Abstract
Objectives
Develop a multifunctional analytics platform for efficient management and analysis of healthcare data.
Materials and Methods
Management, Analysis, and Visualization of Clinical Data (MAV-clic) is a Health Insurance Portability and Accountability Act of 1996 (HIPAA)-compliant framework based on the Butterfly Model. MAV-clic extracts, cleanses, and encrypts data then restructures and aggregates data in a deidentified format. A graphical user interface allows query, analysis, and visualization of clinical data.
Results
MAV-clic manages healthcare data for over 800 000 subjects at UConn Health. Three analytic capabilities of MAV-clic include: creating cohorts based on specific criteria; performing measurement analysis of subjects with a specific diagnosis and medication; and calculating measure outcomes of subjects over time.
Discussion
MAV-clic supports clinicians and healthcare analysts by efficiently stratifying subjects to understand specific scenarios and optimize decision making.
Conclusion
MAV-clic is founded on the scientific premise that to improve the quality and transition of healthcare, integrative platforms are necessary to analyze heterogeneous clinical, epidemiological, metabolomics, proteomics, and genomics data for precision medicine.
Keywords: analysis, database, data mining, healthcare, HIPAA, management
INTRODUCTION
Healthcare data include information about patients’ lifestyles, medical histories, visits to the practices, lab tests, imaging tests, diagnoses, medications, surgical procedures, metabolomics and genomics profiles, consulted providers and claims. Healthcare analytics has the potential to revolutionize the field of medicine by improving the quality and transition of care, improving outcomes by reducing costs, detecting diseases at earlier stages,1 developing a better understanding of biological mechanisms, and modeling complex biological interactions through integration and analysis of data with holistic approach.2 The ability to stratify patients, understand scenarios, and optimize decision-making would consistently improve based on the myriad data obtained during the care-delivery process.
Healthcare data analytics begins with the process of database development: this includes data collection, preparation (extraction, cleansing, discretization, validation, integration, and transformation), modeling, validation, and resulting in the creation of patient knowledge bases. To effectively implement analytic processes, various big data management challenges3–9 must be overcome. These include the inadequacy of clinical data10; existence of multiple data standards, structures and types; rapid growth in heterogeneous data; understanding of analysis algorithms for clinical data interpretation, exploration, and drawing inference; unavailability of effective open source tools that combine various approaches to model biological interactions; integration of clinical and analytic systems; interdisciplinary field barriers; high cost11; and implementation of secure frameworks for the healthcare data collection, simplification, synchronizations, raw to knowledge conversion12–14 management, analysis, reporting etc. Establishment of healthcare data analytics system can help achieve goals for better care and health of populations at lower costs with better work–life balance for clinicians and staff. However, it is not forthright,6 as significant efforts are required from experts in various disciplines (eg data science, biomedical informatics, bioinformatics, biostatistics, genomics, metabolomics, clinical science, etc.) and from within multiple organizational units (eg hospitals, research and bench/wet laboratories, insurance companies, information technology providers, data security, etc.). Another challenge is to establish an efficient and secure workflow that can connect all organizational units to streamline transparent data flow and sharing.
In past decades, various systems1 have been developed both in academic (eg dRiskKB,15 PhenoPredict,16 MeSHDD,17 3D-MICE,18 CRISP,19 TCMRs,20 PhOSCo,21 OpenEMR, GNU Health, ClearHealth, OpenClinica, openCDMS, TrialDB, OpenMRS, FreeMED), and commercial (eg NextGen, Epic, Cerner, GE Healthcare, eClinicalWorks, athenahealth, McKesson, Allscripts, Care360, Practice Fusion, Meditech, Greenway Health, etc.) sectors. Academic systems put significant value on analytics; while commercial systems focus on supporting clinical operations. However, both commercial and academic sectors are unable to identify problems by their effects,5 and significantly help in clinical decision-making, healthcare process implementation, and cost reduction. Promoting significant medical transformation in public health, here, we present a new clinical operation and research based scientific platform, named: Management, Analysis, and Visualization of Clinical Data (MAV-clic). It is based on the vision of allowing research for innovation and sustainability to solve public health problems and challenges, while focusing on high quality research.
