Abstract
Translating CYP2D6 genotype to metabolizer phenotype is not standardized across clinical laboratories offering pharmacogenetic (PGx) testing and PGx clinical practice guidelines, such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG). The genotype to phenotype translation discordance between laboratories and guidelines can cause discordant cytochrome P450 2D6 (CYP2D6) phenotype assignments and, thus lead to inconsistent therapeutic recommendations and confusion among patients and clinicians. A modified‐Delphi method was used to obtain consensus for a uniform system for translating CYP2D6 genotype to phenotype among a panel of international CYP2D6 experts. Experts with diverse involvement in CYP2D6 interpretation (clinicians, researchers, genetic testing laboratorians, and PGx implementers; n = 37) participated in conference calls and surveys. After completion of 7 surveys, a consensus (> 70%) was reached with 82% of the CYP2D6 experts agreeing to the final CYP2D6 genotype to phenotype translation method. Broad adoption of the proposed CYP2D6 genotype to phenotype translation method by guideline developers, such as CPIC and DPWG, and clinical laboratories as well as researchers will result in more consistent interpretation of CYP2D6 genotype.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Translating CYP2D6 genotype to metabolizer phenotype is not standardized across clinical laboratories offering pharmacogenetic (PGx) testing and PGx clinical practice guidelines.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ The purpose of this project was to harmonize the systems used by the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the Dutch Pharmacogenetics Working Group (DPWG) and reach consensus among an international panel of CYP2D6 experts regarding the standardization of how to translate CYP2D6 genotype to phenotype.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ We engaged a diverse group of international CYP2D6 experts to establish a standardized method for translating CYP2D6 genotype to metabolizer phenotype.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Broad adoption of the proposed CYP2D6 genotype to phenotype translation method by guideline developers, such as the CPIC and DPWG, and clinical laboratories as well as researchers will result in more consistent interpretation of CYP2D6 genotype.
Cytochrome P450 2D6 (CYP2D6) is directly involved in the metabolism of ~ 20% of currently approved medications,1 and genetic variation in the CYP2D6 gene has been implicated in the efficacy and/or toxicity of many drugs. Consequently, the highly polymorphic CYP2D6 gene is the focus of several Clinical Pharmacogenetics Implementation Consortium (CPIC) and/or Dutch Pharmacogenetics Working Group (DPWG) clinical practice guidelines on 15 widely used medications, including selective serotonin reuptake inhibitors, tricyclic antidepressants, atomoxetine, codeine, tramadol, tamoxifen, ondansetron, and tropisetron.2, 3, 4, 5, 6, 7, 8 Recently, the CPIC and DPWG reported some discrepancies in their guidelines, primarily related to how certain CYP2D6 genotypes or diplotypes (from here on referred to as “genotype”) were translated into metabolizer phenotype or metabolizer status (from here on referred to as “phenotype”).9 Figure 1 describes the process used to translate identified CYP2D6 genetic variants into phenotypes. Given that the clinical recommendations for CYP2D6 in the CPIC and DPWG guidelines are based on phenotype, the assignment of CYP2D6 phenotype based on genotype is a critical aspect for consistent clinical implementation.
Figure 1.
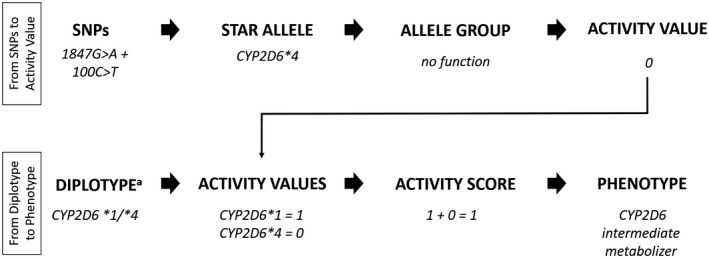
Process for translation of CYP2D6 genotype to phenotype. Diplotype describes the combination of two alleles (or haplotypes), which can involve multiple variants. Diplotype and genotype, a term that technically describes variation at a single nucleotide polymorphism (SNP), are often used interchangeably. Because genotype is the more commonly used term, it is used throughout this report. AS, activity score.
Translating CYP2D6 genotype to phenotype is also not standardized across clinical laboratories offering pharmacogenetic (PGx) testing. Current systems used to translate genotype to phenotype rely on the star (*) allele nomenclature (defining which variant(s) are present in an allele), and the assignment of function to the star alleles (i.e., increased, normal, decreased, or no function) with inferring phenotype based on the identified genotype. Some systems utilize the activity score (AS) system for assignment of phenotype where each allele is assigned an “activity value” ranging from 0−1 (e.g., 0 for no function, 0.5 for decreased function, and 1.0 for normal function).10 In addition, given that the CYP2D6 allele can also have variable copy number, the activity value of an allele is multiplied by the number of gene copies (i.e., ×2, ×3, etc.) if copy number is known. If copy number is not known, a sample maybe defaulted to ×2 assignment or shown as xN. As such, the CYP2D6 AS is the sum of the activity values assigned to each allele10 (Figure 1 ). Within the CPIC guidelines, the CYP2D6 AS is then translated into a phenotype using the following classification system: individuals with an AS of 0 are poor metabolizers (PMs), those with a score of 0.5 are intermediate metabolizers (IMs), those with a score of 1.0, 1.5, and 2.0 are normal metabolizers (NMs), and those with a score > 2 are ultrarapid metabolizers (UMs; later referred to as the “CPIC method”; Table 1 ).
