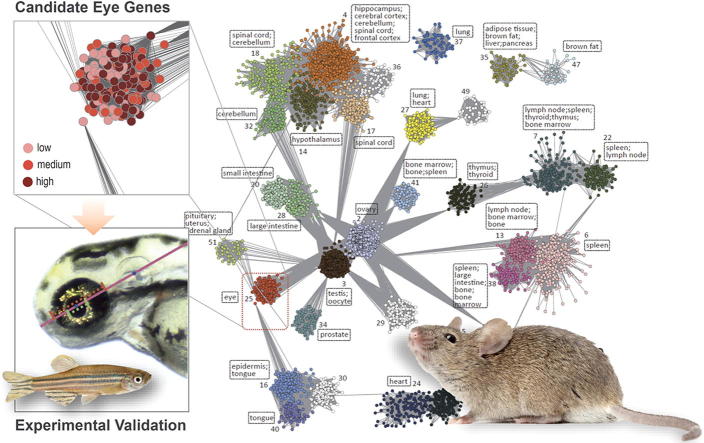
Graphical abstract
Keywords: Mammals, Retina, Expression profile, Modularity, Systems biology, Functional genomics
Abstract
In the last few decades, reverse genetic and high throughput approaches have been frequently applied to the mouse (Mus musculus) to understand how genes function in tissues/organs and during development in a mammalian system. Despite these efforts, the associated phenotypes for the majority of mouse genes remained to be fully characterized. Here, we performed an integrated transcriptome-phenome analysis by identifying coexpressed gene modules based on tissue transcriptomes profiled with each of various platforms and functionally interpreting these modules using the mouse phenotypic data. Consequently, >15,000 mouse genes were linked with at least one of the 47 tissue functions that were examined. Specifically, our approach predicted >50 genes previously unknown to be involved in mice (Mus musculus) visual functions. Fifteen genes were selected for further analysis based on their potential biomedical relevance and compatibility with further experimental validation. Gene-specific morpholinos were introduced into zebrafish (Danio rerio) to target their corresponding orthologs. Quantitative assessments of phenotypes of developing eyes confirmed predicted eye-related functions of 13 out of the 15 genes examined. These novel eye genes include: Adal, Ankrd33, Car14, Ccdc126, Dhx32, Dkk3, Fam169a, Grifin, Kcnj14, Lrit2, Ppef2, Ppm1n, and Wdr17. The results highlighted the potential for this phenome-based approach to assist the experimental design of mutating and phenotyping mouse genes that aims to fully reveal the functional landscape of mammalian genomes.
1. Introduction
Mus musculus is one of the most important mammalian organisms used for biomedical research since it exhibits extensive genetic, physiological, and behavioral similarities with humans. Consequently, over the last two decades, high throughput technologies have been applied to this animal model as part of ongoing efforts to understand how genes function in tissues/organs and during development [1], [2], [3], [4], [5]. Reverse genetics has provided straightforward phenotypic descriptions of gene functions at the organismal level [4], [6]. However, >50% of genes encoded in the mouse genome still lack such data [7]. Furthermore, among the mouse genes which have been phenotyped for various alleles (i.e., spontaneous, targeted, N-ethyl-N-nitrosourea-induced, endonuclease-mediated, etc.), the majority of these mutant alleles have only been phenotyped in a limited subset of tissues/organs without a standardized and comprehensive examination conducted across developmental stages and organs. To address this issue, The International Mouse Phenotyping Consortium (IMPC) has been working to systematically catalog gene functions for mice generated by the International Knockout Mouse Consortium (IKMC) by performing comprehensive phenotyping of using standardized pipelines to measure various phenotypes [8]. However, despite these efforts, prioritization and identification of functions of mammalian genes remain a challenge.
Modularity is an essential property in biology because it allows a living system to function [9] and evolve [10] efficiently. Modular structures are observed in various gene networks, including those inferred from coregulation relationships [11], [12], [13]. In our previous study, we provided genomic evidence that a mutated mouse gene tends to present abnormal phenotype(s) in tissues where it is expressed [14]. Thus, we postulated that gene modules in a coexpression network are enriched with genes which co-function within the same anatomical structure (i.e., tissue/organ), and this anticipated tendency, if exists, may facilitate a prediction of gene function. Therefore, we compiled transcriptome datasets of mouse tissues profiled with various platforms to identify gene modules in coexpression networks inferred from each dataset. Statistically enriched tissue functions for each module were annotated based on mouse phenome data. By using this approach, we linked tissue functions >15,000 mouse genes. Specifically, we predicted 50 mouse genes with potential roles in the development and/or physiological functions of the eye, which is one of the most conserved organs during vertebrate evolution [15]. Among the 50 genes identified, 35 have not had their visual functions elucidated in any animal model. The results of the subsequent experiments based on a subset of candidate eye genes demonstrated that predicted functions of >85% of the candidate eye genes can be validated with functional analyses performed in zebrafish (Danio rerio) embryos. These results indicated the potential for this phenome-based approach to guide the experimental design of mutating and phenotyping mouse genes. We discuss the relevance of the novel eye genes discovered to genetics of several congenital eye diseases in humans.
2. Materials and methods
2.1. Mouse gene modules in coexpression networks
Expression signals of mouse genes in 78 and 96 tissues/cell types were obtained from the bioinformatic portal BioGPS (http://biogps.org/; accessed on September 23, 2015) [16] (see Tables S1–S2). These expression signals were derived from mRNA hybridization data collected with Affymetrix microarray chips, GNF1M and MOE430 2.0, respectively. The downloaded microarray-based expression signals were previously processed/normalized with the GCRMA algorithm [17]. In addition, RNA-seq data for 53 mouse tissues were obtained from data sources listed in Table S3 to provide a third transcriptome dataset for this study. For each set of RNA-seq data, raw reads were mapped to the reference mouse genome, NCBIM37 (Ensembl release 67), with TopHat (v2.0.12) [18]. The read count for each gene was then calculated with HTSeq [19]. Both procedures were performed with default parameters. The expression signals were represented as raw counts and then were transformed by using a variance-stabilizing transformation (VST) procedure available in the DESeq2 R software package [20]. The expression signals were measured multiple times in tissues/cells from biological replicates and these values were averaged after normalization.
The strengths of coexpression (CoExp) between all possible gene pairs were estimated based on the absolute value of Pearson’s correlation coefficient of expression signals of compared genes across all of the tissues/cells examined. As a result, a weighted gene coexpression network was generated for each of the three transcriptome datasets. The R software package WGCNA and Python program PGCNA were subsequently used to construct a coexpression network and to identify the modular structures within it. WGCNA analyses used following settings: networkType = “unsigned”; TOMType = “unsigned”; minModuleSize = 30, maxBlockSize = 1000; reassignThreshold = 0. The soft-thresholding power values (and the corresponding R2 of the scale free topology model fit) were determined by the “pickSoftThreshold” function of WGCNA, and these values for the GNF1M, MOE430 and RNA-seq datasets were 3 (0.82), 3 (0.65), and 9 (0.71), respectively. As for PGCNA analyses, default settings were used except for using “-f 1” to preserved all the nodes and using “–usePearson” to define CoExp as described above. Consequently, WGCNA (or PGCNA) identified 54 (or 45), 64 (or 43), and 22 (or 28) modules of coexpressed genes from the GNF1M, MOE430, and RNA-seq datasets, respectively (see below).
