Abstract
Dravet syndrome (DS) is a severe epileptic encephalopathy caused mainly by heterozygous loss-of-function mutations of the SCN1A gene, indicating haploinsufficiency as the pathogenic mechanism. Here we tested whether catalytically dead Cas9 (dCas9)-mediated Scn1a gene activation can rescue Scn1a haploinsufficiency in a mouse DS model and restore physiological levels of its gene product, the Nav1.1 voltage-gated sodium channel. We screened single guide RNAs (sgRNAs) for their ability to stimulate Scn1a transcription in association with the dCas9 activation system. We identified a specific sgRNA that increases Scn1a gene expression levels in cell lines and primary neurons with high specificity. Nav1.1 protein levels were augmented, as was the ability of wild-type immature GABAergic interneurons to fire action potentials. A similar enhancement of Scn1a transcription was achieved in mature DS interneurons, rescuing their ability to fire. To test the therapeutic potential of this approach, we delivered the Scn1a-dCas9 activation system to DS pups using adeno-associated viruses. Parvalbumin interneurons recovered their firing ability, and febrile seizures were significantly attenuated. Our results pave the way for exploiting dCas9-based gene activation as an effective and targeted approach to DS and other disorders resulting from altered gene dosage.
Keywords: gene therapy, Dravet syndrome, epileptic encephalopathy, activatory CRISPR
Colasante et al. exploit an activatory CRISPR-targeting Scn1a gene promoter as a therapeutic strategy to rescue Scn1a haploinsufficiency in a mouse model of Dravet syndrome and restore physiological levels of its gene product, the Nav1.1 voltage-gated sodium channel.
Introduction
Dravet syndrome (DS) is a severe epileptic encephalopathy beginning in the first year of life with seizures often associated with fever that evolve into frequent, prolonged, and clustered epileptic crises.1, 2, 3 In subsequent years, patients often develop psychomotor delay, behavioral disturbances, and cognitive impairment.4 DS is a genetic condition mainly caused by mutations in the SCN1A gene encoding for the Nav1.1 voltage-gated sodium channel α subunit.5, 6 Over 650 missense and nonsense SCN1A mutations have been described in DS patients. Although most are de novo, some mutations have been found to be inherited in familial cases.7 SCN1A mutations affect only one copy of the gene, typically leading to loss of function and indicating that a haploinsufficient genetic mechanism is responsible for DS. These data suggest that a reduced amount of Nav1.1 channel impairs neuronal activity and function. Scn1a heterozygous mutant mice display similar neurological symptoms, including severe epilepsy, behavioral alterations, and premature death.8, 9, 10, 11 Functional studies revealed that cortical fast-spiking GABAergic inhibitory interneurons exhibit reduced intrinsic excitability and defects in action potential firing.8, 10, 12 In contrast, both excitability and firing of cortical excitatory neurons from Scn1a heterozygous mutant mice appear to be substantially unaltered.8, 13 These findings potentially resolve the paradox that epilepsy arises from loss-of-function mutations in Nav1.1, which contributes to the fast depolarization of neuronal membranes during an action potential. Of note, Nav1.1 has been found to be mainly expressed in inhibitory interneurons by immunohistochemistry analysis, suggesting that this sodium channel isoform has a preponderant function in that neuronal population.10 Accordingly, selective inactivation of Scn1a in cortical interneurons is sufficient to elicit neurological deficits comparable with those described in constitutive mutant mice,14, 15 and, conversely, Scn1a loss restricted to the dorsal-telencephalic (e.g., neocortical, hippocampal) excitatory neurons has ameliorating effects on epileptic seizures and sudden death.13 Scn1a heterozygous mutant mice develop spontaneous and recurrent seizures starting from 3 weeks after birth, often leading to premature and sudden death.8, 9, 10 Remarkably, body temperature elevation triggers myoclonic and generalized seizures in these mice, recapitulating febrile seizures in DS patients.16 Thus, DS mice represent a valuable model of the disease, not only to dissect the pathological mechanisms but also to evaluate the efficacy of innovative therapies. Drug treatment of DS patients, including stiripentol in combination with clobazam and valproate, has limited efficacy and poorly controls convulsive seizures.17, 18 Cannabidiol or serotonin uptake inhibitors have been reported to reduce seizure frequency in some patients, but larger studies are needed to appreciate the exact therapeutic indications for these treatments.19, 20, 21 Nonetheless, complete seizure cessation is rarely obtained with any of these pharmacological anticonvulsants. Gene therapy approaches for neurodevelopmental disorders are in rapid development because of the introduction of novel serotypes of recombinant adeno-associated viruses (AAVs), allowing efficient transduction of neurons.22 However, the SCN1A coding sequence is 6 kb long, exceeding the strict cargo limit for AAVs. Although lentiviruses (LVs) can carry the SCN1A gene sequence, they show limited spread in neural tissue and are therefore inadequate to treat diseases affecting large brain areas.23 These obstacles have prevented substantial advances in gene-based therapies for DS. Given that one copy of the SCN1A gene is still functional in DS, stimulating its endogenous expression over physiological levels might lead to increased availability of the Nav1.1 channel protein, potentially leading to symptomatic improvement. Thus, a system able to induce SCN1A gene expression in neurons in a regulated manner, without significant off-target effects, would be a strong therapeutic candidate tool for DS.
CRISPR-Cas9 technology has become a powerful tool for genome editing, allowing DNA to be targeted with high efficiency and specificity. As demonstrated by pioneering works in several cell types and organisms, the Cas9/single guide RNA (sgRNA) complex can efficiently generate double-strand breaks, which then trigger non-homologous end joining-mediated gene knockout or homology-directed repair-mediated recombination.24, 25, 26 A modified version of the CRISPR-Cas9 system has been developed by generating a nuclease-dead Cas9 (dCas9) fused to effector domains for transcriptional gene regulation. Hence, the dCas9/sgRNA complex has provided a crucial platform for programming diverse types of transcriptional or epigenetic manipulation of the genome without cleaving the target DNA.27, 28, 29 Seminal studies have shown that dCas9-based gene activation is highly specific in DNA binding and gene regulation and promotes chromatin remodeling of the regulatory elements of the gene of interest.30, 31, 32 This system has been successfully implemented to investigate hierarchies in gene regulatory networks, screening for cellular phenotypes and directing somatic cell fate.33, 34, 35, 36 In the activatory CRISPR system, dCas9 is fused to multiple VP16 transcriptional activator domains that robustly boost gene transcription when combined with one or more sgRNAs targeting sequences in the proximal promoters or close to transcription start sites (TSSs).26, 36, 37 As proof of concept, this technology has also been employed to activate endogenous genes in mouse models of disease to ameliorate biomarkers of diabetes, muscular dystrophy, and acute kidney disease.30 More recently, it has been applied to enhance expression of the Sim1 gene in the hypothalamus and to rescue the associated obesity phenotype.38 Here we describe a dCas9-based system that significantly upregulates Scn1a expression and restores Nav1.1 protein levels in both cellular and animal models of DS. This targeted gene activation rescues membrane excitability and action potential firing in DS cortical interneurons and significantly attenuates hyperthermia-induced seizures in DS mice.