Table 1.
Comparison of systems used for CYP2D6 genotype to phenotype translation
| CPIC | DPWG | System 1a (n = 1) | System 2 (n = 1) | System 3 (n = 3) | System 4 (n = 1) | |
|---|---|---|---|---|---|---|
| AS | ||||||
| UM | > 2 | > 2.5 | ≥ 3 | ≥ 3 | > 2 | Not tested |
| NM to UM | 2.25 < x < 3 | 2.5 | ||||
| NM | 1−2 | 1.5−2.5 | 1.75 ≤ x ≤ 2.25 | 2 | 1.5−2 | 1b to 2 |
| IM to NM | 1.25 < x < 1.75 | 1.5 | ||||
| IM | 0.5 | 0.5−1 | 0.75 ≤ x ≤ 1.25 | 1 | 0.5−1 | 0.5−1b |
| PM to IM | 0 < x < 0.75 | 0.5 | ||||
| PM | 0 | 0 | 0 | 0 | 0 | 0 |
AS, activity score; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenomics Working Group; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
AS ranges shown in gray are not reported. n refers to the number of laboratories that reported using the system.
aThis laboratory utilizes a propriety system of values to determine AS. bNM AS = 1 is a combination of a fully functional allele plus a no function allele; IM AS = 1 is a combination of two decreased function alleles.
Although the CPIC guidelines and some clinical laboratories categorize an AS of 1.0 as a CYP2D6 NM, other clinical laboratories and the DPWG guidelines consider an AS of 1.0 as CYP2D6 IMs6, 7, 8, 11 (Table 1 ). Differences also exist for the AS value that separates NMs from UMs. Consequently, the different ways of inferring CYP2D6 phenotype between laboratories and guidelines can cause discordant CYP2D6 phenotype assignments and, thus, lead to inconsistent therapeutic recommendations. To minimize confusion, it is important to maintain standardized CYP2D6 phenotype translation from genotype data. As such, the purpose of this project was to harmonize the systems used by the CPIC and DPWG and reach consensus among an international panel of CYP2D6 experts regarding the standardization of how to translate CYP2D6 genotype to phenotype.
Materials and Methods
The Delphi survey technique is an established approach for seeking expert consensus on a given topic.12, 13, 14 The method uses a series of repeated structured questionnaires or “rounds.” Each round provides written, systematic refinement of expert opinion, where feedback of group opinion is provided after each round.15 Delphi survey technique guidelines proposed by Hasson et al.16 were consulted in the design of the project. The method used is this study is often referred to as a “modified‐Delphi” as a major modification to the Delphi technique consists of beginning the process with a set of carefully selected options vs. an open‐ended questionnaire.
For the Delphi method used (Figure 2 ), CYP2D6 expert members of the CPIC and DPWG were solicited by email invitation, as well as other international investigators with published expertise in CYP2D6 PGx. In addition, experts were solicited by posting a description of the project on the PharmGKB and CPIC websites.
Figure 2.
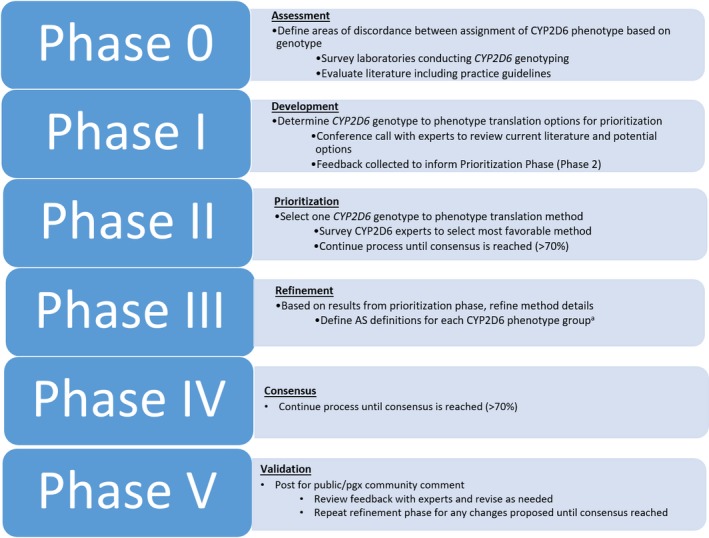
Modified Delphi process. aComments from each round were made available to all experts and discussed on conference calls. AS, activity score.