Zsummary module preservation statistics were obtained by performing a WGCNA. These statistics included four statistics related to density and three statistics related to connectivity. These statistics allow a quantitative assessment to be performed to determine whether the density and connectivity patterns of modules defined in a reference dataset are preserved in a test dataset. A Zsummary value between 2 and 10 indicates moderate module preservation, whereas a Zsummary value >10 indicates strong module preservation. The score of the overlap between two modules from different networks was calculated by the function “overlapTable” with default settings.
2.2. Associating coexpressed gene modules with tissue functions based on phenotypic data
Annotations of mouse genes and their associated phenotypes derived from mutagenesis approaches were obtained from MGI (http://www.informatics.jax.org/, version 5.2; accessed on October 8, 2015). Ensembl IDs of the phenotyped mouse protein-coding genes were found at MRK_ENSEMBL.rpt, while information regarding genotype-phenotype associations (presented as Mammalian Phenotype IDs, MP IDs) was found at MGI_GenePhenoMP.rpt. Phenotypes caused by mutations in multiple genes were discarded. Consequently, there were “phenotyped” 7791 mouse protein coding genes with one or more MP IDs. These data derived from gene knock out, gene knock down, transgenic insertions, and/or point mutation experiments conducted in mouse models. MP IDs are hierarchically structured and a parent MP ID represents a phenotype lineage that may include several child MP IDs to describe a more detailed abnormal phenotype. Genes with a child MP ID were also assigned to the parent MP IDs. As described in a previous study [14], we transformed the MGI annotated MP IDs into records of phenotypic defects at the tissue level (Table S4). MP ID terms used to define abnormal phenotypes in the 47 tissues examined are listed in Dataset S1. There were 4363 genes associated with at least one of the 47 tissue functions in our dataset. For the remaining 3428 genes, although mutant strains have been generated (and phenotyped) for these, no phenotypic defect has been reported in any of the 47 tissues of interest. Furthermore, in the latter cases, the absence of a phenotype profile may partially be explained by incomprehensive phenotyping.
To functionally annotate the gene sets represented as modules in our coexpression network, we performed an enrichment analysis on the mutant phenotypes available for the 47 tissues listed in Table S4. For each module and each tissue to which a gene’s function could be associated, the number of phenotyped genes found to be associated with (and not associated with) the focal tissue function in the module of interest, as well as the number of genes in other non-focal modules, were counted. Tissues with entries overrepresented with P < 0.001 by Fisher’s exact test were assigned to the coexpressed gene module.
2.3. Network properties of module members
To estimate the centrality of genes in a module, only interactions between genes of the same modules were considered in the approximation of centrality of the focal gene in a module. Connectivity (K) was defined as the averaged connection strength between the focal gene and the rest of the genes in the same module. Betweenness centrality (the number of shortest paths between all pairs of genes that pass through the gene) and closeness centrality (the sum of the length of the shortest paths between the genes and all other genes) were calculated with the ‘networkx’ Python package.
2.4. Known and predicted human orthologs associated with eye diseases
To understand if human orthlogs of mouse eye candidate genes were enriched with known human genes associated with eye diseases, we obtained the lists of human genes associated with retina disorder or eye disease from RetNet (http://www.sph.uth.tmc.edu/RetNet/, accessed on May 22, 2017) or HPO (https://hpo.jax.org/, accessed on March 28, 2019), respectively. The file “hp.obo” and “ALL_SOURCES_ALL_FREQUENCIES_genes_to_phenotype.txt” were downloaded as the phenotype ontology data and the gene-phenotype association data, respectively. Genes annotated with any of the HP term “Abnormality of the eye” (HP:0000478) or/and the downstream terms of HP:0000478 were defined as known human genes associated with congenital eye diseases.
2.5. Zebrafish maintenance and injection of morpholinos
Zebrafish Danio rerio AB strains were maintained at 28 °C with a 14 h/10 h dark/light cycle. Two males and four females were set-up for breeding. Embryo collection and microinjections were performed according to a standard protocol [21]. Orthology and sequence information for zebrafish/mouse/human genes were obtained from the portal BioMart of the Ensembl database (v87). MO were designed and synthesized by GeneTools, LLC. To minimize possible off-target effects, a BLAST search was performed against the zebrafish genome (GRCz10). MO which were found to target translational start sites or splicing junctions of a secondary gene were redesigned. The criteria for judging off-target binding of MO were as follows: 1) MO needed to be at least 14 bases in length to target regions between the 5′ cap and within 25 bases of the 3′ translational start site of a mRNA; and 2) MO needed to target splice junctions within 10 bases of an exon and 25 bases of an intron. The specific amount of MO which were injected into embryos with a Nanoject II instrument (Drummond Scientific) (see below). The standard control MO (CCTCTTACCTCAGTTACAATTTATA) which targets a human beta-globin intron mutation were injected into embryos to serve as the negative control group for each experiment. As suggested by GeneTools, the standard control MO is a widely used and frequently reported sequence of negative control [22]. Injected embryos were maintained and raised under the same conditions as those for the adult zebrafish. Observations regarding phenotype were made at 72 hpf and images were captured and measured with a stereomicroscope (Nikon SMZ645) equipped with a CCD camera. The fixation and embedding of zebrafish embryos for paraffin sectioning and H&E staining were according to the standard protocol [23]. The Institutional Animal Care and Use Committee of Tzu Chi University approved all of the animal experiments conducted in this study (IACUC approval number: 106050).
2.6. Statistics
Fisher’s exact test was used to examine if the proportions of samples with a specified characteristic were the same between two compared groups. The Mann-Whitney U test was used to examine if two compared samples were equivalent in their network properties or in regard to parameters used to measure eye phenotypes. Fisher’s exact test (two-tailed) or the Mann-Whitney U test (two-tailed) were applied by using the R function fisher.test or wilcox.test, respectively, to determine P-values.
3. Results and discussion
3.1. Coexpressed mouse gene modules and the associated tissue functions
The procedures of our analysis are summarized as Fig. S1 (see below for details). To identify modular structures in coexpression gene networks generated from transcription profiles of mouse tissues/cells, we compiled three platform-specific datasets from transcriptomes obtained from three sources: 1) 78 tissues/cells of mouse embryos, 8- to 10-week-old mice, or 10- to 12-week-old mice (Table S1) profiled by the GNF1M microarray platform [1], 2) 96 tissues/cells of mouse embryos or 8- to 10-week-old mice (Table S2) profiled by the MOE430 2.0 microarray platform (referred to as MOE430 hereafter) [24], and 3) 53 embryonic and adult mouse tissues profiled by Illumina RNA-seq in the ENCODE project (30 tissues) [25] or other individual studies (23 tissues) (Table S3), respectively. Both the GNF1M and MOE430 datasets were generated and processed from a single study. To integrate the RNA-seq-based transcriptome data produced from various studies, the raw reads of each transcriptome set of data for 53 tissues were remapped, reprocessed, and normalized (see Section 2). In the GNF1M, MOE430, and RNA-seq datasets, mRNA expression levels of 17,384, 16,688, and 20,968 protein-coding genes were estimated, respectively. As a result, a total of 15,521 common genes whose mRNA levels were estimated across the platforms were identified. To construct coexpression gene networks and identify gene modules within each network, we adopted two methods, including Weighted correlation network analysis (WGCNA) [12], [26] and Parsimonious Gene Correlation Network Analysis (PGCNA) [27]. Although WGCNA and PGCNA employ very different algorithms to define modules of coexpressed genes [WGCNA: hierarchical clustering [28]; PGCNA: fast unfolding [29]], the results generated from the two methods are consistent with each other (see below, and Section 2). Because WGCNA is more widely used, we present the data based on WGCNA as the main results as follows.