Results
A Single sgRNA Enhances Scn1a Gene Expression by Targeting Its Proximal Promoter
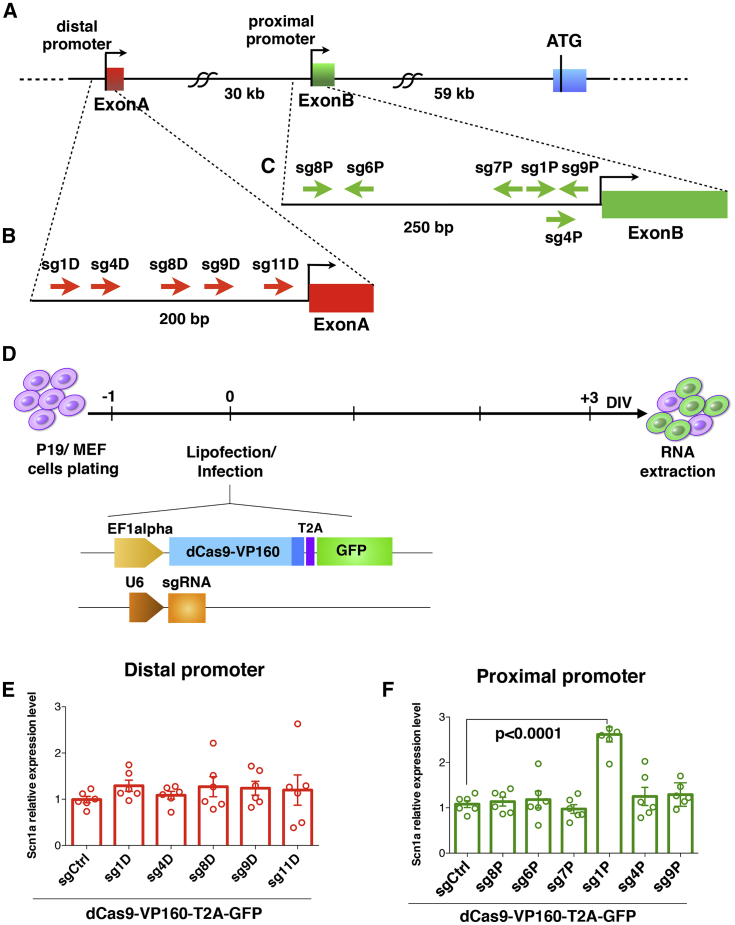
To achieve upregulation of Scn1a gene transcription, we sought to define the necessary dCas9/sgRNA elements by a candidate approach in vitro. Through an extensive bioinformatics analysis, we determined the Scn1a gene promoter regions to focus the sgRNA design. Several studies have pointed out that sgRNAs can transactivate genes of interest more efficiently when localized within 500 bp from the gene TSS.34 We interrogated the Encyclopedia of DNA Elements (ENCODE) and Fantom5 databases for the expression profiling and epigenetic marks of actively transcribed genes in the adult mouse brain. In addition, CAGE-seq and DNase-seq datasets were queried to determine the exact TSSs for Scn1a (Figure S1). We identified two regions in the Scn1a locus where RNA polymerase II (Pol II), mono- and tri-methylation of lys4, and acetylation of lys27 of H3 histone were strongly enriched and revealed DNase I-hypersensitive sites. CAGE-seq peaks were aligned to the same sequences, confirming the existence of two active TSSs (TSS1 and TSS2) located upstream of two non-coding exons (exon A and exon B) (Figure S1) and producing two different mRNA isoforms, both expressed in the adult mouse brain.39 200 bp upstream of the exon A (distal promoter) and 250 bp upstream of exon B (proximal promoter) (Figure 1A) were submitted to the CRISPOR web tool (http://crispor.tefor.net) for sgRNA design. We selected five guides in the distal promoter and six in the proximal one with specificity scores higher than 50% (Figures 1B and 1C; Table S1). Then we determined whether dCas9 fused to VP160 (dCas9-VP160), a transcriptional activator that carries 10 tandem copies of VP16 (a herpes simplex virus type 1 transcription factor), in association with the selected sgRNAs was able to upregulate Scn1a gene expression in the P19 murine teratocarcinoma cell line. sgRNAs specific for Scn1a promoters and one control guide (sgCtrl), targeting the β-galactosidase bacterial sequence, were cloned into the pU6 vector and individually lipofected into P19 cells together with the Ef1a-dCas9-VP160-T2A-GFP (Figure 1D), and 3 days later, after ascertaining GFP expression, cells were harvested for RNA extraction and qRT-PCR (Figure 1D). Interestingly, none of the sgRNAs targeting the distal promoter were able to significantly alter the basal expression of Scn1a (Figure 1E). Conversely, among the guides targeting the proximal promoter, only sg1P was found to significantly increase Scn1a mRNA levels with respect to sgCtrl (Figure 1F). Comparable results were obtained when sgRNAs were lipofected with dCas9 linked to a puromycin resistance cassette (Ef1a-dCas9-VP160-T2A-PuroR) and puromycin was added to the culture medium the day after transfection (Figures S2A–S2C), indicating that antibiotic selection and consequent enrichment of lipofected cells were not strictly necessary to detect sg1P-mediated Scn1a induction. sg1P upregulated Scn1a gene expression to a similar extent in primary mouse embryonic fibroblasts (MEFs) (Figure S2D). In conclusion, we identified sg1P as an sgRNA that is sufficient, when associated with the dCas9 activation system, to stimulate basal transcription of Scn1a consistently in different cell types.
Figure 1.
sgRNA Design and Screening for Stimulating Scn1a Gene Expression with the dCas9 Activation System in P19 Cells
(A–C) Schematic representation of the Scn1a gene (A) with distal (B) and proximal (C) promoter regions; the positions of the sgRNAs selected for this screening are highlighted. (D) Experimental setting for the sgRNA screening in P19 cells and schematic representation of the constructs employed for cell lipofection. One day after plating, P19 cells were lipofected, and the subsequent day, GFP expression was ascertained. At 3 DIV, the cells were processed for RNA extraction. (E and F) qRT-PCRs for Scn1a mRNA levels performed on RNA extracted from P19 cells lipofected with dCas9VP160-T2A-GFP together with sgRNAs targeting the distal (E) or proximal (F) promoter. Data are normalized on the 18S rRNA and relative to sgCtrl-lipofected cells. sg1p induces significant upregulation of Scn1a compared with sgCtrl (n = 6, p < 0.0001, one-way ANOVA followed by Bonferroni multiple comparisons test). Data are shown as mean ± SEM, with dots representing individual samples.
sg1P/dCas9-VP160 Lentiviral Transduction Upregulates Scn1a Expression in Primary Neurons
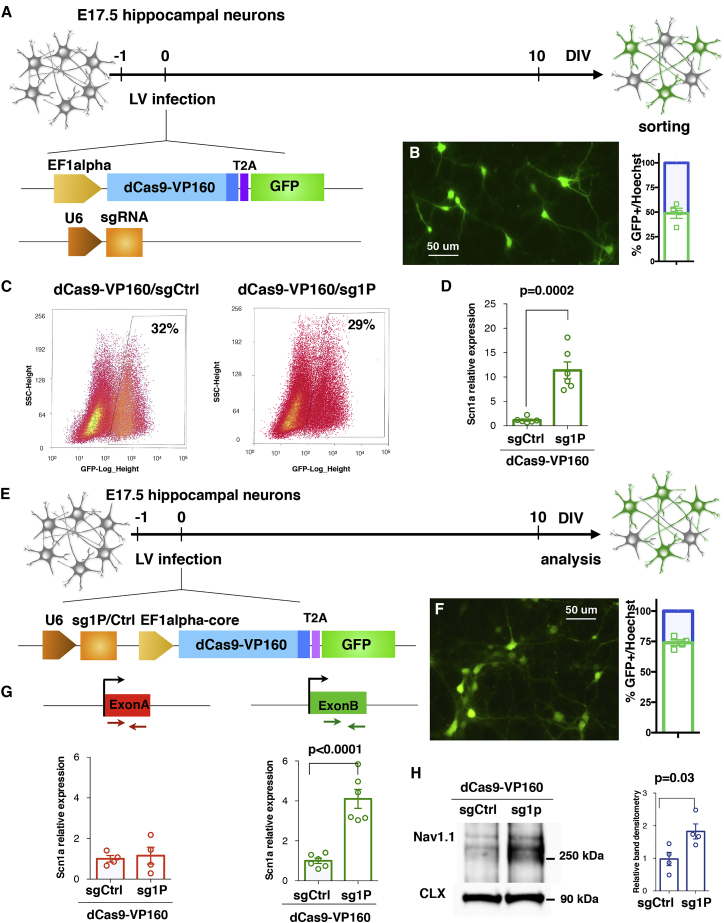
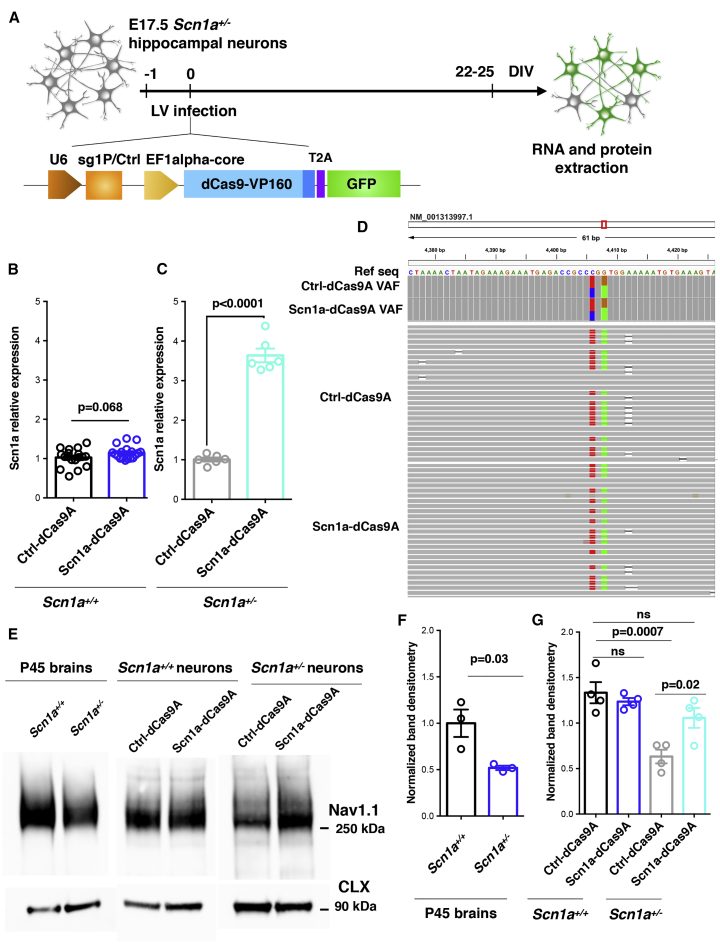
We asked whether the sg1P/dCas9-VP160 system could stimulate Scn1a expression in primary hippocampal neurons. Neurons were co-transduced with Ef1a-dCas9VP160-T2A-GFP and pU6-sg1P or pU6-sgCtrl LVs the day after plating to maximize transduction efficiency (Figure 2A). Immunofluorescence analysis performed at 10 days in vitro (DIV) showed that almost 50% of the plated neurons were transduced (Figure 2B). With this intermediate efficiency, we decided to purify the infected cells to obtain reliable information regarding the regulation of Scn1a expression. Thus, 10 DIV transduced GFP+ neurons from sgCtrl- and sg1P-treated samples were isolated by fluorescence-activated cell sorting (FACS) (Figure 2C), and the RNA was extracted for gene expression analysis. Scn1a expression levels were robustly increased when transduced with sg1P with respect to sgCtrl (Figure 2D). However, FACS is detrimental to neurons and prevents their functional analysis. For this reason, we generated a single lentiviral vector carrying both sg1P and dCas9-VP160 (Figure 2E), which improved transduction efficiency up to ∼75% (Figure 2F). In this setting, a 4-fold increase in Scn1a expression was detected in sg1P with respect to sgCtrl-treated neurons. Moreover, no alteration in the transcriptional levels of the second Scn1a mRNA isoform carrying exon A was detectable (Figure 2G), indicating that the sg1P/dCas9-VP160 activation system does not affect the transcriptional status of the distal promoter. To evaluate whether increased transcription of Scn1a led to higher Nav1.1 protein levels, we performed western blot analysis of membrane lysates isolated from transduced 10 DIV primary neuronal cultures, which showed a 2-fold increase in membrane-associated Nav1.1 protein (Figure 2H).
