recently identified human AXL+ DCs. We name this conserved population transitional DCs (tDCs) to reflect their heterogeneous phenotype and function, which span between pDCs and cDC2s. Our multifaceted analysis of the phenotype, transcriptome, and function of tDCs in mouse and human is a resource to further dissect the role of these cells in different diseases and ultimately move the field forward toward DC-based therapeutics.
Human and mouse tDCs share several characteristics: (1) a conserved gene expression profile; (2) expression of key TFs; (3) cellular heterogeneity showing a range of pDC to cDC phenotypes; (4) lower capacity than pDCs to produce IFNα; and (5) higher capacity than pDCs to promote allogeneic T cell proliferation. Because tDCs appear quite heterogeneous, we found it useful to divide them into two functional groups: CD11clow (pDC-like) tDCs and CD11chigh (cDC-like) tDCs. However, we emphasize that this distinction was created for the purpose of characterizing tDC heterogeneity, and consequently, we do not consider these to be distinct developmental DC subsets.
In humans, tDCs can be distinguished from pDCs by their expression of AXL; however, we could not detect surface or intracellular Axl protein in mouse tDCs, indicating that this is not a shared characteristic. We found that mouse tDCs are best distinguished from pDCs by expression of Cx3cr1, a marker also expressed by human tDCs. A lack of concordance in DC subset-specific surface markers between human and mouse is well documented for other DC subsets (Vu Manh et al., 2015). For example, Pdca1 (BST2) is a pDC-specific marker in mouse at steady state but is widely expressed in human immune cells (Blasius et al., 2006). These differences may reflect divergent evolution, perhaps driven by species-specific pathogens. Because of these differences, it is imperative to consider a combination of phenotypic, functional, and transcriptional assays to establish cell-type homology between species, as we have done here. We found that mouse and human tDCs did share variable expression of CD2, CD5, CD81, and SIGLEC1. Despite these similarities, we could not identify any marker that unequivocally distinguished mouse tDCs from pDCs and cDCs, which is in accordance with their transitional phenotype. These results suggest that tDCs were probably overlooked when using gating strategies based on a few markers and illustrate the importance of using high-dimensional approaches to avoid oversimplification of the DC compartment. As shown here, high-dimensional approaches can be used to design simplified gating strategies for flow cytometry analysis and cell purification.
Although further investigation of the origin of tDCs is required, we report several observations that link tDCs closely to pDCs. First, tDCs expressed the pDC lineage-defining TF TCF4. This TF is necessary for the development of both mouse and human pDCs and mouse tDCs, as demonstrated here (Cisse et al., 2008; Grajkowska et al., 2017; Nagasawa et al., 2008). The developmental dependence of tDCs on Tcf4 is in accordance with a lack of noncanonical DCs in Tcf4−/− mice (Bar-On et al., 2010), cells that we show are included within the broader definition of tDCs. In addition, pDCs and tDCs shared expression of other TFs required for pDC development and function (e.g., BCL11A, IRF8, SPIB, and RUNX2), which are known to be upstream or downstream of the master regulator TCF4 (Ceribelli et al., 2016; Ghosh et al., 2010; Sawai et al., 2013). Similar to pDCs, tDCs do not require IRF8 for their development, which is in contrast to cDC1s (Schiavoni et al., 2002; Sichien et al., 2016). Altogether, the shared TF signature of pDCs and tDCs strongly indicates that their development likely progresses along a shared path.
Another observation that associates tDCs with pDCs is the presence of lymphoid characteristics: rearrangement of the IgH locus, which is a permanent change to the genomic DNA, and expression of PTCRA-EGFP. The presence of these features in pDCs was originally interpreted to be indicative of lymphoid origin. Indeed, analysis at the single-cell level has identified a lymphoid precursor that can give rise to pDCs (Dress et al., 2019; Rodrigues et al., 2018). However, care should be taken when analyzing pDC lymphoid features, because these are also known to be carried by pDCs derived from myeloid progenitors (Corcoran et al., 2003; Sathe et al., 2013; Shigematsu et al., 2004). Regardless of their lymphoid or myeloid origin, IgH rearrangement can be detected in pDCs and tDCs, which suggests that these cells are closely related developmentally. Similar developmental associations can be implied by the expression of PTCRA-EGFP in pDCs and tDCs. These lymphoid characteristics may result from their shared expression of Bcl11a, which controls RAG-1 expression (Lee et al., 2017). Alternatively, they may be a consequence of an ectopic lymphoid program activated by TCF4, as previously suggested (Reizis, 2019). We found that tDCs, especially CD11chigh tDCs, simultaneously express PTCRA-EGFP and Zbtb46, a TF generally associated with cDCs. Zbtb46 expression has also been found in embryonic-derived Langerhans cells (Wu et al., 2016), suggesting that this TF does not necessarily label a pre-DC developmental pathway but perhaps reveals the functional capabilities of the cells, as suggested (Meredith et al., 2012b).
The exact developmental origin of tDCs also requires further study. It is possible that tDCs are generated as a byproduct of pDC development, as suggested for noncanonical DCs (Reizis, 2019). Alternatively, tDCs may arise directly from pDCs. Indeed, pDCs are known to lose IFN-I production potential and increase antigen presentation capacity during stimulation (Palucka et al., 2005), which are characteristics associated with tDCs. This pDC conversion, also called plasticity, has been observed for both human and mouse pDCs (Alcántara-Hernández et al., 2017; Alculumbre et al., 2018; Grouard et al., 1997; Liou et al., 2008; Manh et al., 2013). The identification of mouse and human tDCs creates an opportunity to explore pDC plasticity in vivo.
The observations that tDCs harbor lymphoid characteristics and depend developmentally on Tcf4 do not support the hypothesis that these cells are the traditional cDC precursor or pre-DCs, as previously suggested (See et al., 2017). Using a high-dimensional alignment strategy at the protein level, we show that mouse tDCs have little to no overlap with splenic and BM pre-DCs. Furthermore, splenic tDCs express SiglecH, and SiglecHCRE RFPfl/fl reporter mice have limited (~0.5%) RFP labeling in cDCs (Puttur et al., 2013). Similarly, DT administration to SiglecHDTR/DTR mice does not result in cDC depletion (Takagi et al., 2011). Altogether, these observations support our conclusion that tDCs are a different population of cells distinct from pre-DCs. Future work will aim to trace tDC fate in health and infection.
We found limited numbers of tDCs in lung and skin at steady state. However, similar to pDCs, tDCs accumulate in the lung during infection, suggesting that these two cell types may have cooperative roles during anti-viral responses. Our in vitro data show that complementary IFN-I secretion and antigen presentation functions are enacted by pDCs and tDCs, respectively. Surprisingly, we found that tDCs are present in DT-treated CLEC4CDTR mice, which allowed us to assess their role in the absence of pDCs. We found that a lack of pDCs, but not tDCs, promotes an increase in the frequency of Treg cells during influenza infection. This observation suggests that tDCs may be suited to promote Treg cells in accordance with their superior antigen presentation capabilities. However, the role of pDCs during influenza infection appears to be pro-inflammatory. It remains to be determined why tDCs are not depleted in the CLEC4CDTR mice given that DTR expression is thought to be modulated by TCF4 in this model (Cisse et al., 2008; Swiecki et al., 2010). Nevertheless, this finding points to the need to dissect whether tDCs are affected in other mouse models that have been used to evaluate pDC function. It is conceivable that tDCs, similar to pDCs, would be depleted when using anti-BST2 antibodies or SiglecHDTR models. Finally, to unequivocally identify the role of tDCs during immune responses, it is necessary to develop strategies to specifically deplete these cells without affecting pDCs or cDCs.
In summary, we identify and characterize a tDC population in mouse that mirrors the phenotype, function, and transcriptome of the recently defined human AXL+ DCs (unified under the name tDCs). Although tDCs have been unappreciated until now, these cells most likely serve as the missing link to explain the range of functions previously associated with pDCs. Our in-depth characterization is a resource to understand the role of these cells in the context of infection, cancer, and autoimmunity and ultimately to harness pDCs and tDCs therapeutically.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Juliana Idoyaga (jidoyaga@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Specimens
Blood was obtained from healthy adult volunteers following the guidelines of the Research and Laboratory Environmental Health and Safety program of Stanford University. All donors provided informed consent in accordance with IRB protocols approved by the Stanford University Administrative Panel on Human Subjects in Medical Research (IRB Protocol #37612). Blood donors were healthy, without acute diseases and between 20-45 years old. Males and females were equally represented. We did not identify significant differences between sexes in our analyses. Spleen samples were obtained from the Stanford Tissue Bank and classified by a pathologist as healthy. These spleen samples were from distal pancreatectomy from patients that were not subjected to any chemotherapy before organ extraction.
Mice
Female C57BL/6 (B6) and BALB/c mice were purchased from The Jackson Laboratory. Transgenic PTCRA-EGFP mice have been described previously (Shigematsu et al., 2004). CD11cCRE, Irf8fl/fl and CLEC4CDTR mice have been described and were obtained from The Jackson Laboratory (Caton et al., 2007; Feng et al., 2011; Swiecki et al., 2010). CD11cCRE mice were crossed to Irf8fl/fl in house and screened routinely for the appearance of Irf8 germline deletions. CD11cCRE Tcf4fl/fl mice were obtained from Dr. Reizis (Cisse et al., 2008). Animals were maintained under specific pathogen-free conditions and used at 6-8 weeks of age in accordance with the Stanford University Administrative Panel on Laboratory Animal Care and overseen by the Institutional Animal Care and Use Committee (APLAC Protocol #28926).
Influenza Virus
NR-29029 Influenza A/Puerto Rico/8/1934-WG (H1N1) was obtained from BEI resources, expanded in MDCK cells as described (Kronstad et al., 2018) and generously provided by Drs. C. Blish and L. Kronstad.