There were 54, 64, and 22 modules of coexpressed gene clusters identified among the GNF1M, MOE430, and RNA-seq datasets, respectively (see Section 2; Tables S5–S7) by WGCNA. A Zsummary score, which provides evidence that a module is preserved in another independent network, was calculated for each of the abovementioned modules (see Section 2). Although GNF1M, MOE430, and RNA-seq datasets were generated by different platforms/approaches and include expression profiles of different sets of tissues, a proportion of the modules analyzed [(GNF1M: 31/54 (57.4%), MOE430: 33/64 (51.5%), and RNA-seq: 7/22 (31.8%)] were found to be associated with highly significant preservation scores (Zsummary > 10) in at least one of the two other datasets (Fig. S2). The analysis on overlapped modules between networks indicated that the relatively few GNF1M- or MOE430-based modules found to have a high Zsummary score in the RNA-seq-based network is because multiple GNF1M- or MOE430-based modules often corresponded to the same module in the RNA-seq-based network (Fig. S2). In fact, only a small number of modules in microarray-data-based network were not overlapped with any module in the RNA-seq-based network (Fig. S2). This result suggested the reproducibility of modules detected across the datasets analyzed.
To functionally annotate coexpressed gene modules based on the mouse phenome, we focused on genes that had mutagenesis-derived phenotypes manifested in the 47 focal mouse tissues defined according to annotations of Mouse Genome Informatics (MGI) (see Table S4 and Dataset S1, and Section 2), and looked for enriched tissue-level phenotypes for each module according to Fisher’s exact tests (see Section 2) [14]. Because 47 tissues were tested, a P-value < 10−3 (0.05/47 = 1.06 × 10−3, Bonferroni correction) was considered to be statistically significant to correct for multiple testing. The tissue(s) associated with each of the modules in the coexpression network examined derived from the GNF1M, MOE430, and RNA-seq datasets are shown in Figs. S3–S5, respectively (also see Tables S5–S8).
3.2. Prediction of mouse genes with eye-related functions
Through the abovementioned analyses, 12,832, 15,453 and 5738 genes were associated with at least one tissue function based on their module membership in the GNF1M-, MOE430-, and RNA-seq-networks, respectively. Some of these genes have been phenotyped experimentally, while others have not (Tables S5–S7). Because modular structures can vary across different coexpression networks, a gene that is associated with a tissue function in a given network (e.g., the GNF1M-based network) is not necessarily associated with the same tissue function in another network (e.g., the RNA-seq-based network). Based on the consistency of assigned tissue functions in the three expression datasets used in the current study, we determined confidence scores for each tissue function for each gene. Thus, low, medium, and high confidence scores for a given tissue function of a gene were considered to indicate that the assigned tissue function for that gene was observed in one, two, or three of the datasets, respectively. To estimate the accuracy of predicting associated tissue function in this manner, we calculated Rdcv/phe (the ratio of “the number of genes previously discovered to have mutant phenotypes in a predicted tissue” divided by “the number of total phenotyped genes”) for each set of genes having a confidence score and a predicted tissue function (see Table S9). The two gene numbers used to compute Rdcv/phe for each gene set were based on gene annotations obtained from MGI [30] (see Section 2). The full gene lists of predicted tissue functions with the corresponding confidence score for each gene in the lists are provided as Dataset S2.
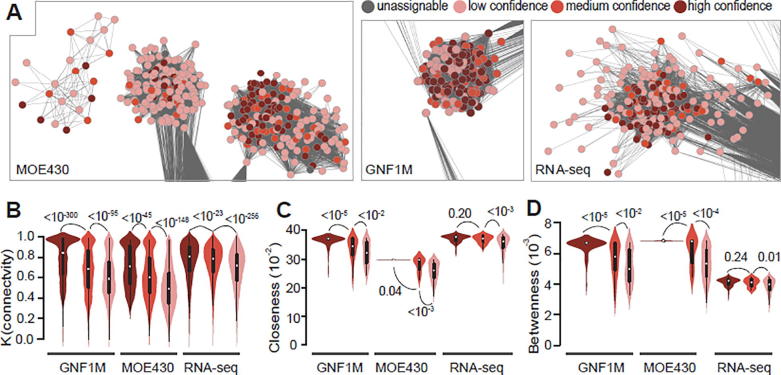
The gene sets predicted to function in “eye” achieved relatively high Rdcv/phe scores (Rdcv/phe = 96.3% [52/54], 77.3% [34/44], and 34.4% [65/189] for high, medium, and low confidence scores, respectively; Rdcv/phe of genes outside the eye module(s) was 19.3% [784/4072]) compared with the Rdcv/phe values of the gene sets predicted to function in other tissues (Table S9). For example, the Rdcv/phe values for genes with predicted functions in “spleen” were only 60.9% (56/92, high confidence), 37.3% (128/343, medium confidence), 22.7% (152/670, low confidence), and 9.3% (301/3254, others) (Table S9). Subnetworks of the eye module in the GNF1M-, MOE430- and RNA-seq-based coexpression networks, as well as the confidence scores of the module members within each network, are presented in Fig. 1A, B, and C, respectively. Next, we focused on this subnetwork structure by only considering intra-modular interactions. Specifically, we computed connectivity (K), closeness, and betweenness for each gene in the largest component (i.e., the module with the greatest number of nodes) of eye modules (and in this module the number of genes of each confidence score has to be at least 10) for each network (see Section 2). In general, the genes with higher confidence scores were associated with greater K, closeness, and betweenness values (Fig. 1D–F). These results indicate that the genes which were more consistently reported to be associated with eye function across the platforms analyzed were also more likely to represent hub genes in the subnetwork of the eye module, and these genes may be more important for eye function or development. Although the algorithm adopted by PGCNA does not clustered genes into modules by considering all edges in a network, the reproduced Fig. 1 based on PGCNA-defined modules showed consistent trends, except for the lack of statistical significance when the analyses were performed based on the RNA-seq-data (Fig. S6).
Fig. 1.
Genes associated with the “eye” module. (A) Genes with different levels of consistency in being located in the eye module across the three platforms are shown according to their confidence scores [e.g., high (dark red), medium (red), low (pink)] for predicted retinal function. (B-D) Genes with higher confidence scores tend to have greater values of (B) connectivity, (C) betweenness, and (D) closeness, thereby indicating that they are more likely to be the central node of the module(s). In (B–D), P-values were determined with the Mann-Whitney U test and are associated with the arched lines that indicate the values that were compared. The corresponding weighted coexpression networks used to generate the data are indicated under each panel. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further verify the predicted eye functions of the mouse genes that were identified, we examined if the human orthologs of these mouse genes tended to be known genes associated with eye diseases in humans. We found that the human orthlogs were enriched with genes cataloged in the Retinal Information Network (RetNet, a database which compiled known retinal disease genes in humans; see Section 2) (Table 2) and enriched with eye disease-related genes annotated by Human Phenotype Ontology (HPO) [31] (see Section 2) (Table 2). More importantly, when we focused on a subgroup of genes which did not have any previously reported retinal phenotype exclusive to mice, the abovementioned enrichment was still observed (Table 2). These results strongly suggest that many mouse genes in the eye modules identified are associated with previously undiscovered visual-related functions.
