Abstract
Objectives
Systematic reviews of clinical trials could be updated faster by automatically monitoring relevant trials as they are registered, completed, and reported. Our aim was to provide a public interface to a database of curated links between systematic reviews and trial registrations.
Materials and Methods
We developed the server-side system components in Python, connected them to a PostgreSQL database, and implemented the web-based user interface using Javascript, HTML, and CSS. All code is available on GitHub under an open source MIT license and registered users can access and download all available data.
Results
The trial2rev system is a web-based interface to a database that collates and augments information from multiple sources including bibliographic databases, the ClinicalTrials.gov registry, and the actions of registered users. Users interact with the system by browsing, searching, or adding systematic reviews, verifying links to trials included in the review, and adding or voting on trials that they would expect to include in an update of the systematic review. The system can trigger the actions of software agents that add or vote on included and relevant trials, in response to user interactions or by scheduling updates from external resources.
Discussion and Conclusion
We designed a publicly-accessible resource to help systematic reviewers make decisions about systematic review updates. Where previous approaches have sought to reactively filter published reports of trials for inclusion in systematic reviews, our approach is to proactively monitor for relevant trials as they are registered and completed.
Keywords: review literature as topic, semi-supervised learning, databases as topic, bibliographic databases
INTRODUCTION
Systematic reviews of clinical trials are a critical resource supporting policy and clinical decision-making, but they are challenging to keep current. Producing and updating systematic reviews is resource-intensive and the volume with which new evidence is produced can outpace our ability to keep up.1,2 Updating systematic reviews to incorporate new evidence has traditionally involved a manual search for new relevant studies and a re-evaluation of the results. Without undertaking a new search for evidence, it is difficult to determine whether an update may be warranted,3 or if the available new evidence is likely to lead to a change in conclusion.
To reduce the effort needed to undertake systematic reviews, researchers have automated some systematic review processes. Much of the effort on automation has been on the most burdensome processes: searching and screening of bibliographic databases.4–9 Investigators attempting to automate the screening of articles rely on private sources of data or need to manually extract information from published systematic reviews. Training data used in the development and evaluation of these methods were limited to between 1 and 24 systematic reviews.5–13
Using bibliographic databases as the primary source of information for creating or updating a systematic review creates challenges related to timeliness and completeness. Around half of all clinical trials remain unpublished after 2 years,14–16 and of those that are published, around half have missing or changed outcomes.17 Safety outcomes in particular tend to be left out of published trial reports, introducing a challenge for systematic reviewers wanting to provide a timely and complete synthesis of clinical trial results.
Compared with bibliographic databases, clinical trial registries may provide an earlier and more complete view of trials that may be relevant to a systematic review. Some investigators have used clinical trial registries to identify publication bias,14,16,18–22 and the selective reporting of results that can lead to bias in the evidence captured by a systematic review.16,23,24 A series of changes in culture and policy now mean that most published trials are registered.25–27 While trial registries are also imperfect sources of information about trials,28,29 they can be a timelier source of evidence than bibliographic databases, and may be especially useful for signaling when new trial evidence is—or is soon to be—available for incorporation into a systematic review update.
Other recent advances in systematic review technologies include the development of living systematic reviews,30,31 which reduce overhead associated with undertaking new systematic reviews and full updates. Another emerging method is based on crowd-sourcing,32–34 which has been effective in reducing reliance on experts for screening. Both approaches suggest a change in focus away from improving processes inside standalone systematic reviews and toward connecting resources to improve the efficiency of the entire systematic review ecosystem.
Our aim was to create a new method for systematic reviewers to monitor when relevant trial evidence becomes available and assess the need for a systematic review update.
METHODS
We created trial2rev, a shared space for humans and software agents to work together to proactively monitor the status of registered trials that are likely to be relevant to a systematic review update.
System description
The central component of the system is a database of structured records linking trials from their registrations to published systematic reviews. The database is populated by extracting information from a range of data sources and from crowd-sourcing. The typical way in which a human user interacts with the system is by browsing or searching for a published systematic review, discovering what the system already knows about the trials included in the review, verifying the assignment of included trials, and reviewing and voting on the trials that the system predicts to be relevant to an update of the systematic review. Users must register an account in the system to carry out actions such as verifying and voting on trials; unregistered users may interact with the system in a read-only fashion. As users interact with the system, software agents work in parallel to retrieve information about links from external data sources or to suggest candidate trials through machine learning methods. The system records all information provided by registered users, and immediately makes it available for software agents and other users.