METHOD DETAILS
Enrichment of DCs from Mouse Spleen
Mouse spleens and LN were digested with 400 U/mL Collagenase D (Millipore Sigma) and 50 μg/mL DNase I (Millipore Sigma) for 30 minutes at 37°C. 10 mM EDTA (Corning) was added for the last 5 minutes of culture. For splenocytes, red blood cells were then lysed with ACK Lysis buffer (Lonza). Both spleen and LN cell suspension were filtered through a 70 mm strainer. For CyTOF analysis, cell suspensions were stained directly or enriched using a Bovine Serum Albumin (BSA, Millipore Sigma) gradient. In brief, splenocytes were re-suspended in 3 mL of 30% BSA, and 1 mL of PBS was overlaid, then centrifuged at 1055 g for 30 minutes. For cell sorting of all DC subsets, whole spleen cell suspensions were incubated with anti-CD3 (KT3) and anti-CD19 (1D6) followed by negative selection with anti-rat Dynabeads (Thermo Fisher Scientific). Cells were sorted using a FACSAria II or FACSAria Fusion (BD Biosciences) at the Stanford Shared FACS Facility. All DC populations were first gated on live cells, singlets, lineage negative (CD3− CD19− CD20− TCRβ−), and CD11c+. cDC1s were sorted as CD11chigh Xcr1+. cDC2s were sorted as Xcr1− CD11chigh CD11b+. For functional assays, the sorting strategy for pDCs and tDCs avoided the use of antibodies with potential inhibitory capacity. pDCs were sorted asXcr1− CD11b− Cx3cr1− B220+ Ly6C+. tDCs were sorted as Xcr1− CD11b− Cx3cr1+ and separated based on CD11clow Ly6Chigh and CD11chigh Ly6Clow. For RNA and DNA extraction, SiglecH was included in the sorting strategy.
Lung, Skin, and Bone Marrow Preparation
Perfused lungs were digested with 0.13 U/mL Liberase TM (Millipore Sigma) and 50 μg/mL DNase I for 25 minutes at 37°C and filtered using a 70 μm strainer. Skin from both ears was split in dorsal and ventral halves and incubated dermis down with 1.3 U/mL Liberase TL (Millipore Sigma) and 50 μg/mL DNase I for 45 minutes. Ears were minced with scissors, incubated for additional 45 minutes and filtered through 70 μm strainer. For bone marrow analysis, femurs and tibias were obtained and flushed using RPMI. The epiphysis was minced with scissors and washed thoroughly with RPMI. Cell suspensions were filtered through a 70 μm strainer.
Isolation of DCs from Human Blood and Spleen
Blood from healthy adult human donors was collected using EDTA-coated tubes (BD Biosciences). Spleens were perfused with 10 mL of digestion medium consisting of RPMI containing 10% FBS, 2 mM L-glutamine (Corning), 100 IU Penicillin, 100 μg/mL Streptomycin (Corning), 0.8 mg/mL Collagenase IV (Worthington) and 0.05 mg/mL DNase I. After perfusion, spleen was cut in small pieces and incubated in digestion medium for 30 min at 37°C with continuous shaking. Digestion was stopped by adding 5 mM EDTA. Cell suspensions were filtered through 100 μm cell strainers. Mononuclear cells were isolated by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare) for both blood and spleen. To sort pure DC subsets from blood, PBMCs were Fc-blocked with human gamma globulin (Thermo Fisher Scientific), then negatively enriched using mAb against CD3 (OKT3), CD19 (HIB19), CD14 (HCD14), and CD335 (9E2) followed by anti-mouse magnetic beads (Dynabeads, Thermo Fisher Scientific) at a concentration of 2-4 beads per target cell. Cells were sorted using a FACSAria II or FACSAria Fusion (BD Biosciences) at the Stanford Shared FACS Facility. All DC populations were first gated on live, singlets, lineage negative (CD3− CD19− CD20− CD335− CD66b−) and monocyte negative (CD14− CD16−). cDC1s were sorted as HLA-DR+ CD123− BDCA3high. cDC2s were sorted as HLA-DR+ CD123− BDCA3-low CD11c+ BDCA1+. pDCs were sorted as HLA-DR+ CD123+ BDCA1− AXL− CD11c−. tDCs were gated as HLA-DR+ CD123+ BDCA1− AXL+ and separated into CD11c− and CD11c+. To allow IL-3 binding in culture, only CD123 clone 6H6 was used for sorting.
Staining Cell Suspensions for Flow Cytometry
Antibodies (Abs) for flow cytometry were purchased from Biolegend, R&D, MBL International Corp. and Thermo Fisher Scientific. Anti-Clec9a (Idoyaga et al., 2011) and anti-TCF4 were labeled using the Alexa 647 Labeling Kit following manufacturer’s instructions (Thermo Fisher Scientific) or detected using anti-Rabbit-Alexa 647 (Jackson ImmunoResearch). Cells were acquired on a 5-laser LSRFortessa X-20 (BD Biosciences), and data analyzed using FlowJo software (Tree Star, Inc). Unstained cells and single-fluoro-chrome-stained compensation beads (BD Biosciences) or cells were used for accurate compensation. Control samples included fluorescence minus one (FMO) for DC markers.
Mouse:
Cell suspensions from spleen, lung, and skin were incubated with supernatant against CD16/CD32 (clone 2.4G2, produced in house) to block non-specific binding for 15 minutes at 4°C. Cell suspensions were incubated in Ab mixes in mouse FACS buffer (2 mM EDTA, 2% FBS in PBS) for 20 minutes at 4°C. For transcription factor staining, cells were stained with surface Abs and LIVE/DEAD Fixable Blue (Thermo Fisher Scientific) in PBS for 20 minutes at 4°C, then fixed with FoxP3 Transcription Factor Fix/Perm Buffer (Thermo Fisher Scientific) for 2 hours to overnight and stained intracellularly for 30 minutes in 1X Permeabilization Buffer (Thermo Fisher Scientific).
Human:
PBMCs were incubated with human gamma-globulin (Thermo Fisher Scientific) to block non-specific binding for 15 minutes at 4°C. Cells were incubated with Ab mixes in human FACS buffer (2 mM EDTA, 2% Donor equine serum in PBS) for 20 minutes at room temperature. For transcription factor staining, cells were stained with surface Abs and LIVE/DEAD Fixable Blue in PBS for 20 minutes at room temperature, then fixed with FoxP3 Transcription Factor Fix/Perm Buffer for 1 hour at 4°C and stained intracellularly for 20 minutes in 1X Permeabilization Buffer.
Staining of Cell Suspensions for CyTOF
Metal-labeled Abs were obtained from Fluidigm or labeled using the MaxPar X8 labeling kit (Fluidigm) according to manufacturer’s instructions (see Table S1). Freshly isolated mouse (BSA- or Dynabead-enriched) splenocytes or thawed human mononuclear cells from blood and spleen were stained with 1 mL of 0.25 μM cisplatin (Fluidigm) for 5 minutes at room temperature to exclude dead cells. Cells were then washed with CyFACS buffer (2 mM EDTA, 1% BSA, 1% in PBS) and stained with heavy-metal-labeled Ab cocktail for 30 minutes on ice. Cells were washed twice with CyFACS then fixed with FoxP3 Transcription Factor Fix/Perm Buffer (Thermo Fisher Scientific) for 2 hours. Human surface CyTOF Abs that were sensitive to FoxP3 buffer in our hands (i.e., CX3CR1, CD123, CD33, CD135, CD172a and CD163), were instead stained after fixation and permeabilization for 30 minutes at 4°C. After staining, samples were washed and incubated with 2% paraformaldehyde (Electron) in PBS containing 125 nM Iridium intercalator (Fluidigm) overnight. Cells were washed with water, filtered, and acquired in a CyTOF2 (Fluidigm) at the Stanford Shared FACS Facility.
In Vivo Mouse DC Stimulation
Two μg of CpG-A (ODN2216; Invivogen) and 12 μL DOTAP (Santa Cruz Biotechnology) were mixed with 100 μL of PBS and administered intravenously (i.v.) by tail vein injection. Control mice received 12 μL DOTAP with PBS. Spleens were harvested and analyzed 6 hours after inoculation.
In Vitro DC Stimulation
Mouse:
For detection of cytokines in supernatant, sorted DC subsets were cultured in 96 well U-bottom plates at 37°C at a concentration of 5,000 cells in 200 μL of complete R10 media, consisting of RPMI (Corning) with 10% FBS, 2 mM L-glutamine (Corning), 100 IU Penicillin, 100 mg/mL Streptomycin (Corning), 25 mM HEPES (Corning), 1 mM Sodium Pyruvate (Corning), 100 mM MEM Nonessential Amino Acids (Corning) and 55 mM 2-Mercaptoethanol (GIBCO). DCs were cultured with 6 μg/mL CpG-A (ODN 2216), 5 μg/mL Imiquimod (Invivogen), or 6 μg/mL CpG-B (ODN1826; Invivogen) for 16-18 hours. Supernatants were frozen and used to detect cytokines by ELISA.
For detection of intracellular IL-12p40, mouse DCs were enriched using CD11c Microbeads (Miltenyi Biotec). CD11c+ cells were cultured at 37°C in 5 mL polypropylene tubes at a concentration of 1×106 cells in 500 μL complete R10 media with 6 μg/mL CpG-A (ODN 2216) for 4 hours in the presence of 5 μg/mL Brefeldin A (Millipore Sigma) for the last 3.5 hours of culture. Cytokine expression was detected by intracellular staining and measured by flow cytometry.
Human:
Sorted DC subsets were cultured in 96 well U-bottom plates at 37°C at a concentration of 3,000 cells in 150 μL complete R10 media. Media was supplemented with 10 ng/mL recombinant human IL-3 (R&D Systems). DCs were cultured with 5 μg/mL CpG-A (ODN 2216) or with “LPR” cocktail consisting of 100 ng/mL LPS (Invivogen), 25 μg/mL Poly(I:C) (Invivogen), and 2.5 μg/mL R848 (Invivogen) for 24 hours. Supernatants were frozen and used to detect cytokines by ELISA; DCs were analyzed for expression of maturation markers by flow cytometry.
Cytokine Detection in Culture Supernatant
Mouse and human IFNα was detected using a VeriKine Mouse Interferon Alpha ELISA Kit and VeriKine Human IFN Alpha Multi-Subtype ELISA Kit, respectively (PBL Assay Science). Mouse IL-12p70 was detected using a Quantikine ELISA kit (R&D Systems). Human IL-12p70 was detected using the CBA Enhanced Sensitivity Flex Set (BD Biosciences).
Mixed Leukocyte Reaction
Mouse:
Spleens from BALB/c animals were disrupted mechanically. CD4+ T cells were obtained by negative selection using a cocktail of Abs containing the following culture supernatants (produced in house): B220 (RA3-6B2), F4/80 (HB-198), MHC-II (TIB 120), NK1.1 (HB-191) and CD8 (2.43). After supernatant incubations, negative selection was performed using anti-rat Dynabeads (Thermo Fisher Scientific). Enriched CD4+ T cells were stained with CellTrace Violet (CTV, Thermo Fisher Scientific) for 10 minutes at 37°C. FACS-purified DC subsets were co-cultured with BALB/c T cells in a 1:5 ratio for 4 days. As a control for homeostatic proliferation, T cells were cultured alone without DCs. Results were expressed as frequency of CTVlow CD4+ T cells.