Table 2.
The human orthologs of mouse genes with predicted eye functions are enriched with genes cataloged in RetNet (dubbed “RetNet”) or HPO-defined eye disease genes (dubbed “HPO”).
| Confidence score of predicted eye functions |
P-valuea |
|||
|---|---|---|---|---|
| All orthologs |
Orthologs without any previously reported eye phenotype in mice |
|||
| RetNet | HPO | RetNet | HPO | |
| High | <10−11 | <10−6 | <10−2 | 0.02 |
| High + Medium | <10−12 | <10−9 | <10−2 | 0.01 |
| High + Medium + Low | <10−4 | <10−10 | 0.12 | n.s. |
P-value was obtained under the null hypothesis of no enrichment by Fisher’s exact test (n.s.: not significant).
Highly expressed genes and tissue-specific genes are more easily to be clustered in modules of a gene coexpression network. To understand if it was coexpression information, or the expression abundance in the eye tissues alone, contributing to the predictive power of our approach, we computed Zeye for each gene for each dataset as Zeye = (Eeye − Eavg)/SDE, where Eeye is the expression signals of the focal gene in the eye tissues, Eavg is its mean expression signals across all the tissues, and SDE is the standard deviation of expression signals in all the tissues. Zeye quantified the relative mRNA abundance of a gene in the eye tissues in comparison with other tissues, and did not incorporate the information of coexpression between genes. We found that genes with a higher confidence score defined in Fig. 1A tend to have a higher Zeye (Fig. S7), suggesting the need to verify the contribution of expression patterns at a single gene level to the predictive power of our approach. We subsequently generated three gene lists by selecting 185, 549 and 306 genes with the highest Zeye values from each of the GNF1M-, MOE430-, and RNA-seq platforms, respectively (the numbers of genes selected were the same as the numbers of candidate genes identified from the eye related modules for each network shown in Fig. 1A). Similar to Fig. 1A, low, medium, and high confidence scores of the candidate eye genes identified through Zeye were determined based on the observed association of a gene in one, two, or three Zeye-defined gene lists, respectively. We found that, when candidate genes were defined based on top ranked Zeye, the enrichment in known genes associated with eye diseases became insignificant, or not as significance as the P-values shown in Table 2 (Table S10). This result, indicated that coexpression information, which emphasizes the relationship between genes, has assisted the gene function prediction at the phenotypic level.
3.3. External data supporting the prediction of genes with eye-related functions
In the present study, a total of 75 mouse genes and 73 mouse genes were found to be associated with the eye module and to have high and medium confidence scores, respectively (Tables S11 and S12). Among these genes, 50 have no previously documented eye phenotype according to the MGI annotations, or they do not have any mutant strains produced for phenotyping (Tables S11–S13, Table 1, and Section 2). When the same approach to identify eye candidate genes with a confidence score was applied to PGCNA-defined coexpressed gene modules, 45 out of these 50 genes were also found to be medium- or high-confident eye candidate genes by the PGCNA method (genes marked with the superscript “e” in Table 1), indicating the consistency of the results when different methods were used to predict gene functions when the phenome-based approach was applied to. It should also be noted that the 50 novel eye genes listed in Table 1 were predicted in December 2016. Subsequently, in 2017 and 2018, eye-related functions for two genes which each received a high-confidence score (i.e., Samd7 and Lrit1 Tspan10, respectively) were confirmed in targeted mutagenesis studies conducted in a mouse model[32], [33] (Table 1). We also found that 17 of the remaining 48 candidate genes have characterized functions related to vision in at least one other vertebrate species, including human, chicken (Gallus gallus), or zebrafish (Danio rerio) (see Table 1) [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55]. For example, while the functions of Cnga1, Pou6f2, and Vit in mouse remain unexplored, nonsense or misense mutations of human CNGA1 is associated with Retinitis pigmentosa [35], [36], a common form of inherited retinal degeneration; knock-down of zebrafish pou6f2 suppresses retina regeneration [52]; mutations in vit in chicken have been linked to weakened vision during the domestication process [55].
Table 1.
List of candidate genes underlying proper eye development and function in mice. Only genes without a previously reported gene deletion phenotype in any of the 47 selected tissues in mice are included in this list.
| MGI Gene symbola | Number of ortholog(s)b |
Characterized vision/eye-related phenotypes due to mutations (or reduced expression, if noted) [gene symbol, speciesc], if available | |
|---|---|---|---|
| Human | Zebrafish | ||
| High confidence genes | |||
| Ankrd33d,e | 1 | 2 | Unreported |
| Arr3e | 1 | 2 | Knockdown caused a sever delay in photoresponse recovery [arr3, zebrafish] [34] |
| Cnga1e | 1 | 2 | Retinitis pigmentosa 49, RP49 [CNGA1, human] [35], [36] |
| Cryba4e | 1 | 1 | Cataracts and microphthalmia, CTRCT23 [CRYBA4, human] [37], [38] |
| Crybb3e | 1 | 1 | Congenital cataracts, CTRCT22 [CRYBB3, human] [39] |
| Gm4792e | 0 | 0 | Unreported |
| Gm9918e | 1 | 0 | Unreported |
| Grifind,e | 1 | 1 | Unreported |
| Kcnj14d,e | 1 | 1 | Unreported |
| Kcnv2e | 1 | 2 | Retinal cone dystrophy 3B, RCD3B [KCNV2, human] [40] |
| Lrit1e | 1 | 2 | Unreported until 2018; visual acuity impairments in optokinetic response [Lrit1, mouse] [32] |
| Lrit2d,e | 1 | 1 | Unreported |
| Mlanae | 1 | 0 | None [ps. no eye abnormality detected in mice [41]] |
| Pde6he | 1 | 1 | Incomplete achromatopsia [43] and Retinal cone dystrophy 3A, RCD3A [42] [PDE6H, human] |
| Ppef2d,e | 1 | 2 | Unreported |
| Ppm1nd,e | 1 | 2 | Unreported |
| Samd7e | 1 | 1 | Unreported until 2017; ectopic expression of nonrod genes and rod photoreceptor cell dysfunction [Samd7, mouse] [33] |
| Slc1a7e | 1 | 2 | Early pathological change in the development of aged-related macular degeneration [EAAT5, human] [44] |
| Stx3e | 1 | 2 | Congenital cataract and intellectual disability [STX3, human] [45] |
| Tmem215e | 1 | 0 | Unreported |
| Tspan10e | 1 | 1 | Unreported until 2019, concomitant strabismus [TSPAN10, human] [46] |
| Medium confidence genes | |||
| 4632404H12Rike | 0 | 0 | Unreported |
| Adald,e | 1 | 1 | Unreported |
| Car14d,e | 1 | 1 | Unreported |
| Ccdc126d,e | 1 | 1 | Unreported |
| Crxose | 0 | 0 | Unreported |
| Crybb1e | 1 | 1 | Pediatric or age-related cataracts CATARACT 17 [CRYBB1, human] [47], [48] |
| Crygne | 1 | 2 | Unreported |
| Defb9e | 0 | 0 | Unreported |
| Dhx32d,e | 1 | 2 | Unreported |
| Dkk3d,e | 1 | 2 | Unreported |
| Fabp12e | 1 | 3 | Unreported |
| Fam169ad | 1 | 2 | Unreported |
| Fam19a3e | 1 | 0 | Unreported |
| Frmpd2d,e | 1 | 1 | Unreported |
| Gzmme | 1 | 0 | Unreported |
| Impg1e | 1 | 2 | Vitelliform macular dystrophies [IMPG1, human] [49] |
| Lyg2e | 1 | 3 | Unreported |
| Make | 1 | 1 | Retinitis pigmentosa 62, RP62 [MAK, human] [50], [51] |
| Mgarpe | 1 | 0 | Unreported |
| Otore | 1 | 0 | Unreported |
| Pdzph1e | 0 | 1 | Unreported |
| Plk5 | 1 | 0 | Unreported |
| Pou6f2 | 1 | 1 | Knockdown suppressed retina regeneration [pou6f2, zebrafish] [52] |
| Rrhe | 1 | 1 | Retinitis punctata albescens [RRH, human] [53] |
| Slco4a1 | 1 | 1 | Expression reduction marks the occurrence of retinal detachment [SLCO4A1, human] [54] |
| Tldc1d,e | 1 | 1 | Unreported |
| Vite | 1 | 1 | Evolutionarily reduced expression linked to weakened vision during domestication [vit, chicken] [55] |
| Wdr174 | 1 | 1 | Unreported |
| Zfp563e | 1 | 0 | Unreported |
See Tables S11 and S12 for the corresponding MGI ID and full gene name.