Experts were invited to participate in a series of surveys using an internet‐based survey tool (SurveyMonkey, Palo Alto, CA; http://www.surveymonkey.com), supplemented with multiple live webinars that were used to explain the survey and solicit feedback. The webinars were designed to facilitate understanding of the survey to encourage completion; toward the end of the process an additional webinar was used to assist in developing consensus. Each survey also included questions regarding the expert's workplace setting and degree of CYP2D6 expertise (i.e., role in clinical PGx, time devoted to CYP2D6). Responses were included in the analysis if the respondent provided their name and contact information, which were necessary to enable follow‐up with the respondent for the subsequent round (but not disclosed). Responses were tabulated as numeric counts and frequencies for each phase to determine whether consensus was reached. Consensus was defined as 70% of experts agreeing; this level of agreement has been considered appropriate in previous Delphi studies.17, 18, 19
Phase 0: Assessment
The objective of the assessment phase was to define areas of discordance among assignments of CYP2D6 phenotype based on genotype. The Genetic Testing Registry20 was queried for laboratories performing clinical CYP2D6 genetic testing, and emails requesting participation in a survey were sent to each laboratory. The survey consisted of 16 questions regarding CYP2D6 genotype interpretation, including questions regarding current methods used to translate CYP2D6 genotype to phenotype (see ref. 21 for the laboratory survey questions). Clinical practice guidelines (CPIC and DPWG) were evaluated for systems used to translate genotype to phenotype. References used in the evidence tables of the CPIC guidelines and additional literature were evaluated for differences in AS between 0.5 vs. 1 and 1 vs. 2, and consequences of CYP2D6*10‐containing genotypes on AS assignment. Results were presented to the CYP2D6 experts on the first conference call.
Phase I: Development
The objective of the development phase was to determine CYP2D6 genotype to phenotype translation options for evaluation and assess initial expert opinions on current systems being used by clinical laboratories and available PGx guidelines. Given that the discordance between genotype to phenotype translation is mostly related to the AS of 0.5, 1, and 2 and disagreements regarding the activity value assigned to CYP2D6*10, the first expert conference call provided examples of pharmacokinetic studies with AS data and additional studies comparing CYP2D6*10 activity (see ref. 21 for this spreadsheet). Experts were required to either attend the live conference call or to listen to the recorded version and asked to provide feedback and additional references if warranted.
Phase II: Prioritization
The expert opinions discussed in the development phase were used to inform the prioritization phase with the final objective to select a genotype to phenotype translation system to which at least 70% of the experts agreed upon. Survey 1 asked specifically if experts thought there was a clinically significant difference between AS of 1 vs. 2 and 0.5 vs. 1, and if there was a rationale to use a lower activity value for AS calculation (i.e., “downgrade” the activity value from 0.5 to 0.25) for some CYP2D6 alleles (e.g., CYP2D6*9, *10, *17, *29, and *41) to more accurately reflect activity relative to other CYP2D6 alleles. Survey 1 also presented five different systems for translating CYP2D6 genotype to phenotype to assess expert opinion of each system. All questions required expert explanation, references, and examples to support the opinion. The results from Survey 1 were presented on a conference call and discussed, and a subsequent call presented two methods for assigning AS (i.e., AS ordinal groups vs. continuous percentage activity values). The results from Surveys 2 and 3 were used to prioritize one method to move into the refinement phase. Results including expert comments from previous surveys were provided with each survey.
Phases III and IV: Refinement and consensus
Based on the results from Survey 3, Surveys 4−6 were used to refine the details of the selected approach. Experts were asked a series of questions related to AS definitions for each CYP2D6 phenotype. A summary of comments from previous surveys was provided and experts were asked to review the comments prior to responding to subsequent questions.
Phase V: Validation
Once consensus was reached, results were presented on a member‐wide CPIC call and posted to the CPIC website for 2 months to allow for public comment. PharmGKB also blogged about the project and solicited feedback. Feedback was presented to the experts on a subsequent conference call and discussed. Survey 7 measured acceptance of incorporation of the feedback into the previous consensus system. The final survey (Survey 8) measured the level of acceptance of the final CYP2D6 genotype to phenotype translation system.
Results
Expert panel composition
A total of 37 CYP2D6 experts participated in the project with 27 completing Survey 1, 28 completing Survey 2, 24 completing Survey 3, 25 completing Survey 4, 27 completing Survey 5, 31 completing Survey 6, 23 completing Survey 7, and 27 completing Survey 8. Not all experts participated in each round and some experts participated in the initial or early rounds but not in the later rounds or vice versa. The participants represented a diverse group of self‐identified experts with varying levels of CYP2D6 expertise (Table 2 ) and an international representation: 59% were from the United States, 27% from Europe, and 11% from other countries (i.e., South Korea, Japan, Canada, and Australia). The study was facilitated by representatives from both the CPIC (n = 5) and the DPWG (n = 3) leadership.
Table 2.
Key demographics of CYP2D6 experts
| No. (%) respondents (n = 37) | |
|---|---|
| Workplace setting | |
| Laboratory test interpretation | 3 (8) |
| Nonprofit or academic hospital | 14 (38) |
| Reference/clinical laboratory | 7 (19) |
| Research or clinical institute | 3 (8) |
| University | 10 (27) |
| Time related to work involving CYP2D6 | |
| 0–5% | 2 (5) |
| 6–25% | 16 (43) |
| 26–50% | 11 (30) |
| 51–75% | 6 (16) |
| 76–100% | 2 (5) |
Phases 0 and I: Assessment and development
Email invitations were sent to 43 clinical testing laboratories who reported performing CYP2D6 genotype testing to the Genetic Testing Registry. A total of 15 laboratories completed a survey regarding how their laboratory translated CYP2D6 genotype to phenotype. Of those, 47% (n = 7) reported using the CPIC method for translating CYP2D6 genotype to phenotype (i.e., AS of 1.0 is classified as NM). Of the eight laboratories (53%) not using the CPIC method (i.e., AS of 1.0 is classified as IM), six disclosed their CYP2D6 genotype to phenotype translation methods (Table 1 ). Full results can be found in ref. 21. Experts participated in an initial conference call during which results from the laboratory survey were reported, evidence supporting differences in AS of 0.5, 1, and 2 was presented, and available information regarding CYP2D6*10 activity was shared. Finally, options for a system for translating genotype to phenotype were discussed.