Human:
PBMCs stained with CFSE (Millipore Sigma) at 37°C in a water bath for 10 minutes before T cell purification using the Pan T Cell Isolation Kit (Miltenyi Biotec) according to manufacturer’s instructions. FACS-purified DC subsets were co-cultured with allogeneic T cells in a 1:20 ratio for 6 days. As a control for homeostatic proliferation, T cells were cultured alone without DCs. Results are expressed as frequency of CFSElow CD4+ T cells.
Quantitative PCR
RNA from sorted DCs was extracted immediately after sorting using the Nucleospin RNA XS kit (Takara Bio) according to manufacturer’s instructions. Total RNA from each DC subset was reverse transcribed using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories). cDNA was amplified in a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories) with the iTaq Universal SYBR® Green Supermix (Bio-Rad Laboratories). Primers are listed in Table S2. Expression was calculated following a ΔΔCq method relative to RPL13A/Rpl13a (human/mouse) and cDC2s and shown as 2-ΔΔCq.
PCR Assay for Genomic IgH D-J Rearrangement
IgH D-J rearrangement PCR assay was adapted from Schlissel et al. (1991) using the primers listed in Table S2. Genomic DNA was isolated with the Nucleospin Tissue XS kit (Takara Bio). To equalize the amount of DNA in PCR reactions, samples were diluted based on relative quantification determined by qPCR using primers against mouse Actin DNA. PCR products were run on a 2% agarose gel and imaged with a ChemiDoc MP (Bio-Rad).
Influenza Infection
B6 mice were anesthetized and inoculated intranasally with 1000 PFU of PR8 influenza virus. For T cell analysis, leukocytes were enriched using a Percoll (GE Healthcare) gradient (45% / 67.5%). Cell suspensions from mediastinal LNs were obtained by digestion with 0.13 U/mL Liberase TL and 50 μg/mL DNase I for 30 minutes at 37°C. 10 mM EDTA was added for the last 5 minutes of culture. CLEC4CDTR mice were treated with 1 μg of diphtheria toxin (DT, Millipore Sigma) i.v. one day before infection. Depletion was maintained by inoculating 500 ng DT i.p. every other day.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments, including statistical tests used and value of n, can be found in figure legends. All statistical tests were run with GraphPad Prism 6. Significance is depicted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All graphs show mean ± SD. gMFI indicates geometric mean fluorescence intensity.
CyTOF Data Analysis
Files in FCS format were normalized with Cytofkit. For both mouse and human, live, single cells were gated using FlowJo. For analysis, mouse splenocytes were gated as CD3− CD19− CD335− Ly6G−. Human PBMC and splenocytes were gated as CD3− CD19− CD335− CD66b− CD14− CD16− HLA-DR+. Events of interest were imported into CYT and transformed using hyperbolic arcsin (asinh x/5). tSNE plots were calculated using all of the markers except lineage markers. Principal Component Analysis (PCA) and CyTOF heatmaps were performed and visualized in R with the ggplot2, prcomp, rgl, and viridis packages. UMAP visualization was performed in Python following the guide found at https://umap-learn.readthedocs.io/en/latest//.
Comparison of Mouse and Human DC Subsets
See workflow in Figure S4A. Human data: Log transformed and filtered TPM counts of scRNA-seq data were downloaded from the Broad Single Cell Portal study “Atlas of human blood dendritic cells and monocytes” (https://singlecell.broadinstitute.org/single_cell/study/SCP43/atlas-of-human-blood-dendritic-cells-and-monocytes#study-download). Significantly differentially expressed genes between each pair of subsets were determined with the limma package in R. The cut-off for significant genes was log2FC > 1 (FC > 2) and adjusted p value < 0.05. For PCA analysis, 30 cells of each subset were randomly selected, and the expression Z-scores were calculated. Mouse data: Raw RNA-seq counts were downloaded from NCBI GEO (GSE76132). Significantly differentially expressed genes between each pair of subsets were determined with the DESeq2 package in R. The cut-off for significant genes was log2FC > 1 (FC > 2) and adjusted p value < 0.05. For PCA analysis, normalized counts were downloaded from NCBI GEO. Two samples of each subset were randomly selected, and expression z-scores were calculated.
DATA AND CODE AVAILABILITY
This study did not generate unique datasets or code. Links to the publicly available transcriptome data analyzed in this study are provided in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Antibodies used for CyTOF are summarized in Table S1. | ||
| Anti-human BDCA1/CD1c APC/Cy7 (clone L161) | Biolegend | Cat# 331520; RRID:AB_10644008 |
| Anti-human BDCA2/CD303 FITC (clone 201A) | Biolegend | Cat# 354208; RRID:AB_2561364 |
| Anti-human BDCA2/CD303 APC (clone 201A) | Biolegend | Cat# 354206; RRID:AB_11150412 |
| Anti-human BDCA3/CD141 PECy7 (clone M80) | Biolegend | Cat# 344110; RRID:AB_2561623 |
| Anti-human BDCA4/CD304 BV510 (clone 12C2) | Biolegend | Cat# 354515; RRID:AB_2563074 |
| Anti-human BDCA4/ CD304 APC (clone 12C2) | Biolegend | Cat# 354506; RRID:AB_11219600 |
| Anti-human CD11c PECy7 (clone Bu15) | Biolegend | Cat# 337216; RRID:AB_2129790 |
| Anti-human CD11c Alexa Fluor 700 (clone Bu15) | Biolegend | Cat# 337220; RRID:AB_2561503 |
| Anti-human CD123 BUV395 (clone 7G3) | BD Biosciences | Cat# 564195; RRID:AB_2714171. |
| Anti-human CD14 APC (clone M5E2) | Biolegend | Cat# 982506; RRID:AB_2650643 |
| Anti-human CD14 BV785 (clone M5E2) | Biolegend | Cat# 301840; RRID:AB_2563425 |
| Anti-human CD14 BV510 (clone M5E2) | Biolegend | Cat# 301842; RRID:AB_2561946 |
| Anti-human CD16 BV650 (clone 3G8) | Biolegend | Cat# 302042; RRID:AB_2563801 |
| Anti-human CD19 PerCP/Cy5.5 (clone HIB19) | Biolegend | Cat# 302230; RRID:AB_2073119 |
| Anti-human CD19 Pacific Blue (clone HIB19) | Biolegend | Cat# 302224; RRID:AB_493653 |
| Anti-human CD20 PerCP/Cy5.5 (clone 2H7) | Biolegend | Cat# 302325; RRID:AB_893285 |
| Anti-human CD20 Pacific Blue (clone 2H7) | Biolegend | Cat# 302320; RRID:AB_493651 |
| Anti-human CD2 APC/Cy7 (clone RPA-2.10) | Biolegend | Cat# 300220; RRID:AB_2571989 |
| Anti-human CD3 PerCP/Cy5.5 (clone UCHT1) | Biolegend | Cat# 300430; RRID:AB_893299 |
| Anti-human CD3 Pacific Blue (clone UCHT1) | Biolegend | Cat# 300431; RRID:AB_1595437 |
| Anti-human CD335 PerCP/Cy5.5 (clone 9E2) | Biolegend | Cat# 331920; RRID:AB_2561665 |
| Anti-human CD335 Pacific Blue (clone 9E2) | Biolegend | Cat# 331912; RRID:AB_2149280 |
| Anti-human CD4 BV785 (clone OKT4) | Biolegend | Cat# 317442; RRID:AB_2563242 |
| Anti-human CD45 BV785 (clone HI30) | Biolegend | Cat# 304048; RRID:AB_2563129 |
| Anti-human CD5 BV737 (clone UCHT2) | Biolegend | Cat#564451; RRID:AB_2714177 |
| Anti-human CD66b PerCP/Cy5.5 (clone G10F5) | Biolegend | Cat# 305108; RRID:AB_2077855 |
| Anti-human CD66b Pacific Blue (clone G10F5) | Biolegend | Cat# 305112; RRID:AB_2563294 |
| Anti-human CD8a APC/Cy7 (clone RPA-T8) | Biolegend | Cat# 301016; RRID:AB_314134 |
| Anti-human CLEC9A/DNGR1 APC (clone 8F9) | Biolegend | Cat# 353806; RRID:AB_2565519 |
| Anti-human HLADR BV605 (clone L243) | Biolegend | Cat# 307640; RRID:AB_2561913 |
| Anti-human Axl AlexaFluor488 (clone 108724) | R&D Systems | Cat# FAB154G; RRID:AB_2714170 |
| Anti-human CD80 BV421 (clone 2D10) | Biolegend | Cat# 305222; RRID:AB_2564407 |
| Anti-human CD81 PerCP/Cy5.5 (clone 5A6) | Biolegend | Cat# 349508; RRID:AB_2564019 |
| Anti-human CD86 BUV737 (clone FUN-1) | BD Biosciences | Cat# 612784; RRID:AB_2814790 |
| Anti-human SIGLEC1 PE (clone 7-239) | Biolegend | Cat# 346004; RRID:AB_2189029 |
| Anti-human/mouse TCF4/E2-2 purified (clone NCI-R159-6) | Abcam | Cat# ab217668; RRID:AB_2714172 |
| Anti-human/mouse IRF8 APC (clone V3GYWCH) | ThermoFisher Scientific | Cat# 17-9852-82; RRID: 17-9852-82 |
| Anti-human/mouse IRF4 PE Antibody (clone IRF4.E4) | Biolegend | Cat# 646404; RRID:AB_2563005 |
| Anti-mouse Axl PECy7 (clone MAX8LDS) | Invitrogen | Cat #: 25-1084-80; RRID: AB_2734851 |
| Anti-mouse Axl PE (clone 175128) | R&D Systems | Cat #: FAB8541P; RRID: AB_2814643 |
| Anti-mouse B220 BV650 (clone RA3-6B2) | Biolegend | Cat #: 103241; RRID: AB_11204069 |
| Anti-mouse Ccr2 PE (clone SA203G11) | Biolegend | Cat #: 150609; RRID: AB_2616981 |
| Anti-mouse Ccr9 PECy7 (clone CW-1.2) | Biolegend | Cat #: 128711; RRID: AB_10901176 |
| Anti-mouse CD103 PerCP/Cy5.5 (clone M290) | BD Biosciences | Cat #: 563637; RRID: AB_2738337 |