Data sources and database structure
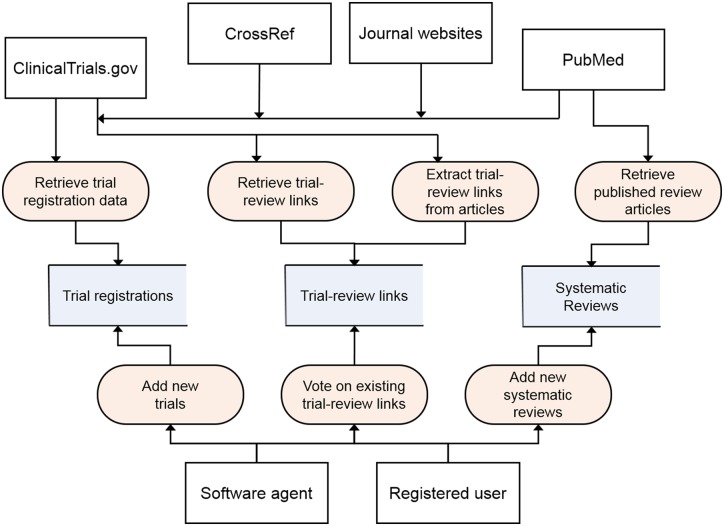
The main sources of data for the system include ClinicalTrials.gov, PubMed, CrossRef, and the websites of journals including the Cochrane Database of Systematic Reviews (Figure 1).
Figure 1.
Major data flows across the trial2rev system, including external data sources and software agents (white boxes), process classes (rounded rectangles), and tables in the database (open rectangles).
We use the Entrez Programming Utilities to access bibliographic information available in PubMed via the application programming interface (API). When the system or a registered user adds a systematic review or trial publication to the system, we store a local copy of the title, abstract, authors, source, publication date, PubMed ID, and Digital Object Identifier (DOI), where available.
We access ClinicalTrials.gov to download a subset of the available information for every registered trial or study. This information is updated daily, and our local copy includes the title, number of expected or actual participants, expected or actual completion date, and the most recent status.
We access the CrossRef database via API when a registered user or scheduled search adds a new systematic review to the database. Using the systematic review’s DOI, we use CrossRef to find articles included in its references. If these data are available via CrossRef, we then search for each returned article on PubMed to check if any have metadata links to a trial registration on ClinicalTrials.gov. Not all cited articles with links to ClinicalTrials.gov represent included trials (systematic reviews often cite relevant but excluded studies), so we add these with an unverified status.
To facilitate further research in the area, we make the complete set of trial-review links available for direct download as a single large sparse matrix linking the set of all available trials with the set of all available systematic reviews. For each trial, the dataset includes information about their verification (for potentially included trials) and the patterns of voting (for potentially relevant trials). We designed the downloadable dataset so that the trial-review matrix can be used directly in the training and testing of new methods aimed at supporting screening, and can be transformed to construct a review–review network of similarity where the connections between any 2 systematic reviews is a function of the number of shared trials.
Software agents and machine learning methods
The software agents interact with the system to add and vote on included and relevant trials (Table 1). Basic software agents that use document similarity methods rank relevant trials based on their similarity to either the systematic review text or the text of already-included trials. Other software agents are designed to crawl or access external data sources including CrossRef and the websites of individual journals to mine for references that are likely to capture some of the included trials. Software agents that use machine learning methods include one based on a matrix factorization method that used a shared latent space to rank relevant trials.35
Table 1.