Phase II:Prioritization
In Survey 1, 93% (n = 25) of the experts agreed that there is a clinically significant difference between a CYP2D6 AS of 1 and 2, and 78% (n = 21) agreed that there also is a significant difference between an AS of 0.5 and 1. Among the experts agreeing to the need to downgrade some alleles to an activity value of 0.25 (53%; n = 14), 85% selected CYP2D6*10 and 50% selected CYP2D6*41. Based on the first conference call discussion, Survey 1 included five potential options to move forward ( Supplemental Figure S1 ). However, no method reached consensus (> 70%). Comments and Survey 1 results were made available to all participants and discussed on the second conference call. Based on feedback provided after the second call, a third call was held to discuss using a “percentage activity system” vs. the AS system (see Discussion). After receiving feedback from several of the experts, the CPIC and the DPWG recommended to the experts to proceed with the use of the AS system to which 94% (n = 29) agreed in Survey 2. Also in Survey 2, 42% (n = 12) preferred a system that classifies AS of 0.5 and 1 as IMs, 38% (n = 11) preferred to create a new phenotype group for AS of 1, 7% (n = 2) thought both methods would be acceptable, and 14% (n = 4) recommended another method. Experts were asked to provide their rationale for their responses and Survey 2 results were discussed on a conference call. Survey 2 results can be found in ref. 22. Based on the conference call discussion and Survey 2 results, CPIC and DPWG representatives recommended to proceed with the use of AS and to downgrade the activity value of some alleles (currently limited to CYP2D6*10). Using an activity value of 0.25 for AS calculation to more accurately reflect the considerably decreased activity of CYP2D6*10 results in the introduction of additional AS groups. In Survey 3, the majority of experts (96%) agreed to create an activity value of 0.25 category, and 88% agreed to the assignment of AS 0.5−1 as an IM.
Phases III and IV: Refinement and consensus
Surveys 4 and 5 were used to refine the new system discussed above. Specifically, the experts discussed how to integrate the new AS groups (i.e., 0.25, 0.75, 1.25, 1.75, and 2.25) that are introduced by the addition of an activity value of 0.25 for AS calculations into the four phenotype categories (i.e., PM, IM, NM, and UM). As shown in Supplemental Table S1 , Survey 5 included two options. Fourteen (52%) of the experts chose option 1, 11 (41%) chose option 2, and 2 (7%) disagreed with both options. Because the experts favored option 1 (52% vs. 41%), the CPIC and DPWG representatives recommended option 1 on Survey 6; this decision was supported by expert comments regarding the small contribution of an AS of 0.25 to clinically appreciable activity to the overall function. Experts agreed with the option and consensus was reached (27 (87%) agreed, whereas 4 experts (13%) disagreed; Table 3 ).
Table 3.
Final consensus CYP2D6 genotype to phenotype translation compared to previously reported CPIC and DPWG methods
| Inferred CYP2D6 phenotype | Previous CPIC definition (AS) | Previous DPWG definition (AS) | Consensus definition (AS) | Consensus contiguous definition (AS) | Examples of CYP2D6 diplotypes for consensus translation method |
|---|---|---|---|---|---|
| UM | > 2 | > 2.5 | > 2.25 | > 2.25 | *1/*1xN, *1/*2xN b , *2 a /*2xN b , *1x2/*9 |
| NM | 1–2 | 1.5–2.5 | 1.25 | 1.25 ≤ x ≤ 2.25 | *1/*10 |
| 1.5 | *1/*41, *1/*9 | ||||
| 2.0 | *1/*1, *1/*2 | ||||
| 2.25 | *2x2/*10 | ||||
| IM | 0.5 | 0.5–1 | 0.25 | 0 < x < 1.25 | *4/*10 |
| 0.5 | *4/*41, *10/*10 | ||||
| 0.75 | *10/*41 | ||||
| 1 | *41/*41, *1/*5 | ||||
| PM | 0 | 0 | 0 | 0 | *3/*4, *4/*4, *5/*5, *5/*6 |
AS, activity score; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenomics Working Group; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
a CYP2D6*2 is currently considered to be a normal function allele by CPIC and DPWG; however, this function assignment has been challenged32 and some laboratories report CYP2D6*2 function differently. Function of this allele will be reassessed as additional data become available. bN is categorical and indicates the number of copy variants (e.g., *1x2, *1x3, etc).