Figure 2.
dCas9-VP160/sg1P Potentiates Scn1a Gene Transcription in Primary Hippocampal Neurons
(A) Schematic drawing depicting the experimental setting to deliver the Ctrl- and Scn1a-dCas9A system in primary neurons. Hippocampal neurons were derived from E17.5 embryos and, the day after plating, were co-transduced with two distinct lentiviruses (LVs) carrying Ef1a-dCas9-VP160- T2A-GFP and pU6-sg1P or pU6-sgCtrl guides, respectively. (B) Representative image of anti-GFP immunofluorescence at 10 DIV and quantification of GFP+ transduced neurons over the total cell number. Scale bar, 50 μm. (C) Representative FACS images of GFP+ neurons transduced with either the sgCtrl- or sg1P-dCas9 activation system. (D) qRT-PCR reveals the increased Scn1a transcriptional levels in hippocampal neurons infected with sg1P with respect to sgCtrl conditions (n = 6, p = 0.0002, Student’s t test). Data are shown as mean ± SEM, with dots representing individual samples. (E) Schematic setting of E17.5 neurons transduced with a LV carrying both the pU6-sgRNA cassette and dCas9-VP160-T2A-GFP sequence under control of the EF1alpha core promoter. (F) Anti-GFP immunofluorescence at 10 DIV and relative quantification of transduced GFP+ cells over total, showing that a single LV reaches a transduction efficiency of 75%. Scale bar, 50 μm. (G) qRT-PCR for Scn1a with primers amplifying the first (exon A) and second 5′ UTR exon (exon B). Data are normalized on 18S rRNA and expressed as relative to sgCtrl. Exon A, sg1P versus sgCtrl: p = 0.7353; exon B, sg1P versus sgCtrl: p < 0.0001; Student’s t test). (H) Left: western blot for Nav1.1 and Calnexin on protein lysates from Ctrl-dCas9A- and Scn1a-dCas9A-treated neurons at 10 DIV. Right: quantification obtained through densitometry and normalized on Calnexin levels and expressed as Scn1a-dCas9A relative to Ctrl-dCas9A (data are shown as mean ± SEM, with dots representing individual samples); n = 4; sg1P versus sgCtrl: p = 0.03, Student’s t test.
Taken together, these results indicate that sg1P associated with the dCas9 activation system can modulate Scn1a gene activity in primary neurons and, accordingly, increase the levels of the Nav1.1 channel protein.
dCas9-Based Gene Activation Is Highly Specific for Scn1a in Primary Neurons
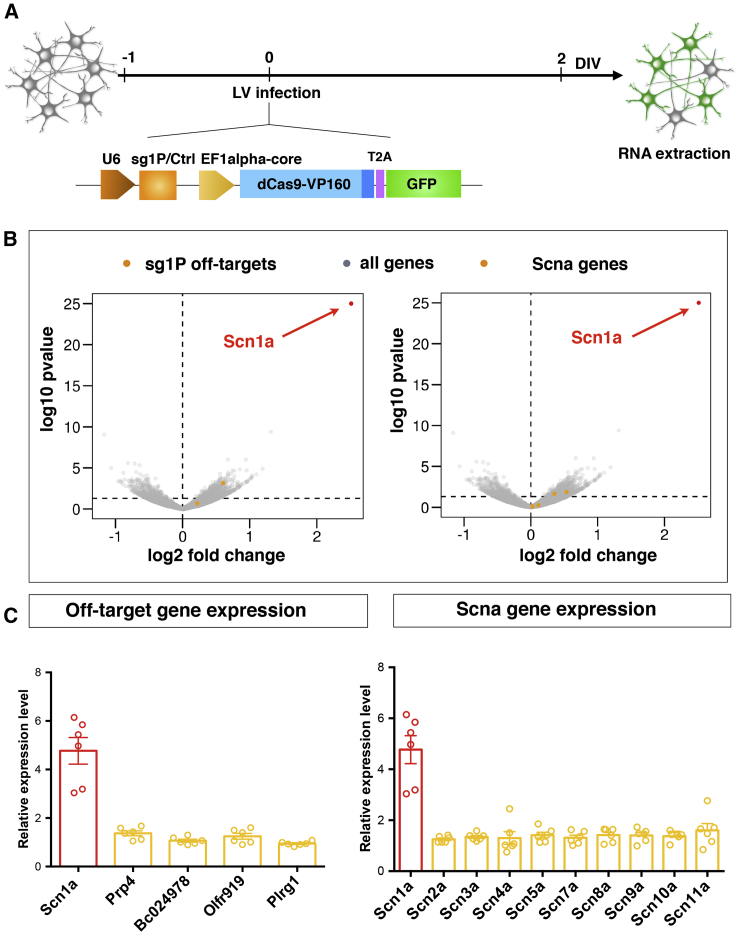
We examined the specificity of the dCas9 activation system by assessing global gene expression in primary neurons transduced with dCas9-VP160 together with either sg1P or sgCtrl. RNA sequencing (RNA-seq) was performed on 3 DIV neurons 2 days after lentiviral transduction with either sg1P or the sgCtrl (Figure 3A). Notably, the only gene with significantly increased expression relative to the control was Scn1a (log2 fold change > 1.5, p < 0.005), indicating the high specificity of the dCas9 activation system in primary neuronal cells (Figures 3B, red dots). CRISPOR provided a list of 195 putative off-target genes associated with sg1P. Because dCas9A is nuclease defective, we reasoned that aspecific transcription activation could occur only when off-target sequences were in close proximity to TSSs of genes. Therefore, we filtered the list using the web tool Galaxy to investigate which of those putative off-targets were located within 500 bp of TSSs of any annotated gene. Only 4 of 195 putative off-target genes were identified in the putative promoter regions upstream of the Prp4, BC02, Olfr919, and Plrg1 genes. However, as shown by both sequencing and qRT-PCR analyses, the expression of these genes was not altered after delivery of the dCas9 activation system (Figures 3B and 3C, left panel, yellow dots).
Figure 3.
Global Gene Expression analysis of Transduced Neurons Confirms the High Specificity Profile of the Scn1a-dCas9A System
(A) Schematic view of the experimental setting to perform gene expression profiling of Ctrl-dCas9A- and Scn1a-dCas9A-treated primary neurons. E17.5 embryo-derived neurons were transduced with single LVs at DIV 1 expressing either the Ctrl-dCas9A or Scn1a-dCas9A elements and processed for RNA extraction 48 h later (DIV 3). (B) Volcano plots showing the log10 p value as a function of log2 fold changes in gene expression in Scn1a-dCas9A-treated neurons with respect to Ctrl-dCas9A. Scn1a is shown as a red dot. Yellow dots represent off-target genes in the left panel and other Scna genes in the right panel. All other genes are shown as gray dots. (C) qRT-PCRs for profiling the expression of predicted off-targets (genes Prp4, BC024978, Olfr919, and Plrg1; left panel) or other Scna genes (Scn2a, Scn3a, Scn4a, Scn5a, Scn7a, Scn8a, Scn9a, and Scn11a; right panel). Plotted values are normalized on 18S rRNA and expressed as relative to sgCtrl-treated samples (value = 1, data not shown). n = 6; sg1p versus sgCtrl p < 0.0004, Student’s t test. Data are shown as mean ± SEM, with dots representing individual samples.
