Abstract
Our objective was to build a mock pharmacogenomic (PGx) patient portal and assess its ability to disseminate test results and information to patients. The YourPGx Portal delivered four sample PGx results (omeprazole, simvastatin, clopidogrel, and codeine). We hosted two study groups to assess patient knowledge and perceptions of PGx before and after accessing the portal. Ten PGx‐tested and 10 traditional care participants were included (average 61 years, 60% women, 50% African American, and 55% had a bachelor's/advanced degree). Participants scored significantly higher on the post‐test compared with the pre‐test, with no significant differences between baseline scores or score change between the groups. Patient perceptions also improved after accessing the portal—more patients wanted their providers to have access to test results, and more patients would encourage family/friends to get PGx testing. Patients would share their test results with their healthcare providers, spouse/partner, and family; none would share results with their friends or social media. Almost all patients (95%) said the portal was easy to use and 65% said it was easy to understand. In this pilot study, patients’ knowledge and perceptions of PGx improved after accessing the YourPGx Portal.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THIS TOPIC?
☑ There is very little current knowledge on the effect of pharmacogenomic (PGx) patient portals on patients’ knowledge and perceptions of PGx.
WHAT QUESTIONS DID THIS STUDY ADDRESS?
☑ This study assessed patient knowledge and perceptions of PGx before and after assessing a patient results portal, and showed that the YourPGx Portal improved patient knowledge and perceptions of PGx.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ The results of this study indicate that a patient‐friendly portal can effectively deliver PGx results and information to patients, and that patients may act as advocates for their own personalized care when empowered with PGx information by sharing their results with different members of their healthcare team.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The results of this study justify further exploration of the role of allowing patients to have direct access to their own PGx test results and suggest a potentially positive impact of patient educational engagement within PGx implementation programs.
Patients today have access to numerous educational materials relating to their health care, however, these materials need to be readable, understandable, and applicable for patients to use this knowledge therein to actively engage in their own health care.1 Advances in health information technologies have increased the development of patient web portals to support patient self‐management of their health care and improve communication between patients and their providers.2, 3 However, patient portals pose additional barriers, such as complex visual layout and poor usability features.4 Nonetheless, if carefully constructed to overcome these barriers, a patient portal can better equip patients to participate more fully in their care: to manage their conditions and medications, coordinate care across multiple providers, and improve communication with different members of their healthcare team.3
Advances in next‐generation sequencing technologies and bioinformatics are driving the integration of genetics‐centered health care into clinical practice. Pharmacogenomics (PGx) is the study of how genetic variations affect individual responses to medications. Within the past decade, PGx approaches have been successfully implemented in several clinical trials, including The University of Chicago “The 1200 Patients Project,” which has demonstrated utility and positive outcomes in the outpatient care setting.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 In our previous institutional PGx implementation studies, PGx results have been primarily disseminated to providers who may or may not share the results with their patients; however, patients are often not provided direct access to their PGx results. The results of a single PGx test can reveal a patient's predisposition to many medications for the patient's entire lifetime, and, hence, understanding one's results could lead to improved outcomes, medication adherence, and patient satisfaction for decades.17 Therefore, there is a pressing need to develop additional tools to safely, effectively, and responsibly deliver PGx test results and information directly to patients with varying levels of health and genetic literacy.
Methods
Development of the YourPGx portal
The patient portal was modeled after the design of our institutional PGx result delivery tool used for providers, the Genomic Prescribing System (GPS).18 Like the GPS, the portal was designed to be user‐friendly by allowing patients to access different pages with as few clicks as possible. The portal used our traffic light iconography—red “warning,” yellow “caution,” and green “favorable” lights—to help patients quickly and easily identify their risk for each medication. A sample set of results were delivered from a simplified mock database. Similar to the GPS, the portal contained an autocomplete drug search functionality allowing for suggested search terms in the case of misspelling. Enlarged text, elements of responsive web design, and visual hierarchy were implemented across the site in order to make the portal accessible and navigable to individuals with varying degrees of web literacy. Results were presented in two formats:
A broad overview of medications listed under their “traffic light” recommendations, and
An accordion listing which allows the user to click and expand to learn more about the details of each result without navigating to a separate page.
Other features of the patient portal included a background tab to introduce basic concepts of genetics and PGx ( Supplementary Figures ), information on how a PGx test is performed and how to interpret test results, and a help tab with a discussion on privacy as well as contact information. Model‐View‐Controller architecture was implemented to allow easy future extension of the site from a demo format to a working prototype.