Phase V: Validation
After 2 months of accepting public comments, 2 issues were revisited for consideration: (i) inclusion of a rapid metabolizer (RM) phenotype group between NM and UM; and (ii) use of contiguous AS values to define each phenotype (i.e., no gaps between AS categories). After discussion on a conference call and Survey 6, 87% (n = 20) of the experts rejected the introduction of an RM phenotype group, whereas 70% agreed to use contiguous AS ranges to define CYP2D6 phenotype based on genotype. Experts were also asked regarding the range for PMs: the majority (61%; n = 14) favored to define PMs as having two no function alleles (AS = 0), 30% (n = 7) favored defining AS ≤0.25 as PMs, and 9% indicated “I do not know.” Results were discussed on a subsequent conference call on which the experts also discussed and agreed on the contiguous ranges for the other phenotype groups (Survey 7; 82% of the participants (n = 22) agreeing to the final assignments shown in Table 3 ).
Discussion
We engaged a diverse group of international CYP2D6 experts to establish a standardized method for translating CYP2D6 genotype to metabolizer phenotype. The major focus of this working group was to harmonize how to translate CYP2D6 genotype into phenotype; a secondary aim was to explore how currently used systems could be improved. This international group of experts consisted of representatives of academia and industry, including clinical genetic testing laboratories. In addition, individuals with experience in implementing CYP2D6 PGx into clinical practice and electronic health records at large hospitals were included to assess the impact of the project on past or ongoing CYP2D6 implementation efforts. Importantly, the final CYP2D6 translation method presented in Table 3 will be incorporated into the CYP2D6 tables on http://www.cpicpgx.org and used in all new and updated CPIC and DPWG guidelines. We also recommend that this system be considered as standard practice across all areas of clinical PGx, including clinical genetic testing laboratories. We also strongly encourage PGx researchers to use this standardized method to report their findings as this will greatly facilitate future data collection from the literature and comparison of data.
Throughout the project several issues and challenges were identified and discussed in detail (Table 4 ) as follows. (i) Lengthy discussions entailed the possibility of generating a new phenotype group for AS = 0.5 as patients with genotypes consisting of one decreased and one nonfunctional allele seem to have lower activity compared with those with genotypes giving rise to an AS of 1. Concerns were raised that combining AS of 0.5 and 1.0 in research studies may mask potentially significant differences among these AS groups because there are considerably fewer subjects with an AS of 0.5. The introduction of a new phenotype group describing patients between PM and IM was, however, rejected by the CPIC and DPWG representatives based on the CPIC term standardization project, which determined that five phenotype groups are sufficient18; the majority of experts also rejected the introduction of an additional phenotype group. (ii) A number of factors weighed into the decision to reclassify AS of 1 from NM to IM. Because published studies vary on how subjects with an AS of 1 are grouped (NM vs. IM), it is difficult to compare AS of 0.5–1 vs. 2 or AS of 1 vs. 2 with confidence to support differences in outcomes between these groups. In addition, more laboratories also currently classify an AS of 1 as IMs and not NMs (Table 1 ), indicating that classifying an AS of 1 as IM may be minimally disruptive to most research and clinical laboratories. (iii) Although the goal is to have a translation system that is agnostic to the drug used, the experts realized that certain genotypes may need recommendations that differ from their “drug‐agnostic” phenotype group assignment. To address this challenge, recommendations from the CPIC and the DPWG can be different for certain drugs (see CYP2D6*10‐containing genotypes in CPIC tamoxifen guideline for example3), or for a particular AS group if warranted. In other words, a recommendation can be based on the AS vs. the phenotype group. Therefore, it is extremely important that clinical laboratories not only report phenotype but also detail the patient's genotype and sequence variations tested (see Bousman et al.22 for guidance of how to select a PGx test).
Table 4.
Discussion points raised by CYP2D6 experts during Delphi process
| Discussion points | Pros | Cons |
|---|---|---|
| Addition of CYP2D6 RM phenotype |
|
|
| Addition of new phenotype group between IM and PM |
|
|
| Changing AS of 1 from NM to IM |
|
|
| Creation of new activity value (0.25) |
|
|
| Contiguous AS scale |
|
|
| Percentage activity system |
|
|
AS, activity score; CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Dutch Pharmacogenetics Working Group; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; RM, rapid metabolizer; UM, ultrarapid metabolizer.
A secondary goal of the project was to re‐evaluate the activity values assigned to alleles with decreased function. Currently, a value of 0.5 bins decreased function alleles together regardless of the percentage activity they retain compared to the CYP2D6.1 (wild‐type) protein product. The majority of values used today for AS calculation are based on the original report by Gaedigk et al.10 that was published 11 years ago. The prospect of lowering the value assigned to CYP2D6*10, an allele that is anecdotally known to have “little” activity was reviewed earlier, but the authors did not find sufficient evidence to downgrade this allele based on evidence available 6 years ago.23 Since then, there was mounting evidence suggesting that CYP2D6*10 not only consistently conveys decreased function across substrates, but also seems to be, on average, considerably lower compared with other decreased function alleles. Thus, using an activity value of 0.5 for AS calculation for CYP2D6*10‐containing genotypes may overestimate the metabolic capacity of patients with CYP2D6*10/*10 or *10/no function genotypes. Assigning an activity value of 0.25 to the CYP2D6*10 allele for AS calculation will group CYP2D6*10/*10 as AS = 0.5 and *10/no function as AS = 0.25 (opposed to AS = 1 and AS = 0.5, respectively), which more precisely aligns with the level of reduction of enzyme activity.