Absolute and relative levels of the various Nav α subunits are strictly regulated, allowing a fine balance of neuronal membrane excitability.40 Therefore, we examined the expression of other Nav α subunit-encoding genes. The global transcriptional analysis revealed no significant changes in their expression levels (Figure 3B, right panel, yellow dots). These results were confirmed by qRT-PCR assays of independent cellular replicates (Figure 3C, right panel). In conclusion, the global and targeted gene expression analyses of primary neurons transduced with the sg1P-dCas9 activation system (hereafter called Scn1a-dCas9A) confirmed the high specificity for Scn1a gene transactivation at a genome-wide level.
dCas9-Based Scn1a Gene Activation Enhances Neuronal Activity in Immature Wild-Type (WT) Cortical Interneurons
To evaluate whether the increased levels of Nav1.1 protein in primary neurons were sufficient to alter neuronal excitability, whole-cell patch-clamp experiments were carried out in dCas9A-transduced neurons. We conceived a dual LV-inducible system designed with a first lentivector carrying dCas9-VP160 with the tdTomato reporter regulated by the reverse tetracycline-controlled transactivator (rtTA)-responsive element (TRE) and a second lentivector carrying the transactivator rtTA and sgRNAs (sg1P or sgCtrl) to explore a setting that would be relevant in in vivo experiments (Figure 4A; Figure S3A). Indeed, the split of Scn1a-dCas9A in two vectors is required for in vivo delivery mediated by AAVs, characterized by a limited cargo capacity. In this setting, upon doxycycline (dox) administration, about 60% of neurons were transduced (Figure S3B). A 2-fold increase in the basal level of Scn1a gene expression (Scn1a-dCas9A versus Ctrl-dCas9A) in WT neurons at 7 DIV was observed upon dox administration (Figure S3C, +dox), but not under dox-absent conditions (−dox), although some leaky expression of dCas9A could be detected (Figure S3D).
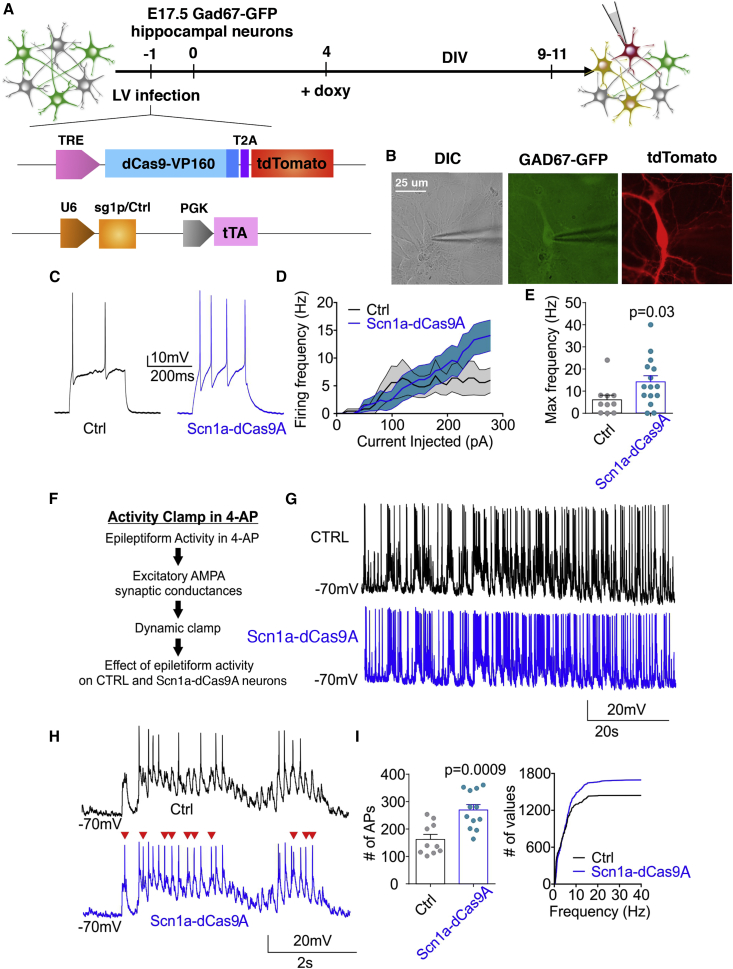
Figure 4.
Scn1a-dCas9A Increases Neuronal Excitability in Cortical Immature Wild-Type Interneurons
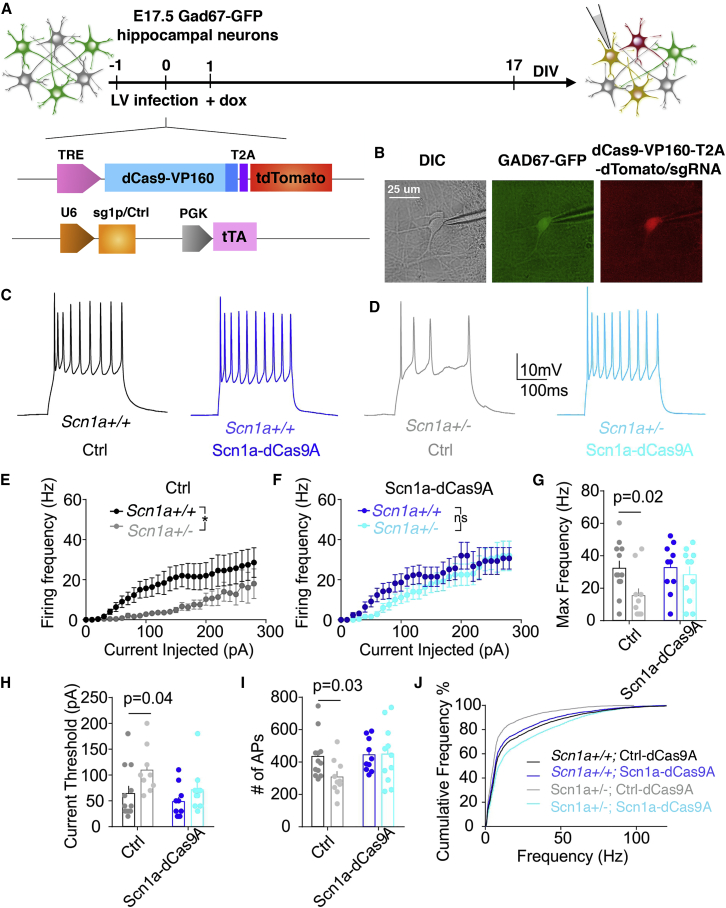
(A) Schematic drawing showing the timeline of transduction with LVs expressing the dCas9A systems on primary wild-type GAD67-GFP neurons and their subsequent functional analysis. (B) Representative images of a patch-clamp-recorded interneuron expressing both GFP under the GAD67 promoter and tdTomato, reflecting the active Scn1a-dCas9A system. Scale bar, 25 μm. (C) Representative current-clamp traces of APs induced by a single current step in dCas9A (black trace, sgCtrl) or Scn1a-dCas9A interneurons (blue trace, sg1P). (D) Firing frequency versus injected current for Ctrl- and Scn1a-dCas9A-transduced interneurons (Ctrl-dCas9A, n = 11; Scn1a-dCas9A, n = 15). (E) Histogram of the maximum frequency reached by interneurons during the current step protocol (p = 0.03, Mann-Whitney U test). (F) Experimental design of activity clamp in primary neuronal cultures in the presence of 4AP (Materials and Methods). (G and H) Representative full traces (G) and magnified traces (H) for the activity clamp protocol in Ctrl-dCas9A (black trace, sgCtrl) and Scn1a-dCas9A (blue trace, sg1P) interneurons. (I) Activity clamp analysis for the number of events during the full traces (left) and cumulative plot for AP frequency (right) (Ctrl, n = 10; Scn1a-dCas9A, n = 12; p = 0.0009, unpaired Student`s t test).
Because Nav1.1 channel loss mainly affects GABAergic interneurons, we established primary neuronal cultures from GAD67-GFP mouse embryos that were transduced with either the Ctrl-dCas9A or Scn1a-dCas9A system and analyzed when double-positive for GFP and tdTomato (Figures 4A and 4B).
First recordings were performed on 9–11 DIV primary neurons before their achievement of full functional maturation. Current step injections showed a significant increase in firing rate in interneurons transduced with the Scn1a- compared with the Ctrl-dCas9 activation system (Ctrl-dCas9A, n = 11; Scn1a-dCas9A, n = 15; p = 0.03, Mann-Whitney non-parametric t test) (Figures 4C–4E; Figure S4). No alteration of neuronal firing rate was mediated by Scn1a-dCas9A in the absence of dox (Figure S3E). These results underline the potential efficiency of Scn1a-dCas9A to increase interneuron excitability upon alteration of Scn1a gene dosage, at least in an immature network.
Recently, we developed a new electrophysiological approach (“activity clamp”) to analyze how a neuron in a given epileptic network responds to antiepileptic drugs.41 Here we modified this method to adapt it for primary neuronal cultures to compare interneurons transduced with either the Ctrl-dCas9A or Scn1a-dCas9A system. First, we recorded in interneurons the barrage of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor-mediated excitatory synaptic currents that occurs in the presence of the chemoconvulsant potassium channel blocker 4-aminopyridine (4-AP). Then we converted the recorded currents into a conductance waveform, and, by applying dynamic current clamp, we fed them back to interneurons pharmacologically isolated from the network. We then compared neurons transduced with either the Scn1a- or Ctrl-dCas9A system (Figures 4F–4I). Activity clamp showed that GABAergic interneurons transduced with Scn1a-dCas9A exhibited an increase in the number of action potentials (APs) evoked by the same epileptic inputs as well as higher firing frequencies reached during the protocol (Ctrl-dCas9A, n = 10; Scn1a-dCas9A, n = 12; p = 0.0009, parametric Student’s t test) (Figure 4I). Altogether, these results indicate that increased expression of Scn1a obtained by the dCas9A system is sufficient to increase interneuron excitability in response to epileptiform barrages of synaptic excitation.
dCas9-Based Scn1a Activation Increases Nav1.1 Protein Levels and Rescues Excitability in Scn1a+/− Mutant Cortical Interneurons
The experiments described above show that the Scn1a-dCas9A system upregulates Scn1a expression in WT interneurons, increasing Nav1.1 protein levels and enhancing their excitability. We asked whether this system could also boost transcription of the single WT Scn1a allele in DS mice to reach sufficient Nav1.1 protein levels to compensate for haploinsufficiency and attenuate the pathology.