Participants
Study participants were recruited from our existing institutional, prospective PGx implementation study called “The 1200 Patients Project” (http://clinicaltrials.gov NCT01280825).15, 16 We prespecified a target sample size of 10–12 participants per group for the 2 separate days of testing (anticipating that there would be dropout among those who agreed by telephone before the actual participation day). This planned sample size was in accordance with other literature, which demonstrated that detectable increases in knowledge and changes in perceptions were measurable in pilot groups of this size.19, 20 We contacted patients by telephone for potential participation in one of two study sessions: one group consisted of patients who had previously received broad pre‐emptive genomic testing and whose actual results had been previously made available to their treating physicians (PGx), and a second control group consisted of patients who had not received genomic testing within the context of “The 1200 Patients Project,” reflecting traditional care (TC). Purposeful sampling of both cohorts was carried out by continuous assessment of the demographics of confirmed participants to guide subsequent telephone call invitations in order to ensure that the demographic characteristics of participants in each study session were not significantly different. A recruitment transcript was used to explain the purpose of study sessions—to assess patients’ knowledge and perceptions of PGx before and after having access to a mock PGx patient web portal. Potential participants were informed that they would be given a $50 Visa gift card, complimentary parking, and would be provided refreshments at the session as incentives for participation. A confirmation letter with the study session details was mailed to all individuals who agreed to participate. This study was approved by The University of Chicago Institutional Review Board.
Study design and data collection
The two groups convened separately in August 2018. At the start of each session, participants were asked to complete a short demographic questionnaire and a pre‐test, and then participants were given 15–20 minutes to navigate the mock PGx patient web portal, and finally complete a post‐test. In order to ensure the clarity of questions, the pre‐test and post‐test instruments were first pre‐tested and then refined before formal use in this study based on feedback from a group of ~ 10 stakeholders, including physicians, pharmacists, informatics personnel, and laypersons. The instruments ( Supplementary Materials ) consisted of multiple‐choice questions to assess PGx knowledge as well as Likert‐scale questions to assess perceptions of PGx and the patient portal. Responses from the demographic questionnaires and tests were collected to compare responses and scores within and between groups, and to investigate response changes between the pre‐tests and post‐tests.
Data analysis
Demographic data were compared using Fisher's exact test for categorical variables and a t‐test for continuous variables, as appropriate. For multiple‐choice questions to assess general knowledge of PGx, data were reported as a mean and SD of total % correct, and differences were compared using a t‐test. For Likert‐scale questions to assess general PGx perceptions, “strongly agree” and “somewhat agree” were combined and compared with combined responses from those who reported “not sure,” “somewhat disagree,” and “strongly disagree.” A P value < 0.05 was considered statistically significant for all comparisons.
Results
The YourPGx Portal
The YourPGx Portal contained four sample drug results with varying levels of information: omeprazole (Figure 1 a), clopidogrel (Figure 1 b), simvastatin (Figure 1 c), and codeine (Figure 1 d). The mock portal displayed the list of all four sample PGx‐associated medications in a compact accordion view, which could be expanded to provide more information for each medication by clicking on its name without leaving the page. In addition to the traffic light signal, each drug result expanded to include a quick sentence to summarize the drug‐specific patient risk. The summaries also included additional information, such as what typical condition(s) the medication is used to treat, population and patient risks for adverse drug events, and graphics and iconography to illustrate side effects, population risk, and/or mechanism of drug metabolism. We purposely provided varying levels of information detail and depth, and different display formats for each of the four summaries in order to test different ways to disseminate information to patients and assess patient preferences for each of the summaries.
Figure 1.
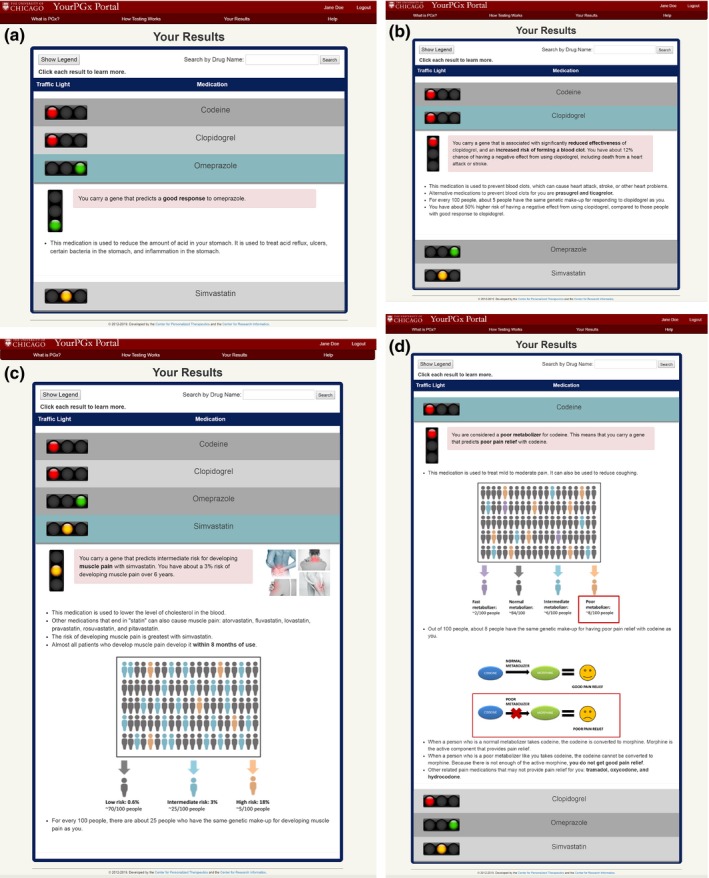
The YourPGx patient web portal contained mockup results for (a) omeprazole, (b) clopidogrel, (c) simvastatin, and (d) codeine. The summaries used our traffic light symbolism (red—“warning,” yellow—“caution,” and green—“favorable”) to help the patient quickly identify their risk for each medication. The summaries also included additional information such as what typical condition(s) the medication is used to treat, population and patient risks for adverse drug events, and graphics and iconography to illustrate side effects, population risk, and/or mechanism of drug metabolism. PGx, pharmacogenomic.