See Table S15 for the corresponding gene IDs of the orthologs.
Human: Homo sapiens; mouse: M. musculus; chicken: Gallus gallus; zebrafish: Danio rerio.
Indicates genes which were functionally validated in zebrafish in the present study.
Eye candidate genes with a medium or a high confidence score when coexpressed gene modules were defined by PGCNA.
However, while few of the other candidate genes (i.e., Mlana, Pde6h, and Ppef2) with potential eye-related functions have been examined in mammals, eye-related functions have not been discovered for these genes in mice. For example, no visible morphological abnormality was observed in the eyes of Mlana-deleted mice [41]. While mutations in PDE6H in humans causes total color blindness [43] or retinal cone dystrophy [42], Pde6h knockout in mice exhibited normal retinal morphology and no visual functional impairment [56]. Meanwhile, loss of rdgC, the ortholog of mouse Ppef2, results in light-induced retinal degeneration in Drosophila [57], yet mouse double knockouts of Ppef2 and Ppef1revealed no visual defect [58]. Also, IMPC has examined and unable to discover vision/eye phenotypes for a subset of candidate eye genes, including Kcnj14, Kcnv2, Adal, Fabp12, Plk5, and Wdr17 (Table S14), yet the ortholog of Kcnv2 in humans is associated with retinal cone dystrophy [40]. For these cases, it remains unclear whether the functions of the abovementioned genes in mice are truly unrelated to vision, or whether vision-related phenotypes can only be observed under previously unconsidered conditions. One such an example is that Cabp5 (a gene with a high confidence score, see Table S11) knockout mouse exhibit no significant retinal abnormalities. In spite of this, reduced sensitivity of rod-mediated light responses of retinal ganglion cells is observed [59]. Similarly, IMPC has only examined gross morphological/physiological alternations in mutant strains (see Table S14); more comprehensive phenotypic assays might be needed to definitively conclude if any of these genes have eye-related functions.
Molecular experiments have been conducted for some of the genes listed in Table 1. The results from these experiments indirectly support the retinal functions predicted for these genes. For example, mRNA of mouse Gzmm localizes exclusively to photoreceptor cells in the retina and is only expressed after the eye opens. These results imply that Gzmm transcripts are related to maintenance of the retinal structure or functions of mature photoreceptor cells [60]. Another example is the protein product of mouse Mgarp which specifically localizes to mitochondria in retina cells. When MGARP is overexpressed without its N-terminal region, severe aggregation of mitochondria occurs. Taken together, these results imply that Mgarp may have a role in retinal-energetic metabolism [61]. Furthermore, mouse Slc1a7 has been identified as a target of the light-regulated microRNA, miR-183/96/182, in photoreceptor cells [62]. While all of these molecular data are valuable, we only considered phenotypes as direct evidence of the gene functions predicted in the present study, since gene expression activities or interactions could, in part, be spurious and not all of them produce phenotypic outcomes [7], [63].
3.4. Functional validation of candidate mouse eye genes in zebrafish
The anatomy, histology, and biochemistry of the eye in zebrafish (Danio rerio) are comparable to mammals. Concerning the rapid embryonic development, zebrafish could be excellent in vivo model for efficiently validating eye-related gene functions predicted from mouse -omics data [64]. Accordingly, zebrafish orthologs of candidate mouse genes that met the following criteria were selected and examined for their requirement in eye development. First, non-crystalline genes with corresponding human orthologs that are not linked to eye-related diseases according to RetNet and Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/) databases. Second, genes without any reported visual function in the non-human vertebrate model. Third, to reduce the complexity of the zebrafish experiments, the number of orthologs in the zebrafish genome for each of the selected genes was limited to two. Accordingly, 15 out of 50 mouse genes corresponding to 21 zebrafish orthologs were selected for further study (Table 1, asterisks, and Table S15).
We individually knocked down these zebrafish orthologs with use of morpholino oligos (MO) by injecting them into the yolk of 1-cell stage embryos to block the translation of the targeted mRNAs (Table S16; see Section 2). Embryos of the same brood were also injected with an equal amount of standard control oligo (see Table S16, Section 2) as negative controls. Since the role of pax6b in eye development in zebrafish is well-known [65], MO targeting on pax6b was used as a positive control.
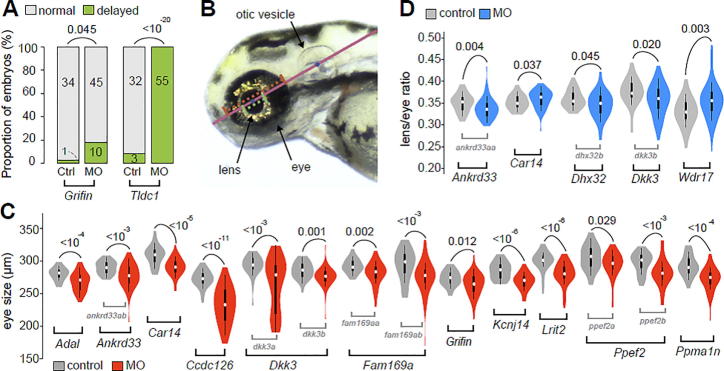
According to the gross phenotypic consequences of these experiments, we categorized the eye phenotypes of MO-injected embryos into three categories: the global development delay, small eye, and abnormal lens/eye ratio. The developmental delay phenotype is defined by the gross morphology of MO-injected embryos (Fig. 2A; Figs. S8–S23). When zebrafish orthologs of Grifin and Tldc1 mouse genes were targeted, a significant increase in the proportion of developmentally delayed embryos (embryos arrested at 48 hpf, hours post-fertilization, or earlier) was observed (Fig. 2A). For examination of the phenotype of eye development, the gross phenotype of the embryonic eye at 72 hpf was characterized based on the size of the eye, as well as the relative size of the lens by the lens/eye ratio (Fig. 2B) (see Section 2, Figs. S8–S23). The lens/eye ratio is to describe the potential knockdown effect which specifically interferes the lens development. To control for the confounding effect of late embryonic development in our measurements of eye phenotype, we excluded the data from developmentally delayed embryos from our analyses. As a positive control, the embryos injected with pax6b MO exhibited small eye phenotype, yet no change in their lens/eye ratio (Fig. S23), which is consistent with that described in a previous report [65].