We tested the efficacy of the Scn1a-dCas9A system in Scn1a+/− neurons derived from a DS mouse model (Figure 5A).10 Considering that the Scn1a gene starts to be expressed around post-natal day 10 (P10),10, 42 the analysis was done at DIV 22–25 to ensure display of the characteristic DS phenotype. Interestingly, at this time point, WT neurons seemed to be unresponsive to Scn1a-dCas9A treatment (Figure 5B), whereas we observed a 3.5-fold increase in levels of Scn1a expression in Scn1a-dCas9A- compared with Ctrl-dCas9A-treated DS neurons (Figure 5C). To assess the nature of Scn1a-dCas9A-induced mRNA, we performed deep sequencing of PCR amplicons spanning the mutation region and found out that, in Ctrl-dCas9A-transduced neurons corresponding to basal conditions, WT and mutant transcripts were approximately equally abundant (almost 50% each) (Figure 5D). The same relative proportion was maintained in Scn1a-dCas9A-treated neurons (Figure 5D). These data indicate that mutant mRNA is stable in DS neurons and, consequently, that Scn1a-dCas9A treatment induces upregulation of both transcripts. However, at the protein level, untreated Scn1a+/− postnatal brains showed an ∼50% reduction in Nav1.1 protein levels with respect to the control counterparts (Figures 5E and 5F). In accordance with mRNA data, the levels of Nav1.1 protein did not change upon Scn1a-dCas9A treatment in Scn1a+/+ neurons, whereas, remarkably, Scn1a-dCas9A-treated Scn1a+/− neurons exhibited almost doubled Nav1.1 protein levels compared with Ctrl-dCas9A-treated Scn1a+/− neurons at 25 DIV (Figures 5E and 5F). An immunoblot signal lower than 170 kDa and corresponding to truncated Nav1.1 protein was not observed, neither in Scn1a+/− adult brains nor in Scn1a+/− neurons under basal conditions (Ctrl-dCas9A) or treated with Scn1a-dCas9A (Figure 5E). These results imply that the mutant protein is likely degraded and not targeted to the plasma membrane. To assess whether rescue of Nav1.1 protein levels has a functional effect on DS neurons, electrophysiological experiments were repeated in Scn1a+/−;GAD67-GFP GABAergic interneurons transduced either with the Ctrl-dCas9A or Scn1a-dCas9A system (Figures 6A and 6B). Recordings were performed on 18–20 DIV neuronal cultures to treat fully mature and functional interneurons. Current step injections showed a decreased frequency-current relationship and maximum AP frequency in Scn1a+/− interneurons compared with the WT when both were transduced with the Ctrl-dCas9A system (Figures 6C–6E). Accordingly, an increased current threshold to trigger a single AP was observed (Figure 6G). These defects in Scn1a+/− interneurons were completely rescued by transducing the Scn1a-dCas9A system (Ctrl WT, n = 12; Ctrl Scn1a+/−, n = 10; Scn1a-dCas9A WT, n = 10; Scn1a-dCas9A Scn1a+/−, n = 11; p = 0.02, p = 0.04, 2-way ANOVA followed by Bonferroni’s multiple comparisons test) (Figures 6D–6H). Activity clamp confirmed rescue of DS mutant interneuron firing in the face of epileptiform activity following Scn1a-dCas9A treatment compared with Ctrl-dCas9A treatment (Ctrl-dCas9A WT, n = 12; Ctrl-dCas9A Scn1a+/−, n = 10; Scn1a-dCas9A WT, n = 10; Scn1a-dCas9A Scn1a+/−, n = 11; p = 0.03, 2-way ANOVA followed by Bonferroni’s multiple comparisons test) (Figure 6I). As expected from the molecular data, the increase in excitability in Scn1a-dCas9A-treated WT interneurons observed at 9–10 DIV was no longer observed at 18–20 DIV (Figures 6C–6J).
Figure 5.
Scn1a-dCas9A Stimulates Scn1a Basal Expression and Nav1.1 Protein Levels in Scn1a+/− Hippocampal Neurons
(A) Schematic drawing of the dCas9A treatments in E17.5 Scn1a+/− primary neurons. Neurons were transduced with either the Ctrl-dCas9A or Scn1a-dCas9A system 1 day after plating and processed for RNA and protein extraction at 22–25 DIV. (B and C) qRT-PCRs for Scn1a transcriptional levels performed on RNA extracted from Ctrl-dCas9A- or Scn1a-dCasA-treated Scn1a+/+(B) and Scn1a+/− (C) primary neurons. Plotted data are expressed as relative to Ctrl-dCas9A. Scn1a+/+: n = 18, p = 0.068; Scn1a+/−: n = 4, p < 0.0001; Student’s t test. (D) Binary alignment map (BAM) within the mouse Scn1a transcript (NM_001313997.1). The red box indicates the region amplified and sequenced with high coverage. Ctrl-dCas9A and Scn1a-dCas9A variant allele frequency (VAF) tracks show the observed VAF. Ctrl-dCas9A and Scn1a-dCas9A read tracks display a sample of about 30 different sequencing reads per sample; nucleotides diverging from the reference genome are highlighted. (E) Western blot for Nav1.1 and Calnexin on protein lysates from adult (P45) Scn1a+/+ and Scn1a+/− mice (left panel) and from Ctrl-dCas9A- and Scn1a-dCasA-treated Scn1a+/+ and Scn1a+/− neurons at 22–25 DIV (center and right panels). (F) Densitometric quantification of immunoreactive bands in the western blots of adult mouse brains. Values corresponding to the Nav1.1 band were normalized to Calnexin levels (n = 3, p = 0.03, Student’s t test). (G) Densitometric quantification of immunoreactive bands in the western blots of Scn1a+/+ and Scn1a+/− neurons transduced with Ctrl-and Scn1a-dCas9A.Values corresponding to the Nav1.1 band were normalized to Calnexin levels (n = 4, one-way ANOVA followed by Turkey’s multiple comparisons test). Data are shown as mean ± SEM, with dots representing individual samples.
Figure 6.
Scn1a-dCas9A Rescues Neuronal Excitability Defects in Cortical Mature Scn1a+/− Interneurons
(A) Schematic drawing showing the experimental time frame for lentiviral transductions and functional analysis of Scn1a+/+;GAD67-GFP+ or Scn1a+/−;GAD67-GFP+ primary hippocampal neurons transduced with the two depicted lentiviruses. (B) Representative images of a patch-clamp-recorded Scn1a+/− interneuron expressing both GFP under the GAD67 promoter and tdTomato reflecting the active Scn1a-dCas9A system (Materials and Methods). (C and D) Representative current-clamp traces of APs induced by single current steps administered to Ctrl-dCas9A-transduced WT interneurons (black trace, C), Scn1a-dCas9A-transduced Scn1a+/+interneurons (blue trace, C), Ctrl-dCas9A-transduced Scn1a+/− interneurons (gray trace, D), and Scn1a-dCas9A-transduced Scn1a+/− interneurons (cyan trace, D). (E and F) Firing frequency versus injected current for Ctrl-dCas9A-transduced (E) and Scn1a-dCas9A-transduced (F) Scn1a+/+ and Scn1a+/− interneurons. Ctrl-dCas9A wild-type, n = 12; Ctrl-dCas9A Scn1a+/−, n = 10; Scn1a-dCas9A wild-type, n = 10; Scn1a-dCas9A Scn1a+/−, n = 1 (p < 0.05, 2-way ANOVA). (G and H), Histogram plots of the maximum frequency (G) and current threshold (H) reached by interneurons during the current step protocol (p = 0.02, p = 0.04, 2-way ANOVA/Bonferroni’s multiple comparisons tests). (I and J) Activity clamp analysis for the number of events during the full traces (I) and cumulative plot for AP frequency (J) (p = 0.03, 2-way ANOVA/Bonferroni’s multiple comparisons tests).
AAV-Mediated Scn1a-dCas9A Transduction of Cortical Interneurons Rescues Parvalbumin (PV)+ Interneuron Deficiency and Protects Scn1a+/− Mutant Mice from Hyperthermia-Induced Seizures
Given the encouraging results with Scn1a-dCas9A treatment obtained on neuronal cultures, we sought to test its efficacy in rescuing the epileptic phenotype in a DS mouse model.