OBJECTIVES
Innovative healthcare data analytic platforms are necessary to improve the quality and transition of healthcare by analyzing heterogeneous healthcare data of huge volume, velocity, variety, and veracity, and to obtain actionable care gap-based information about patients, developing communication and co-ordination across healthcare units, providers, nurses, quality inspectors, researchers, analysts, and administration. The overall concept of this research was to help support and implement a new healthcare data analytic and research process that can connect people from different backgrounds and specialists with electronic health records (EHR) to facilitate analytical queries and informative outputs. Another goal was to design and implement a proficient Extract-Transform-Load (ETL) strategy to derive meaningful information across different EHR systems. With vast differences across EHRs, the goal is to unite them on a single platform. The ability to deliver these analytics services are increasingly compromised by tight fiscal conditions, therefore, we aimed to build the capacity of our institution to innovate and invent solutions to complex and previously intractable healthcare problems at an affordable price.
METHODS
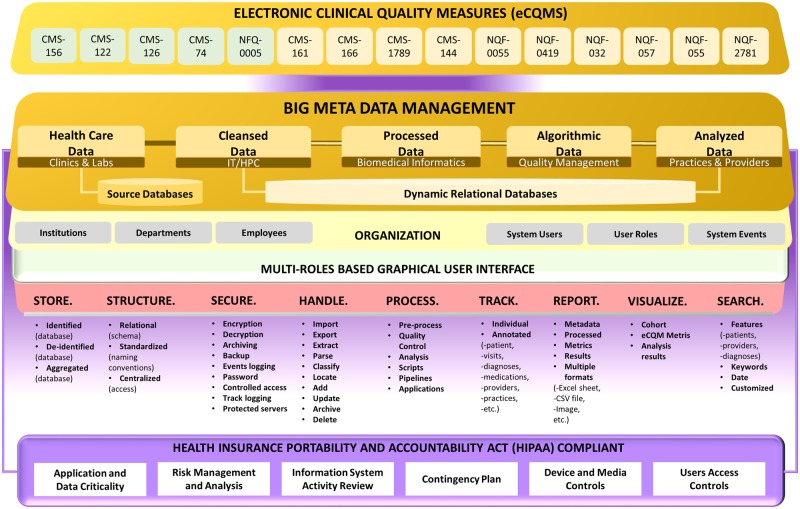
MAV-clic is Health Insurance Portability and Accountability Act of 1996 (HIPAA)22–25 compliant platform, which implements healthcare data analytic processes. Its product line architecture (Figure 1) is based on the Butterfly model26–28 and developed using JAVA programming language, where all major modules are capable of performing individual key roles and can integrate with each other. The data oriented functions of MAV-clic are represented by different components: Store, Structure, Secure, Handle, Process, Track, Visualize, and Search. It implements healthcare and users data security, which includes: Application and Data Criticality; Risk Management and Analysis; Information System Activity Review; Contingency Plan; Device and Media Controls; and Access Controls.22–25 The user-friendly graphical interface of MAV-clic offers multi-role-based operation, which is divided in to six different modules: Main, Users, Analysis, Measures, Databases, and ETL (details are provided in attached Supplementary Material S1).
Figure 1.
MAV-clic HIPAA-compliant product line architecture for clinical data handling, extraction, cleansing, transfer, load, store, structure, standardization, management, processing, analysis, quality assessment, visualization, security, tracking, searching, and reporting.