Notably, there are other star (*) alleles that harbor the CYP2D6*10 defining variant (100C>T; rs1065852) in combination with other variants that, to the best of current knowledge, do not impact function or have decreased function on their own (e.g., 1023C>T), which are currently classified by the CPIC as “decreased” function and, thus, receive a value of 0.5 for AS calculation (e.g., CYP2D6*49, *54, *65, and *72). Note that positions are provided according to the genomic CYP2D6 RefSeq NG_008376.3, the numbering system recommended by PharmVar.24 There are also a number of alleles with g.100C>T that are currently labeled as “uncertain” function (e.g., *37, *52, *64, *87, *94, and *95). Most experts recommended that these alleles should also receive an activity value of 0.25; however, concerns were raised by some of the experts regarding the lack of evidence (i.e., in vitro or in vivo studies) for most of these alleles (e.g., 100C>T in combination with other SNP(s) may obliterate function, or compensate for the decreased function caused by 100C>T).10, 25 Thus, other CYP2D6 alleles containing the 100C>T variant besides *10 will be assessed as part of future CPIC guideline development. At that time, functional status and values for AS calculations will be assigned for these alleles; other alleles will also be reviewed and re‐assessed during this process.
It was also discussed whether genotype to phenotype translation should be standardized across all CYP450 enzymes. Currently, the AS is applied to CYP2D6 for which it was originally devised to accommodate a large (and growing) number of alleles with varying activity and was widely adopted after being published10; hence, it was a natural decision for the CPIC to adopt this system. The AS was eventually also adopted for DPYD gentoype to phenotype translation to accommodate the vast number of sequence variants that emerged for this gene. As shown in Supplemental Table S2 , other CYP genes have their distinct systems to translate genotype to phenotype. There was no consensus among the group whether this would be a desirable goal because a major revision toward a CYP‐wide system may pose a major challenge for clinical reporting and implementation with unclear benefits.
Feedback included the suggestion to add an RM phenotype group. One argument for having an RM phenotype group was that certain genotypes may have increased activity compared with NMs (e.g., *1x2/*41), but less than UMs (e.g., *1x2/*2); it was also argued that the introduction of an RM group would be in alignment with CYP2C19. However, the experts felt that there was not enough evidence to differentiate between two “increased function” phenotypes (rapid and ultrarapid) for CYP2D6 and, thus, these groups would not be clinically useful.
In 2016, the CPIC published a consensus project aimed to standardize terms describing allele function and phenotype.18 Prior to this project, various terms were used for allele function and phenotype, which impeded reporting and sharing of test results across clinical laboratories and electronic health records. Based on the results of this project, Systematized Nomenclature of Medicine‐Clinical Terms (SNOMED‐CT) and Logical Observation Identifiers Names and Codes (LOINC) terms were created for use in the electronic health record to facilitate efficient reporting of PGx results. Although the 2016 project did not address standardization of the translation of genotype to phenotype, PGx experts were asked whether they favor a four or five major category phenotype system. The majority of participants (91%; n = 48) agreed to four categories (see all survey results in ref. 21). The CYP2D6 experts and CPIC and DPWG representatives considered this result suggesting that adding an additional phenotype category may not be widely accepted by the PG community.
The use of a contiguous AS scale for defining metabolizer phenotype was addressed at two stages during the Delphi process. Early in the process (call #3, Survey 2), a group of experts advocated for an alternative system referred to as the “percentage activity” system. Similar to the AS, in the percentage activity system, each allele is assigned a value on the scale of 0 (no activity) to 1 (normal activity); however, in the percentage activity system, values are assigned in increments of 0.1 instead of 0.5 (now 0.25; Figure 3 ). In addition, instead of calculating the sum of the two activity values to calculate the AS, the values would be averaged and multiplied by 100 for the percentage activity system so that each patient's CYP2D6 metabolic capacity is described on a percentage activity scale of 0% (analogous to AS = 0) to 100% (AS = 2.0) or higher. It was argued that the percentage activity system may be more intuitive to clinicians. Although such a system may ultimately be more precise, there are a number of hurdles. For example, the determination of activity for an allele is difficult as is and to discriminate activity on a scale of 10% increments seems impossible as there are no data for the vast majority of alleles at this point in time. Second, there is a broad range of interindividual variability among subjects within the same genotype group10, 25 and third, even if activity could be determined on a 10% scale, percent activities may still need to be translated into a limited number of phenotyping categories for feasibility of clinical implementation.
Figure 3.
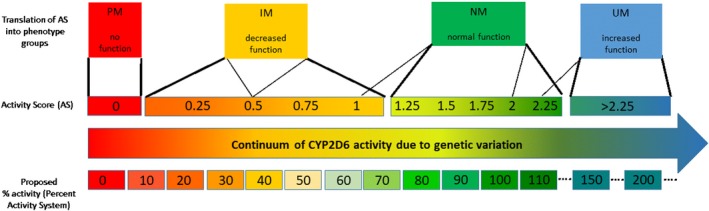
Comparison of the Clinical Pharmacogenetics Implementation Consortium method and percentage activity method for translating CYP2D6 genotype to phenotype. Thin lines represent different ways to translate activity score (AS) into phenotype and the bold lines represent the recommended CYP2D6 genotype to phenotype translation consensus system. IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
Given these challenges, the experts came to consensus on Survey 2 to move forward with the AS system mainly due to limited data for estimating percentage activity of individual alleles, with a general interest in future work that moves the field in the direction of more precise activity estimates as well as the prospect of developing more sophisticated dosing algorithms that are based on population pharmacokinetic and dynamic models taking genotype along other pertinent factors into account.