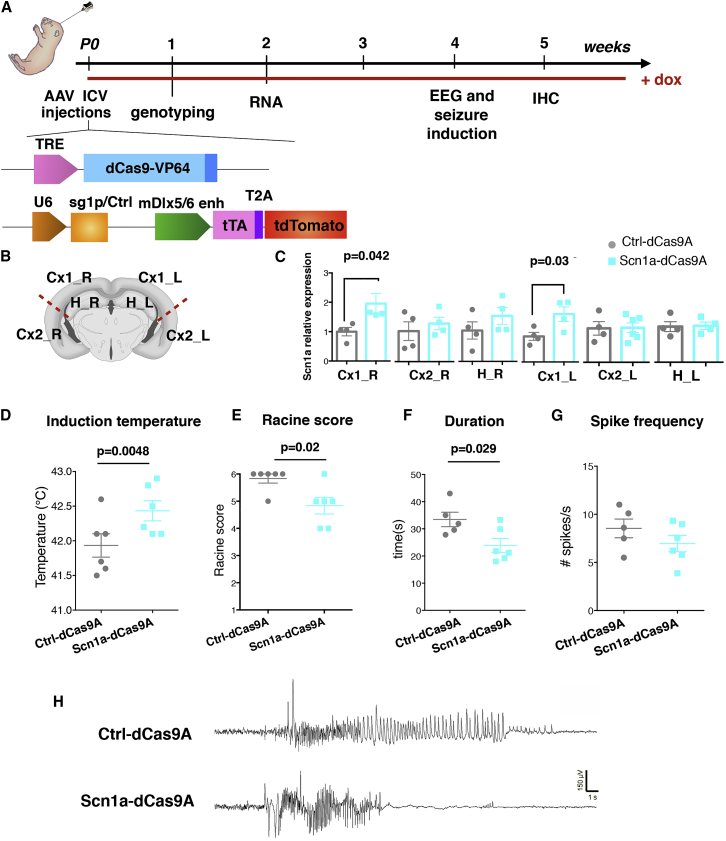
To exploit the Scn1a-dCas9A system in vivo, we sought to stimulate Scn1a expression selectively in forebrain GABAergic interneurons. We used a dual AAV9-based system because these viral particles diffuse efficiently in the brain parenchyma after intracerebroventricular (i.c.v.) injections in neonatal mouse pups.43 The VP64 activator domain, carrying four tandem copies of VP16, was chosen for in vivo delivery because its smaller size allows this dCas9A together with the TRE promoter to fit in an AAV vector. Similar to the aforementioned dual lentiviral system, a second AAV9 was packaged with the sg1P cassette, followed by the mDlx5/6 promoter driving selective expression of the rtTA-T2A-Tomato cassette in forebrain GABAergic interneurons (Figure 7A).44, 45
Figure 7.
In Vivo Scn1a-dCas9A Delivery through Intracerebroventricular Brain Injections Attenuates Seizures in the Scn1a+/− Mice
(A) Schematic illustration showing the experimental setting for in vivo delivery of the Scn1a-dCas9A system through intracerebroventricular injections into P0 pups of AAVs (2.9) carrying the Ctrl-dCas9A and Scn1a-dCas9A system. After 1 week, treated mice were genotyped, and then Scn1a+/− animals then selected for implantation of EEG electrodes and analysis of the epileptic phenotype. Wild-type (WT) litters were processed for molecular (2 weeks) and histological (5 weeks) characterization of in vivo AAV targeting. Doxycycline (dox) was administered in drinking water or food until final analysis. (B) Scheme of cerebral cortex dissection in treated mice for Scn1a expression at the mRNA level (Cx1, medial cortex; Cx2, lateral cortex; R, right; L, left). (C) qRT-PCRs performed on dissected areas of the brains in Ctrl-dCas9A- and Scn1a-dCas9A-treated wild-type mice (n = 6 for each group, p = 0.042 for Cx1_R, p = 0.03 for Cx1_L, Student’s t test). (D and E) Mean (± SEM) threshold temperatures (D) for the occurrence of myoclonic seizures (n = 6 for each group, p = 0.048, Student’s t test) and severity of the epileptic seizures, evaluated by a modified Racine score (E) in Ctrl-dCas9A-and Scn1a-dCas9A-treated Scn1a+/− mice (n = 6 for each group, p = 0.02, chi-square test). (F and G) Duration (F) and spike frequency (G) of temperature-induced seizures in Ctrl-dCas9A-or Scn1a-dCas9A-treated Scn1a+/− mice (n = 5 for Ctrl-dCas9A and n = 6 for Scn1a-dCas9A treated mice, p = 0.029 for duration and p = 0.2 for spike frequency, Student’s t test). (H) Representative EEG traces of hyperthermia-induced seizures in Ctrl-dCas9A-and Scn1a-dCas9A-treated Scn1a+/− mice.
When the mDlx5/6-promoter driven dCas9A elements were virally transduced in GAD67-GFP transgenic pups, we estimated that about 85% of the viral reporter tdTomato+ cells in the cerebral cortex also expressed the GFP transgene (Figures S7A–S7C and S7F). Furthermore, different interneuron subtypes could be targeted specifically by our system, as shown by co-labeling of tdTomato with parvalbumin, somatostatin, neuropeptide Y (NPY), or vasoactive intestinal peptide (VIP) (Figures S6B–S6F). Histological characterization of WT litters injected with the dCas9A elements revealed discrete transduction efficiency along the antero-posterior axis, with about 20% of total GABAergic interneurons transduced in cortical areas close to injection sites (Cx1_L, left, and Cx1_R, right; Figure S6G). Neatly, in these same areas, a significant increase in Scn1a gene expression was detected in 2-week-old mice treated with the Scn1a-dCas9A system compared with the Ctrl-dCas9A system (Figures 7B and 7C). To assess whether Scn1a-dCas9A has a functional effect on PV interneurons in vivo, we performed patch-clamp analysis on treated Scn1a+/+ and Scn1a+/− mice crossed with PV-Cre;Ai9-tdTomato mice at P21–P28 (Figure S7A).
Current step injections highlighted a decreased frequency-current relationship (Figure S7C) and maximum AP frequency (Figure S7D) in Scn1a+/− PV interneurons compared with Scn1a+/+ when injected with Ctrl-dCas9A. These defects in Scn1a+/− PV interneurons were rescued by transducing the Scn1a-dCas9A system (Scn1a+/+;Ctrl-dCas9A, n = 9; Scn1a+/−;Ctrl-dCas9A, n = 12; Scn1a+/−;Scn1a-dCas9A, n = 11; input/output (I/O), p = 0.003; two-way ANOVA (Figures S7C and S7D).
At 1 month, Scn1a+/− mice were implanted with electrodes, and an electroencephalogram (EEG) was recorded after subjecting the mice to hyperthermia-induced seizures. When Scn1a+/− mice were exposed to hyperthermia, we observed that the seizure threshold temperature was increased in Scn1a-dCas9A- compared with Ctrl-dCas9A-treated mice (Ctrl-dCas9A: 41.93 ± 0.1687, n = 6; Scn1a-dCas9A: 42.343 ± 0.1453, n = 6; p = 0.0048, Student’s t test) (Figure 7F). Furthermore, Scn1a-dCas9A-treated Scn1a+/− mice displayed seizures with a generally lower average clinical severity score than Ctrl-dCas9A-treated mice (Ctrl-dCas9A: 5.83 ± 0.17, n = 6; Scn1a-dCas9A: 4.83 ± 0.31, n = 6; p = 0.02, chi-square test) (Figure 7G). The average seizure duration defined by EEG recordings was also shorter (Ctrl-dCas9A: 33.5 ± 2.7 s, n = 5; Scn1a-dCas9A: 23.9 ± 2.6 s, n = 6; p = 0.029, Student’s t test) (Figure 7H). Finally, we observed a non-significant trend for the spike frequency to be lower in Scn1a-dCas9A-treated mice compared with Ctrl-dCas9A-treated mice (Figures 7I and 7J).
The upregulation of Nav1.1 during development in vivo may be protective against epileptic insults because loss of Nav1.1, in the inverse scenario, is epileptogenic. To test this hypothesis, we performed intraventricular injections of P0 pups with either Ctrl-dCas9A or Scn1a-dCas9A viruses using a pan-neuronal promoter. At P14–P17, pups were injected with lipopolysaccharide (LPS) to elicit fever by infection,46, 47 followed by repeated low-dose kainic acid (KA) injections every 30 min 2 h later. Epileptic seizures were scored according to the Racine scale, and pups were observed every 10 min until they reached grade 5, marked by tonic-clonic convulsive seizures (Figure S8A). The time taken to reach grade 5 was used to define susceptibility to epileptic insults. Pups injected with Scn1a-dCas9A had a higher seizure threshold compared with sham and Ctrl-dCas9A injected animals, confirming the hypothesis that upregulation of Nav1.1 during development is protective against seizures (Figure S8B).
In conclusion, these results show that the Scn1a-dCas9A system can be efficiently delivered in vivo by AAV-mediated gene transfer in Scn1a+/− mice to ameliorate temperature-induced seizures characteristic of this DS mouse strain.