| Anti-mouse CD115 APC (clone AFS98) | ThermoFisher Scientific | Cat #: 17-1152-80; RRID: AB_ 1210790 |
| Anti-mouse CD11b BV785 (clone M1/70) | Biolegend | Cat #: 101243; RRID: AB_2561373 |
| Anti-mouse CD11b BV650 (clone M1/70) | Biolegend | Cat #: 101259; RRID: AB_ 2566568 |
| Anti-mouse CD11c PECy7 (clone N418) | ThermoFisher Scientific | Cat #: 25-0114-81; RRID: AB_ 469589 |
| Anti-mouse CD11c APC-Alexa780 (clone N418) | ThermoFisher Scientific | Cat #: 47-0114-80; RRID: AB_1548663 |
| Anti-mouse CD135 Biotin (clone A2F10) | Biolegend | Cat #: 135307; RRID: AB_1953266 |
| Anti-mouse CD172a APC-Fire750 (clone P84) | Biolegend | Cat #: 144029; RRID: AB_2721316 |
| Anti-mouse CD19 FITC (clone 6D5) | Biolegend | Cat #: 115505; RRID: AB_313640 |
| Anti-mouse CD19 APC-Alexa700 (clone 1D3) | BD Biosciences | Cat #: 557958; RRID: AB_396958 |
| Anti-mouse CD19 eFluor450 (clone 1D3) | ThermoFisher Scientific | Cat #: 48-0193-82; RRID: AB_2734905 |
| Anti-mouse CD2 PE (clone RM2-5) | Biolegend | Cat #: 100107; RRID: AB_2073691 |
| Anti-mouse CD20 FITC (clone SA275A11) | Biolegend | Cat #: 150407; RRID: AB_2566776 |
| Anti-mouse CD24 APC-eFlour780 (clone M1/69) | ThermoFisher Scientific | Cat #: 47-0242-82; RRID: AB_10853172 |
| Anti-mouse CD25 PerCP/Cy5.5 (clone PC61.5) | ThermoFisher Scientific | Cat #: 45-0251-82; RRID: AB_914324 |
| Anti-mouse CD3 APC-Alexa700 (clone 17A2) | Biolegend | Cat #: 100215; RRID: AB_493696 |
| Anti-mouse CD3 eFluor 450 (clone 17A2) | ThermoFisher Scientific | Cat #: 48-0032-82; RRID: AB_1272193 |
| Anti-mouse CD4 APC-Alexa780 (clone RM4-5) | ThermoFisher Scientific | Cat #: 47-0042-82; RRID: AB_1272183 |
| Anti-mouse CD4 BV785 (clone RM4-5) | Biolegend | Cat #: 100552; RRID: AB_2563053 |
| Anti-mouse CD45 BV510 (clone 30-F11) | Biolegend | Cat #: 103138; RRID: AB_2563061 |
| Anti-mouse CD45 BV785 (clone 30-F11) | Biolegend | Cat #: 103149; RRID: AB_2564590 |
| Anti-mouse CD45RA PerCP/Cy5.5 (clone 14.8) | BD Biosciences | Cat #: 564359; RRID: AB_2738767 |
| Anti-mouse CD5 APC (clone 53-7.3) | BD Biosciences | Cat #: 561895; RRID: AB_10895562 |
| Anti-mouse CD69 PE (clone H1.2F3) | Biolegend | Cat #: 104507; RRID: AB_313110 |
| Anti-mouse CD8 BV510 (clone 53-6.7) | Biolegend | Cat #: 100752; RRID: AB_2563057 |
| Anti-mouse CD8 APC-A780 (clone 53-6.7) | ThermoFisher Scientific | Cat #: 47-0081-82; RRID: AB_1272185 |
| Anti-mouse CD8 BUV737 (clone 53-6.7) | BD Biosciences | Cat #: 564297; RRID: AB_2722580 |
| Anti-mouse CD81 PerCP-Cy5.5 (clone Eat-2) | Biolegend | Cat #: 104911; RRID: AB_2562994 |
| Anti-mouse CD80 PE (clone 16-10A1) | BD Biosciences | Cat #: 553769; RRID: AB_395039 |
| Anti-mouse CD86 APC-Alexa700 (clone GL1) | Biolegend | Cat #: 105024; RRID: AB_493721 |
| Anti-mouse CD86 PE (clone GL1) | BD Biosciences | Cat #: 561963; RRID: AB_10896971 |
| Anti-mouse Clec9a Alexa647 (clone 10B4) | Idoyaga et al., 2011 | N/A |
| Anti-mouse Cx3cr1 FITC (clone SA011F11) | Biolegend | Cat #: 149019; RRID: AB_2565702 |
| Anti-mouse Cx3cr1 PE (clone SA011F11) | Biolegend | Cat #: 149005; RRID: AB_2564314 |
| Anti-mouse Cx3cr1 APC (clone SA011F11) | Biolegend | Cat #: 149007; RRID: AB_2564491 |
| Anti-mouse Cx3cr1 Biotin (clone SA011F11) | Biolegend | Cat #: 149018; RRID: AB_2565701 |
| Anti-mouse DEC205 (CD205) PECy7 (clone Yekta) | Invitrogen | Cat #: 25-2051-42; RRID: AB_1834473 |
| Anti-mouse ESAM PE (clone1G8) | ThermoFisher Scientific | Cat #: 12-5852-81; RRID: AB_891539 |
| Anti-mouse F4/80 PerCP/Cy5.5 (clone BM8) | ThermoFisher Scientific | Cat #: 45-4801-82; RRID: AB_914345 |
| Anti-mouse F4/80 eFluor450 (clone BM8) | ThermoFisher Scientific | Cat #: 48-4801-42; RRID: AB_1548747 |
| Anti-mouse FoxP3 APC (clone FJK-16 s) | ThermoFisher Scientific | Cat #: 17-5773-80; RRID: AB_469456 |
| Anti-mouse IL-12p40 PE (clone C15.6) | BD Biosciences | Cat #: 562038; RRID: AB_10895571 |
| Anti-mouse Ly6C BV510 (clone HK1.4) | Biolegend | Cat #: 128033; RRID: AB_2562351 |
| Anti-mouse Ly6C APC-Alexa700 (clone HK1.4) | Biolegend | Cat #: 128023; RRID: AB_10640119 |
| Anti-mouse Ly6C eFluor450 (clone HK1.4) | ThermoFisher Scientific | Cat #: 48-5932-82; RRID: AB_10805519 |
| Anti-mouse Ly6D PE (clone 49-H4) | Biolegend | Cat #: 138603; RRID: AB_2281362 |
| Anti-mouse Ly6G BUV395 (clone 1A8) | BD Biosciences | Cat #: 563978; RRID: AB_2716852 |
| Anti-mouse MHC-II PE (clone AF6-120.1) | BD Biosciences | Cat #: 553552; RRID: AB_394919 |
| Anti-mouse MHC-II BV510 (clone M5/114.15.2) | Biolegend | Cat #: 107635; RRID: AB_2561397 |
| Anti-mouse MHC-II APC-Alexa700 (clone M5/114.15.2) | Biolegend | Cat #: 107622; RRID: AB_493727 |
| Anti-mouse NK1.1 APC-Alexa700 (clone PK136) | Biolegend | Cat #: 108729; RRID: AB_2074426 |
| Anti-mouse PD-L1 SuperBright780 (clone MIH5) | ThermoFisher Scientific | Cat #: 78-5982-82; RRID: AB_2724081 |
| Anti-mouse Pdca-1 FITC (clone 927) | ThermoFisher Scientific | Cat #: 11-3172-82; RRID: AB_763416 |
| Anti-mouse Siglec1 (CD169) PE (clone 3D6.112) | Biolegend | Cat #: 142403; RRID: AB_10915470 |
| Anti-mouse SiglecF PE (clone E50-2440) | BD Biosciences | Cat #: 552126; RRID: AB_394341 |
| Anti-mouse SiglecF BV421 (clone E50-2440) | BD Biosciences | Cat #: 565934; RRID: AB_2739398 |
| Anti-mouse SiglecH PerCP/Cy5.5 (clone 551) | Biolegend | Cat #: 129613; RRID: AB_10639936 |
| Anti-mouse SiglecH FITC (clone 551) | Biolegend | Cat #: 129604; RRID: AB_1227761 |
| Anti-mouse SiglecH PE (clone 551) | Biolegend | Cat #: 129605; RRID: AB_1227763 |
| Anti-mouse SiglecH APC (clone 551) | Biolegend | Cat #: 129612; RRID: AB_10641134 |
| Anti-mouse Xcr1 FITC (clone ZET) | Biolegend | Cat #: 148210; RRID: AB_2564366 |
| Anti-mouse Xcr1 PE(clone ZET) | Biolegend | Cat #: 148204; RRID: AB_2563843 |
| Anti-mouse Xcr1 PerCP/Cy5.5 (clone ZET) | Biolegend | Cat #: 148208; RRID: AB_2564364 |
| Anti-mouse Xcr1 APC (clone ZET) | Biolegend | Cat #: 148206; RRID: AB_2563932 |
| Anti-mouse Zbtb46 PE (clone U4-1374) | BD Bioscience | Cat #: 565832; RRID: AB_2739372 |
| Bacterial and Virus Strains | ||
| NR-29029 Influenza A/Puerto Rico/8/1934-WG (H1N1) | BEI resources | https://www.beiresources.org/Catalog/animalViruses/NR-29029.aspx |
| Biological Samples | ||
| Whole Blood from healthy donors | Obtained from donors with informed consent. IRB approved by Stanford University Research Compliance Office. | N/A |
| Human Spleen | Normal spleen from distal pancreatectomy from patients not subjected to any chemotherapy. | Stanford Tissue Bank http://med.stanford.edu/cancer/research/shared-resources/tissue-procurement.html |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ficoll-Paque PLUS | GE Healthcare | Cat # 300-25 |
| Percoll | GE Healthcare Life Sciences | Cat# 17089101 |
| Fetal Bovine Serum, qualified, US origin | GIBCO | Cat # 26140079 |
| RPMI 1640 with L- Glutamine | Corning | Cat# 10040CV |
| L-glutamine Solution | Corning | Cat # 25005CI |
| Sodium Pyruvate Solution | Corning | Cat # 25000CI |
| Penicillin-Streptomycin | Corning | Cat# 30002CI |
| HEPES solution | Corning | Cat # 25060CI |
| MEM Nonessential Amino Acid Solution | Corning | Cat # 25025CI |
| Recombinant Human IL-3 | R&D Systems | Cat # 203IL010CF |
| EDTA 0.5M pH 8.0 | Corning | Cat # 46034CI |
| ACK Lysis Buffer | Lonza | Cat# 10-548E |
| Benzonase’ Nuclease | Millipore-Sigma | Cat # E1014-25KU |
| Cell-ID Intercalator-Ir | Fluidigm | Cat#: 201192A |
| Cell-ID Cisplatin | Fluidigm | Cat # 201064 |
| 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester | Millipore-Sigma | Cat # 21888-25MG-F |
| CellTrace Violet Cell Proliferation Kit | ThermoFisher Scientific | Cat# C34557 |
| Bovine Serum Albumin solution 30% ± 2% in 0.85% sodium chloride, aseptically filled | Millipore-Sigma | Cat # A7284-50ML |
| Dimethyl sulfoxide > 95% | Millipore-Sigma | Cat# D4540 |
| Collagenase D from Clostridium histolyticum | Millipore-Sigma | Cat #11088858001 |
| DNase I from bovine pancreas | Millipore-Sigma | Cat# 10104159001 |
| Collagenase IV | Worthington Biochemical Corporation | Cat# LS004189 |
| Liberase TL | Millipore-Sigma | Cat# 5401020001 |
| Liberase TM | Millipore-Sigma | Cat# 5401119001 |
| Paraformaldehyde 16% aqueous solution | Electron Microscopy Sciences | Cat# 15710 |
| Indium 113 metal chloride | Trace Sciences International | Cat# In-113 |
| Indium 115 metal chloride | Trace Sciences International | Cat# In-115 |
| LPS | Invivogen | Cat# tlrl-eblps |
| Poly(I:C) | Invivogen | Cat# tlrl-pic |
| R848 | Invivogen | Cat# tlrl-r848 |
| CpGA 2216 | Invivogen | Cat# tlrl-2216-1 |
| CpGB 1826 | Invivogen | Cat# tlrl-1826-1 |