Fig. 2.
Consequences of knocking down zebrafish orthologs of mouse candidate eye genes with MO. (A) Groups of MO for two candidate genes (Grifin, Tldc1) exhibited a greater proportion of developmentally delayed embryos than the control groups. The numbers of embryos used to calculate the proportions are indicated for each bar. (B) The left side of each embryo was imaged and a line was drawn across the centroids of the lens and the otic vesicle to measure gross eye morphology. Based on this line, eye (or lens) size was defined as the length (in pixels) of the red (or green) dashed line that starts from the anterior boundary and extends to the posterior boundary of the eye (or lens). The lens/eye ratio was calculated by lens size divided by eye size. (C) Groups of MO for 11 candidate genes induced a significant reduction in eye size. (D) Groups of MO for 5 candidate genes induced aberrant lens/eye ratios. In (C–D), violin plots present the values for (C) eye size and (D) lens/eye ratios. In (A, C, D), P-values were determined with Fisher’s exact test (A) or the Mann-Whitney U test (C, D). These values are associated with arched gray lines at the top of each panel which indicate the values that were compared. At the bottom of the plots, names of the corresponding mouse genes (black font) which were validated with MO are indicated at the bottom of the plots. The names of the MO groups (grey font) are specified only when multiple MO were designed for a focal gene. Only comparisons that differed significantly are shown (Figs. S8–S22 present the complete set of data). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
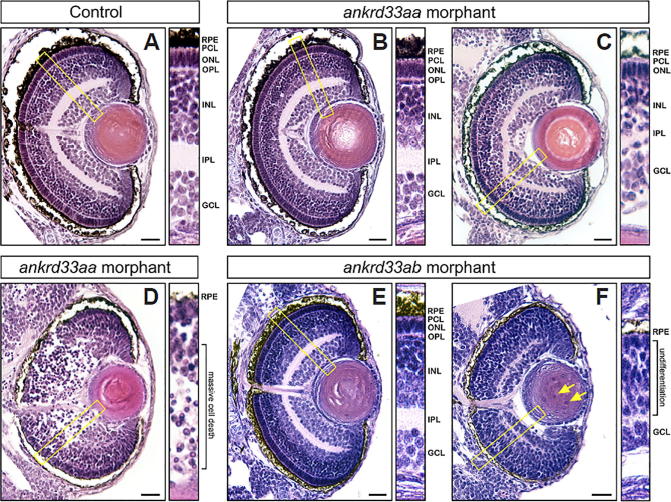
As a result, 14 zebrafish genes, corresponding to mouse candidate genes Adal, Ankrd33, Car14, Ccdc126, Dkk3, Fam169a, Grifin, Kcnj14, Lrit2, Ppef2, and Ppm1n, were found to exhibit reduced eye size phenotype (Fig. 2C). The other five zebrafish genes, corresponding to mouse candidate genes, Ankrd33, Car14, Dhx32, Dkk3, and Wdr17, exhibited an aberrant lens/eye ratio phenotype (Fig. 2D) (Figs. S8–S22). Strikingly, knockdown of zebrafish orthologs of mouse genes Ankrd33, Car14, and Dkk3 exhibited both of these eye abnormalities. Therefore, the predicted eye-related functions were validated for the majority (13 out of 15) of mouse candidate genes in zebrafish. Strikingly, knockdown of zebrafish orthologs of mouse genes Ankrd33, Car14, and Dkk3 exhibited both of these eye abnormalities. Among them, Ankrd33 has been found to be expressed in the mouse retina and acts as a transcriptional repressor that suppresses CRX-regulated photoreceptor genes [66]. However, the phenotypic analysis of Ankrd33 in retina is lacking. In zebrafish, there are two orthologs of mouse Ankrd33, ankrd33aa and ankrd33ab. The knockdown of zebrafish ankrd33aa showed the phenotype of aberrant lens/eye ratio (P = 0.004, Fig. 2D), whereas knockdown of zebrafish ankrd33ab caused the reduced eye size (P < 10−3, Fig. 2C). To further investigate whether the gross phenotypes presented in ankrd33aa and ankrd33ab morphants could reflect specific eye abnormalities in tissue level, we performed the subsequent histological analysis. In control MO-injected embryos, the retinal lamination was well structured, and the crescent-shaped inner plexiform layer (IPL) and outer plexiform layer (ONL) can be clearly observed (Fig. 3A) for the all the eyes examined. Among the analyzed thirteen eyes from randomly selected ankrd33aa morphants, we observed retinal abnormalities such as disorganized plexiform layers (either IPL or OPL) accompanying mild cell death (9/13) (Fig. 3C) and massive retinal degenerations in the ganglion cell layer (GCL) and inner nuclear layer (INL) (2/13) (Fig. 3D). In ankrd33ab morphants, other than normal retinal lamination (7/16) (Fig. 3E), we observed the poor differentiation of retinal layers and the enucleation defect in lens fiber cells (9/16) (Fig. 3F). These results phenotypically verified visual related functions of Ankrd33, and suggested that the gross morphological defects could correlate with specific structural defects in the retina.
Fig. 3.
Histological examination of retinal structures in morphants of ankrd33aa and ankrd33ab. (A) The eye section of control MO-injected embryos. The retina of all control embryos (13/13) showed well-structured lamination. (B,C,D) Eye sections of ankrd33aa morphant embryos. The retina of ankrd33aa morphant embryos could be (B) normal (2/13), (C) disorganized in plexiform layers (9/13), or (D) severely degenerated (2/13). (E,F) Eye sections of ankrd33ab morphant embryos. The retina of ankrd33ab morphant embryos could be (E) normal (7/16) or (F) underdeveloped with an enucleation defect in lens fiber cells (yellow arrows) (9/16). In (A–F), coronal sections of the eye were performed to reveal retinal lamination by hematoxylin and eosin staining. The area within yellow rectangle is enlarged to show the retina structure. The anterior is to the top. Scale bars: 20 μm. RPE: retinal pigment epithelium; PCL: photoreceptor cell layer; ONL: outer nuclear layer; OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Final remarks
In the present study, an integrated transcriptome-phenome analysis based on the mouse data was used to predict tissue functions for mammalian genes. In particular, we focused on a subset of eye candidate genes with potential biomedical relevance to validate the results of gene function prediction. For 86.7% (13/15) of these genes, eye-related functions were experimentally validated with targeted MO in zebrafish. These 13 genes include genes whose previous targeted deletion experiment did not lead to any observable eye abnormality in mice, such as Ppef2 [58] (Fig. 2C and Fig. S19). In addition to the previously mentioned issues regarding the comprehensiveness of phenotyping, it remains to be determined if the absence of an eye phenotype in knockout mice of some of these genes is due to genetic compensation mechanisms activated by a gene deletion approach [67]. For some of the candidate genes, their human orthologs were found to be located in genomic regions where inherited eye diseases have been mapped to (see Table 3) [68], [69], [70], [71], [72], [73], [74], [75], [76]. However, each of these mapped regions harbor multiple genes. Our study thus could assist researchers to pin down the causal genes underlying these diseases.