Discussion
DS poses severe challenges for developing an effective therapeutic strategy to control epileptic seizures and associated neurodevelopmental dysfunctions. Currently available antiepileptic drugs are inadequate to suppress recurrent seizures. Furthermore, novel gene therapy approaches for Scn1a gene replacement are hampered by the relatively large size of the Scn1a gene, which exceeds the packaging cargo size of AAV particles. Finally, Scn1a transcriptional levels need to be carefully gauged to maintain an Nav1.1 protein level compatible with physiological membrane excitability in mature neurons. In light of these significant hurdles, we propose that the dCas9A-guided approach for Scn1a gene regulation has invaluable advantages for developing an effective and safe gene therapy strategy for this disease. We identified the sg1P guide, targeting a sequence close to the Scn1a proximal promoter, capable of significantly stimulating Scn1a expression. A preliminary genomic analysis confirmed that this promoter region is transcriptionally active in the adult mouse and human neurons, showing the exact transcriptional start site (TSS) by CAGE-seq and the crucial epigenetic modifications associated with its functional state. Rapid advances regarding our knowledge of the transcriptional and epigenetic state of the regulatory elements across the genome of neurons will improve the design of effective guides. Importantly, our data provide evidence that this dCas9-based activation system can further stimulate Scn1a, increasing its basal expression in young post-mitotic neurons. Surprisingly, we could not detect a significant increase in Scn1a expression in mature WT neurons, whereas it was evident in Scn1a+/− neurons at both the mRNA and protein levels upon Scn1a-dCas9A treatment. Considering that approximately 70% of the newly synthesized Nav1.1 constitutes a metabolically stable intracellular pool of protein and only 30% is trafficked to the plasma membrane and axon initiation segment (AIS),48 we can speculate that excessive accumulation of Nav1.1 induces negative regulation of the Scn1a transcript that cannot be overcome even by Scn1a-dCas9A treatment in a WT situation. Conversely under Scn1a+/− conditions, only half of the protein is produced, and this “saturation” is not achieved. In light of this, we propose that the total amount of Nav1.1 channel available in the cell can exert control by repressing transcription or destabilizing Scn1a mRNA. Further studies are required to determine the details of this regulation and to assess whether stimulation of Scn1a gene expression alters the chromatin marks within the promoter region with specific histone modifications associated with this particular transcriptional state. These results have valuable implications for manipulation of gene expression in the adult brain, providing a tool for targeted and tunable transcriptional regulation of potentially any genetic element. We achieved good gene transcriptional activation using dCas9 fused with the effector domain VP64 or VP160. However, recent studies have identified novel transactivators that can elicit higher levels of gene activation.29, 34, 49 Establishment of different Cas9 activator systems, each with its own advantages, can provide an invaluable toolbox for obtaining the right fine-tuning of transcriptional levels adequate for each specific application. Using global RNA-seq, we showed that targeted Scn1a gene activation was exquisitely specific, with no detectable off-target gene activation in primary neurons. These data reveal the high specificity of this approach, which will contribute to the high safety level for its future therapeutic applications. In fact, both the new models for accurate prediction of sgRNA off-targets and the strict requirement for targeting promoter regions close to the TSS contribute to elevating the level of specificity of this approach. Additionally, use of dCas9 eliminates the risks of DNA cleavage and its consequences in post-mitotic neurons that have lost the ability to activate homology-directed repair mechanisms to resolve DNA damage.50, 51 Importantly, dCas9-based stimulation of Scn1a expression led to a significant increase in membrane-associated Nav1.1 protein levels that restored correct functioning of DS mutant inhibitory interneurons in vitro. Our approach did not distinguish between the two Scn1a alleles and also stimulated expression of the mutant Scn1a allele (R1407X). Multiple studies have confirmed that the great majority of SCN1A mutations are loss-of-function ones and have a negligible effect because they do not produce any stable protein capable of functioning at the neuronal membrane.10 However, few mutations in SCN1A have been hypothesized from in vitro studies to cause the disease through a gain-of-function mechanism.52 We anticipate that, in these particular cases, our approach would not be of any advantage. Neuropathological studies have shown that, even in advanced stages of the disease, there is no evident sign of neuronal cell loss in patients.53 These observations strongly imply that dysfunctional interneurons can potentially recover their activity whenever a sufficient amount of Nav1.1 channel is available and indicate that at least some DS pathological defects are reversible. Altogether, these results raise the prospect of a cure for this disease even when pathological manifestations are already evident.
In this study, the different elements of the dCas9 activation system were packaged in two different AAVs but designed to provide interdependent expression of the different genetic elements. Considering that DS globally affects forebrain interneurons, we carried out AAV i.c.v. injections in neonatal mouse pups to cover the entire forebrain structures with a single treatment. In patients, because symptom onset generally occurs within the first two years of life and some more time is required to ascertain Scn1a gene mutation by exon sequencing, later delivery of gene therapy will be required. Nevertheless, the recent discovery of new AAV synthetic serotypes capable of crossing the blood-brain barrier from the bloodstream might open new opportunities for delivery of therapeutic AAVs for treating CNS disorders. In this respect, peripheral injections of AAV9 in infants with spinal muscular atrophy (SMA), a devastating infantile neurological disorder affecting spinal motor neurons, have recently shown substantial and long-term clinical benefits.54 In fact, a single intravenous infusion of the AAV9 expressing the corrected gene resulted in wide protection of motor skills for an extensive period of time and longer survival.54 This unprecedented clinical success regarding SMA with a systemic AAV gene therapy approach might facilitate the introduction of a similar strategy to treat other incurable neurological infantile disorders and DS in particular.
Our gene therapy strategy was targeted selectively to forebrain interneurons using the small Dlx5/6 enhancer, which has been shown to reliably deliver reporter genes within these neuronal classes.44, 45 Similarly, we reported that this regulatory element also ensured restricted expression of the transgenes in the GAD67-GFP neuronal sub-population in DS adult mice. Even though Scn1a is also expressed in subpopulations of cortical excitatory neurons, our strategy almost completely avoided Scn1a gene activation in these cells. Nevertheless, Scn1a deletion in this neuronal population does not induce noticeable abnormalities in mice, whereas it ameliorates the pathological phenotype of mice with Scn1a deletion in GABAergic neurons.13 Scn1a expression levels are likely different in cortical excitatory and inhibitory neurons, and our approach does not allow us to deliver different levels of gene activation in different neuronal subtypes. Thus, we considered it safer to employ the Dlx5/6 enhancer to exclusively target the neuronal population, whose dysfunction leads to pathological manifestations. The possibility that transduction of extra-cortical interneurons may affect treatment efficacy needs to be considered. Alternatively, a more selective promoter driving expression of the Scn1a-dCas9A system only in cortical interneurons and not in other interneurons (i.e., striatal) could be exploited. The Scn1a gene activation system attenuated induced epileptic seizures in terms of threshold temperature, total duration, overall clinical severity, and recovery period. However, seizures were not completely suppressed. The results can be explained by the relatively low co-infection efficiency of the two separate AAVs in the interneuron population, reaching around 20% in the injected area. In fact, the considerable size of SpCas9 requires use of two independent AAVs to assemble all elements of the activation system. Thus, future work is necessary to improve this strategy to package all of the system in a unique AAV vector by using significantly smaller Cas9 orthologs, such as SaCas9,55 GeoCas9,56 CjCas9.57 When the AAV vector for Scn1a-dCas9A treatment is optimized, it would be interesting to also test its effect on the survival rate and spontaneous seizure number and severity in Dravet mice. In conclusion, we showed that the dCas9 activation system can be tailored to obtain a robust and highly specific activation of the Scn1a gene both in cultured neurons and in brain tissue. Moreover, the dCas9 activation system can be packaged into AAVs to establish a gene therapy approach for treating DS mice and obtaining protection from temperature-induced epileptic seizures. A similar approach can then be considered for other haploinsufficient genetic disorders where stimulation of the WT allele can rescue molecular dysfunction and lead to a clinical benefit.
Materials and Methods
Bioinformatics Analysis
Transcriptomics and epigenetics next-generation sequencing (NGS) data were downloaded from the ENCODE58 and Functional Annotation of the Mammalian Genome (FANTOM)59 databases. Tracks are visualized along the mm10 mouse reference genome with the Integrative Genome Viewer (IGV).60
Molecular Cloning
sgRNAs were cloned in a LV-U6 vector as described previously. Ef1alpha-dCas9VP160-T2A- PuroR was generated from pAC94-pmax-dCas9VP160-2A-puro, a gift from R. Jaenisch (Addgene plasmid 48226).61 The dCas9VP160-2A-puro cassette was cut with AgeI and inserted into the TetO-FUW vector digested with AgeI. The dCas9VP160-2A-puro cassette was restriction digested with HpaI/AfeI and blunt-cloned into the Ef1alpha-GFP promoter, where GFP was removed by SmaI/EcoRV digestion. Ef1alpha-dCas9VP160-T2A-GFP was obtained by restriction digestion of Ef1alpha-dCas9VP160-T2A-PuroR with AscI/XbaI; the VP160-T2A fragment was obtained by AscI/XhoI digestion from Ef1alpha-dCas9VP160-T2A-PuroR, whereas the GFP fragment was PCR amplified with primers containing XhoI/XbaI restriction sites; the vector and the two fragments were ligated together.