Patient demographics
We contacted 324 participants by telephone: 167 did not answer, 118 refused or were unable to participate, and 39 agreed to participate. The most commonly cited reasons for declining participation (in descending order) were schedule conflict, lack of interest, and no means of transportation to the sessions. Nineteen participants who initially agreed to participate by telephone did not come to the group on the assigned day (10 in the PGx group and 9 in the TC group), leaving 10 and 10 participants in the PGx and TC groups, respectively. The purposeful sampling method proved successful as there were no significant differences in demographic characteristics between the two groups (Table 1 ). The average age was 61 years (38–74), 60% were women, 50% were African American, and 55% had a bachelor's or advanced degree. Almost all patients (95%) reported currently taking at least one prescription medication and 90% reported self‐management of their medications. Notably, 40% of participants in both groups self‐reported a history of medication‐related side effects and 40% recalled stopping a medication due to a side effect or because they felt like it was ineffective. Only 30% of the PGx group participants recalled having a genetic test done, and 30% recalled receiving a PGx‐determined prescription medication, compared with 10% and 0% in the TC group, respectively.
Table 1.
Demographics of the participants in the TC and PGx test groups
| Column 1 | Total (N = 20) | TC (N = 10) | PGx (N = 10) | P valuea |
|---|---|---|---|---|
| Age (years), mean (SD) | 61.2 (12.3) | 64.7 (11.0) | 57.7 (13.7) | 0.22 |
| Male, N (%) | 8 (40) | 5 (50) | 3 (30) | 0.65 |
| Race, N (%) | 0.37 | |||
| White | 7 (35) | 2 (20) | 5 (50) | |
| African American | 10 (50) | 7 (70) | 3 (30) | |
| Other | 3 (15) | 1 (10) | 2 (20) | |
| Education, N (%) | 1.00 | |||
| High school or less | 4 (20) | 2 (20) | 2 (20) | |
| Some college | 5 (25) | 3 (30) | 2 (20) | |
| Bachelor's degree | 3 (15) | 1 (10) | 2 (20) | |
| Advanced degree | 8 (40) | 4 (40) | 4 (40) | |
| Number of prescription medications, N (%) | 1.00 | |||
| 0 | 1 (5) | 0 (0) | 1 (10) | |
| 1–4 | 13 (65) | 7 (70) | 6 (60) | |
| 5–8 | 4 (20) | 2 (20) | 2 (20) | |
| 9 or more | 2 (10) | 1 (10) | 1 (10) | |
| Self‐reported history of medication‐related side effects, N (%) | 8 (40) | 3 (30) | 5 (50) | 0.65 |
| Self‐reported medication management, N (%) | 1.00 | |||
| Self | 18 (90) | 9 (90) | 9 (90) | |
| Family | 1 (5) | 0 (0) | 1 (10) | |
| Caretaker | 1 (5) | 1 (10) | 0 (0) | |
| Other | 1 (5) | 0 (0) | 0 (0) | |
| Recalls stopping a medication due to a side effect or because felt like it was ineffective, N (%) | 8 (40) | 3 (30) | 5 (50) | 0.65 |
| Recalls having a genetic test done, N (%) | 4 (20) | 1 (10) | 3 (30) | 0.58 |
| Recalls receiving a PGx‐determined prescription, N (%) | 3 (15) | 0 (0) | 3 (30) | 0.21 |
PGx, pharmacogenomics group; TC, traditional care group.
Fisher's exact P value presented for categorical variables and t‐test P value is presented for age.
Pre‐test and post‐test PGx knowledge scores
Patients’ knowledge of PGx significantly improved after accessing the YourPGx Portal for both groups (Figure 2 ). The average pre‐test scores were 40.0 ± 23.9% and 51.8 ± 20.6% for the TC and PGx groups, respectively (P = 0.25). The average post‐test scores were 67.3 ± 23.2% and 64.5 ± 21.2% for the TC and PGx groups, respectively (P = 0.79). Patients scored 20% higher in the post‐test compared with the pre‐test and there was a significant increase in the average total score (% correct) from the pre‐test (45.9 ± 22.6) to post‐test (65.9 ± 21.6; P < 0.05) for all 20 patients.
Figure 2.
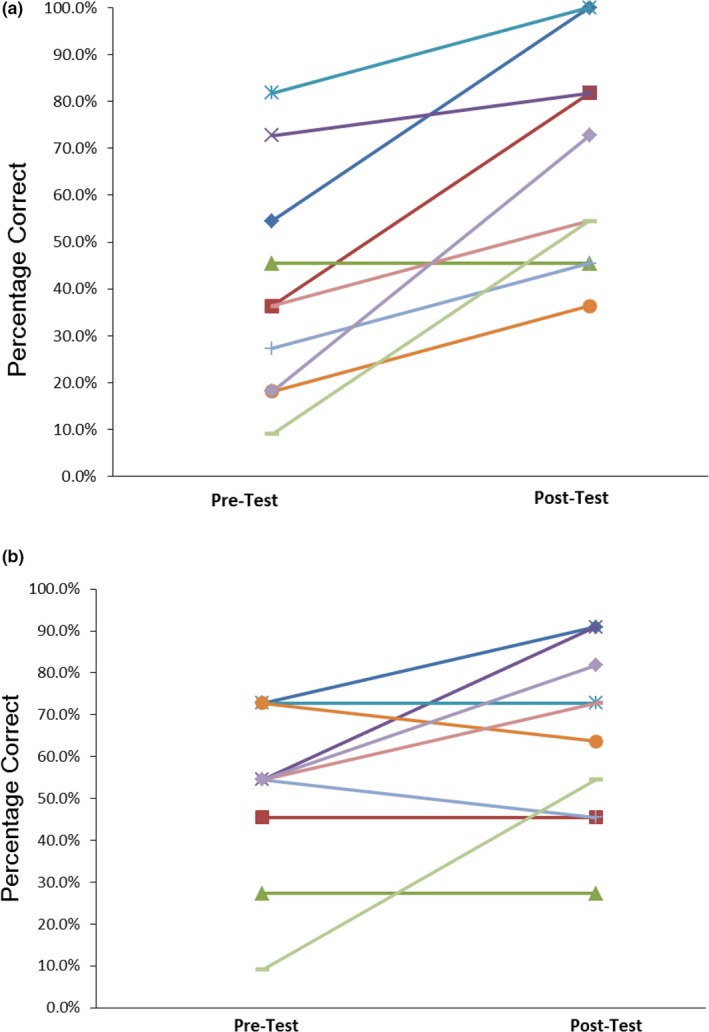
Pre‐test and post‐test pharmacogenomic (PGx) knowledge scores for the (a) traditional care and (b) PGx groups. Patients overall scored 20% higher in the post‐test compared with the pre‐test. There were no significant differences in baseline PGx knowledge scores or score change between the groups.
Pre‐test and post‐test PGx perceptions
Likert‐scale questions were used to assess patient perceptions of PGx before and after accessing the YourPGx Portal. Patients were asked to respond to the statement “based on my current knowledge, I wish my doctor had PGx information before prescribing [drug name]” for the four sample PGx drug results. Overall, fewer patients in the TC group answered “somewhat” or “strongly agree” in the pre‐test compared with patients in the PGx group across all medications except simvastatin (Table 2 ). However, there was an increase in the number of patients who responded “somewhat” or “strongly agree” in both groups across all medications in the post‐test—ranging from 60–90% across the four medications in the pre‐test compared with 89‐100% in the post‐test.
Table 2.
Number of patients in the TC and PGx groups who said they somewhat or strongly wished their physician had sample PGx results for four drugs before and after accessing the YourPGx Portal
| Pre‐test | Post‐test | |
|---|---|---|
| Omeprazole | ||
| TC | 6/10 (60%) | 9/10 (90%) |
| PGx | 9/10 (90%) | 10/10 (100%) |
| Clopidogrel | ||
| TCa | 6/10 (60%) | 8/9 (89%) |
| PGx | 9/10 (90%) | 10/10 (100%) |
| Simvastatin | ||
| TC | 7/10 (70%) | 10/10 (100%) |
| PGx | 7/10 (70%) | 10/10 (100%) |
| Codeine | ||
| TC | 7/10 (70%) | 10/10 (100%) |
| PGx | 9/10 (90%) | 10/10 (100%) |
PGx, pharmacogenomics group; TC, traditional care group.
One patient in the TC group did not respond to the post‐test questions for clopidogrel.
When asked to respond to the statement “It is important that I can control who has access to my PGx information,” almost all patients in both groups somewhat or strongly agreed in both the pre‐test and post‐test (90–100%). With regard to the statement “If I have access to my PGx test results, I would encourage a friend or family member to consider PGx testing,” 80% and then 90% (pre/post‐test) of patients in both the TC and PGx groups responded “somewhat” or “strongly agree” (Figure 3 ). Notably, there were more patients who responded “strongly agree” in the post‐test compared with the pre‐test for both groups (60−80% in the TC group, and 40−80% in the PGx group).
Figure 3.
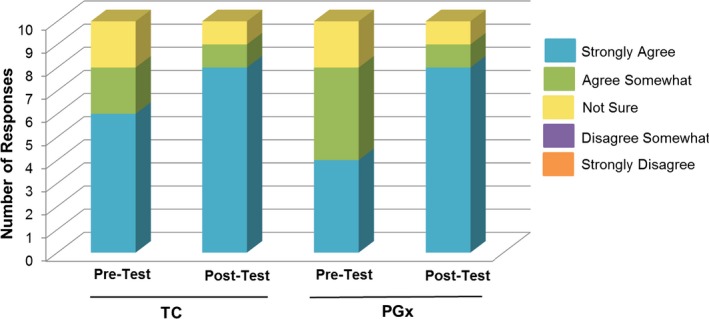
Patient responses to the Likert‐scale question “If I have access to my pharmacogenomics test results, I would encourage a friend or family member to consider getting pharmacogenomics testing.” PGx, pharmacogenomics group; TC, traditional care group.
Patients were asked to choose all that applied to the statement “If I have access to my PGx test results, I would share my results with…” (Figure 4 ). Almost all patients (90%) said they would share their results with their healthcare provider, and this did not change from pre‐test to post‐test. Both groups also stated the desire to share their results with their pharmacist, spouse/partner, parents, siblings, and children. However, no patients said they would share their results with friends or social media. Interestingly, fewer patients in the TC group wanted to share their results with other individuals besides their healthcare provider in the post‐test, and one patient even changed their respond to “I would not share my results.” However, more patients in the PGx group wanted to share their results with their pharmacist, spouse/partner, and parents by the time of the post‐test, but fewer wanted to share results with their siblings and children in the post‐test, and one patient changed their response from “I would not share my results” in the pre‐test to the post‐test.
Figure 4.
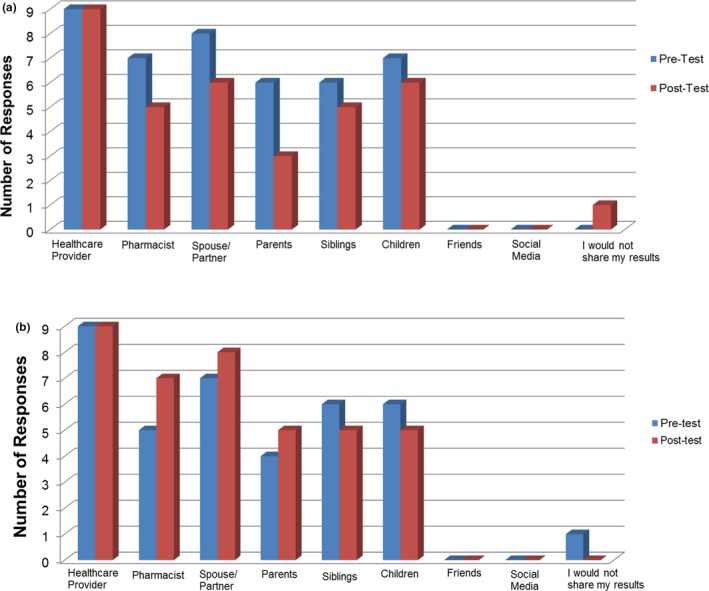
Patient responses to the question “If I have access to my pharmacogenomics test results, I would share my results with…” for the traditional care (TC) (a) and pharmacogenomics (PGx) (b) groups. *One patient from the TC and the PGx group did not respond.
Post‐test patient perceptions on the usability and understandability of the PGx patient portal
Almost all patients somewhat or strongly agreed that the patient portal was easy to use—90% in the TC group, and 100% in the PGx group (Figure 5 a). When asked about the statement “the patient portal contained a lot of technical or complicated words I did not understand,” almost all patients in the PGx group (80%) somewhat or strongly disagreed compared with only 50% of patients in the TC group (Figure 5 b). All patients in the PGx group (100%) and 80% in the TC group thought that 15–20 minutes was enough time to evaluate the contents of the portal (Figure 5 c).
Figure 5.
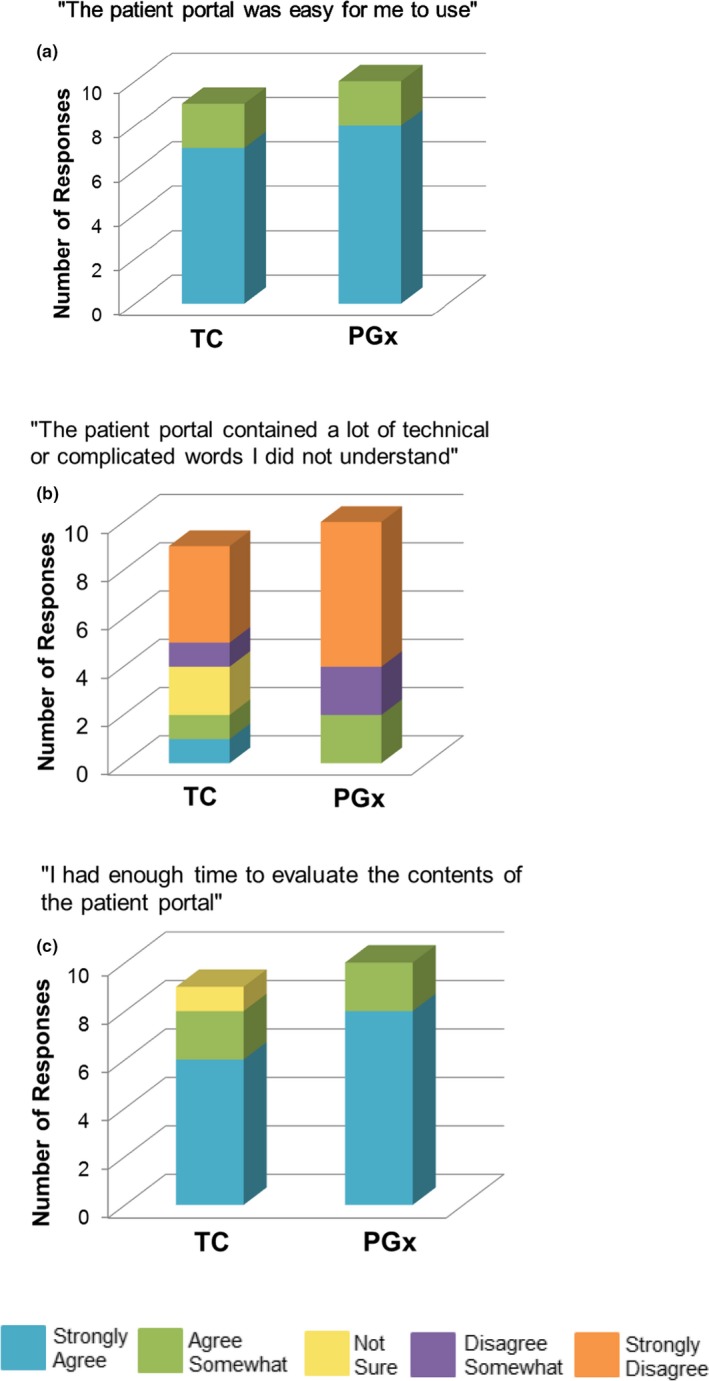
Likert‐scale questions to assess patients’ perceptions of the usability and understandability of the YourPGx Portal. TC, traditional care; PGx, pharmacogenomics. *One patient in the TC group did not respond.
Discussion and Conclusion
Discussion
In this study, we built a mock PGx patient portal, the YourPGx Portal, and assessed its ability to disseminate PGx test results and information to patients, regardless of whether patients had previously received PGx testing or not. PGx knowledge and perceptions improved overall for the patients in both groups. Additionally, after reviewing the mock results, more patients expressed that they wished their provider had access to PGx information before prescribing the four sample drugs, and more patients would encourage a friend or family member to get PGx testing. The positive measures detected in this pilot will now be utilized in a follow‐on study to test a live, functional version of the YourPGx Portal in a large cohort of patients receiving PGx‐informed care.
Although the patient groups receiving the PGx educational intervention were small in this pilot study, the ~ 20% improvements in knowledge about PGx are on par with other similar educational interventions outside of the realm of genomics.19, 20, 21 Interestingly, after accessing the patient portal, almost all patients (90%) would share their results with their healthcare providers and 60% would share their results with pharmacists. These findings alone may justify further exploration of the true impact of allowing patients to have direct access to their own PGx test results (whether willingness to share PGx results might be medication‐specific is a topic that was underpowered for our current study but we plan to explore this in our follow‐up study). One salient prior study previously explored questions about PGx result education in a similar fashion to our current study. The prior study by Olson et al.22 examined patients’ knowledge and perceptions of PGx after having received a mailed letter with their individual CYP2D6 results. Unlike our study, however, only post‐result disclosure knowledge/attitudes were assessed (not pre‐result), and patients were not required to access results via an online portal (in the Olson et al.22 study, the primary mode of delivery of results was a paper letter). Nevertheless, many of the patient responses to similar questions were comparable with our current findings.
Interestingly, in our study, only 30% of participants in the PGx group recalled having genetic testing done. This may be because our institutional PGx implementation projects have previously only returned PGx test results to providers, not patients. Providers may or may not share these results with patients. Our current findings suggest a potentially positive impact of patient educational engagement regarding results delivery within PGx implementation programs. In line with this idea, we previously showed that patient‐provider communications about PGx results increased patient recall of medication changes, a finding that may have direct impacts on medication adherence.23 Additionally, Olson et al.22 found that 91% of patients reported being “much more likely” or “somewhat likely” to take their medication as prescribed if PGx information was used to help select their medications.22
There were some potential improvements that were learned from this assessment, which will be incorporated in the next phase of portal testing and implementation. Both patient groups expressed the importance of being able to control who had access to their PGx test results. To address this concern, the functional version of the YourPGx Portal will be password‐protected and will require patients to choose their own authentication credentials, which will allow them to control with whom they want to share PGx test results. The implemented version will also store patient genomic data and patient demographics on a Health Insurance Portability and Accountability Act (HIPAA) compliant server farm at The University of Chicago Center for Research Informatics to protect all patient information. Means for portability of results for patients who do wish to share results with other healthcare providers outside of the University hospital system is being explored, but can currently be accomplished through pdfs of individual pages printed by the patient. Patients from this study indicated that the mock YourPGx Portal was easy to use and easy to understand. We will leverage the learnings from this study to build‐out a secured, protected‐access, live version of the YourPGx Portal with expanded content beyond the four initial medications, to deliver actual patient‐specific PGx results within our healthcare system. We will initially test this live portal in a larger cohort of patients by allowing participants from our existing institutional PGx implementation programs to access the YourPGx portal once assent is given by their treating provider(s).
There were some limitations to this study. First, half of the participants had been previously genotyped, and the results may not be generalizable to the overall population. Second, the readability, understandability, and actionability testing of the patient portal were beyond the scope of our initial pilot study described here. We will formally test the readability of the portal (for the full, expanded set of medication summaries) by using both the Fry and SMOG methods to produce an average score for the reading grade level of the content in our next‐step expanded implementation pilot phase.24, 25 In the next‐phase pilot, we will also test the understandability and actionability of the portal by using the Patient Education Materials Assessment Tool (PEMAT).26 Our eventual objective is to provide all patients who enroll in our various PGx implementation programs access to the portal and to their PGx test results. Test results will first be delivered to participating providers enrolled in our studies, who will give permission for the results to be shared with patients via the YourPGx Portal. This will address possible safety concerns of directly providing PGx results to patients without proper initial education and consultation with a healthcare professional.
Conclusion
A patient‐friendly portal can effectively deliver PGx results and information directly to patients. Patients’ knowledge and perceptions of PGx improved after accessing the YourPGx Portal. Our results show that patients may act as advocates for their own personalized care when empowered with PGx information by sharing their results with different members of their healthcare team.
Funding
This research was supported by National Institutes of Health (NIH) training grant 5T32GM007019‐41 (T.M.T.); NIH grant 1R01HG009938‐01A1 (P.H.O.); and the Benjamin McAllister Research Fellowship (T.M.T.). The funding sources had no involvement in the conduct of this research and/or preparation of this article.
Conflicts of Interest
Dr. Ratain is a co‐inventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping. No royalties were received from this research. All other authors declared no competing interests for this work.
Author Contributions
P.H.O. wrote manuscript, designed research, performed research, analyzed data, contributed new reagents/analytical tools; T.M.T. wrote manuscript, designed research, performed research, analyzed data, contributed new reagents/analytical tools; E.L. performed research, contributed new reagents/analytical tools; K.D. contributed new reagents/analytical tools; E.S. performed research; M.J.R. contributed new reagents/analytical tools.
Supporting information
Figure S1. The YourPGx Portal (a) background tab to introduce basic concepts of genetics and PGx, and (b) information on how to interpret results and other warning and precautionary statements.
Pre‐Test and Post‐Test Questionnaires
References
- 1. Wilson, M. Readability and patient education materials used for low‐income populations. Clin. Nurse. Spec. 23, 33–40 (2009). [DOI] [PubMed] [Google Scholar]
- 2. Coughlin, S.S. , Stewart, J.L. , Young, L. , Heboyan, V. & De Leo, G. Health literacy and patient web portals. Int. J. Med. Inform. 113, 43–48 (2018). [DOI] [PubMed] [Google Scholar]
- 3. Ricciardi, L. , Mostashari, F. , Murphy, J. , Daniel, J.G. & Siminerio, E.P. A national action plan to support consumer engagement via E‐health. Health Aff. 32, 376–384 (2013). [DOI] [PubMed] [Google Scholar]
- 4. Zarcadoolas, C. , Vaughon, W.L. , Czaja, S.J. , Levy, J. & Rockoff, M.L. Consumers’ perceptions of patient‐accessible electronic medical records. J. Med. Internet. Res. 15, e168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weitzel, K.W. et al Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 56–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dunnenberger, H.M. et al Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luzum, J.A. et al The Pharmacogenomics Research Network Translational Pharmacogenetics Program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin. Pharmacol. Ther. 102, 502–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van der Wouden, C.H. et al Implementing pharmacogenomics in Europe: design and implementation strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 101, 341–358 (2017). [DOI] [PubMed] [Google Scholar]
- 9. Friedman, P.N. et al The ACCOuNT Consortium: a model for the discovery, translation, and implementation of precision medicine in African Americans. Clin. Transl. Sci. 12, 209–217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramsey, L.B. et al Implementation of pharmacogenetics at Cincinnati Children's Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin. Pharmacol. Ther. 105, 49–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hinderer, M. et al Implementing pharmacogenomic clinical decision support into German hospitals. Stud. Health Technol. Inform. 247, 870–874 (2018). [PubMed] [Google Scholar]
- 12. Borobia, A.M. et al Clinical implementation of pharmacogenetics testing in a hospital of the Spanish National Health System: strategy and experience over 3 years. Clin. Transl. Sci. 11, 189–199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sissung, T.M. et al Pharmacogenomics implementation at the National Institutes of Health Clinical Center. J. Clin. Pharmacol. 57(suppl. 10), S67–S77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harada, S. et al Precision Medicine at the University of Alabama at Birmingham: laying the foundational processes through implementation of genotype‐guided antiplatelet therapy. Clin. Pharmacol. Ther. 102, 493–501 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Donnell, P.H. et al Adoption of a clinical pharmacogenomics implementation program during outpatient care‐initial results of the University of Chicago “1,200 Patients Project”. Am. J. Med. Genet. 166c(1), 68–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Donnell, P.H. et al Pharmacogenomics‐based point‐of‐care clinical decision support significantly alters drug prescribing. Clin. Pharmacol. Ther. 102, 859–869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haga, S.B. & LaPoint, N.M. The potential impact of pharmacogenomics testing on medication adherence. Pharmacogen. J. 13, 481–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Danahey, K. et al Simplifying the use of pharmacogenomics in clinical practice: building the genomic prescribing system. J. Biomed. Inform. 75, 110–121 (2017). [DOI] [PubMed] [Google Scholar]
- 19. Finley, K. , Giannamore, M. , Bennett, M. & Hall, L. Assessing the impact of lifestyle modification education on knowledge and behavior changes in gastroesophageal reflux disease patients on proton pump inhibitors. J. Am. Pharm. Assoc. 49, 544–548 (2009). [DOI] [PubMed] [Google Scholar]
- 20. Klein, C.J. , Riggenbach‐Hays, J.J. , Sollenberger, L.M. , Harney, D.M. & McGarvey, J.S. Quality of life and compassion satisfaction in clinicians: a pilot intervention study for reducing compassion fatigue. Am. J. Hosp. Palliat. Care 35, 882–888 (2018). [DOI] [PubMed] [Google Scholar]
- 21. Giuliano, C. , Nofar, T. & Edwin, S.B. Can a short video improve Apixaban knowledge in an inpatient setting? P T. 42, 256–260 (2017). [PMC free article] [PubMed] [Google Scholar]
- 22. Olson, J.E. et al Participant‐perceived understanding and perspectives toward pharmacogenomics: Mayo Clinic Right Drug, Right Dose, Right Time (RIGHT) Protocol. Genet. Med. 19, 819–825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borden, B.A. et al Patient‐provider communications about pharmacogenomic results increase patient recall of medication changes. Pharmacogen. J. 19, 528–537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedman, D.B. & Hoffman‐Goetz, L. A systematic review of readability and comprehension instruments used for print and web‐based cancer information. Health Educ. Behav. 33, 352–373 (2006). [DOI] [PubMed] [Google Scholar]
- 25. Centers for Medicare & Medicaid Services . The toolkit for making written material clear and effective. <https://www.cms.gov/Outreach-and-Education/Outreach/WrittenMaterialsToolkit/index.html> (2012). Accessed April 4, 2019.
- 26. Shoemaker, S.J. , Wolf, M.S. & Brach, C. Development of the Patient Education Materials Assessment Tool (PEMAT): a new measure of understandability and actionability for print and audiovisual patient information. Pat. Educ. Couns. 96, 395–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The YourPGx Portal (a) background tab to introduce basic concepts of genetics and PGx, and (b) information on how to interpret results and other warning and precautionary statements.
Pre‐Test and Post‐Test Questionnaires