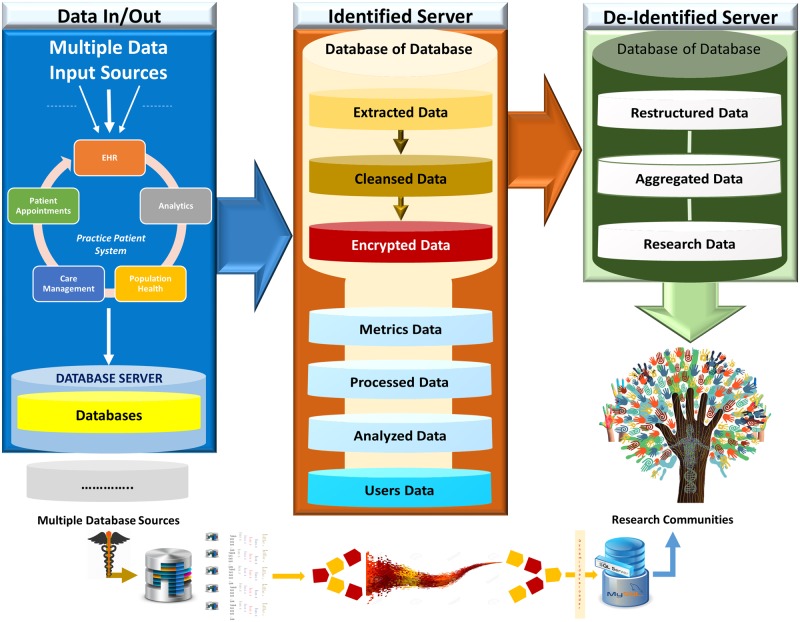
One of the most difficult and complex tasks of implementing healthcare data analytics, is the extraction of data from multiple databases, as each database has large volume and a complex structure that includes different numbers of relations attributed with different numbers of fields and data types. MAV-clic interfaces multiple databases for a streamline data flow with secured and controlled accessibility, transparency, and full audit trail. It provides a dynamic data classification module to organize all integrated databases, which is capable of automatically understanding the structure of a source databases, removing data abnormalities, performing data simplification, and efficiently establishing direct sequential and parallel High Performance Computing (HPC) -based data transfer (Figure 2). The data are structured (normalized relational schemas, standardized naming conventions) and secured in terms of the encryption, archiving, backup, event logging, limited access, and password management in identified and deidentified formats. In order to turn these data into actionable intelligence, MAV-clic provides a holistic view of the data by applying our proposed approach to consolidate healthcare data into a secure data warehouse to perform big data analytics.
Figure 2.
MAV-clic big data workflow, handling input from multiple data sources, dynamic transfer of data using high-performance computing, partitioning of extracted heterogeneous data into different databases, and sharing of restructured and aggregated data.
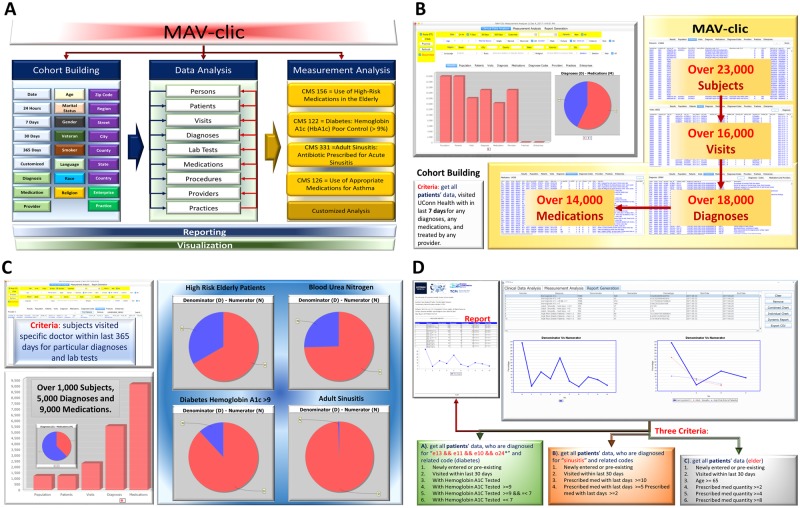
MAV-clic consists of three main modules: Cohort building, Data analysis, and Measurement analysis (Figure 3A). In the cohort building module, multiple features are offered to build groups and ontologies based on patients’ personal details (eg gender, age, marital status, language, race, religion, smoking, and other habits), regional information (eg zip code, street, city, county, state, country, etc.), and medical history. Once the cohort is built, care providers can analyze their patients’ data based on customized date and time, visit to practices, diagnoses, lab tests, prescribed medications, surgical procedures, and consulted providers in the data analysis module. It is capable of tracking one specific patient in the selected cohort as well as the cohort itself. In the measurement analysis module, customized functions are offered to report electronic clinical quality measures (eCQMs) that the Center for Medicare and Medicaid Services (CMS) has proposed to help uncover insights from patient data that demonstrate value in evidence-based medicine, such as improving outcomes and reducing costs (details are provided in attached Supplementary Material S2). The confluence of MAV-clic modules can create space for a new era of open data and discovery in public healthcare by making the most of the new opportunities, utilizing resources maximally and sensibly, and identifying our capabilities to build capacity.