The second discussion of a “contiguous scale” system was held after the public comment period (Survey 7), after thresholds for each phenotype had already been agreed upon. Given the possibility of future allelic re‐estimates or percentage activities, the experts defined the consensus scale contiguously, such that all potential values of AS have a consensus phenotype translation. For example, given that an AS = 0 is PM and an AS = 0.25 is IM, would an AS = 0.2 be a PM or IM? Thus, the contiguous consensus scale (Table 3 ) can accommodate any future scores regardless of the number of groups or system used.
A central aim of this project was to continue the previously reported and ongoing efforts dedicated to standardizing inconsistent components related to clinical PGx, including genetic testing, interpretation, recommendations, and implementation.18, 26, 27, 28, 29 Importantly, we strongly encourage all PGx stakeholders to adopt the consent CYP2D6 translation system that has emerged from this project. Broad adoption of the proposed CYP2D6 translation system by clinical laboratories as well as researchers will ultimately lead to reduced interlaboratory discrepancies, increased consistency in CYP2D6 reporting, thus more consistent test interpretation. The performance of this system will also be measurable over time based on the metrics from the College of American Pathologists Pharmacogenetic Proficiency Survey, as CYP2D6 genotyping/phenotyping has historically had the greatest interlaboratory variability among the commonly tested PGx genes.30 However, we acknowledge that adopting this process, if distinct from a previous reporting protocol, may also result in laboratory cost and effort to modify workflows and reconcile previously reported CYP2D6 results based on prior translation systems.
Healthcare institutions that have already implemented CYP2D6 genotyping using the CPIC method will be affected by this new system as follows: (i) patients with a CYP2D6 AS of 1.0 who were previously assigned an NM phenotype will now have to be reassigned an IM phenotype and patients with a CYP2D6 AS of 2.25 who were previously assigned a UM phenotype assigned as NM; and (ii) CYP2D6 interpretive reports as well as all applicable educational materials pertaining to an AS of 1.0 (or 2.25) will need to be updated. Because the former change will necessitate substantial efforts in order to back‐track patients and inform them of their new phenotype assignment, some institutions may elect not to inform previously tested patients of their new re‐assigned CYP2D6 phenotype.
The Delphi method is a powerful tool that was developed to build consensus among and to develop standards across different disciplines.12, 13, 15 Key risks to the validity of a Delphi study include overestimating the expertise of participants and attrition across the consensus rounds. Given that each participant had some CYP2D6 PGx expertise and 51% of survey respondents indicated that they spend > 26% of their time devoted to work related to CYP2D6, we believe to have had adequate CYP2D6 expertise among our survey participants. Although attrition rates were not defined a priori, 76% of the experts participated in Survey 7 (participation averaged 74% for Surveys 1–6) and relative to other Delphi panels and the recommended minimum panel size, our final consensus panel was relatively large (suggested minimum for expert panels is 10 participants), which reinforces the validity of our results.31 To reduce bias, especially the authority or reputation of specific individuals, Delphi panel participants are often kept anonymous throughout the process. Although survey creators and analysts were not blinded to participants, identifying information was not shared among survey participants. The only occasions of participant identification were in between surveys when nonblinded email invitations were sent to participants in conference calls and webinars during which interim results were discussed. Because many survey results were close, the CPIC and DPWG representatives discussed options for the next survey based on previous results and comments from the experts, which resulted in recommendations of limited choices to move forward. However, experts still had to agree to the option.
In conclusion, consensus among an international panel of CYP2D6 experts regarding the standardization of translating CYP2D6 genotype to phenotype was achieved. Moving forward, the CPIC and DPWG will use this system in their practice guidelines. As most PGx clinical recommendations are based on phenotype, we anticipate that broad adoption of the proposed CYP2D6 genotype to phenotype translation framework will minimize discrepant CYP2D6 test results and inconsistent therapeutic recommendations.
Funding
This work was funded by the National Institutes of Health (NIH) for CPIC (R24GM115264; U24HG010135‐01), PharmGKB (R24GM61374), PharmVar (R24GM123930), and by the European Union's Horizon 2020 Research and Innovation Programme (U‐PGx: 668353).
Conflicts of Interest
The authors declared no competing interests for this work.
Author Contributions
K.E.C., K.S., M.W.‐C., J.J.S., C.E.H., T.E.K., R.S.G., M.V.R., S.A.S., D.L.H., H.‐J.G., and A.G. wrote the manuscript. K.E.C., K.S., M.W.‐C., J.J.S., R.S.G., M.V.R., H.‐J.G., and A.G. designed the research. K.E.C., K.S., M.W.‐C., and A.G. performed the research. K.E.C., K.S., M.W.‐C., and A.G. analyzed the data.