| Imiquimod | Invivogen | Cat# tlrl-imqs |
| DOTAP transfection reagent | Santa Cruz Biotechnologies | Cat# sc362166 |
| Critical Commercial Assays | ||
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | ThermoFisher Scientific | Cat#: 00-5521-00 |
| Permeabilization Buffer (10X) | ThermoFisher Scientific | Cat#: 00-8333-56 |
| Maxpar X8 Multimetal Labeling Kit Fluidigm | Fluidigm | Cat#: 201300 |
| LIVE/DEAD Fixable Dead Cell Stain Sampler Kit | ThermoFisher Scientific | Cat# L34960 |
| Dynabeads Sheep Anti-Rat IgG | ThermoFisher Scientific | Cat# 11035 |
| Dynabeads Pan Mouse IgG | ThermoFisher Scientific | Cat# 11042 |
| Pan T cell Isolation Kit, human | MACS, Miltenyi Biotec | Cat# 130-096-535 |
| Mouse IFN Alpha All Subtype ELISA Kit, High Sensitivity | PBL Assay Science | Cat# 42115-1 |
| Human IFN Alpha Multi-Subtype ELISA Kit (TCM) | PBL Assay Science | Cat# 41105-1 |
| Mouse IL-12 p70 Quantikine ELISA Kit | R&D Systems | Cat# M1270 |
| Cytometric Bead Array Human Enhanced Sensitivity Master Buffer Kit | BD Biosciences | Cat# 561521 |
| Cytometric Bead Array Human IL-12p70 Enhanced Sensitivity Flex Set | BD Biosciences | Cat# 561518 |
| BD CompBead Anti-Mouse Ig, κ/Negative Control Compensation Particles Set | BD Biosciences | Cat# 552843 |
| Alexa Fluor 647 Antibody Labeling Kit | ThermoFisher Scientific | Cat# A20186 |
| NucleoSpin RNA XS | Takara Bio | Cat# 740902.10 |
| NucleoSpin Tissue XS | Takara Bio | Cat# 740901.10 |
| Phusion Hot Start II Master Mix | ThermoFisher Scientific | Cat# F565S |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat# 1708840 |
| iTaq Universal SYBRGreen Supermix | Bio-Rad Laboratories | Cat# 1725120 |
| Deposited Data | ||
| Human data single cell RNaseq | Broad Single Cell Portal study “Atlas of human blood dendritic cells and monocytes” | https://singlecell.broadinstitute.org/single_cell/study/SCP43/atlas-of-human-blood-dendritic-cells-and-monocytes#study-download |
| Mouse Raw RNA-Seq | NCBI GEO | GSE76132 |
| Software and Algorithms | ||
| GraphPad Prism 6 | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| MATLAB | N/A | https://www.mathworks.com/products/matlab.html |
| Cytofkit | N/A | https://bioconductor.riken.jp/packages/3.7/bioc/html/cytofkit.html |
| ViSNE (CYT) | Amir et al., 2013. | https://dpeerlab.github.io/dpeerlab-website/cyt-download.html |
| FlowJo Software v10.0.8 | TreeStar, Inc. | https://www.flowjo.com/solutions/flowjo |
| UMAP | Becht et al., 2018 | https://github.com/lmcinnes/umap |
| R | N/A | https://www.R-project.org/ |
| rgl v0.98.1 | N/A | https://r-forge.r-project.org/projects/rgl/ |
| ggplot2 v2.2.1 | N/A | https://github.com/tidyverse/ggplot2 |
| viridis | N/A | https://github.com/sjmgarnier/viridis |
| limma | N/A | https://bioconductor.org/packages/release/bioc/html/limma.html |
| DESeq2 | N/A | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Supplementary Material
Highlights.
We identify the murine homolog of human AXL+ DCs, now called transitional DCs (tDCs)
tDC development depends on Tcf4, similar to pDCs
tDCs are inefficient at IFN-I production but can efficiently activate T cells
During influenza infection, tDCs and pDCs accumulate in the lung
ACKNOWLEDGMENTS
R.L. is the recipient of an NSF GRFP fellowship (DGE-1147470). M.A.-H. is the recipient of Stanford Medicine Dean’s Office and Katharine McCormick fellowships. This work was supported by NIH grant DP2AR069953 and Baxter and Freidenrich Foundation awards to J.I., and NIH grants AI072571 and AI128949 to B.R. and AI100853 and CA232666 to O.A.P. Cell sorting was performed on instruments in the Stanford Shared FACS Facility funded through S10RR025518 and S10OD016318 and the Parker Institute for Cancer Immunotherapy. We thank the blood donors for their participation; Idoyaga lab members for technical support and discussions; Drs. Moore, Lauvau, Zengel, Blish, and Kronstad for providing reagents; Dr. Carette for fruitful discussion; and Dr. Gottfried for critical reading of the manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.042.
REFERENCES
- Alcántara-Hernández M, Leylek R, Wagar LE, Engleman EG, Keler T, Marinkovich MP, Davis MM, Nolan GP, and Idoyaga J (2017). High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 47, 1037–1050.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alculumbre SG, Saint-André V, Di Domizio J, Vargas P, Sirven P, Bost P, Maurin M, Maiuri P, Wery M, Roman MS, et al. (2018). Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol 19, 63–75. [DOI] [PubMed] [Google Scholar]
- Alculumbre S, Raieli S, Hoffmann C, Chelbi R, Danlos F-X, and Soumelis V (2019). Plasmacytoid pre-dendritic cells (pDC): from molecular pathways to function and disease association. Semin. Cell Dev. Biol 86, 24–35. [DOI] [PubMed] [Google Scholar]
- Amir AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, and Pe’er D (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Paturel C, Boonstra A, Dalod M, Durand I, Yessaad N, Dezutter-Dambuyant C, Vicari A, O’Garra A, Biron C, Brière F, and Trinchieri G (2001). Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol 2, 1144–1150. [DOI] [PubMed] [Google Scholar]
- Banchereau J, and Steinman RM (1998). Dendritic cells and the control of immunity. Nature 392, 245–252. [DOI] [PubMed] [Google Scholar]
- Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, Buer J, Murphy KM, Reizis B, and Jung S (2010). CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA 107, 14745–14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. [DOI] [PubMed] [Google Scholar]
- Björck P, Leong HX, and Engleman EG (2011). Plasmacytoid dendritic cell dichotomy: identification of IFN-α producing cells as a phenotypically and functionally distinct subset. J. Immunol 186, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Giurisato E, Cella M, Schreiber RD, Shaw AS, and Colonna M (2006). Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol 177, 3260–3265. [DOI] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, and Reizis B (2007). Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med 204, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceribelli M, Hou ZE, Kelly PN, Huang DW, Wright G, Ganapathi K, Evbuomwan MO, Pittaluga S, Shaffer AL, Marcucci G, et al. (2016).A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell 30, 764–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, and Ludewig B (2007). Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109, 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Lewis KL, Firner S, Thiel V, Hugues S, Reith W, Ludewig B, and Reizis B (2012). Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci. USA 109, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M, Preston SP, Lun ATL, Tellier J, Smyth GK, Pellegrini M, Belz GT, Corcoran LM, Visvader JE, Wu L, and Nutt SL (2016). RUNX2 Mediates Plasmacytoid Dendritic Cell Egress from the Bone Marrow and Controls Viral Immunity. Cell Rep. 15, 866–878. [DOI] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, et al. (2008). Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L, Ferrero I, Vremec D, Lucas K, Waithman J, O’Keeffe M, Wu L, Wilson A, and Shortman K (2003). The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J. Immunol 170, 4926–4932. [DOI] [PubMed] [Google Scholar]
- Dress RJ, Dutertre C-A, Giladi A, Schlitzer A, Low I, Shadan NB, Tay A, Lum J, Kairi MFBM, Hwang YY, et al. (2019). Plasmacytoid dendritic cells develop from Ly6D+ lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol 20, 852–864. [DOI] [PubMed] [Google Scholar]
- Feng J, Wang H, Shin D-M, Masiuk M, Qi C-F, and Morse HC 3rd. (2011). IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol 186, 1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Cisse B, Bunin A, Lewis KL, and Reizis B (2010). Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 33, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajkowska LT, Ceribelli M, Lau CM, Warren ME, Tiniakou I, Nakandakari Higa S, Bunin A, Haecker H, Mirny LA, Staudt LM, and Reizis B (2017). Isoform-Specific Expression and Feedback Regulation of E Protein TCF4 Control Dendritic Cell Lineage Specification. Immunity 46, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson MH, Ramos MS, Rohlfing MM, Kitchens R, Wang HD, Gould A, Agapov E, and Holtzman MJ (2007). Controls for lung dendritic cell maturation and migration during respiratory viral infection. J. Immunol 179, 1438–1448. [DOI] [PubMed] [Google Scholar]
- Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, and Liu YJ (1997). The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med 185, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, and Yona S (2014). Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol 14, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadeiba H, Sato T, Habtezion A, Oderup C, Pan J, and Butcher EC (2008). CCR9 expression defines tolerogenic plasmacytoid dendritic cells able to suppress acute graft-versus-host disease. Nat. Immunol 9, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, et al. (2012). Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest 122, 4037–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, and Taniguchi T (2005). IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777. [DOI] [PubMed] [Google Scholar]
- Idoyaga J, Lubkin A, Fiorese C, Lahoud MH, Caminschi I, Huang Y, Rodriguez A, Clausen BE, Park CG, Trumpfheller C, and Steinman RM (2011). Comparable T helper 1 (Th1) and CD8 T-cell immunity by targeting HIV gag p24 to CD8 dendritic cells within antibodies to Langerin, DEC205, and Clec9A. Proc. Natl. Acad. Sci. USA 108, 2384–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito GC, Dekker JD, Wang Y-H, Lee B-K, Shaffer AL 3rd, Lin J, Wall JK, Lee B-S, Staudt LM, Liu Y-J, et al. (2014). Dendritic cell fate is determined by BCL11A. Proc. Natl. Acad. Sci. USA 111, E998–E1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre A, Barnes BJ, Amrute S, Yeow W-S, Megjugorac N, Dai J, Feng D, Chung E, Pitha PM, and Fitzgerald-Bocarsly P (2003). Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol 74, 1125–1138. [DOI] [PubMed] [Google Scholar]
- Kronstad LM, Seiler C, Vergara R, Holmes SP, and Blish CA (2018). Differential Induction of IFN-α and Modulation of CD112 and CD54 Expression Govern the Magnitude of NK Cell IFN-γ Response to Influenza A Viruses. J. Immunol 201, 2117–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CM, Nish SA, Yogev N, Waisman A, Reiner SL, and Reizis B (2016). Leukemia-associated activating mutation of Flt3 expands dendritic cells and alters T cell responses. J. Exp. Med 213, 415–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BS, Lee BK, Iyer VR, Sleckman BP, Shaffer AL III, Ippolito GC, Tucker HO, and Dekker JD (2017). Corrected and Republished from: BCL11A Is a Critical Component of a Transcriptional Network That Activates RAG Expression and V(D)J Recombination. Mol. Cell. Biol 38, e00362–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leylek R, and Idoyaga J (2019). The versatile plasmacytoid dendritic cell: function, heterogeneity, and plasticity. Int. Rev. Cell Mol. Biol 349, 177–211. [DOI] [PubMed] [Google Scholar]
- Liou L-Y, Blasius AL, Welch MJ, Colonna M, Oldstone MBA, and Zúñiga EI (2008). In vivo conversion of BM plasmacytoid DC into CD11b+ conventional DC during virus infection. Eur. J. Immunol 38, 3388–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu F-F, Randolph GJ, Rudensky AY, and Nussenzweig M (2009). In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manh T-PV, Alexandre Y, Baranek T, Crozat K, and Dalod M (2013). Plasmacytoid, conventional, and monocyte-derived dendritic cells undergo a profound and convergent genetic reprogramming during their maturation. Eur. J. Immunol 43, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Connolly JE, Michnevitz M, Chaussabel D, Yu C-I, Glaser C, Tindle S, Pypaert M, Freitas H, Piqueras B, et al. (2009). CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J. Immunol 182, 6815–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, and Mortha A (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol 31, 563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao K-H, Niec RE, and Nussenzweig MC (2012a). Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med 209, 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Kamphorst AO, Idoyaga J, Yamane A, Guermonprez P, Rihn S, Yao K-H, Silva IT, Oliveira TY, et al. (2012b). Zinc finger transcription factor zDC is a negative regulator required to prevent activation of classical dendritic cells in the steady state. J. Exp. Med 209, 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GRS, and Perlman H (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol 49, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M, Schmidlin H, Hazekamp MG, Schotte R, and Blom B (2008). Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix-loop-helix factor E2-2 and the Ets factor Spi-B. Eur. J. Immunol 38, 2389–2400. [DOI] [PubMed] [Google Scholar]
- Naik SH, Sathe P, Park H-Y, Metcalf D, Proietto AI, Dakic A, Carotta S, O’Keeffe M, Bahlo M, Papenfuss A, et al. (2007). Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol 8, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, and Manz MG (2007). Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol 8, 1207–1216. [DOI] [PubMed] [Google Scholar]
- Palucka AK, Blanck J-P, Bennett L, Pascual V, and Banchereau J (2005). Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc. Natl. Acad. Sci. USA 102, 3372–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttur F, Arnold-Schrauf C, Lahl K, Solmaz G, Lindenberg M, Mayer CT, Gohmert M, Swallow M, van Helt C, Schmitt H, et al. (2013). Absence of Siglec-H in MCMV infection elevates interferon alpha production but does not enhance viral clearance. PLoS Pathog. 9, e1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B (2019). Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 50, 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues PF, Alberti-Servera L, Eremin A, Grajales-Reyes GE, Ivanek R, and Tussiwand R (2018). Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol 19, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe P, Vremec D, Wu L, Corcoran L, and Shortman K (2013). Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 121, 11–19. [DOI] [PubMed] [Google Scholar]
- Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, and Murphy KM (2012). Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med 209, 1135–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai CM, Sisirak V, Ghosh HS, Hou EZ, Ceribelli M, Staudt LM, and Reizis B (2013). Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med 210, 2151–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, Morse HC 3rd, Belardelli F, and Gabriele L (2002). ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J. Exp. Med 196, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS, Corcoran LM, and Baltimore D (1991). Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med 173, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HRB, Schreuder J, Lum J, Malleret B, Zhang S, Larbi A, Zolezzi F, et al. (2015). Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol 16, 718–728. [DOI] [PubMed] [Google Scholar]
- Schotte R, Nagasawa M, Weijer K, Spits H, and Blom B (2004). The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J. Exp. Med 200, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P, Dutertre CA, Chen J, Günther P, McGovern N, Irac SE, Gunawan M, Beyer M, Händler K, Duan K, et al. (2017). Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D, Traver D, Leder P, Sakaguchi N, and Akashi K (2004). Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective oftheircellular origin. Immunity 21, 43–53. [DOI] [PubMed] [Google Scholar]
- Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M, Joeris T, De Prijck S, Vanhoutte L, Vanheerswynghels M, et al. (2016). IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity 45, 626–640. [DOI] [PubMed] [Google Scholar]
- Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, and Liu YJ (1999). The nature of the principal type 1 interferon-producing cells in human blood. Science 284, 1835–1837. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, and Kumatori A (2004). Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha-dendritic cell development. Proc. Natl. Acad. Sci. USA 101, 8981–8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Gilfillan S, Vermi W, Wang Y, and Colonna M (2010). Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Wang Y, Gilfillan S, and Colonna M (2013). Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 9, e1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Fukaya T, Eizumi K, Sato Y, Sato K, Shibazaki A, Otsuka H, Hijikata A, Watanabe T, Ohara O, et al. (2011). Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity 35, 958–971. [DOI] [PubMed] [Google Scholar]
- Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu Manh T-P, Bertho N, Hosmalin A, Schwartz-Cornil I, and Dalod M (2015). Investigating Evolutionary Conservation of Dendritic Cell Subset Identity and Functions. Front. Immunol 6, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm TR, Taddeo A, Winter O, Schulz AR, Mälzer J-N, Domingo C, Biesen R, Alexander T, Thiel A, Radbruch A, et al. (2016). Siglec-1-positive plasmacytoid dendritic cells (pDCs) in human peripheral blood: A semi-mature and myeloid-like subset imbalanced during protective and autoimmune responses. Clin. Immunol 163, 42–51. [DOI] [PubMed] [Google Scholar]
- Wolf AI, Buehler D, Hensley SE, Cavanagh LL, Wherry EJ, Kastner P, Chan S, and Weninger W (2009). Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J. Immunol 182, 871–879. [DOI] [PubMed] [Google Scholar]
- Wu X, Satpathy AT, Kc W, Liu P, Murphy TL, and Murphy KM (2013). Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS ONE 8, e64800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Briseño CG, Durai V, Albring JC, Haldar M, Bagadia P, Kim K-W, Randolph GJ, Murphy TL, and Murphy KM (2016). Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med 213, 2553–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gregorio JD, Iwahori T, Zhang X, Choi O, Tolentino LL, Prestwood T, Carmi Y, and Engleman EG (2017). A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc. Natl. Acad. Sci. USA 114, 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate unique datasets or code. Links to the publicly available transcriptome data analyzed in this study are provided in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Antibodies used for CyTOF are summarized in Table S1. | ||
| Anti-human BDCA1/CD1c APC/Cy7 (clone L161) | Biolegend | Cat# 331520; RRID:AB_10644008 |
| Anti-human BDCA2/CD303 FITC (clone 201A) | Biolegend | Cat# 354208; RRID:AB_2561364 |
| Anti-human BDCA2/CD303 APC (clone 201A) | Biolegend | Cat# 354206; RRID:AB_11150412 |
| Anti-human BDCA3/CD141 PECy7 (clone M80) | Biolegend | Cat# 344110; RRID:AB_2561623 |
| Anti-human BDCA4/CD304 BV510 (clone 12C2) | Biolegend | Cat# 354515; RRID:AB_2563074 |
| Anti-human BDCA4/ CD304 APC (clone 12C2) | Biolegend | Cat# 354506; RRID:AB_11219600 |
| Anti-human CD11c PECy7 (clone Bu15) | Biolegend | Cat# 337216; RRID:AB_2129790 |
| Anti-human CD11c Alexa Fluor 700 (clone Bu15) | Biolegend | Cat# 337220; RRID:AB_2561503 |
| Anti-human CD123 BUV395 (clone 7G3) | BD Biosciences | Cat# 564195; RRID:AB_2714171. |
| Anti-human CD14 APC (clone M5E2) | Biolegend | Cat# 982506; RRID:AB_2650643 |
| Anti-human CD14 BV785 (clone M5E2) | Biolegend | Cat# 301840; RRID:AB_2563425 |
| Anti-human CD14 BV510 (clone M5E2) | Biolegend | Cat# 301842; RRID:AB_2561946 |
| Anti-human CD16 BV650 (clone 3G8) | Biolegend | Cat# 302042; RRID:AB_2563801 |
| Anti-human CD19 PerCP/Cy5.5 (clone HIB19) | Biolegend | Cat# 302230; RRID:AB_2073119 |
| Anti-human CD19 Pacific Blue (clone HIB19) | Biolegend | Cat# 302224; RRID:AB_493653 |
| Anti-human CD20 PerCP/Cy5.5 (clone 2H7) | Biolegend | Cat# 302325; RRID:AB_893285 |
| Anti-human CD20 Pacific Blue (clone 2H7) | Biolegend | Cat# 302320; RRID:AB_493651 |
| Anti-human CD2 APC/Cy7 (clone RPA-2.10) | Biolegend | Cat# 300220; RRID:AB_2571989 |
| Anti-human CD3 PerCP/Cy5.5 (clone UCHT1) | Biolegend | Cat# 300430; RRID:AB_893299 |
| Anti-human CD3 Pacific Blue (clone UCHT1) | Biolegend | Cat# 300431; RRID:AB_1595437 |
| Anti-human CD335 PerCP/Cy5.5 (clone 9E2) | Biolegend | Cat# 331920; RRID:AB_2561665 |
| Anti-human CD335 Pacific Blue (clone 9E2) | Biolegend | Cat# 331912; RRID:AB_2149280 |
| Anti-human CD4 BV785 (clone OKT4) | Biolegend | Cat# 317442; RRID:AB_2563242 |
| Anti-human CD45 BV785 (clone HI30) | Biolegend | Cat# 304048; RRID:AB_2563129 |
| Anti-human CD5 BV737 (clone UCHT2) | Biolegend | Cat#564451; RRID:AB_2714177 |
| Anti-human CD66b PerCP/Cy5.5 (clone G10F5) | Biolegend | Cat# 305108; RRID:AB_2077855 |
| Anti-human CD66b Pacific Blue (clone G10F5) | Biolegend | Cat# 305112; RRID:AB_2563294 |
| Anti-human CD8a APC/Cy7 (clone RPA-T8) | Biolegend | Cat# 301016; RRID:AB_314134 |
| Anti-human CLEC9A/DNGR1 APC (clone 8F9) | Biolegend | Cat# 353806; RRID:AB_2565519 |
| Anti-human HLADR BV605 (clone L243) | Biolegend | Cat# 307640; RRID:AB_2561913 |
| Anti-human Axl AlexaFluor488 (clone 108724) | R&D Systems | Cat# FAB154G; RRID:AB_2714170 |
| Anti-human CD80 BV421 (clone 2D10) | Biolegend | Cat# 305222; RRID:AB_2564407 |
| Anti-human CD81 PerCP/Cy5.5 (clone 5A6) | Biolegend | Cat# 349508; RRID:AB_2564019 |
| Anti-human CD86 BUV737 (clone FUN-1) | BD Biosciences | Cat# 612784; RRID:AB_2814790 |
| Anti-human SIGLEC1 PE (clone 7-239) | Biolegend | Cat# 346004; RRID:AB_2189029 |
| Anti-human/mouse TCF4/E2-2 purified (clone NCI-R159-6) | Abcam | Cat# ab217668; RRID:AB_2714172 |
| Anti-human/mouse IRF8 APC (clone V3GYWCH) | ThermoFisher Scientific | Cat# 17-9852-82; RRID: 17-9852-82 |
| Anti-human/mouse IRF4 PE Antibody (clone IRF4.E4) | Biolegend | Cat# 646404; RRID:AB_2563005 |
| Anti-mouse Axl PECy7 (clone MAX8LDS) | Invitrogen | Cat #: 25-1084-80; RRID: AB_2734851 |
| Anti-mouse Axl PE (clone 175128) | R&D Systems | Cat #: FAB8541P; RRID: AB_2814643 |
| Anti-mouse B220 BV650 (clone RA3-6B2) | Biolegend | Cat #: 103241; RRID: AB_11204069 |
| Anti-mouse Ccr2 PE (clone SA203G11) | Biolegend | Cat #: 150609; RRID: AB_2616981 |
| Anti-mouse Ccr9 PECy7 (clone CW-1.2) | Biolegend | Cat #: 128711; RRID: AB_10901176 |
| Anti-mouse CD103 PerCP/Cy5.5 (clone M290) | BD Biosciences | Cat #: 563637; RRID: AB_2738337 |
| Anti-mouse CD115 APC (clone AFS98) | ThermoFisher Scientific | Cat #: 17-1152-80; RRID: AB_ 1210790 |
| Anti-mouse CD11b BV785 (clone M1/70) | Biolegend | Cat #: 101243; RRID: AB_2561373 |
| Anti-mouse CD11b BV650 (clone M1/70) | Biolegend | Cat #: 101259; RRID: AB_ 2566568 |
| Anti-mouse CD11c PECy7 (clone N418) | ThermoFisher Scientific | Cat #: 25-0114-81; RRID: AB_ 469589 |
| Anti-mouse CD11c APC-Alexa780 (clone N418) | ThermoFisher Scientific | Cat #: 47-0114-80; RRID: AB_1548663 |
| Anti-mouse CD135 Biotin (clone A2F10) | Biolegend | Cat #: 135307; RRID: AB_1953266 |
| Anti-mouse CD172a APC-Fire750 (clone P84) | Biolegend | Cat #: 144029; RRID: AB_2721316 |
| Anti-mouse CD19 FITC (clone 6D5) | Biolegend | Cat #: 115505; RRID: AB_313640 |
| Anti-mouse CD19 APC-Alexa700 (clone 1D3) | BD Biosciences | Cat #: 557958; RRID: AB_396958 |
| Anti-mouse CD19 eFluor450 (clone 1D3) | ThermoFisher Scientific | Cat #: 48-0193-82; RRID: AB_2734905 |
| Anti-mouse CD2 PE (clone RM2-5) | Biolegend | Cat #: 100107; RRID: AB_2073691 |
| Anti-mouse CD20 FITC (clone SA275A11) | Biolegend | Cat #: 150407; RRID: AB_2566776 |
| Anti-mouse CD24 APC-eFlour780 (clone M1/69) | ThermoFisher Scientific | Cat #: 47-0242-82; RRID: AB_10853172 |
| Anti-mouse CD25 PerCP/Cy5.5 (clone PC61.5) | ThermoFisher Scientific | Cat #: 45-0251-82; RRID: AB_914324 |
| Anti-mouse CD3 APC-Alexa700 (clone 17A2) | Biolegend | Cat #: 100215; RRID: AB_493696 |
| Anti-mouse CD3 eFluor 450 (clone 17A2) | ThermoFisher Scientific | Cat #: 48-0032-82; RRID: AB_1272193 |
| Anti-mouse CD4 APC-Alexa780 (clone RM4-5) | ThermoFisher Scientific | Cat #: 47-0042-82; RRID: AB_1272183 |
| Anti-mouse CD4 BV785 (clone RM4-5) | Biolegend | Cat #: 100552; RRID: AB_2563053 |
| Anti-mouse CD45 BV510 (clone 30-F11) | Biolegend | Cat #: 103138; RRID: AB_2563061 |
| Anti-mouse CD45 BV785 (clone 30-F11) | Biolegend | Cat #: 103149; RRID: AB_2564590 |
| Anti-mouse CD45RA PerCP/Cy5.5 (clone 14.8) | BD Biosciences | Cat #: 564359; RRID: AB_2738767 |
| Anti-mouse CD5 APC (clone 53-7.3) | BD Biosciences | Cat #: 561895; RRID: AB_10895562 |
| Anti-mouse CD69 PE (clone H1.2F3) | Biolegend | Cat #: 104507; RRID: AB_313110 |
| Anti-mouse CD8 BV510 (clone 53-6.7) | Biolegend | Cat #: 100752; RRID: AB_2563057 |
| Anti-mouse CD8 APC-A780 (clone 53-6.7) | ThermoFisher Scientific | Cat #: 47-0081-82; RRID: AB_1272185 |
| Anti-mouse CD8 BUV737 (clone 53-6.7) | BD Biosciences | Cat #: 564297; RRID: AB_2722580 |
| Anti-mouse CD81 PerCP-Cy5.5 (clone Eat-2) | Biolegend | Cat #: 104911; RRID: AB_2562994 |
| Anti-mouse CD80 PE (clone 16-10A1) | BD Biosciences | Cat #: 553769; RRID: AB_395039 |
| Anti-mouse CD86 APC-Alexa700 (clone GL1) | Biolegend | Cat #: 105024; RRID: AB_493721 |
| Anti-mouse CD86 PE (clone GL1) | BD Biosciences | Cat #: 561963; RRID: AB_10896971 |
| Anti-mouse Clec9a Alexa647 (clone 10B4) | Idoyaga et al., 2011 | N/A |
| Anti-mouse Cx3cr1 FITC (clone SA011F11) | Biolegend | Cat #: 149019; RRID: AB_2565702 |