Figure 3.
(A) MAV-clic features for cohort building and ontology making for patient data analysis using CMS proposed and customized measures. (B) Cohort building example: patients who visited UConn Health within the last 7 days for any diagnosis, any medication, and with any provider. (C) Measurement analysis example: patients visited a specific doctor within the last 365 days for particular diagnoses and lab tests. (D) Calculated measure outcome example: customized measurement analysis of patients diagnosed for diabetes, sinusitis, and aged over 65 years.
RESULTS
MAV-clic has developed for healthcare data analytics and research at UConn Health, fulfilling the requirements of data owners and users in implementing the health information system. It efficiently extracts data from the NextGen (deployed EHR system) having over 7000 relations and other inhouse databases, then cleanses and stores the extracted data in Microsoft SQL and MySQL data clusters. It is managing dataset of over 800 000 subjects, which includes demographics, medical visits, diagnoses, lab tests, prescribed medications, and procedures. Furthermore, it includes detailed information about all associated providers to practices at UConn Health, diagnosis (International Classification of Diseases) and medications (National Drug Codes) codes.
In this manuscript, we present three examples to help potential users better understand the analytic capabilities of the MAV-clic system. Example 1 (Figure 3B) explains the cohort building of subjects, who visited UConn Health within the last 7 days for any diagnosis, medication, and treated by any provider. Aggregated, approximate results show that over 23 000 subjects were entered and updated in the system. Out of those, over 16 000 physically appeared and were diagnosed with over 18 000 diagnoses and 14 000 medications were prescribed.
Example 2 (Figure 3C) presents a measurement analysis, which includes the information about subjects’ visits to a specific doctor within the last 365 days for particular diagnoses and lab tests. The selected cohort of subjects are used as an input to evaluate the quality measures of “High Risk Elderly Patients,” “Blood Urea Nitrogen,” “Diabetes HbA1c > 9%,” and “Adults Sinusitis,” Aggregated results show that over 1000 subjects have visited a selected doctor during last 365 days and were prescribed over 9000 medications for over 5000 diagnoses.
The current version of MAV-clic supports 3 eCQMs (CMS156v6, CMS122v5, and PQRS331) for measurement analysis. Example 3 (Figure 3D) shows a calculated measure outcome for each, which includes 3 aggregated cases reported in a deidentified form. Each report includes subjects (newly entered or pre-existing) who have visited UConn Health within the last 30 days and satisfy the following clinical conditions:
-
A.
Diagnosed with “diabetes” with ICD-9 Codes E13, E11, E10, O24 and/or related codes (diabetes) with Hemoglobin A1C Tested: 1) with Hemoglobin A1C Tested ≥ 9, 2) with Hemoglobin A1C Tested ≥ 9 && ≤ 7, and 3) with Hemoglobin A1C Tested ≤ 7.
-
B.
Diagnosed for “sinusitis” and related codes: 1) prescribed medicine within last days ≥ 10, 2) prescribed medicine within last days ≥ 5, and 3) prescribed med with last days ≥ 2.
-
C.
Elder Age ≥ 65 and: 1) prescribed med quantity ≥2, 2) prescribed med quantity ≥4, and 3) prescribed med quantity ≥8.