Table 3.
Candidate genes whose human ortholog were found to be located in the genomic regions mapped by congenital eye diseases in humans.
| Gene symbol | Mapped congenital eye diseases (OMIM ID) | References |
|---|---|---|
| High confidence genes | ||
| Kcnj14 | Cataract 35 (OMIM: 609376) | [68] |
| Ppm1n | Cataract 35 (OMIM: 609376) | [68] |
| Medium confidence genes | ||
| Adal | Microphthalmia with coloboma 2 (OMIM: 605738) |
[69], [70] |
| Ccdc126 | Dominant cystoid macular dystrophy (OMIM: 153880) | [72] |
| Dhx32 | Cone-rod dystrophy 17 (OMIM: 615163) | [73] |
| Frmpd2 | Usher syndrome, type IK (OMIM: 614990) | [74] |
| Lyg2 | Glaucoma 1B, primary open angle, adult onset; (OMIM: 606689) | [75] |
| Otor | Glaucoma 1 K, primary open angle, juvenile-onset (OMIM 608696) | [76] |
| Wdr17 | Retinitis pigmentosa 29 (OMIM: 612165) | [71] |
It should be noted that except for Ankrd33 (Fig. 3), a more detailed phenotyping had not been performed for the rest of 12 genes whose predicted functions were validated by observing the gross phenotype of developing eyes in the zebrafish system (Fig. 2). In the future, a more careful examinations of the consequences of gain- or loss-of-functions mutations of these genes, especially in mammals, would be desired. In addition to the identification of eye candidate genes, our combined transcriptome-phenome approach could be used to identify genes related with functions of non-eye tissues. With improvements and greater availability of both expression and phenotype data in mouse tissues in the future, utilization of our presented approach is expected to accelerate the elucidation of functional profiles of a substantial proportion of mammalian coding genes, and potentially noncoding genes when their target information becomes available.
Funding
This work was supported by intramural funding from the National Health Research Institutes and research grants from the Ministry of Science and Technology [grant numbers 104-2311-B-400-002-MY3, 105-2314-B-400-021-MY4] to B.-Y.L.
Author contributions
Y-H.C. and B-Y.L. conceived the study. C-Y.C. and B-Y.L. designed computational experiments. Y-H.C., M.-D.L. and B-Y.L. designed and supervised validation experiments and data analyses. C-Y.C. and T-Y.C. performed bioinformatic analyses. C-Y.C., L-S.H., Y-S.Y., P-Y.K, I.M. performed/assisted validation experiments. C-Y.C., Y-H.C., T-Y.C., L-S.H., M.-D.L. and B-Y.L. analyzed data. C-Y.C., Y-H.C., M.-D.L. and B-Y.L. wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Meng-Pin Weng for assisting the bioinformatic analysis on mouse phenotypes.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.12.009.
Contributor Information
Ming-Der Lin, Email: mingder@gms.tcu.edu.tw.
Ben-Yang Liao, Email: liaoby@nhri.org.tw.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Su A.I. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrest A.R.R. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huttlin E.L. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hrabe de Angelis M.H. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25(4):444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 5.Diez-Roux G. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9(1) doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams M.D., Sekelsky J.J. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat Rev Genet. 2002;3(3):189–198. doi: 10.1038/nrg752. [DOI] [PubMed] [Google Scholar]
- 7.Liao B.-Y., Weng M.P. Functionalities of expressed messenger RNAs revealed from mutant phenotypes. Wiley Interdiscip Rev RNA. 2016;7(4):416–427. doi: 10.1002/wrna.1329. [DOI] [PubMed] [Google Scholar]
- 8.Meehan T.F. Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nat Genet. 2017;49(8):1231–1238. doi: 10.1038/ng.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tosh C.R., McNally L. The relative efficiency of modular and non-modular networks of different size. Proc Biol Sci. 2015;282(1802) doi: 10.1098/rspb.2014.2568. pii: 20142568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner G.P. Homologues, natural kinds and the evolution of modularity. Am Zool. 1996;36(1):36–43. [Google Scholar]
- 11.Stuart J.M. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 13.Oldham M.C. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao B.-Y., Weng M.P. Unraveling the association between mRNA expressions and mutant phenotypes in a genome-wide assessment of mice. Proc Natl Acad Sci USA. 2015;112(15):4707–4712. doi: 10.1073/pnas.1415046112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb T.D. Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci. 2007;8(12):960–976. doi: 10.1038/nrn2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gharaibeh R.Z. Background correction using dinucleotide affinities improves the performance of GCRMA. BMC Bioinf. 2008;9:452. doi: 10.1186/1471-2105-9-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerfield M. 4 ed. Univ. of Oregon Press; 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 22.Eisen J.S., Smith J.C. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135(10):1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 23.Sabaliauskas N.A. High-throughput zebrafish histology. Methods. 2006;39(3):246–254. doi: 10.1016/j.ymeth.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Lattin J.E. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue F. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Care M.A. Parsimonious Gene Correlation Network Analysis (PGCNA): a tool to define modular gene co-expression for refined molecular stratification in cancer. NPJ Syst Biol Appl. 2019;5:13. doi: 10.1038/s41540-019-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P., Horvath S. Fast R functions for robust correlations and hierarchical clustering. J Stat Softw. 2012;46(11):1–17. [PMC free article] [PubMed] [Google Scholar]
- 29.Blondel V.D. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008:P10008. [Google Scholar]
- 30.Bello S.M. Allele, phenotype and disease data at Mouse Genome Informatics: improving access and analysis. Mamm Genome. 2015;26(7–8):285–294. doi: 10.1007/s00335-015-9582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson P.N. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83(5):610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueno A. Lrit1, a retinal transmembrane protein, regulates selective synapse formation in cone photoreceptor cells and visual acuity. Cell Rep. 2018;22(13):3548–3561. doi: 10.1016/j.celrep.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Omori Y. Samd7 is a cell type-specific PRC1 component essential for establishing retinal rod photoreceptor identity. Proc Natl Acad Sci USA. 2017;114(39):E8264–E8273. doi: 10.1073/pnas.1707021114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renninger S.L. Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur J Neurosci. 2011;33(4):658–667. doi: 10.1111/j.1460-9568.2010.07574.x. [DOI] [PubMed] [Google Scholar]
- 35.Jin X. Novel compound heterozygous mutation in the CNGA1 gene underlie autosomal recessive retinitis pigmentosa in a Chinese family. Biosci Rep. 2016;36(1) doi: 10.1042/BSR20150131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dryja T.P. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA. 1995;92(22):10177–10181. doi: 10.1073/pnas.92.22.10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billingsley G. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79(4):702–709. doi: 10.1086/507712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J. Molecular genetic analysis of Pakistani families with autosomal recessive congenital cataracts by homozygosity screening. Invest Ophthalmol Vis Sci. 2017;58(4):2207–2217. doi: 10.1167/iovs.17-21469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiao X. A common ancestral mutation in CRYBB3 identified in multiple consanguineous families with congenital cataracts. PLoS ONE. 2016;11(6) doi: 10.1371/journal.pone.0157005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiagalingam S. Novel mutations in the KCNV2 gene in patients with cone dystrophy and a supernormal rod electroretinogram. Ophthalmic Genet. 2007;28(3):135–142. doi: 10.1080/13816810701503681. [DOI] [PubMed] [Google Scholar]
- 41.Aydin I.T. Coat color dilution in mice because of inactivation of the melanoma antigen MART-1. Pigment Cell Melanoma Res. 2012;25(1):37–46. doi: 10.1111/j.1755-148X.2011.00910.x. [DOI] [PubMed] [Google Scholar]
- 42.Piri N. A substitution of G to C in the cone cGMP-phosphodiesterase gamma subunit gene found in a distinctive form of cone dystrophy. Ophthalmology. 2005;112(1):159–166. doi: 10.1016/j.ophtha.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Kohl S. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012;91(3):527–532. doi: 10.1016/j.ajhg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee A. A novel splice variant of the Excitatory Amino Acid Transporter 5: cloning, immunolocalization and functional characterization of hEAAT5v in human retina. Neurochem Int. 2016 doi: 10.1016/j.neuint.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Chograni M. Autosomal recessive congenital cataract, intellectual disability phenotype linked to STX3 in a consanguineous Tunisian family. Clin Genet. 2015;88(3):283–287. doi: 10.1111/cge.12489. [DOI] [PubMed] [Google Scholar]
- 46.Plotnikov D. A commonly occurring genetic variant within the NPLOC4-TSPAN10-PDE6G gene cluster is associated with the risk of strabismus. Hum Genet. 2019;138(7):723–737. doi: 10.1007/s00439-019-02022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel R. Novel mutations in the crystallin gene in age-related cataract patients from a north indian population. Mol Syndromol. 2017;8(4):179–186. doi: 10.1159/000471992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan A.O. Phenotypes of recessive pediatric cataract in a cohort of children with identified homozygous gene mutations (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2015;113:T7. [PMC free article] [PubMed] [Google Scholar]
- 49.Manes G. Mutations in IMPG1 cause vitelliform macular dystrophies. Am J Hum Genet. 2013;93(3):571–578. doi: 10.1016/j.ajhg.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tucker B.A. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108(34):E569–E576. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozgul R.K. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89(2):253–264. doi: 10.1016/j.ajhg.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powell C. Zinc-binding domain-dependent, deaminase-independent actions of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 2 (Apobec2), mediate its effect on zebrafish retina regeneration. J Biol Chem. 2014;289(42):28924–28941. doi: 10.1074/jbc.M114.603043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivolta C. Mutation screening of the peropsin gene, a retinal pigment epithelium specific rhodopsin homolog, in patients with retinitis pigmentosa and allied diseases. Mol Vis. 2006;12:1511–1515. [PubMed] [Google Scholar]
- 54.Delyfer M.N. Transcriptomic analysis of human retinal detachment reveals both inflammatory response and photoreceptor death. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M.S. Positive selection rather than relaxation of functional constraint drives the evolution of vision during chicken domestication. Cell Res. 2016;26(5):556–573. doi: 10.1038/cr.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennenstuhl C. Targeted ablation of the Pde6h gene in mice reveals cross-species differences in cone and rod phototransduction protein isoform inventory. J Biol Chem. 2015;290(16):10242–10255. doi: 10.1074/jbc.M114.611921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vinos J. A G protein-coupled receptor phosphatase required for rhodopsin function. Science. 1997;277(5326):687–690. doi: 10.1126/science.277.5326.687. [DOI] [PubMed] [Google Scholar]
- 58.Ramulu P. Normal light response, photoreceptor integrity, and rhodopsin dephosphorylation in mice lacking both protein phosphatases with EF hands (PPEF-1 and PPEF-2) Mol Cell Biol. 2001;21(24):8605–8614. doi: 10.1128/MCB.21.24.8605-8614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rieke F. Characterization of Ca2+-binding protein 5 knockout mouse retina. Invest Ophthalmol Vis Sci. 2008;49(11):5126–5135. doi: 10.1167/iovs.08-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniguchi M. High expression of alternative transcript of granzyme M in the mouse retina. Neurosci Res. 1999;34(2):115–123. doi: 10.1016/s0168-0102(99)00036-x. [DOI] [PubMed] [Google Scholar]
- 61.Qi S. A mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is highly expressed in retina that exhibits a large area of intrinsic disorder. Mol Biol Rep. 2011;38(5):2869–2877. doi: 10.1007/s11033-010-9948-x. [DOI] [PubMed] [Google Scholar]
- 62.Krol J. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141(4):618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 63.Graur D. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 2013;5(3):578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fadool J.M., Dowling J.E. Zebrafish: a model system for the study of eye genetics. Prog Retin Eye Res. 2008;27(1):89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kleinjan D.A. Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 2008;4(2) doi: 10.1371/journal.pgen.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanuki R. Panky, a novel photoreceptor-specific ankyrin repeat protein, is a transcriptional cofactor that suppresses CRX-regulated photoreceptor genes. FEBS Lett. 2010;584(4):753–758. doi: 10.1016/j.febslet.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 67.El-Brolosy M.A., Stainier D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017;13(7) doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riazuddin S.A. A new locus for autosomal recessive nuclear cataract mapped to chromosome 19q13 in a Pakistani family. Invest Ophthalmol Vis Sci. 2005;46(2):623–626. doi: 10.1167/iovs.04-0955. [DOI] [PubMed] [Google Scholar]
- 69.Morle L. A locus for autosomal dominant colobomatous microphthalmia maps to chromosome 15q12-q15. Am J Hum Genet. 2000;67(6):1592–1597. doi: 10.1086/316894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michon L. Physical and transcript map of the autosomal dominant colobomatous microphthalmia locus on chromosome 15q12-q15 and refinement to a 4.4 Mb region. Eur J Hum Genet. 2004;12(7):574–578. doi: 10.1038/sj.ejhg.5201197. [DOI] [PubMed] [Google Scholar]
- 71.Hameed A. A new locus for autosomal recessive RP (RP29) mapping to chromosome 4q32-q34 in a Pakistani family. Invest Ophthalmol Vis Sci. 2001;42(7):1436–1438. [PubMed] [Google Scholar]
- 72.Saksens N.T. Dominant cystoid macular dystrophy. Ophthalmology. 2015;122(1):180–191. doi: 10.1016/j.ophtha.2014.07.053. [DOI] [PubMed] [Google Scholar]
- 73.Kamenarova K. A novel locus for autosomal dominant cone-rod dystrophy maps to chromosome 10q. Eur J Hum Genet. 2013;21(3):338–342. doi: 10.1038/ejhg.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaworek T.J. USH1K, a novel locus for type I Usher syndrome, maps to chromosome 10p11.21-q21.1. J Hum Genet. 2012;57(10):633–637. doi: 10.1038/jhg.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stoilova D. Localization of a locus (GLC1B) for adult-onset primary open angle glaucoma to the 2cen-q13 region. Genomics. 1996;36(1):142–150. doi: 10.1006/geno.1996.0434. [DOI] [PubMed] [Google Scholar]
- 76.Wiggs J.L. A genomewide scan identifies novel early-onset primary open-angle glaucoma loci on 9q22 and 20p12. Am J Hum Genet. 2004;74(6):1314–1320. doi: 10.1086/421533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.