Supporting information
(Supplement to standardizing CYP2D6 genotype to phenotype translation: Consensus recommendations from the clinical pharmacogenetics implementation consortium (CPIC) and Dutch pharmacogenetics working Group)
Supplementary Materials. Figure S1, Tables S1‐S2.
Acknowledgments
The members of the CPIC are acknowledged for their support in this project and all the CYP2D6 experts that participated in the Delphi process. We would also like to acknowledge Dr. John Black at the Mayo Clinic for his insights during this project and his thoughtful review of this manuscript. All CPIC members are listed at https://cpicpgx.org/members/.
References
- 1. Saravanakumar, A. , Sadighi, A. , Ryu, R. & Akhlaghi, F. Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: a systematic review of the top 200 most prescribed drugs vs. the FDA‐approved drugs between 2005 and 2016. Clin. Pharmacokinet. 58, 1281–1294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown, J.T. et al Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 genotype and atomoxetine therapy. Clin. Pharmacol. Ther. 106, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goetz, M.P. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 103, 770–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell, G.C. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 102, 213–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crews, K.R. et al Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swen, J.J. et al Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 9. Bank, P.C.D. et al Comparison of the guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 103, 599–618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaedigk, A. et al The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 83, 234–242 (2008). [DOI] [PubMed] [Google Scholar]
- 11. AmpliChip™ CYP450 Package Insert. 2005.
- 12. Dalkey, N. & Helmer, O. An experimental application of the Delphi method to the use of experts. Manage. Sci. 9, 458–467 (1963). [Google Scholar]
- 13. Beretta, R. A critical review of the Delphi technique. Nurse Res. 3, 79–89 (1996). [DOI] [PubMed] [Google Scholar]
- 14. Green, B. , Jones, M. , Hughes, D. & Williams, A. Applying the Delphi technique in a study of GPs’ information requirements. Health Soc. Care Community 7, 198–205 (1999). [DOI] [PubMed] [Google Scholar]
- 15. von der Gracht, H.A. Consensus measurement in Delphi studies: review and implications for future quality assurance. Technol. Forecast. Soc. Chang. 79, 125–1536 (2012). [Google Scholar]
- 16. Hasson, F. , Keeney, S. & McKenna, H. Research guidelines for the Delphi survey technique. J. Adv. Nurs. 32, 1008–1015 (2000). [PubMed] [Google Scholar]
- 17. Slade, S.C. , Dionne, C.E. , Underwood, M. & Buchbinder, R. Standardised method for reporting exercise programmes: protocol for a modified Delphi study. BMJ Open 4, e006682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caudle, K.E. et al Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med. 19, 215–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henderson, E.J. & Rubin, G.P. Development of a community‐based model for respiratory care services. BMC Health Serv. Res. 12, 193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubinstein, W.S. et al The NIH genetic testing registry: a new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res. 41, D925–D935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clinical Pharmacogenetics Implementation Consortium (CPIC) . CYP2D6 genotype to phenotype standardization project. <https://cpicpgx.org/resources/cyp2d6-genotype-to-phenotype-standardization-project/>. (2019).
- 22. Bousman, C.A. , Zierhut, H. & Muller, D.J. Navigating the labyrinth of pharmacogenetic testing: a guide to test selection. Clin. Pharmacol. Ther. 106, 309–312 (2019). [DOI] [PubMed] [Google Scholar]
- 23. Hicks, J.K. , Swen, J.J. & Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metab. 15, 218–232 (2014). [DOI] [PubMed] [Google Scholar]
- 24. Gaedigk, A. et al The evolution of PharmVar. Clin. Pharmacol. Ther. 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montane Jaime, L.K. , Lalla, A. , Steimer, W. & Gaedigk, A. Characterization of the CYP2D6 gene locus and metabolic activity in Indo‐ and Afro‐Trinidadians: discovery of novel allelic variants. Pharmacogenomics 14, 261–276 (2013). [DOI] [PubMed] [Google Scholar]
- 26. Caudle, K.E. et al Standardization can accelerate the adoption of pharmacogenomics: current status and the path forward. Pharmacogenomics 19, 847–860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalman, L.V. et al Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin. Pharmacol. Ther. 99, 172–185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pratt, V.M. et al Recommendations for clinical CYP2C19 genotyping allele selection: a report of the association for molecular pathology. J. Mol. Diagn. 20, 269–276 (2018). [DOI] [PubMed] [Google Scholar]
- 29. Moriyama, B. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. 102, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu, A.H. Genotype and phenotype concordance for pharmacogenetic tests through proficiency survey testing. Arch. Pathol. Lab. Med. 137, 1232–1236 (2013). [DOI] [PubMed] [Google Scholar]
- 31. Okoli, C. & Pawlowski, S. The Delphi method as a research tool: an example, design considerations and applications. Inform. Manage. 42, 15–29 (2004). [Google Scholar]
- 32. Wang, D. et al Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long‐range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 23, 268–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Supplement to standardizing CYP2D6 genotype to phenotype translation: Consensus recommendations from the clinical pharmacogenetics implementation consortium (CPIC) and Dutch pharmacogenetics working Group)
Supplementary Materials. Figure S1, Tables S1‐S2.