MAV-clic reports include information about customized eCQMs, denominators, numerators, and percentages drawn in chronological line charts (Figure 3D). These reports can be exported and shared in different file formats, including PDF and CSV (details are provided in attached Supplementary Material S3).
DISCUSSION
To facilitate and improve public sector clinical research and practice, there is a critical need for pure academic frameworks that can connect operational and analytical systems in a way that experts from multiple domains can perform measurement and descriptive analysis. MAV-clic offers numerous conceptual and technological innovations as it addresses a major gap in the field by establishing a state-of-the-art application. It is a new platform at UConn Health, supporting comparative research to determine clinically relevant quality improvements and to evaluate cost-effectiveness in the healthcare system.29 It is a multidatabase management system, capable of handling different levels and types of healthcare data. Programmed analytic processes in MAV-clic help build cohorts by collecting patients’ demographics and medical histories, which can facilitate application of different quality measures, visualization of patterns, and reporting of summaries in an automated and timely manner.
The well-organized data management features in MAV-clic allow users to analyze complex and disparate healthcare data. Its potential benefits include: best strategies to diagnose and treat patients especially at risk for medical complications; patient recruitment candidates matching to the treatments; analyzing clinical trials and patient records to identify follow-on indications1; analyzing large volumes of clinical data to predict and prevent crisis29; and analyzing patient profiles and medical history to perform proactive care. All the combinations from thousands of diagnosis codes, millions of medications, and dozens of lab tests create vast numbers of scenarios that can now be efficiently managed and analyzed by MAV-clic.
CONCLUSION
MAV-clic is a pure academic application to facilitate and improve public sector clinical research and practice. It is based on the vision of allowing research for innovation and sustainability to solve public health problems and challenges, while focusing on high quality research. It has the potential for not only solving many complex healthcare data oriented problems of implementing secure networking, management, pervasive computing, advanced analytics, process modeling, data representation, integrity, privacy, reliability, and exchange but establishing collaborative research environment that can lead to new fundamental insights and advancements in healthcare by analyzing original as well as aggregated healthcare data.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONTRIBUTORS
ZA perceived the idea and did all work on the software and infrastructure design and implementation and related aspects of MAV-clic. ZA and MK did analysis and performance evaluation of MAV-clic. BL guided study. ZA drafted manuscript and all authors participated in writing and review.
Supplementary Material
ACKNOWLEDGMENTS
We would like to give special thanks to Dr Christopher Bonin for stylistic and native speaker corrections. We are grateful to the University of Connecticut Health Center (UConn Health), School of Medicine, Department of Genetics and Genome Sciences, Institute for Systems Genomics, The Pat and Jim Calhoun Cardiology Center, Cardiovascular Biology and Medicine, SNE-PTN, and Ahmed Lab. We appreciate all colleagues, who provided insight and expertise that greatly assisted the research and development.
FUNDING
This work was supported by Ahmed lab, Genetics and Genome Sciences, School of Medicine, University of Connecticut Health Center.
Conflict of interest statement. None declared.
REFERENCES
- 1. Raghupathi W, Raghupathi V.. Big data analytics in healthcare: promise and potential. Health Information Science and Systems 2014; 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alyass A, Turcotte M, Meyre D.. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genomics 2015; 8: 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration predictors in clinical trials: explanation and elaboration. Nature 2013; 5027451: 317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berger B, Peng J, Singh M.. Computational solutions for omics data. Nat Rev Genet 2013; 145: 333–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim MO, Coiera E, Magrabi F.. Problems with health information technology and their effects on care delivery and patient outcomes: a systematic review. J Am Med Inform Assoc 2017; 24: 246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sligo J, Gauld R, Roberts V, et al. A literature review for large-scale health information system project planning, implementation and evaluation. Int J Med Inf 2017; 97: 86–97. [DOI] [PubMed] [Google Scholar]
- 7. Lu Z, Su J.. Clinical data management: current status, challenges, and future directions from industry perspectives. Open Access J Clin Trials 2010; 2: 93–105. [Google Scholar]
- 8. Haux R, Knaup P, Leiner F.. On educating about medical data management the other side of the electronic health record. Methods Inf Med 2007; 461: 74–9. [PubMed] [Google Scholar]
- 9. Rumsfeld JS, Joynt KE, Maddox TM.. Big data analytics to improve cardiovascular care: promise and challenges. Nat Rev Cardiol 2016; 136: 350–9. [DOI] [PubMed] [Google Scholar]
- 10. van Panhuis WG, Paul P, Emerson C, et al. A systematic review of barriers to data sharing in public health. BMC Public Health 2014; 14: 1144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fegan GW, Lang TA.. Could an open-source clinical trial data-management system be what we have all been looking for? PLoS Med 2008; 53: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Williams C, Liu ZH, et al. Big data management challenges in health research—a literature review. Brief Bioinform 2017; doi: 10.1093/bib/bbx086. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duffy DJ. Problems, challenges and promises: perspectives on precision medicine. Brief Bioinformatics 2016; 173: 494–504. [DOI] [PubMed] [Google Scholar]
- 14. Frey LJ, Bernstam EV, Denny JC.. Precision medicine informatics. J Am Med Inform Assoc 2016; 234: 668–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu R, Li L, Wang Q.. dRiskKB: a large-scale disease-disease risk relationship knowledge base constructed from biomedical text. BMC Bioinformatics 2014; 15: 105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu R, Wang Q.. PhenoPredict: a disease phenome-wide drug reposition-ing approach towards schizophrenia drug discovery. J Biomed Inform 2015; 56: 348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown AS, Patel CJ.. MeSHDD: literature-based drug-drug similarity for drug repositioning. J Am Med Inform Assoc 2017; 243: 614–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo Y, Szolovits P, Dighe AS, et al. 3D-MICE: integration of cross-sectional and longitudinal imputation for multi-analyte longitudinal clinical data. J Am Med Inform Assoc 2017; 25(6): 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker JG, Bickerstaffe A, Hewabandu N, et al. The CRISP colorectal cancer risk prediction tool: an exploratory study using simulated consultations in Australian primary care. BMC Med Inform Decis Mak 2017; 1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L, Liu L, Fu X, et al. A cloud-based framework for large-scale traditional Chinese medical record retrieval. J Biomed Inform 2018; 77: 21–33. [DOI] [PubMed] [Google Scholar]
- 21. Krishnankutty B, Bellary S, Kumar NBR, et al. Data management in clinical research: An over-view. Indian J Pharmacol 2012; 442: 168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turner S, Foong S.. Navigating the road to implementation of the Health Insurance Portability and Accountability Act. Am J Public Health 2003; 9311: 1806–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller JD. Sharing clinical research data in the United States under the health insurance portability and accountability act and the privacy rule. Trials 2010; 11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldstein MM. Health information privacy and health information technology in the US correctional setting. Am J Public Health 2014; 1045: 803–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bradford W, Hurdle JF, LaSalle B, et al. Development of a HIPAA-compliant environment for translational research data and analytics. J Am Med Inform Assoc 2014; 211: 185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ahmed Z, Zeeshan S, Dandekar T.. Developing sustainable software solutions for bioinformatics by the “Butterfly” paradigm. F1000Research 2014; 7: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmed Z, Zeeshan S.. Cultivating software solutions development in the scientific academia. Cseng 2014; 71: 54–66. [Google Scholar]
- 28. Ahmed Z. Designing flexible GUI to increase the acceptance rate of product data management systems in industry. Int J Comput Sci Emerg Technol 2011; 2: 100–9. [Google Scholar]
- 29. Manyika J, Chui M, Brown B, et al. Big data: the next frontier for innovation, competition, and productivity. USA: McKinsey Global Institute; 2011. https://www.mckinsey.com/business-functions/digital-mckinsey/our-insights/big-data-the-next-frontier-for-innovation. Last accessed on 20 Nov, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.