| Anti-mouse Cx3cr1 PE (clone SA011F11) | Biolegend | Cat #: 149005; RRID: AB_2564314 |
| Anti-mouse Cx3cr1 APC (clone SA011F11) | Biolegend | Cat #: 149007; RRID: AB_2564491 |
| Anti-mouse Cx3cr1 Biotin (clone SA011F11) | Biolegend | Cat #: 149018; RRID: AB_2565701 |
| Anti-mouse DEC205 (CD205) PECy7 (clone Yekta) | Invitrogen | Cat #: 25-2051-42; RRID: AB_1834473 |
| Anti-mouse ESAM PE (clone1G8) | ThermoFisher Scientific | Cat #: 12-5852-81; RRID: AB_891539 |
| Anti-mouse F4/80 PerCP/Cy5.5 (clone BM8) | ThermoFisher Scientific | Cat #: 45-4801-82; RRID: AB_914345 |
| Anti-mouse F4/80 eFluor450 (clone BM8) | ThermoFisher Scientific | Cat #: 48-4801-42; RRID: AB_1548747 |
| Anti-mouse FoxP3 APC (clone FJK-16 s) | ThermoFisher Scientific | Cat #: 17-5773-80; RRID: AB_469456 |
| Anti-mouse IL-12p40 PE (clone C15.6) | BD Biosciences | Cat #: 562038; RRID: AB_10895571 |
| Anti-mouse Ly6C BV510 (clone HK1.4) | Biolegend | Cat #: 128033; RRID: AB_2562351 |
| Anti-mouse Ly6C APC-Alexa700 (clone HK1.4) | Biolegend | Cat #: 128023; RRID: AB_10640119 |
| Anti-mouse Ly6C eFluor450 (clone HK1.4) | ThermoFisher Scientific | Cat #: 48-5932-82; RRID: AB_10805519 |
| Anti-mouse Ly6D PE (clone 49-H4) | Biolegend | Cat #: 138603; RRID: AB_2281362 |
| Anti-mouse Ly6G BUV395 (clone 1A8) | BD Biosciences | Cat #: 563978; RRID: AB_2716852 |
| Anti-mouse MHC-II PE (clone AF6-120.1) | BD Biosciences | Cat #: 553552; RRID: AB_394919 |
| Anti-mouse MHC-II BV510 (clone M5/114.15.2) | Biolegend | Cat #: 107635; RRID: AB_2561397 |
| Anti-mouse MHC-II APC-Alexa700 (clone M5/114.15.2) | Biolegend | Cat #: 107622; RRID: AB_493727 |
| Anti-mouse NK1.1 APC-Alexa700 (clone PK136) | Biolegend | Cat #: 108729; RRID: AB_2074426 |
| Anti-mouse PD-L1 SuperBright780 (clone MIH5) | ThermoFisher Scientific | Cat #: 78-5982-82; RRID: AB_2724081 |
| Anti-mouse Pdca-1 FITC (clone 927) | ThermoFisher Scientific | Cat #: 11-3172-82; RRID: AB_763416 |
| Anti-mouse Siglec1 (CD169) PE (clone 3D6.112) | Biolegend | Cat #: 142403; RRID: AB_10915470 |
| Anti-mouse SiglecF PE (clone E50-2440) | BD Biosciences | Cat #: 552126; RRID: AB_394341 |
| Anti-mouse SiglecF BV421 (clone E50-2440) | BD Biosciences | Cat #: 565934; RRID: AB_2739398 |
| Anti-mouse SiglecH PerCP/Cy5.5 (clone 551) | Biolegend | Cat #: 129613; RRID: AB_10639936 |
| Anti-mouse SiglecH FITC (clone 551) | Biolegend | Cat #: 129604; RRID: AB_1227761 |
| Anti-mouse SiglecH PE (clone 551) | Biolegend | Cat #: 129605; RRID: AB_1227763 |
| Anti-mouse SiglecH APC (clone 551) | Biolegend | Cat #: 129612; RRID: AB_10641134 |
| Anti-mouse Xcr1 FITC (clone ZET) | Biolegend | Cat #: 148210; RRID: AB_2564366 |
| Anti-mouse Xcr1 PE(clone ZET) | Biolegend | Cat #: 148204; RRID: AB_2563843 |
| Anti-mouse Xcr1 PerCP/Cy5.5 (clone ZET) | Biolegend | Cat #: 148208; RRID: AB_2564364 |
| Anti-mouse Xcr1 APC (clone ZET) | Biolegend | Cat #: 148206; RRID: AB_2563932 |
| Anti-mouse Zbtb46 PE (clone U4-1374) | BD Bioscience | Cat #: 565832; RRID: AB_2739372 |
| Bacterial and Virus Strains | ||
| NR-29029 Influenza A/Puerto Rico/8/1934-WG (H1N1) | BEI resources | https://www.beiresources.org/Catalog/animalViruses/NR-29029.aspx |
| Biological Samples | ||
| Whole Blood from healthy donors | Obtained from donors with informed consent. IRB approved by Stanford University Research Compliance Office. | N/A |
| Human Spleen | Normal spleen from distal pancreatectomy from patients not subjected to any chemotherapy. | Stanford Tissue Bank http://med.stanford.edu/cancer/research/shared-resources/tissue-procurement.html |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ficoll-Paque PLUS | GE Healthcare | Cat # 300-25 |
| Percoll | GE Healthcare Life Sciences | Cat# 17089101 |
| Fetal Bovine Serum, qualified, US origin | GIBCO | Cat # 26140079 |
| RPMI 1640 with L- Glutamine | Corning | Cat# 10040CV |
| L-glutamine Solution | Corning | Cat # 25005CI |
| Sodium Pyruvate Solution | Corning | Cat # 25000CI |
| Penicillin-Streptomycin | Corning | Cat# 30002CI |
| HEPES solution | Corning | Cat # 25060CI |
| MEM Nonessential Amino Acid Solution | Corning | Cat # 25025CI |
| Recombinant Human IL-3 | R&D Systems | Cat # 203IL010CF |
| EDTA 0.5M pH 8.0 | Corning | Cat # 46034CI |
| ACK Lysis Buffer | Lonza | Cat# 10-548E |
| Benzonase’ Nuclease | Millipore-Sigma | Cat # E1014-25KU |
| Cell-ID Intercalator-Ir | Fluidigm | Cat#: 201192A |
| Cell-ID Cisplatin | Fluidigm | Cat # 201064 |
| 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester | Millipore-Sigma | Cat # 21888-25MG-F |
| CellTrace Violet Cell Proliferation Kit | ThermoFisher Scientific | Cat# C34557 |
| Bovine Serum Albumin solution 30% ± 2% in 0.85% sodium chloride, aseptically filled | Millipore-Sigma | Cat # A7284-50ML |
| Dimethyl sulfoxide > 95% | Millipore-Sigma | Cat# D4540 |
| Collagenase D from Clostridium histolyticum | Millipore-Sigma | Cat #11088858001 |
| DNase I from bovine pancreas | Millipore-Sigma | Cat# 10104159001 |
| Collagenase IV | Worthington Biochemical Corporation | Cat# LS004189 |
| Liberase TL | Millipore-Sigma | Cat# 5401020001 |
| Liberase TM | Millipore-Sigma | Cat# 5401119001 |
| Paraformaldehyde 16% aqueous solution | Electron Microscopy Sciences | Cat# 15710 |
| Indium 113 metal chloride | Trace Sciences International | Cat# In-113 |
| Indium 115 metal chloride | Trace Sciences International | Cat# In-115 |
| LPS | Invivogen | Cat# tlrl-eblps |
| Poly(I:C) | Invivogen | Cat# tlrl-pic |
| R848 | Invivogen | Cat# tlrl-r848 |
| CpGA 2216 | Invivogen | Cat# tlrl-2216-1 |
| CpGB 1826 | Invivogen | Cat# tlrl-1826-1 |
| Imiquimod | Invivogen | Cat# tlrl-imqs |
| DOTAP transfection reagent | Santa Cruz Biotechnologies | Cat# sc362166 |
| Critical Commercial Assays | ||
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | ThermoFisher Scientific | Cat#: 00-5521-00 |
| Permeabilization Buffer (10X) | ThermoFisher Scientific | Cat#: 00-8333-56 |
| Maxpar X8 Multimetal Labeling Kit Fluidigm | Fluidigm | Cat#: 201300 |
| LIVE/DEAD Fixable Dead Cell Stain Sampler Kit | ThermoFisher Scientific | Cat# L34960 |
| Dynabeads Sheep Anti-Rat IgG | ThermoFisher Scientific | Cat# 11035 |
| Dynabeads Pan Mouse IgG | ThermoFisher Scientific | Cat# 11042 |
| Pan T cell Isolation Kit, human | MACS, Miltenyi Biotec | Cat# 130-096-535 |
| Mouse IFN Alpha All Subtype ELISA Kit, High Sensitivity | PBL Assay Science | Cat# 42115-1 |
| Human IFN Alpha Multi-Subtype ELISA Kit (TCM) | PBL Assay Science | Cat# 41105-1 |
| Mouse IL-12 p70 Quantikine ELISA Kit | R&D Systems | Cat# M1270 |
| Cytometric Bead Array Human Enhanced Sensitivity Master Buffer Kit | BD Biosciences | Cat# 561521 |
| Cytometric Bead Array Human IL-12p70 Enhanced Sensitivity Flex Set | BD Biosciences | Cat# 561518 |
| BD CompBead Anti-Mouse Ig, κ/Negative Control Compensation Particles Set | BD Biosciences | Cat# 552843 |
| Alexa Fluor 647 Antibody Labeling Kit | ThermoFisher Scientific | Cat# A20186 |
| NucleoSpin RNA XS | Takara Bio | Cat# 740902.10 |
| NucleoSpin Tissue XS | Takara Bio | Cat# 740901.10 |
| Phusion Hot Start II Master Mix | ThermoFisher Scientific | Cat# F565S |
| iScript Reverse Transcription Supermix | Bio-Rad Laboratories | Cat# 1708840 |
| iTaq Universal SYBRGreen Supermix | Bio-Rad Laboratories | Cat# 1725120 |
| Deposited Data | ||
| Human data single cell RNaseq | Broad Single Cell Portal study “Atlas of human blood dendritic cells and monocytes” | https://singlecell.broadinstitute.org/single_cell/study/SCP43/atlas-of-human-blood-dendritic-cells-and-monocytes#study-download |
| Mouse Raw RNA-Seq | NCBI GEO | GSE76132 |
| Software and Algorithms | ||
| GraphPad Prism 6 | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| MATLAB | N/A | https://www.mathworks.com/products/matlab.html |
| Cytofkit | N/A | https://bioconductor.riken.jp/packages/3.7/bioc/html/cytofkit.html |
| ViSNE (CYT) | Amir et al., 2013. | https://dpeerlab.github.io/dpeerlab-website/cyt-download.html |
| FlowJo Software v10.0.8 | TreeStar, Inc. | https://www.flowjo.com/solutions/flowjo |
| UMAP | Becht et al., 2018 | https://github.com/lmcinnes/umap |
| R | N/A | https://www.R-project.org/ |
| rgl v0.98.1 | N/A | https://r-forge.r-project.org/projects/rgl/ |
| ggplot2 v2.2.1 | N/A | https://github.com/tidyverse/ggplot2 |
| viridis | N/A | https://github.com/sjmgarnier/viridis |
| limma | N/A | https://bioconductor.org/packages/release/bioc/html/limma.html |
| DESeq2 | N/A | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |