Abstract
Objectives
Chronic obstructive pulmonary disease (COPD) phenotypes cover a range of lung abnormalities. To allow text mining methods to identify pertinent and potentially complex information about these phenotypes from textual data, we have developed a novel annotated corpus, which we use to train a neural network-based named entity recognizer to detect fine-grained COPD phenotypic information.
Materials and methods
Since COPD phenotype descriptions often mention other concepts within them (proteins, treatments, etc.), our corpus annotations include both outermost phenotype descriptions and concepts nested within them. Our neural layered bidirectional long short-term memory conditional random field (BiLSTM-CRF) network firstly recognizes nested mentions, which are fed into subsequent BiLSTM-CRF layers, to help to recognize enclosing phenotype mentions.
Results
Our corpus of 30 full papers (available at: http://www.nactem.ac.uk/COPD) is annotated by experts with 27 030 phenotype-related concept mentions, most of which are automatically linked to UMLS Metathesaurus concepts. When trained using the corpus, our BiLSTM-CRF network outperforms other popular approaches in recognizing detailed phenotypic information.
Discussion
Information extracted by our method can facilitate efficient location and exploration of detailed information about phenotypes, for example, those specifically concerning reactions to treatments.
Conclusion
The importance of our corpus for developing methods to extract fine-grained information about COPD phenotypes is demonstrated through its successful use to train a layered BiLSTM-CRF network to extract phenotypic information at various levels of granularity. The minimal human intervention needed for training should permit ready adaption to extracting phenotypic information about other diseases.
Keywords: chronic obstructive pulmonary disease, text mining, natural language processing, phenotype, information extraction
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is “a common, preventable, and treatable disease that is characterized by persistent respiratory symptoms and airflow limitation that is due to airway and/or alveolar abnormalities usually caused by significant exposure to noxious particular gases.”1 It is rapidly becoming one of the major causes of morbidity and mortality worldwide.2 COPD is a multifactorial and heterogeneous disease and not every patient responds to all available drugs.3–5 Due to the high prevalence and heterogeneity of COPD, improved deep phenotyping strategies are required. Such in-depth phenotyping can pave the way for personalized treatment regimens,6 ensuring that the most suitable therapies are provided.7,8 A phenotype can be broadly defined as “any observable characteristic of an organism,”9 while a COPD phenotype can be more specifically defined as “a single or combination of disease attributes that describe differences between individuals with COPD as they relate to meaningful outcomes (symptoms, exacerbations, response to therapy, rate of disease progression, or death).”10 Identifying such phenotypes (also described as phenotypic traits) allows grouping of patients according to their prognostic and therapeutic characteristics.10 Early classification of the COPD subtype will facilitate superior healthcare provision and early intervention where it is most required—for example, patients with rapid disease progression or frequent exacerbations.
Various textual sources constitute vital sources of COPD evidence, by providing information about phenotypes, characteristics, and treatment regimens. Although pinpointing relevant information in large, heterogeneous text repositories can be time-consuming, applying text mining (TM) techniques to semantically analyze these repositories11 can significantly reduce the time needed by clinicians and researchers for tasks such as finding relationships amongst concepts (eg, genotype-phenotype,12,13 gene-disease,14–16 and disease-phenotype17,18), diagnosis categorization19 or recruiting patients for trials and studies.20,21 To enhance automatic semantic analysis of COPD-related text, the contributions of this article are two-fold:
We have created a novel corpus of 30 full-text articles, annotated by experts with named entities relating to COPD phenotypes. The fine-grained annotation scheme aims to account for the potentially complex, nested nature of phenotype descriptions. We automatically enrich the annotations with links to UMLS Metathesaurus concepts. The corpus is freely available (http://www.nactem.ac.uk/COPD) to stimulate development of named entity recognition (NER) tools for COPD phenotypic information.
We demonstrate the utility of the corpus by using the expert-added annotations to train a high-performance neural network-based entity recognizer, which exploits nested annotations to accurately detect detailed information relating to COPD phenotypes.
The potential complexity of COPD phenotype descriptions, and how our annotation scheme handles them, is exemplified in Figure 1, where the phrase elevation of pulmonary arterial pressures is identified as a phenotype, and is assigned the category TestOrMeasureResult, since it describes the outcome of a measurement. Analyzing the internal structure of this phenotype reveals the specific measurement undertaken (pulmonary arterial pressures) and anatomical entity involved (pulmonary artery). Our annotations correspond to both complete phrases that constitute COPD phenotypes and other types of concepts frequently mentioned within them, and/or within their context. Such embedding (nesting) of shorter entity mentions within longer (outermost) phenotype descriptions is fairly common (29% of our corpus annotations are embedded).
Figure 1.

Example of a phenotype that includes other concepts nested within it.
The detailed nature of our annotations aims to facilitate the development of automated tools supporting the exploration of COPD phenotypic information in text from multiple perspectives. This will allow not only the location and categorization of COPD phenotypes, including those identified through tests, or those constituting risk-raising individual behaviors (eg, smoking) but will also permit detailed investigations about the nature of these phenotypes, including finding those affecting specific anatomical locations, or those concerning different results of specific tests. Furthermore, our enrichment of the annotations by applying an automatic normalization method helps to link different ways of mentioning the same concept. This can facilitate search at the concept level, such that searching for the condition dyspnea would also retrieve documents mentioning shortness of breath.
To demonstrate the full potential of the corpus for developing NER tools, our neural network-based method is specifically designed to recognize nested and outermost entities. In particular, information about nested mentions is used to improve the accuracy of outermost phenotype recognition, without external knowledge resources. To our knowledge, this is the first attempt to apply such an approach to detecting phenotypic information.
RELATED WORK
Annotated corpora
Several existing annotated corpora contain entity annotations relevant to phenotype recognition, including biomedical abstracts or articles,22–24 medical case reports,25 and clinical records.26,27 Certain corpora are also annotated with relations between disorders and other types of concepts.28,29 For example, the phenotype phrase upper lobe emphysema may be split into Condition (emphysema) and Locus (upper lobe), linked by a has_location relation.28 Such fine-grained analyses allow the potentially complex structure of phenotypes to be exploited to perform more targeted queries, for example, to locate all phenotypes affecting a particular body part. While in most cases, annotations corresponding to phenotypes have rather coarse-grained labels, like Disease, Disorder, or Problem, a more fine-grained annotation scheme for phenotypes of congestive heart failure (CHF)30 distinguishes Causes, Risk Factors, Non-traditional risk factors, and Signs and symptoms.
The annotations in several corpora23,26,31,32 link each annotated entity to a unique concept identifier in a domain-specific terminological resource. Some such resources cover a wide range of medical and biomedical concepts,33,34 while others are specialized for diseases and/or phenotypes.35–37 These links can facilitate the development of normalization methods,31,38–41 which automatically assign a concept identifier in a given terminological resource to each entity, to link together variant concept mentions.
Named entity recognition for COPD
Previous approaches to phenotype NER have included dictionary-based lookup,42–45 possibly coupled with rules to improve accuracy and/or to handle the potentially complex structure of phenotype descriptions.46–50 Whilst some such approaches perform poorly on phenotype recognition,51 an optimized combination of the outputs of these methods can be beneficial.52 However, combining or replacing rules with machine learning (ML) tends to achieve superior performance.53–55
Conventional ML approaches such as conditional random fields (CRFs) have been applied to many NER tasks, including detecting CHF phenotypes30 and recognizing nested entities.56–58 CRF-based models generally require humans to perform feature engineering for each new task, to determine the optimal set of textual features for predicting entities. Features include semantic information from domain-specific terminological resources or the output of linguistic processing tools, which can be time-consuming to apply to huge document collections.
Recently, however, representational methods have improved phenotype extraction performance59–61 by using word embeddings, which remove the need for hand-crafted feature engineering, linguistic processing or terminological resources,62,63 and character embeddings, which encode word morphology information.
Combined with embeddings, advanced deep learning methods can produce high-performance NER systems.64–66 Recurrent neural networks (RNNs)67 are effective for various natural language processing tasks,68 while specializations such as long short-term memory networks (LSTMs)69 and gated recurrent units (GRUs)70 are particularly effective, since they introduce gating mechanisms to handle textual contexts with long dependencies, which can be highly important for NER.71 Bidirectional versions (eg, bidirectional long short-term memory [BiLSTMs]) use information from both left and right contexts, to further boost performance.72,73
In addition to “standard” NER tasks, neural network methods have been applied to nested entity recognition.74,75 Multilayered approaches76,77 use information about entities at a given level of nesting to improve recognition of entities at other levels of nesting. One of these77 uses no linguistic features, and outperforms other methods in detecting nested entities for general language and molecular biology.
METHODS
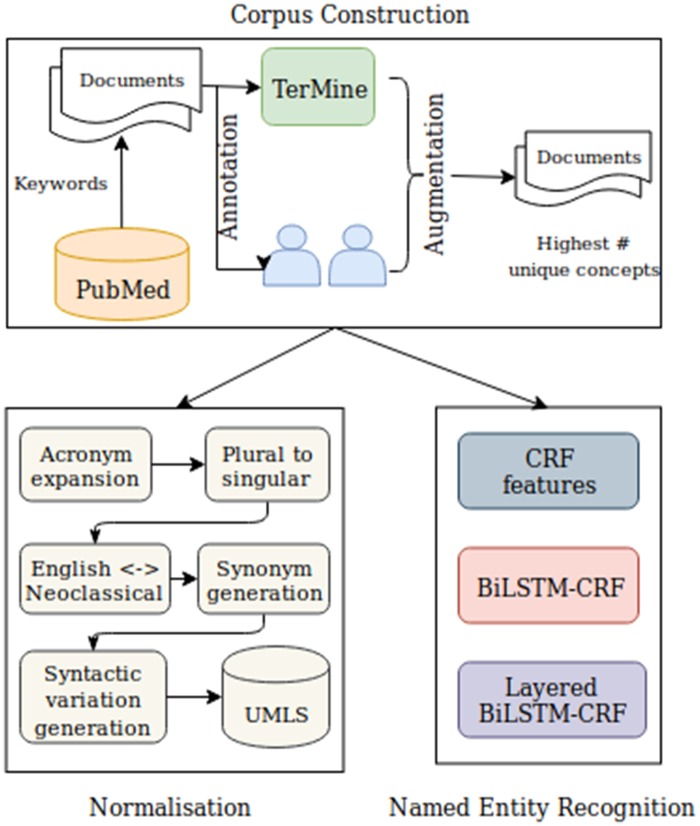
In this section, we explain the methods used in the various steps of our work (see Figure 2 for an overview). Firstly, we describe the construction and annotation of the corpus. We subsequently explain how the expert-added annotations were enriched using an automatic normalization method to link them to UMLS concepts. Finally, we describe the NER methods that were applied to create a named entity recognizer for COPD.
Figure 2.
Workflow for annotation and detection of information relating to COPD phenotypes. COPD: chronic obstructive pulmonary disease; CRF: conditional random field.
Corpus construction
Information about COPD phenotypes may occur in various documents, including clinical records and academic articles. However, the availability of clinical record corpora is restricted, and they tend to be US-centric.78,79 To avoid bias toward practices of a particular country, we decided to create a corpus of scientific articles from various COPD-relevant journals. As previous work has shown that TM tools trained on one text type can be applied to texts with different characteristics,30,80 it is intended that tools trained on our corpus may be adapted for phenotype extraction from clinical records.
We firstly selected COPD-relevant journals in the PubMed Central Open Access Subset, whose titles contain the following keywords: (chronic, obstructive, pulmonary, disease, respiratory, and lung); this resulted in the 10 journals shown in Supplementary Appendix S1. We then retrieved all articles within these journals mentioning either chronic obstructive pulmonary disease or COPD (974 articles). According to limited resources and time, only a subset of these documents could be annotated by our domain experts. We thus attempted to select documents containing the richest and widest COPD phenotype evidence. We firstly applied the automatic term recognition system TerMine81 to the set of the COPD guidelines published jointly by the American Thoracic Society and the European Respiratory Society.82 The automatically extracted terms were augmented with expert-provided terms to create 1925 different terms representing COPD phenotypes. We then selected the 30 full-text articles with the highest numbers of unique COPD phenotype terms. The number of unique terms in each selected document is shown in Supplementary Appendix S2.
Annotation scheme
Our annotation scheme83 (guidelines available at: http://www.nactem.ac.uk/COPD/download.php) aims to balance simplicity of application with the ability to capture fine details about phenotypes. Only simple text spans, rather than relationships, are annotated, since the latter task can considerably increase annotation burden. However, by using a detailed hierarchy of semantic labels, and allowing entities to be nested within each other, we can capture potential relationships between entities. For example, if a treatment is mentioned within a phenotype statement (Steroid-induced skeletal muscle atrophy), then it is likely that the phenotypic manifestation is a side effect of the nested treatment.
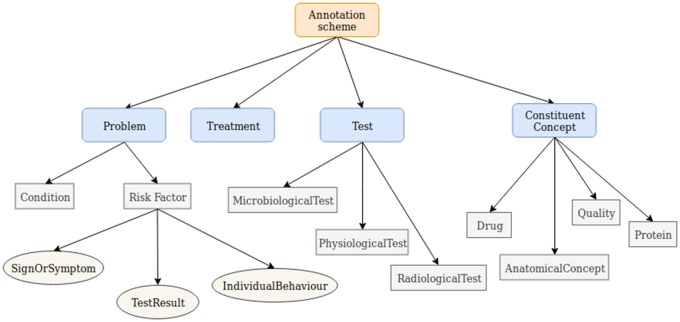
Our scheme (see Table 1 and Figure 3) is inspired by 2 existing schemes. The categories defined in 1 scheme,29 that is, Problem, Treatment, or Test, form the core of the scheme, to identify information about COPD phenotypes, their treatment and discovery. Inspired by the fine-grained labels used for CHF phenotypes,30 we introduce a hierarchy of more detailed labels under these top-level categories; the most specific labels possible are assigned by annotators. Since phenotype descriptions are typically formed from a combination of different types of concepts, our scheme includes the most common of these, for example, anatomical concepts (chronic airways obstruction), proteins (alpha1 antitrypsin deficiency), qualities (eg, decreased COPD exacerbations), and test results (eg, reduced FEV1). These are mainly organized under an additional top-level category, ConstituentConcept.
Table 1.
Descriptions, examples, and counts of each category in the COPD annotation scheme
| Type | Description | Examples | Number of concepts |
|---|---|---|---|
| Problem | An overall category for any COPD indicates of concern | COPD exacerbations; past pulmonary TB | 2556 |
| Condition | Any disease or medical condition includes COPD comorbidities | emphysema; pulmonary vascular disease; asthma | 5119 |
| RiskFactor | A phrase signifying a patient’s increased chances of having COPD | increased levels of the C-reactive protein; alpha1 antitrypsin deficiency | 1211 |
| SignOrSymptom | An observable irregularity manifested by a COPD patient | chronic cough; shortness of breath | 2065 |
| IndividualBehaviour | A patient’s habits leading to susceptibility of having COPD | smoking for 25 years; exercise-limited patients | 194 |
| TestResult | Findings based on COPD-relevant examinations | decrease in rate of lung function; FEV1 45% predicted | 685 |
| Treatment | Any medication, therapy, or treatment program | inhaled corticosteroids; oxygen therapy; pulmonary rehabilitation | 4337 |
| Test | An overall category for any COPD-relevant examinations or measures/parameters | spirometry, respiratory frequency, FEV1 | 3576 |
| RadiologicalTest | Any of the radiological tests for detecting COPD | computed tomography scanning; high resolution computed tomography | 29 |
| MicrobiologicalTest | An examination of a COPD-relevant specimen | complete blood count; bacterial isolates | 11 |
| PhysiologicalTest | A measurement of a COPD patient’s capacity to exercise | 6-min walking distance; incremental cardiopulmonary exercise testing | 17 |
| ConstituentConcept | An umbrella type for elementary concepts that may form part of a phenotype description; should only be chosen if none of the subtypes below apply | bronchodilation; enhancement of skeletal muscle contractility | 5 |
| AnatomicalConcept | A mention pertaining to anatomical entities | lung; heart; pulmonary; hepatic; respiratory airway | 2616 |
| Drug | Any drug name; will mostly overlap with treatment | corticosteroids; short-acting bronchodilators | 2593 |
| Protein | Any protein name | alpha1 antitrypsin; pro-inflammatory cytokines | 820 |
| Quality | Expressions which modify or qualify any of the concepts above | chronic; obstructed; damaged; decreased rate; enhanced; decreased amount | 1153 |
Abbreviations: COPD: chronic obstructive pulmonary disease; FEV1: Forced Expiratory Volume.
Figure 3.
Hierarchical entity annotation scheme for COPD phenotypic information. COPD: chronic obstructive pulmonary disease.
To increase annotation ease and efficiency, we used Argo,84 an interoperable TM platform, to apply a pipeline of pre-existing NER tools to preannotate the corpus with several entity types typically mentioned within phenotypes. The annotators’ task was then limited to reviewing and editing automatically added annotations, or adding longer, spanning annotations corresponding to more complex phenotypes.
To ensure annotation quality and consistency, 6 full-text papers were firstly annotated independently by 2 annotators with medical expertise, and inter-annotator agreement (IAA) rates were calculated. The widely used Cohen's kappa is not suitable here, because it requires the total number of annotated items to be known in advance. Hence, we followed a number of other related efforts85–87 by calculating IAA in terms of F-score. The micro-averaged IAA rate was 80.49% F-score, using strict conditions (ie, requiring both annotators’ annotations to match exactly in terms of text span chosen and semantic category). The main areas of disagreement concerned some fine-grained categories within the Problem branch of the scheme. In consultation with the annotators, the definitions of these categories were reviewed, and disagreements were discussed and resolved. Taking into account the decisions made, one of the annotators annotated the remaining 24 papers.
Entity normalization
We automatically normalized annotated entities to unique concept identifiers in the UMLS Metathesaurus,33 which covers all entity types in our scheme. We chose the HYPHEN method41 because of its flexibility, that is, it can normalize different entity types in documents with varying characteristics to different target terminological resources.33,88
HYPHEN uses a pipeline of different techniques to generate semantically consistent variations of the original entity mention and tries to match these generated variants against existing variants listed in the target terminological resource. The 6 techniques are:
Acronym/abbreviation expansion (eg, Type 2 DM → Type 2 diabetes mellitus).
Plural to singular conversion (eg, alveolar septa → alveolar septum).
Generation of English equivalents of Neoclassical compounds (eg, elevated blood leukocyte counts → elevated white blood cell count).
Generation of Neoclassical equivalents of English terms (eg, pleural inflammation → pleuritis).
Syntactic variation generation (eg, supplemental oxygen → oxygen supplementation).
Synonym generation (eg, worsening pulmonary function → deterioration of lung function).
Table 2 reports on the number and percentage of entities belonging to each category in our corpus that are automatically normalized. For each category, normalization dictionaries were created by filtering the concepts belonging to different UMLS semantic types; these are detailed in Supplementary Appendix S3.
Table 2.
Number of entities normalized by HYPHEN
| Category | Total entities | Number of entities normalized | Percentage of entities normalized |
|---|---|---|---|
| Problem | 2556 | 2151 | 83.15 |
| Condition | 5119 | 4969 | 97.07 |
| RiskFactor | 1211 | 942 | 77.79 |
| SignOrSymptom | 2065 | 1140 | 55.21 |
| IndividualBehaviour | 194 | 124 | 63.92 |
| TestOrMesureResult | 685 | 259 | 37.81 |
| Treatment | 4337 | 3775 | 87.04 |
| TestOrMeasure | 3576 | 2609 | 72.96 |
| AnatomicalConcept | 2616 | 2372 | 90.67 |
| Drug | 2593 | 2368 | 91.32 |
| Protein | 820 | 727 | 87.66 |
| Quality | 1153 | 1015 | 88.03 |
| Total | 26 925 | 22 451 | 83.38 |
As shown in Table 2, HYPHEN normalized a high percentage (83.38%) of entity annotations in the corpus to UMLS concept identifiers. Some examples of successful normalizations are shown in Table 3.
Table 3.
Sample normalization results
| Entity annotation | Semantic category | Mapped UMLS concept |
|---|---|---|
| increased PVR | Problem | Increased pulmonary vascular resistance (C1867423) |
| lung failure | Condition | Pulmonary failure (C0948755) |
| left atrial | AnatomicalConcept | Left atrium (C0225860) |
| arm training | Treatment | Upper limb training (C0556501) |
| spirometric test | TestOrMeasure | Spirometry test (C0037981) |
| genetic predisposition | RiskFactor | Genetic susceptibility to disease (C1455997) |
HYPHEN works well in normalizing entities describing single, straightforward concepts. Although most entity annotations possess such characteristics, performance is lower for categories whose annotations exhibit divergent characteristics. These include SignOrSymptom, whose annotations include long, detailed phrases, for example, daily productive cough for a minimum of 3 months for a minimum of 2 consecutive years or those mentioning multiple concepts, for example, coughing and/or corticosteroid-induced osteoporosis. The most problematic category, TestOrMeasureResult, includes mentions with no corresponding UMLS concepts (eg, negative pleural pressure), or those including numeric values (eg, oxygen saturation level 90%), which cannot be mapped to high oxygen saturation (C0852710) without additional processing.
Named Entity Recognition methods
We used the COPD corpus to train a named entity recognizer which can handle multiple levels of entity nesting.77 We adopted an existing neural network architecture64 for recognizing “flat” (ie, nonnested) named entities, to form the “building blocks” of our layered model for nested entity recognition. In this architecture, rich representations of word properties were obtained by combining word embeddings89 and character-level embeddings. A combination of BiLSTM and CRF was used to detect and classify entities.
Our approach builds upon this architecture, using a stack of multiple BiLSTM-CRF layers, each intended to detect a subset of entities. The input to each layer depends on the output of the previous layer. The input to the first layer consists of word and character-level embeddings for each individual word. The information about all words in each entity detected by this layer is merged into a single unit, whose representation combines information about each individual word in the entity. The merged information is passed to the next layer to aid in recognizing entities with higher levels of nesting. This key feature of our approach aims to account for potential dependencies between entities with different levels of nesting, that is, information about entities with lower nesting levels may provide clues about the presence of higher-level entities that include the nested entities within them.
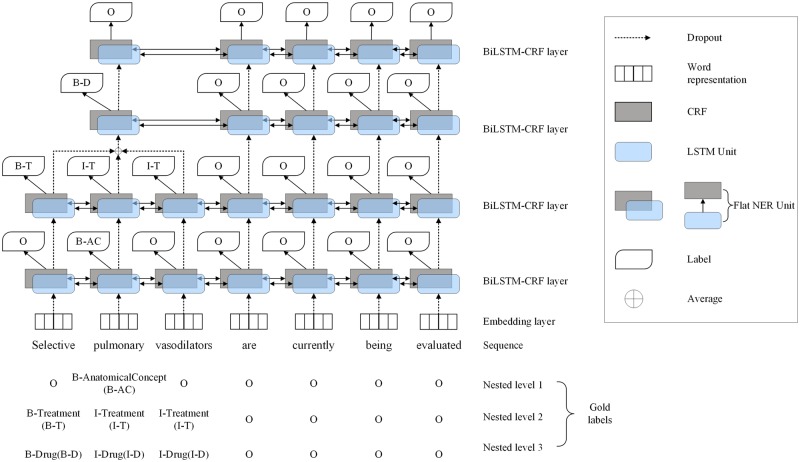
The method is dynamic—it stacks as many new layers as are necessary to allow all nested entities to be recognized; the method terminates when no entities are discovered by a newly stacked layer. Figure 4 illustrates the model architecture, where annotations are transformed into BIO tagging scheme labels to allow the model to be trained. These labels identify whether each word comes at the (B)eginning, I(nside), or (O)uside of an entity annotation. Although more complex tagging schemes may be used, for example, BIOES, which distinguishes words that constitute S(ingle) word entities, or which come at the E(nd) of multi-word entities, we chose to use BIO to avoid data sparsity problems, since some of our categories include relatively few annotations.
Figure 4.
Overview of the layered-BiLSTM-CRF model architecture. B-AC: B-AnatomicalConcept; B-T: B-treatment; I-T: I-treatment; B-D: B-drug; I-D: I-drug.
Baseline models
We firstly randomly split the corpus into 3 different parts—four-fifths for training, one-tenth for development (to tune parameters used by the models using Bayesian optimization90), and one-tenth for testing.
Based on previous studies,73,91 deciding on an optimal deep learning model, and whether to combine it with CRF, appears to be influenced by the task at hand. Using the layered architecture outlined above, we trained and evaluated different deep learning models using different algorithms (BiRNN, BiGRU, and BiLSTM), both in isolation and in combination with CRF; we found that the BiLSTM-CRF model attains the best results (see Supplementary Appendix S4 for performance statistics and tuned hyperparameter values).
We also compared our layered BiLSTM-CRF model to a CRF model and a “flat” (non-layered) BiLSTM-CRF model; the results of these experiments are shown in Table 4. We used NERSuite92 to implement the CRF model, whose features include contextual information, such as n-grams (ie, up to 3 words either side of the entity), parts-of-speech, syntactic chunks, and word base forms.92 In contrast, the non-layered BiLSTM-CRF uses only word and character-level embeddings instead of features, as described above.
Table 4.
Performance of different NER models at different levels of entity nesting
| Level | Model | P (%) | R (%) | F (%) |
|---|---|---|---|---|
| Innermost | CRF | 77.19 | 68.78 | 72.74a |
| BiLSTM-CRF | 73.93 | 73.38 | 73.56 a | |
| Layered BiLSTM-CRF | 69.79 | 70.41 | 70.10 | |
| Outermost | CRF | 73.63 | 66.41 | 69.83a |
| BiLSTM-CRF | 75.61 | 67.35 | 71.24a | |
| Layered BiLSTM-CRF | 74.00 | 74.54 | 74.27 | |
| All | CRF | 75.44 | 67.61 | 71.31a |
| BiLSTM-CRF | 74.71 | 70.42 | 72.50a | |
| Layered BiLSTM-CRF | 77.02 | 75.45 | 76.23 |
Note: For each different level, the best precision (P), recall (R), and F-score (F) amongst the 3 models is shown in bold.
Abbreviations: NER: named entity recognition; CRF: conditional random field.
A significant difference between CRF and (flat) BiLSTM-CRF models at P < .05. Since the layered BiLSTM-CRF takes as input different entities than the baseline models (ie, all entities vs innermost or outermost entities), we did not apply significance testing between layered and flat models.
Experimental settings
We conducted experiments in a single run rather than using cross-validation, in order to minimize overfitting to the training corpus. Our experiments evaluate performance variations of each model when entities with different levels of nesting are considered. We consider innermost entities, outermost entities, and all entities in the test dataset. Innermost entities are the most deeply nested entities, while outermost entities are non-nested entities. In Figure 1, elevation of pulmonary arterial pressures is the outermost entity, while pulmonary arterial is the innermost entity. Entities without nesting (eg, dyspnea) are included in both the innermost and outermost sets.
For the CRF and non-layered BiLSTM-CRF, we train separate models to recognize only innermost and outermost entities. In contrast, our layered BiLSTM-CRF is trained to recognize entities at all levels of nesting; we evaluate its performance in recognizing different levels of entities by considering outputs of different model layers.
RESULTS
Table 4 shows the performance of each model. The non-layered BiLSTM-CRF performs best for innermost entities, demonstrating how embeddings can successfully replace the multiple linguistic features used by the CRF. At this level, however, the layered BiLSTM-CRF has lower performance than the non-layered BiLSTM-CRF. For the layered model, we consider only the output of its first layer, which is expected to recognize only innermost entities. However, error analysis revealed that there is actually not a one-to-one correspondence between model layers and entity nesting levels, that is, the first layer sometimes detects entities belonging to other (ie, not innermost) entity levels. Conversely, higher layers of the model may detect entities that belong to the innermost nesting level.
For outermost entities, the non-layered BiLSTM-CRF still outperforms the CRF, reinforcing the advantages of deep learning. However, in contrast to innermost entities, the layered BiLSTM-CRF outperforms the non-layered model in detecting outermost entities. This clearly demonstrates how the layered model’s use of information about lower-level entities improves recognition of higher-level entities.
The higher performance of the layered BiLSTM-CRF for outermost entities also provides evidence that innermost entities are successfully recognized by lower levels of the model. This is confirmed by its superior performance to the other models in detecting all entities in the test dataset. Although there is no exact correspondence between the recognition of specific levels of entities and layers of the model, the complete model is still able to exploit the output of previous layers to achieve a high level of performance in detecting both outermost and nested entities. Detailed performance statistics for the layered BiLSTM-CRF by entity type are provided in Supplementary Appendix S5.
DISCUSSION
The results achieved by our layered BiLSTM-CRF in recognizing COPD-related information are superior to those achieved by applying the same model to nested entity recognition in well-used corpora from other domains.77 This provides evidence that our corpus is suitable for training high-performance ML-based tools, and that automatic recognition of COPD phenotypic information is a feasible task. Moreover, we have shown that detecting COPD phenotype information using deep learning models, which require minimal human intervention for training, can achieve superior performance to more traditional methods requiring time-consuming feature engineering, linguistic processing, and terminological resources. We have furthermore demonstrated that our layered model can achieve superior performance to a “flat” model, by exploiting information about nested entities when detecting the longer entities in which they are embedded.
These outcomes have important implications, in terms of improving the ease of locating phenotypic information in text. In particular, our nested entity detection method not only allows efficient location of COPD phenotype descriptions hidden in large text collections, but it also detects the internal structure of these descriptions. This provides scope to explore and categorize COPD phenotypes in a fine-grained manner. Since our method can be rapidly adapted to detect different types of information, it could be readily applied to find phenotypic information relating to other diseases, given suitably annotated corpora.
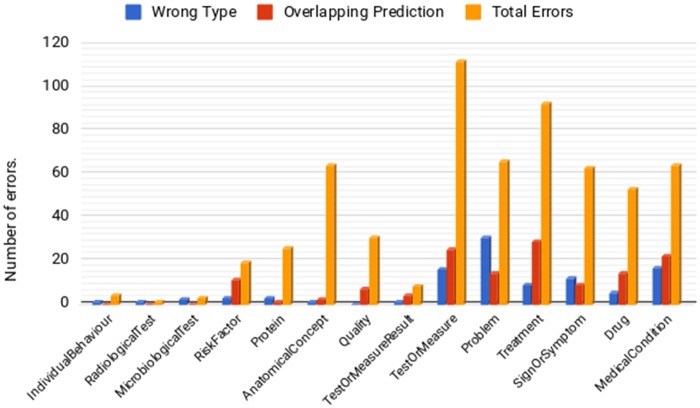
Error analysis of our NER results reveals that about 17% of erroneous entities have the correct text span, but the wrong semantic category. Figure 5 provides detailed error statistics for each semantic type, revealing that Problem is the most frequently misclassified category; these entities are mainly misclassified as either MedicalCondition or SignOrSymptom. Conversely, MedicalCondition entities are mostly misclassified as Problem. Such errors are possibly due to the fine-grained, hierarchical structure of our annotation scheme; the often subtle differences between similar categories may be difficult for the computer to distinguish. A further 23% of errors (most frequently Treatment and TestOrMeasure entities) concern cases where the model assigns the correct category, but the wrong text span (ie, it partially overlaps with the correct span). This may be due to the heterogeneity of phenotype descriptions, which can include mentions of various concept types, and which may or may not include modifier phrases. However, it is significant that in around 40% of the erroneous cases, the model can successfully detect the presence of entities, and categorize them correctly. Thus, even if the span is not completely correct, the model can find documents mentioning relevant entities, and allow examination of the context surrounding these entities.
Figure 5.
Counts of different types of errors for each semantic type.
CONCLUSION
We have described the construction of a novel corpus of full-text articles about COPD, annotated using a scheme that identifies pertinent information about COPD phenotypes, in which nested entity annotations make explicit the internal structure of potentially complex phenotype descriptions. The corpus is intended to assist the development of novel NER approaches to COPD phenotype recognition. The annotations were enriched using a high-performance normalization method to link the majority of them to UMLS Metathesaurus concepts.
We demonstrated the utility of the corpus by using it to train a deep learning-based NER model, which is designed to recognize entities with different levels of nesting and, in contrast to many ML-based models, relies on neither linguistic features nor external knowledge resources.
The detailed, fine-grained information about COPD phenotypes output by our model will facilitate development of semantic search systems for textual repositories, to pinpoint phenotype-relevant information, for example, to identify treatment regimens and investigate their relative effectiveness in different disease phenotypes. The ease of applying the NER model to newly available data will facilitate repeated interrogation of relevant data sources, allowing tracking of disease progression in individuals, and alerting clinicians to changes in disease pattern. Resolving entities to UMLS Metathesaurus concepts will facilitate concept-level search, in which all mentions of a concept of interest can be found automatically, regardless of the actual words or phrases used to describe them.
As future work, we will extend our framework to increase the complexity of the information extracted, inspired by recent work93,94 applying deep neural network models to medical relationship extraction. We will also apply our method to clinical records and to the detection of phenotypes of other diseases. This will reinforce the importance of our method in helping to enhance clinical phenotyping and early classification of disease subtype, providing a means of early, accurate diagnosis, and personalized treatment regimens for patients.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONTRIBUTORS
M.J. ran set up, ran the named entity recognition experiments, and analyzed their output. A.D.S., P.T., N.D.B., L.T., and G.V.G. contributed toward designing and refining the annotation scheme. A.D.S. and L.T. performed the annotation of the corpus. P.T. ran the normalization method and analyzed the output. P.T., M.J., and S.A. drafted the manuscript. N.D.B., L.T., and G.V.G. revised the manuscript. All authors read and approved the final version of the manuscript. S.A. supervised all steps of the work.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank John McNaught for his helpful comments on the manuscript. They also thank Piotr Przybyła for his help with the visualization of the annotations in the corpus.
FUNDING
This work was supported by the Manchester Molecular Pathology Innovation Centre (MMPathIC) [grant id: MR/N00583X/1]. G.V.G acknowledges support from H2020-EINFRA (731075) and the National Science Foundation (IOS:1340112) as well as support from the NIHR Birmingham ECMC, NIHR Birmingham SRMRC and the NIHR Birmingham Biomedical Research Centre and the MRC HDR UK. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, the Medical Research Council or the Department of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Global Strategy for Prevention, Diagnosis and Management of COPD 2018 Report. 2018. https://goldcopd.org/wp-content/uploads/2017/11/GOLD-2018-v6.0-FINAL-revised-20-Nov_WMS.pdf. Accessed October 23, 2018.
- 2. Naghavi M, Abajobir AA, Abbafati C, et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390 (10100): 1151–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miravitlles M, Soler-Cataluña JJ, Calle M, et al. Treatment of COPD by clinical phenotypes: putting old evidence into clinical practice. Eur Respir J 2013; 41 (6): 1252–6. [DOI] [PubMed] [Google Scholar]
- 4. Segreti A, Stirpe E, Rogliani P, et al. Defining phenotypes in COPD: an aid to personalized healthcare. Mol Diagn Ther 2014; 18 (4): 381–8. [DOI] [PubMed] [Google Scholar]
- 5. Cazzola M, Calzetta L, Rogliani P, et al. The challenges of precision medicine in COPD. Mol Diagn Ther 2017; 21 (4): 345–55. [DOI] [PubMed] [Google Scholar]
- 6. Miravitlles M, Calle M, Soler CJJ.. Clinical phenotypes of COPD: identification, definition and implications for guidelines. Arch Bronconeumol 2012; 48 (3): 86–98. [DOI] [PubMed] [Google Scholar]
- 7. Wouters E, Wouters B, Augustin I, et al. Personalized medicine and chronic obstructive pulmonary disease. Curr Opin Pulm Med 2017; 23 (3): 241–6. [DOI] [PubMed] [Google Scholar]
- 8. Heaney LG, McGarvey LP.. Personalised medicine for asthma and chronic obstructive pulmonary disease. Respiration 2017; 93 (3): 153–61. [DOI] [PubMed] [Google Scholar]
- 9. Gkoutos GV, Schofield PN, Hoehndorf R.. The anatomy of phenotype ontologies: principles, properties and applications. Brief Bioinfom 2018; 19 (5): 1008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han MK, Agusti A, Calverley PM, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med 2010; 182 (5): 598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeng Z, Deng Y, Li X, et al. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans Comput Biol Bioinform 2019; 16 (1): 139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Driel MA, Bruggeman J, Vriend G, et al. A text-mining analysis of the human phenome. Eur J Hum Genet 2006; 14 (5): 535.. [DOI] [PubMed] [Google Scholar]
- 13. Singhal A, Simmons M, Lu Z.. Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PLoS Comput Biol 2016; 12 (11): e1005017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017; 45 (D1): D833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson P, Ananiadou S.. Extracting gene-disease relations from text to support biomarker discovery In: proceedings of the 2017 International Conference on Digital Health; July 02–05, 2017; London, United Kingdom: ACM: 180–9. [Google Scholar]
- 16. Bundschus M, Dejori M, Stetter M, et al. Extraction of semantic biomedical relations from text using conditional random fields. BMC Bioinformatics 2008; 9 (1): 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kocbek S, Groza T.. Extracting disease-phenotype relations from text with disease-phenotype concept recognisers and association rule mining In: Proceedings of the 2017 IEEE 30th International Symposium on Computer-Based Medical Systems; June 22–24, 2017; Thessaloniki, Greece: IEEE: 358–63. [Google Scholar]
- 18. Sarntivijai S, Vasant D, Jupp S, et al. Linking rare and common disease: mapping clinical disease-phenotypes to ontologies in therapeutic target validation. J Biomed Semantics 2016; 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carroll RJ, Eyler AE, Denny JC.. Naïve electronic health record phenotype identification for rheumatoid arthritis In: Proceedings of the AMIA Annual Symposium; October 2011; Washington, DC: American Medical Informatics Association: 189–96. [PMC free article] [PubMed] [Google Scholar]
- 20. Wu H, Toti G, Morley KI, et al. SemEHR: a general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research. J Am Med Inform Assoc 2018; 25 (5): 530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ni Y, Kennebeck S, Dexheimer JW, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc 2015; 22 (1): 166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doğan RI, Leaman R, Lu Z.. NCBI disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform 2014; 47: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li J, Sun Y, Johnson RJ, et al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database (Oxford) 2016; 2016: baw068.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Mulligen EM, Fourrier-Reglat A, Gurwitz D, et al. The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships. J Biomed Inform 2012; 45 (5): 879–84. [DOI] [PubMed] [Google Scholar]
- 25. Gurulingappa H, Rajput AM, Roberts A, et al. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J Biomed Inform 2012; 45 (5): 885–92. [DOI] [PubMed] [Google Scholar]
- 26. Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013 In: Forner P, Müller H, Paredes R, Rosso P, Stein B, eds. International Conference of the Cross-Language Evaluation Forum for European Languages. Berlin: Springer, 2013: 212–31. [Google Scholar]
- 27. Ogren PV, Savova GK, Chute CG.. Constructing evaluation corpora for automated clinical named entity recognition In: Proceedings of the 6th international conference on Language Resources and Evaluation; May 2008; Marrakesh, Morocco: 2008: 3143–50. [Google Scholar]
- 28. Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009; 42 (5): 950–66. [DOI] [PubMed] [Google Scholar]
- 29. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alnazzawi N, Thompson P, Batista-Navarro R, et al. Using text mining techniques to extract phenotypic information from the PhenoCHF corpus. BMC Med Inform Decis Mak 2015; 15 (Suppl 2): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alnazzawi N, Thompson P, Ananiadou S.. Mapping phenotypic information in heterogeneous textual sources to a domain-specific terminological resource. PLoS One 2016; 11 (9): e0162287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang L, Bray BE, Shi J, et al. A method for the development of disease-specific reference standards vocabularies from textual biomedical literature resources. Artif Intell Med 2016; 68: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud Health Technol Inform 2006; 121: 279–90. [PubMed] [Google Scholar]
- 35. Kibbe WA, Arze C, Felix V, et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 2015; 43 (D1): D1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015; 43 (D1): D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Köhler S, Vasilevsky NA, Engelstad M, et al. The human phenotype ontology in 2017. Nucleic Acids Res 2017; 45 (D1): D865–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leaman R, Khare R, Lu Z.. Challenges in clinical natural language processing for automated disorder normalization. J Biomed Inform 2015; 57: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan J-W, Sood N, Huang Y.. Disorder concept identification from clinical notes: an experience with the ShARe/CLEF 2013 challenge In: CLEF 2013 Working Notes; 2013. http://ceur-ws.org/Vol-1179/CLEF2013wn-CLEFeHealth-FanEt2013.pdf. Accessed October 23, 2018. [Google Scholar]
- 40. Lee H-C, Hsu Y-Y, Kao H-Y.. AuDis: an automatic CRF-enhanced disease normalization in biomedical text. Database (Oxford) 2016; 2016: baw091.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thompson P, Ananiadou S.. HYPHEN: a flexible, hybrid method to map phenotype concept mentions to terminological resources. Terminology 2018; 24 (1): 91–121. [Google Scholar]
- 42. Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994; 1 (2): 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friedman C, Hripcsak G.. Evaluating natural language processors in the clinical domain. Methods Inf Med 1998; 37 (4-5): 334–44. [PubMed] [Google Scholar]
- 44. Savova GK, Tseytlin E, Finan S, et al. DeepPhe: a natural language processing system for extracting cancer phenotypes from clinical records. Cancer Res 2017; 77 (21): e115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Groza T, Köhler S, Doelken S, et al. Automatic concept recognition using the human phenotype ontology reference and test suite corpora. Database (Oxford) 2015; 2015: bav005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khordad M, Mercer RE, Rogan P.. Improving Phenotype Name Recognition. In: Butz C, Lingras P, eds. Advances in Artificial Intelligence. Berlin: Springer, 2011: 246–57. [Google Scholar]
- 47. Afzal N, Mallipeddi VP, Sohn S, et al. Natural language processing of clinical notes for identification of critical limb ischemia. Int J Med Inform 2018; 111: 83–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Breitenstein MK, Liu H, Maxwell KN, et al. Electronic health record phenotypes for precision medicine: perspectives and caveats from treatment of breast cancer at a single institution. Clin Transl Sci 2018; 11 (1): 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao J, Moore LR, Blank CE, et al. Microbial phenomics information extractor (MicroPIE): a natural language processing tool for the automated acquisition of prokaryotic phenotypic characters from text sources. BMC Bioinformatics 2016; 17: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Collier N, Groza T, Smedley D, et al. PhenoMiner: from text to a database of phenotypes associated with OMIM diseases. Database (Oxford) 2015; 2015: bav104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oellrich A, Collier N, Smedley D, et al. Generation of silver standard concept annotations from biomedical texts with special relevance to phenotypes. PLoS One 2015; 10 (1): e0116040.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collier N, Oellrich A, Groza T.. Concept selection for phenotypes and diseases using learn to rank. J Biomed Semantics 2015; 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Khordad M, Mercer RE, Rogan P.. A machine learning approach for phenotype name recognition. Proceedings of the International Conference on Computational Linguistics. 2012; 2012: 1425–40. [Google Scholar]
- 54. Collier N, Tran M-V, Le H-Q, et al. Learning to recognize phenotype candidates in the auto-immune literature using SVM re-ranking. PLoS One 2013; 8 (10): e72965.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brbić M, Piškorec M, Vidulin V, et al. Phenotype inference from text and genomic data In: Altun Y, Das K, Mielikäinen T, Malerba D, Stefanowski J, Read J, Žitnik M, Ceci M, Džeroski S, eds. Mach Learn Knowl Discov Databases. Cham, Switzerland: Springer, 2017: 373–7. [Google Scholar]
- 56. Finkel JR, Manning CD.. Nested named entity recognition In: Proceedings of the conference on Empirical Methods in Natural Language Processing; August 2009; Singapore: Association for Computational Linguistics: 141–50. [Google Scholar]
- 57. Lu W, Roth D.. Joint mention extraction and classification with mention hypergraphs. In: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; September 2015; Lisbon, Portugal: 857–67. [Google Scholar]
- 58. Muis AO, Lu W.. Labeling gaps between words: recognizing overlapping mentions with mention separators. In: Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing; September 2017; Copenhagen, Denmark: 2608–18. [Google Scholar]
- 59. Gehrmann S, Dernoncourt F, Li Y, et al. Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PLoS One 2018; 13 (2): e0192360.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beaulieu-Jones BK, Greene CS.. Semi-supervised learning of the electronic health record for phenotype stratification. J Biomed Inform 2016; 64: 168–78. [DOI] [PubMed] [Google Scholar]
- 61. Che Z, Kale D, Li W, et al. Deep computational phenotyping In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 2015; Sydney, NSW, Australia: ACM: 507–16. [Google Scholar]
- 62. Collobert R, Weston J.. A unified architecture for natural language processing: deep neural networks with multitask learning In: proceedings of the 25th International Conference on Machine Learning; July 05–09, 2008; Helsinki, Finland: ACM: 160–7. [Google Scholar]
- 63. Collobert R, Weston J, Bottou L, et al. Natural language processing (almost) from scratch. J Mach Learn Res 2011; 12: 2493–537. [Google Scholar]
- 64. Lample G, Ballesteros M, Subramanian S, et al. Neural architectures for named entity recognition In: Proceedings of the Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2016. San Diego, CA: Association for Computational Linguistics: 260–70. [Google Scholar]
- 65. Gridach M. Character-level neural network for biomedical named entity recognition. J Biomed Inform 2017; 70: 85–91. [DOI] [PubMed] [Google Scholar]
- 66. Xing W, Qi J, Yuan X, et al. A gene–phenotype relationship extraction pipeline from the biomedical literature using a representation learning approach. Bioinformatics 2018; 34 (13): i386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goller C, Kuchler A.. Learning task-dependent distributed representations by back propagation through structure. In: Proceedings of the International Conference on Neural Networks; June 1996; Washington DC: 347–52. [Google Scholar]
- 68. Chiu JP, Nichols E.. Named entity recognition with bidirectional LSTM-CNNs. Trans Assoc Comput Linguist 2016; 4: 357–70. [Google Scholar]
- 69. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 70. Cho K, van Merriënboer B, Gulcehre C, et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing; October 2014; Doha, Qatar: 1724–34. [Google Scholar]
- 71. Ratinov L, Roth D.. Design challenges and misconceptions in named entity recognition In: proceedings of the Thirteenth Conference on Computational Natural Language Learning; June 2009; Boulder, Colorado: Association for Computational Linguistics: 147–55. [Google Scholar]
- 72. Ma X, Hovy E.. End-to-end sequence labeling via bi-directional LSTM-CNNS-CRF In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics; August 2016; Berlin, Germany: 1064–74. [Google Scholar]
- 73. Manda P, Beasley L, Mohanty S.. Taking a dive: experiments in deep learning for automatic ontology-based annotation of scientific literature In: Proceedings of the International Conference on Biomedical Ontology; 2018. http://ceur-ws.org/Vol-2285/ICBO_2018_paper_18.pdf. Accessed February 16, 2019. [Google Scholar]
- 74. Xu M, Jiang H, Watcharawittayakul S.. A local detection approach for named entity recognition and mention detection In: Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics; July 2017; Vancouver, Canada: 1237–47. [Google Scholar]
- 75. Li F, Zhang M, Tian B, et al. Recognizing irregular entities in biomedical text via deep neural networks. Pattern Recognit Lett 2018; 105: 105–13. [Google Scholar]
- 76. Nguyen T-S, Nguyen L-M.. Nested named entity recognition using multilayer recurrent neural networks In: Hasida K, Pa Pa W, eds. International Conference of the Pacific Association for Computational Linguistics. Singapore: Springer; 2017: 233–46. [Google Scholar]
- 77. Ju M, Miwa M, Ananiadou S.. A neural layered model for nested named entity recognition In: Proceedings of the Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; June 2018; New Orleans, Louisiana: 1446–59. [Google Scholar]
- 78. Saeed M, Villarroel M, Reisner AT, et al. Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II): a public-access intensive care unit database. Crit Care Med 2011; 39 (5): 952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Uzuner Ö, Luo Y, Szolovits P.. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007; 14 (5): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 17 (1): 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Frantzi K, Ananiadou S, Mima H.. Automatic recognition of multi-word terms: the C-value/NC-value method. Int J Digit Libr 2000; 3 (2): 115–30. [Google Scholar]
- 82. Celli BR, MacNee W, Agusti A, et al. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J 2004; 23 (6): 932–46. [DOI] [PubMed] [Google Scholar]
- 83. Fu X, Batista-Navarro R, Rak R, et al. Supporting the annotation of chronic obstructive pulmonary disease (COPD) phenotypes with text mining workflows. J Biomed Semantics 2015; 6 (1): 8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rak R, Rowley A, Black W, et al. Argo: an integrative, interactive, text mining-based workbench supporting curation. Database (Oxford) 2012; 2012: bas010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thompson P, Daikou S, Ueno K, et al. Annotation and detection of drug effects in text for pharmacovigilance. J Cheminform 2018; 10 (1): 37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hripcsak G, Rothschild AS.. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005; 12 (3): 296–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Thompson P, Iqbal SA, McNaught J, et al. Construction of an annotated corpus to support biomedical information extraction. BMC Bioinformatics 2009; 10 (1): 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Medical Subject Headings (MeSH). https://http://www.nlm.nih.gov/mesh/. Accessed December 19, 2018.
- 89. Chiu B, Crichton G, Korhonen A, et al. How to train good word embeddings for biomedical NLP In: proceedings of the 15th Workshop on Biomedical Natural Language Processing; August 2016; Berlin, Germany: 166–74. [Google Scholar]
- 90. Snoek J, Larochelle H, Adams RP.. Practical Bayesian optimization of machine learning algorithms In: Proceedings of the 25th International Conference on Neural Information Processing Systems; December 03–06, 2012; Lake Tahoe, Nevada: 2951–9. [Google Scholar]
- 91. Yang J, Liang S, Zhang Y.. Design challenges and misconceptions in neural sequence labeling In: Proceedings of the International Conference on Computational Linguistics; August 2018; Santa Fe, New Mexico, USA: 3879–89. [Google Scholar]
- 92. Cho H, Okazaki N, Miwa M, et al. NERsuite: a named entity recognition toolkit. 2010http://nersuite.nlplab.org/. Accessed October 23, 2018.
- 93. Luo Y. Recurrent neural networks for classifying relations in clinical notes. J Biomed Inform 2017; 72: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jagannatha AN, Yu H.. Bidirectional RNN for medical event detection in electronic health records. In: Proceedings of the Annual Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; June 2016; San Diego, California: 473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.