Descriptions of software agents used to add or vote on trials
| Name | Data sources | Schedules and triggers | Descriptions |
|---|---|---|---|
| basicbot1 | PubMed, ClinicalTrials.gov | Triggered when a review is added; updated weekly | Uses document similarity methods to rank and recommend trials for each review based on the cosine similarity between the text of the title and abstract of the systematic review from PubMed, and free text sections of trial registrations from ClinicalTrials.gov |
| basicbot2 | ClinicalTrials.gov | Triggered when a review receives a new included trial; updated weekly | Uses document similarity methods to rank and recommend new trials for a systematic review based on the cosine similarity between free text sections of registry entries of verified and unverified included trials and the free text sections of trial registrations from ClinicalTrials.gov |
| crossrefbot | CrossRef, PubMed | Triggered once when a review is first added | Resolves review-trial links by extracting available reference lists, resolving each citation to a PubMed article, and checking each resolved article for metadata links to ClinicalTrials.gov |
| matfacbot | ClinicalTrials.gov | Scheduled to run once a week | Uses a matrix factorization-based approach to predict missing links between systematic reviews and trial registrations using a small number of positive labeled examples. A computationally expensive method that leverages information from many systematic reviews to learn how to predict missing links |
| cochranebot | Cochrane Database of Systematic Reviews (CDSR); PubMed; ClinicalTrials.gov | Triggered once when a CDSR review is first added | Extracts lists of included, excluded, and ongoing studies from reference lists available on the CDSR journal website. Any ClinicalTrials.gov NCT Numbers are used directly, and references to articles are reconciled against PubMed information and checked for metadata links to trial registrations |
We designed the system to activate software agents in 2 ways—scheduled or triggered by the actions of registered users or other software agents. We use scheduled actions to update the database where we check for updates en masse in external resources or where the methods are computationally expensive (eg the matrix factorization method). We use triggered actions for when registered users might expect real-time or near real-time responses to queries, such as adding a new systematic review, checking external sources for included studies, and for prediction algorithms that only need to access a limited amount of information and take seconds to execute.
Human users and crowdsourcing
We created a web-based interface for the trial2rev system to allow all users to monitor the number and status of new and relevant trials over time, and to facilitate crowdsourcing of information about included and relevant trials by registered users. All users can navigate to systematic reviews already included in the database to monitor the status of relevant trials. Registered users can interact with the system by voting on trials predicted by software agents and other registered users, verifying the correctness and completeness of included trials, and by adding trial registrations not already predicted or added by other registered users and software agents. We implemented user registration for the system because votes and added trials are used as training data, so the impact of introduced errors may propagate to affect the quality of recommendations in other reviews.
Voting is a mechanism used by crowdsourced websites such as Reddit and StackOverflow to outsource the curation, filtering, and ordering of content displayed on their websites. A requirement of trial2rev was to enable registered users to vote on trials based on their expected relevance to an update. This is the main way in which human users and software agents can cooperate to improve the accuracy and robustness of the machine learning methods that underpin the software agents, which in turn reduces the amount of work that users need to do to screen for trials that are expected to be included in any update. Registered users can vote once per trial for each systematic review and can change their vote (eg from voting up on relevance to voting down on relevance) or remove their vote entirely. All users can choose to order the list of relevant trials according to the sum of votes (upvotes plus downvotes) or by the net votes (upvotes minus downvotes). The former allows users to quickly scan through the most relevant trials, while the latter also gives prominence to trials where registered users and software agents disagree on their relevance.
A further requirement of the system was to allow registered users the ability to directly submit relevant or included trials. It is unlikely that computational methods will achieve a recall of 100% for all trials included in or relevant to a review, and our system therefore relies on human users to identify trials that software agents may have missed and to make them available for voting. Users submit trials using the trial’s ClinicalTrials.gov identifier (ie NCT Number). When a user submits a trial, the trial also automatically receives an upvote from the user. Cases where trials have already been added to the included or relevant lists are handled as expected; for example, by moving between relevant and included.
RESULTS
Implementation
We implemented the prototype of the trial2rev system using an agile development approach, adding and expanding on features iteratively. When deciding on features to implement, we were guided by the main goals of the system related to updating published systematic reviews and avoiding duplication of effort. For example, we prioritized the inclusion of lists of related reviews and the ability to search and browse known reviews because it directly addressed the goal of avoiding duplication of effort.
We used a PostgreSQL relational database to store information about systematic reviews, trial registrations, trial articles, and the links connecting them. We used Python to implement the interface to the database, execute trial prediction methods, and handle the backend application logic for the website. The website uses the Flask web framework and the client-side code for the user interface is in JavaScript and HTML using the Bootstrap framework. We host both the database and web servers using Amazon Web Services (AWS). Source code for the system is available on GitHub (https://github.com/evidence-surveillance/trial2rev). To verify that the system supports its intended use cases, we validated it against a set of test cases. We additionally carried out unit testing on individual components of the server-side Python code.
We initially populated the database by searching PubMed for systematic reviews. We were deliberately conservative with our approach to identifying systematic reviews, requiring the use of the terms “systematic review” or “meta-analysis” in the title, and of those, we added systematic reviews to the database if they included at least one known link to a trial in ClinicalTrials.gov. We additionally added all systematic reviews published in the Cochrane Database of Systematic Reviews if they met the same criterion: they must reference at least one PubMed article with a metadata link to ClinicalTrials.gov or have included a reference to an NCT Number in their list of included or ongoing studies. We implemented weekly updates, which identify any newly published systematic reviews with links to trial registrations, daily updates that check for changes in the status of candidate trials (eg trial completion or termination, as well as new links to published reports), and these scheduled updates then trigger software agents that may identify new relevant trial registrations.
The database now includes over 11 500 systematic reviews. In the period between August 2017 and August 2018, an average of 425 new systematic reviews were indexed by PubMed each week. Of these, an average of 61 systematic reviews per week had identifiable links to one or more trial registrations.
Web-based interface
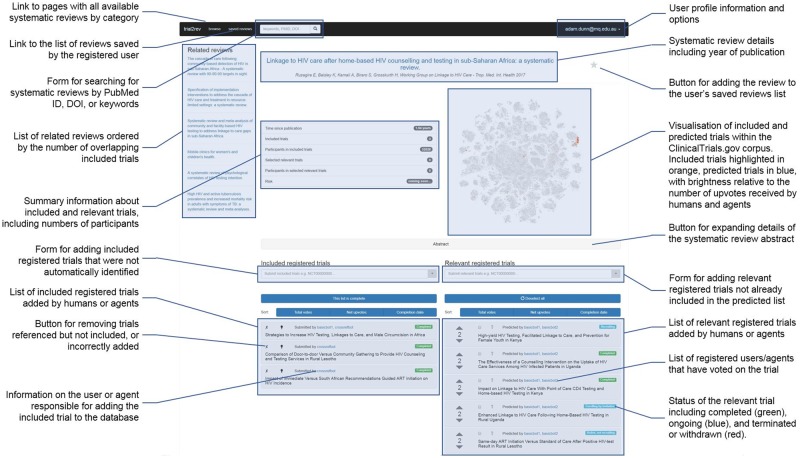
The web-based interface to the system allows users to view and modify what the system knows about a systematic review (Figure 2). We have made it publicly available at https://surveillance-chi.mq.edu.au.
Figure 2.
Design of the prototype web interface for an individual systematic review in the trial2rev system.
After a user finds a systematic review by searching, browsing, or adding it by PubMed identifier or DOI, the system presents users with a view of what is known about the systematic review. This includes a list of included trials (on the left), a list of candidate trials that may be relevant for inclusion in an update (on the right), and related systematic reviews (far left). All users have access to this information and can examine additional details about included and candidate trials, including status, linked articles, numbers of participants, or by using the links to navigate to pages on ClinicalTrials.gov and PubMed. The webpage for each systematic review also displays a visualization of ClinicalTrials.gov trial registrations based on document similarity, which helps users assess the heterogeneity of trials included or predicted for inclusion in a systematic review.
Once they have logged in to the system, registered users are able to modify what is known about a systematic review via the web interface. Registered users can add new systematic reviews; add included and candidate trial registrations; verify the completeness of a list of included trial registrations; upvote candidate trial registrations they believe are likely to be included in an update; downvote trial registrations they believe are likely to be excluded from an update; or move candidate trial registrations between the included and candidate lists if they have been miscategorized. Registered users are also able to see which software agents and registered users have voted on candidate trial registrations or added included trial registrations.
Use cases for systematic reviewers and systematic review researchers
The trial2rev system supports several use cases related to making decisions about updating systematic reviews. For systematic reviewers, the first use case is related to monitoring the availability of new evidence relevant to a review. Systematic reviewers navigate to a systematic review; verify and add to the set of included trial registrations; work together with the software agents to identify trial registration candidates that are likely to be relevant to an update; and add the systematic review to their personal list of saved reviews to receive updates whenever a relevant candidate trial changes status. This allows systematic reviewers to proactively monitor for the need to update a systematic review as trials are completed and their results are made available.
The second class of use cases for systematic reviewers is related to avoiding duplication of effort among systematic review groups. The trial2rev system supports this use case in 2 ways—via the public sharing of information curated by software agents and registered users, and the construction of a network of related systematic reviews. For example, when a systematic reviewer navigates to a systematic review page, the system presents them with a list of related reviews (including the year they were published), which can be used to help decide whether an update or a new systematic review is warranted even when new trial evidence is available.
Use cases for systematic review researchers relate to the training of new machine learning methods for automating trial screening and evaluating the efficiency of the systematic review ecosystem. For example, registered users of the system can download an up-to-date version of what is known about the complete set of systematic reviews known to the system, including information about the registrations of included trials and trials that have been predicted to be relevant. While these data are a mix of verified and noisy information, they may be used in the training and evaluation of new machine learning methods that aim to reduce the manual effort associated with screening trials for inclusion in systematic reviews. By transforming the dataset to produce a similarity network describing the overlapping of systematic reviews, clinical epidemiology researchers may be able to use these data to conduct large-scale assessments of levels of redundancy across systematic reviews or examine differences in conclusion across systematic reviews that include the same trials.
DISCUSSION
Systematic reviewers and systematic review researchers can use the trial2rev system to proactively monitor the accumulation of new trial evidence relative to a published systematic review. The primary function of the system is to help systematic reviewers decide when to update a systematic review. The system may also be used to support the development of tools for automating screening or monitoring, and to undertake large-scale analyses of systematic reviews within and across disciplines.
Other systems have been developed to utilize crowdsourcing and curation of external data sources to improve access to trial information and systematic reviews. Cochrane Cloud is a platform that allows users to screen citations and identify randomized controlled trials.32,34 Cochrane’s Central Register of Controlled Trials stores the crowdsourced citations, which they integrate into their search engine as filters. Vivli,36 aims to improve access to clinical results data through curation, and currently has information about more than 3000 studies, encouraging their use in systematic reviews. TrialsTracker,37 which uses data from ClinicalTrials.gov and metadata links from PubMed (equivalent to one of the steps used in our base system and CrossRef software agent) to detect a biased subset of the trials that have reported results. Our approach differs in that we focus on curating information rather than relying only on recorded links to draw conclusions,38 and we store a large sample of links between systematic reviews and relevant trials.
Our system is also related to research that uses machine learning methods to support the searching and screening processes undertaken by systematic reviewers. Most of the current approaches for reducing manual effort in trial screening use both positive (included studies) and negative (excluded studies) labeled examples to train classifiers.7,11 The matrix factorization approach is different to most other previous approaches not only because it screens trial registrations rather than trial articles, but also because it is a learning to rank method that only requires positive labeled examples.35 However, we expect trial2rev to make a greater number of downvotes available via crowdsourcing and machine-readable lists of excluded studies. As the number of downvotes available in the system increases, it will provide a source of training data for approaches that require positive and negative labeled examples as training data.
Future work
We deliberately chose to represent trials by their registrations on ClinicalTrials.gov rather than their published reports because our aim was to monitor ongoing and completed trials proactively. Bibliographic databases and trial registries represent incomplete but complementary sources of data and we could improve the completeness of the data available in the system by augmenting the sets of included and relevant trials to include trials that have been reported but were not registered. To do this, we would allow extra trials to be represented using one or more DOIs or PubMed identifiers without necessarily having a corresponding NCT Number.
The system currently only accepts published systematic reviews, but could be extended to store information about systematic review protocols in order to support the automation of screening in new systematic reviews rather than just in updates. While some of the machine learning methods that rely on known included trials would not be applicable, other methods that predict relevant trials from the text of the protocol could be used for this purpose. Systematic reviewers may also wish to start with a seeding list of trials that they know match a set of inclusion criteria, and these could be used to identify published reviews to decide whether a new review is warranted. As a systematic reviewer starts to select seeding trials or additional trials for inclusion in a new systematic review, this could be implemented in a process that is equivalent to a cold start followed by an active learning approach.13
Since our focus was on providing information relative to systematic reviews, we did not implement separate pages detailing what is known about individual trials. Though we store information about trials in the database, including links between trial registrations and their published reports, the information is not searchable and only appears on systematic review pages. These could in turn be supported by methods we have developed to identify missing links between trial registrations and their published reports.39 Similarly, we did not provide functionality for previously unpublished systematic reviews.
Methods are available for estimating the likelihood that a systematic review will change conclusion if it is updated, learning from previous examples of updated systematic reviews and using characteristics of the systematic reviews, timing, primary meta-analyses, and included and relevant trials.40,41 We could use the information stored by trial2rev as inputs for similar methods and present an estimate of the risk to users on each systematic review page. However, these estimates may depend critically on information about unregistered trials, and we would need to critically assess whether the approach would work using only registered trials.
Once the system has had time to mature, it will need a formal utility and usability evaluation involving end users. An evaluation of the system will need to assess how the system influences the decisions of expert systematic reviewers about when to update a systematic review and the amount of work required to reach that decision. One approach would be to ask systematic reviewers with and without access to trial2rev to rank previously published systematic reviews according to their need for an update (excluding other factors that might suggest an update is warranted). We would then measure the utility of the system by comparing where the 2 groups of systematic reviewers ranked systematic reviews that had updates reporting a change in results and conclusions; usability would be measured by having the group who accessed trial2rev complete the System Usability Scale questionnaire.42
Individual machine learning methods incorporated into the system have already been assessed for their performance in identifying relevant trial registrations.35 However, it is not yet clear how additional training data produced through crowdsourcing might lead to improvements in the performance of methods aimed at predicting relevant trial registrations. It is also not known how additional machine learning methods might interact and combine with crowdsourcing to change how the system recommends relevant trial registrations. The system is currently new, does not have a critical mass of registered users, and will be updated to include new machine learning based software agents over time. As the number of human user contributions grow, and new software agents are added to the trial2rev system, we recommend that they be evaluated in experiments that are independent of the system (using a snapshot of the available trial-review link matrix) and for their effect on the quality of trial recommendations within the system.
Limitations
First, the choice to utilize crowdsourcing to improve the amount and quality of the data in the system is a limitation. There is no guarantee that the system will have enough contributions from registered users to ensure that new systematic reviews are verified as having listed all included trials with registrations on ClinicalTrials.gov. This may limit the utility of the system for unregistered users who want to view systematic reviews without verifying the available information. Second, there is also the potential for registered users to act maliciously by intentionally providing incorrect information. This is important for a system like trial2rev where some software agents utilize voting patterns as training data to learn how to predict relevant trials, because incorrect information may propagate through the system to affect predictions in other systematic reviews. To balance the potential for abuse with utility we allow only registered users to make changes to the database. Third, the system relies on several external data sources such as PubMed, ClinicalTrials.gov, CrossRef, and journal websites including Cochrane Database of Systematic Reviews. Changes to the APIs or website structures would affect the reliability and maintainability of the system.
CONCLUSION
The trial2rev system is a shared space for humans and software agents to work together to improve access to structured information about systematic reviews and monitor the accumulation of trial evidence relevant to published systematic reviews. The potential benefits of such a system include facilitating more timely updates of systematic reviews, and reducing the workload involved in searching and screening articles for systematic reviews. The system automatically grows with the publication of systematic reviews and includes methods to augment and correct the information available about each systematic review.
CONTRIBUTORS
PM designed and implemented the methods, implemented the system, and drafted the manuscript. RB evaluated the system and critically reviewed the manuscript. DS designed and implemented methods and critically reviewed the manuscript. FB and AD designed the research, evaluated the system, and critically reviewed the manuscript.
FUNDING
This work was supported by the Agency for Healthcare Research and Quality (R03HS024798 to FTB).
Conflict of interest statement. None declared.
REFERENCES
- 1. Garner P, Hopewell S, Chandler J, et al. When and how to update systematic reviews: consensus and checklist. BMJ 2016; 354: i3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastian H, Glasziou P, Chalmers I.. Seventy-five trials and eleven systematic reviews a day: how will we ever keep op? PLoS Med 2010; 79: e1000326.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garritty C, Tsertsvadze A, Tricco AC, Sampson M, Moher D.. Updating systematic reviews: an international survey. PLoS One 2010; 54: e9914.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen AM, Hersh WR, Peterson K, Yen PY.. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc 2006; 132: 206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen AM, Ambert K, McDonagh M.. Cross-topic learning for work prioritization in systematic review creation and update. J Am Med Inform Assoc 2009; 165: 690–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsafnat G, Glasziou P, Choong MK, Dunn A, Galgani F, Coiera E.. Systematic review automation technologies. Syst Rev 2014; 31: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Mara-Eves A, Thomas J, McNaught J, Miwa M, Ananiadou S.. Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev 2015; 41: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shemilt I, Simon A, Hollands GJ, et al. Pinpointing needles in giant haystacks: use of text mining to reduce impractical screening workload in extremely large scoping reviews. Res Synth Methods 2014; 51: 31–49. [DOI] [PubMed] [Google Scholar]
- 9. Thomas J, McNaught J, Ananiadou S.. Applications of text mining within systematic reviews. Res Synth Methods 2011; 21: 1–14. [DOI] [PubMed] [Google Scholar]
- 10. Miwa M, Thomas J, O’Mara-Eves A, Ananiadou S.. Reducing systematic review workload through certainty-based screening. J Biomed Inform 2014; 51: 242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shekelle PG, Shetty K, Newberry S, Maglione M, Motala A.. Machine learning versus standard techniques for updating searches for systematic reviews: a diagnostic accuracy study. Ann Intern Med 2017; 1673: 213–5. [DOI] [PubMed] [Google Scholar]
- 12. Ji X, Ritter A, Yen P-Y.. Using ontology-based semantic similarity to facilitate the article screening process for systematic reviews. J Biomed Inform 2017; 69: 33–42. [DOI] [PubMed] [Google Scholar]
- 13. Wallace BC, Trikalinos TA, Lau J, Brodley C, Schmid CH.. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics 2010; 111: 55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmucker C, Schell LK, Portalupi S, et al. Extent of non-publication in cohorts of studies approved by research ethics committees or included in trial registries. PLoS One 2014; 912: e114023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones CW, Handler L, Crowell KE, Keil LG, Weaver MA, Platts-Mills TF.. Non-publication of large randomized clinical trials: cross sectional analysis. BMJ 2013; 347: f6104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bourgeois FT, Murthy S, Mandl KD.. Outcome reporting among drug trials registered in ClinicalTrials.gov. Ann Intern Med 2010; 1533: 158–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dwan K, Gamble C, Williamson PR, Kirkham JJ.. Systematic review of the empirical evidence of study publication bias and outcome reporting bias—an updated review. PLoS One 2013; 87: e66844.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saito H, Gill CJ.. How frequently do the results from completed US clinical trials enter the public domain? A statistical analysis of the ClinicalTrials.gov database. PLoS One 2014; 97: e101826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song F, Parekh S, Hooper L, et al. Dissemination and publication of research findings: an updated review of related biases. Health Technol Assess 2010; 148: 1–193. [DOI] [PubMed] [Google Scholar]
- 20. Vawdrey DK, Hripcsak G.. Publication bias in clinical trials of electronic health records. J Biomed Inform 2013; 461: 139–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turner EH, Knoepflmacher D, Shapley L.. Publication bias in antipsychotic trials: an analysis of efficacy comparing the published literature to the US Food and Drug Administration database. PLoS Med 2012; 93: e1001189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan A-W, Hróbjartsson A, Haahr MT, Gøtzsche PC, Altman DG.. Empirical evidence for selective reporting of outcomes in randomized trials: comparison of protocols to published articles. JAMA 2004; 29120: 2457–65. [DOI] [PubMed] [Google Scholar]
- 23. Page MJ, McKenzie JE, Forbes A.. Many scenarios exist for selective inclusion and reporting of results in randomized trials and systematic reviews. J Clin Epidemiol 2013; 665: 524–37. [DOI] [PubMed] [Google Scholar]
- 24. Kirkham JJ, Dwan KM, Altman DG, et al. The impact of outcome reporting bias in randomised controlled trials on a cohort of systematic reviews. BMJ 2010; 340: c365. [DOI] [PubMed] [Google Scholar]
- 25. Dickersin K, Rennie D.. Registering clinical trials. JAMA 2003; 2904: 516–23. [DOI] [PubMed] [Google Scholar]
- 26. De Angelis C, Drazen JM, Frizelle FA, et al. Clinical trial registration: a statement from the International Committee of Medical Journal Editors. N Engl J Med 2004; 35112: 1250–1. [DOI] [PubMed] [Google Scholar]
- 27. Trinquart L, Dunn AG, Bourgeois FT.. Registration of published randomized trials: a systematic review and meta-analysis. BMC Med 161: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen R, Desai NR, Ross JS, et al. Publication and reporting of clinical trial results: cross sectional analysis across academic medical centers. BMJ 2016; 352: i637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shamliyan TA, Kane RL.. Availability of results from clinical research: failing policy efforts. J Epidemiol Glob Health 2014; 41: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elliott JH, Synnot A, Turner T, et al. Living systematic review: 1. Introduction—the why, what, when, and how. J Clin Epidemiol 2017; 91: 23–30. [DOI] [PubMed] [Google Scholar]
- 31. Elliott JH, Turner T, Clavisi O, et al. Living systematic reviews: an emerging opportunity to narrow the evidence-practice gap. PLoS Med 2014; 112: e1001603.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mortensen ML, Adam GP, Trikalinos TA, Kraska T, Wallace BC.. An exploration of crowdsourcing citation screening for systematic reviews. Res Synth Methods 2017; 83: 366–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krivosheev E, Casati F, Caforio V, Benatallah B. Crowdsourcing paper screening in systematic literature reviews. arXiv: 2017; 170905168.
- 34. Wallace BC, Noel-Storr A, Marshall IJ, Cohen AM, Smalheiser NR, Thomas J.. Identifying reports of randomized controlled trials (RCTs) via a hybrid machine learning and crowdsourcing approach. J Am Med Inform Assoc 2017; 246: 1165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Surian D, Dunn AG, Orenstein L, Bashir R, Coiera E, Bourgeois FT.. A shared latent space matrix factorisation method for recommending new trial evidence for systematic review updates. J Biomed Inform 2018; 79: 32–40. [DOI] [PubMed] [Google Scholar]
- 36. Bierer BE, Li R, Barnes M, Sim I.. A global, neutral platform for sharing trial data. N Engl J Med 2016; 37425: 2411–3. [DOI] [PubMed] [Google Scholar]
- 37. DeVito NJ, Bacon S, Goldacre B.. FDAAA TrialsTracker: a live informatics tool to monitor compliance with FDA requirements to report clinical trial results. bioRxiv 2018: 266452. [Google Scholar]
- 38. Coens C, Bogaerts J, Collette L.. Comment on the TrialsTracker: Automated ongoing monitoring of failure to share clinical trial results by all major companies and research institutions. F1000Res 2017; 6: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunn AG, Coiera E, Bourgeois FT.. Unreported links between trial registrations and published articles were identified using document similarity measures in a cross-sectional analysis of ClinicalTrials.gov. J Clin Epidemiol 2018; 95: 94–101. [DOI] [PubMed] [Google Scholar]
- 40. Takwoingi Y, Hopewell S, Tovey D, Sutton AJ.. A multicomponent decision tool for prioritising the updating of systematic reviews. BMJ 2013; 347: f7191. [DOI] [PubMed] [Google Scholar]
- 41. Bashir R, Surian D, Dunn A.. An empirically-defined decision tree to predict systematic reviews at risk of change in conclusion In: Cochrane Colloquium. Edinburgh, Scotland: Cochrane; 2018. [Google Scholar]
- 42. Bangor A, Kortum PT, Miller JT.. An empirical evaluation of the system usability scale. Int J Hum Comput Interact 2008; 246: 574–94. [Google Scholar]