LV-TRE-dCas9VP160-T2A-tdTomato was obtained from TetO-FUW dCas9VP160-2A-puro digested with AscI/XbaI; the VP160-T2A fragment was obtained by AscI/XhoI digestion from Ef1alpha-dCas9VP160-T2A-PuroR, whereas the tdTomato fragment was PCR amplified with primers containing XhoI/XbaI restriction sites; the vector and the two fragments were ligated together. The LV-sgRNA-hPGK-rtTA vector was obtained by digesting LV-U6-sgRNA with BamHI and cloning rtTA fragment BamHI digested from LV-hPGK-rtTA. The intermediate LV-U6-rtTA was ClaI-XhoI digested, and the PGK promoter was PCR amplified with primers with ClaI/XhoI and then cloned. AAV-TRE-dCas9-VP64 was obtained by restriction digestion of AAV-SpCas9 (a kind gift from F. Zhang, Addgene PX551),51 where the Mecp2 promoter was removed by XbaI/AgeI digestion and the TRE promoter was amplified with the following primers: FW XbaI (5′-GCTCTAGACCAGTTTGGTTAGATCTC-3′) and RV AgeI (5′-GCACCGGTGCGATCTGACGGTTCACT-3′). SpCas9 was removed with AgeI/EcoRI and Cas9m4-VP64 (a kind gift from G. Church, Addgene 47319)26 was digested with AgeI EcoRI. The VP64 fragment was PCR amplified with the following primers with EcoRI sites: FW: 5′-GATCATCGAGCAAATAAGCGAATTCTC-3′ and RV: 5′-gctaaGAATTCTTATCTAGAGTTAATCAGCATG-3′.
Virus Production
LVs were produced as described previously.62 For AAV production, replication- incompetent, recombinant viral particles were produced in 293T cells by polyethylenimine (PEI) (Polyscience) co-transfection of three different plasmids: a transgene-containing plasmid, a packaging plasmid for rep and cap genes, and pAdDeltaF6 for the three adenoviral helper genes. The cells and supernatant were harvested at 120 h. Cells were lysed in Tris buffer (50 mM Tris (pH 8.5), and 150 mM NaCl; Sigma-Aldrich) by repetitive freezing-thawing cycles (3 times), lysed in Tris buffer, and combined with correspondent cell lysates. To clarify the lysate, benzonase treatment was performed (250 U/mL, 37°C for 30 min; Sigma-Aldrich) in the presence of 1 mM MglCl2 (Sigma-Aldrich), and cellular debris was separated by centrifugation (2,000 × g, 30 min). The viral phase was isolated by an iodixanol step gradient (15%, 25%, 40%, and 60% Optiprep; Sigma-Aldrich) in the 40% fraction and concentrated in PBS with a 100,000 molecular weight cutoff concentrator (Vivaspin 20, Sartorius Stedim). Virus titers were determined by measuring the number of DNase I-resistant viral particles, using qPCR with a linearized genome plasmid as a standard. TRE-dCas9-VP64 was produced by VectorBuilder (CA, USA).
Mice
Mice were maintained at the San Raffaele Scientific Institute institutional mouse facility (Milan, Italy). Scn1a+/−10 mice were backcrossed with 129Sv mice, whereas GAD67-GFP,63 Pvalbtm1(cre)Arbr (JAX 017320, PV-Cre), and Rosa26LSL-tdTomato (JAX 007909, Ai9) mice were backcrossed with C57BL/6N mice. PV-Cre+/+ mice were crossed to Ai9+/+ mice to generate PV-Cre+/+ Ai9+/+ mice. Those mice were then crossed with 129Sv.Scn1a+/− mice to generate Scn1a +/−;PV-Cre+/− Ai9+/− mice and Scn1a+/− ;PV-Cre+/− Ai9+/−. All procedures were performed according to protocols approved by the internal institutional animal care and use committee (IACUC) and reported to the Italian Ministry of Health according to European Commission Council Directive 2010/63/EU and in accordance with the UK Animals (Scientific Procedures) Act of 1986.
Cell Cultures and Primary Neuron Derivation
P19 cells were cultured in alpha-MEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Sigma- Aldrich), 1% non-essential amino acids (Gibco), 1% sodium pyruvate (Sigma-Aldrich), 1% glutamine (Sigma-Aldrich), and 1% penicillin/streptomycin (Sigma-Aldrich). Cells were split every 2–3 days using 0.25% trypsin (Sigma-Aldrich). For transfection, Lipofectamine 300 (Thermo Fisher Scientific) was used according to the manufacturer’s protocol. Primary cultures of mouse embryonic hippocampal neurons were prepared from embryonic day 17.5 (E17.5) embryos or P0 pups derived from GAD67-GFP knockin and Scn1a+/−;GAD67-GFP pregnant females. In the latter case, each brain was processed separately, and a skin biopsy was used for genotyping. Briefly, after dissection, hippocampi were enzymatically digested with 0.025% trypsin (Gibco) in Hank’s balanced salt solution (HBSS; Euroclone) for 20 min at 37°C.Then HBSS with trypsin was removed, and the hippocampi were washed with plating medium (neurobasal medium [Gibco] supplemented with 2% B27, 3.3 mM glucose, 1% glutamine, and penicillin/streptomycin) and mechanically dissociated with a P1000 pipette to obtain a homogeneous cell suspension. Cells were then plated on plates coated with poly-L-lysine (PLL; 0.1 mg/mL) and coverslips. LV infection was performed at DIV 1, and neurons were used at DIV 10 or 21 for electrophysiology, western blot analysis, and immunofluorescence. For recordings from interneurons, we patched cells that showed co-localization of both green (indicating interneurons) and red (indicating successful lentiviral transduction) fluorescent signals.
Western Blotting
Total cerebral cortices from Scn1a+/− mice, WT mice, and primary neurons were homogenized using the Mem-PER Plus Membrane Protein Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions to enrich for the membrane-bound proteins. Western blot analysis was performed on Nupage 4%–12% gradient gels (Thermo Fisher Scientific) using primary antibodies against the following proteins: anti-Nav1.1 (1:200, Millipore) and anti-Calnexin (1:5,000, Sigma).
RNA Isolation qRT-PCR
RNA was extracted using TRI reagent (Merck) according to the manufacturer’s instructions. For qRT-PCR, cDNA synthesis was obtained using the ImProm-II Reverse Transcription System (Promega), and then qRT-PCR was performed in triplicate with custom-designed oligos (Table S2) using Titan HotTaq EvaGreen qPCR Mix (no ROX) (BIOATLAS). Analysis of relative expression was performed using the ΔΔCt method.
RNA-Seq
RNA libraries were generated starting from 1 μg of total RNA extracted from Ctrl-dCas9A and Scn1a-dCas9A neurons. RNA quality was assessed by using a Tape Station instrument (Agilent). To avoid over-representation of 3′ ends, only high-quality RNA with a RNA integrity number (RIN) of 8 or higher was used. RNA was processed according to the QuantSeq 3′ mRNA-Seq Library Prep Kit protocol. The libraries were sequenced on an Illumina HiSeq 2500 with 50-bp stranded reads using Illumina TruSeq technology. Image processing and basecall were performed using the Illumina real-time analysis software. Fastq files were aligned to the mouse genome (NCBI37/mm9) with Bowtie2.64 Differential gene expression and functional enrichment analyses were performed with DESeq2.65. Statistical analysis was performed with the SPSS statistical package (IBM). Data were deposited in NCBI GEO: GSE111436.
Deep Sequencing Data Analysis
Indexed paired-end libraries were generated starting from 1 μg of PCR amplicon spanning the Scn1aRX gene mutation10 performed on cDNA obtained from RT RNA using Illumina TruSeq Nano DNA Library Prep Kits according to the manufacturer’s instructions. Libraries were sequenced on an Illumina MiSeq. FASTQ reads were aligned to the hg38 human reference genome with Bowtie2. Alignments were visualized and quantified with the IGV genome browser.
In Vitro Electrophysiology
Current Steps
For current-clamp recordings, the internal solution contained 126 mM K-gluconate, 4 mM NaCl, 1 mM MgSO4, 0.02 mM CaCl2, 0.1 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA), 15 mM glucose, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 3 mM ATP-Na2, and 0.1 mM GTP-Na (pH 7.3). The extracellular (bath) solution contained 2 mM CaCl2, 140 mM NaCl, 1 mM MgCl2, 10 mM HEPES, 4 mM KCl, and 10 mM glucose (pH 7.3). D-(−)-2-amino-5-phosphonopentanoic acid (D-AP5; 50 μM), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; 10 μM), and picrotoxin (PTX; 30 μM) were added to block synaptic transmission. Experiments were performed at room temperature (22°C–24°C). Transduced cortical and hippocampal interneurons were identified because of GAD67-GFP and tdTomato (dCas9A system) expression. Neurons with unstable resting potential (or more than −50 mV), bridge balance of more than 15 MΩ, and/or holding current of more than 200 pA at −70 mV were discarded. Bridge balance compensation was applied, and the resting membrane potential was held at −70 mV. Current step protocols were used to evoke APs, injecting 250-ms-long depolarizing current steps of increasing amplitude (Δ 10 pA, max 280 pA). Recordings were acquired using a Multiclamp 700A amplifier (Axon Instruments, Molecular Devices) and a Power3 1401 (Cambridge Electron Design [CED]) interface combined with Signal software (CED), filtered at 10 kHz, and digitized at 50 kHz. Passive properties were calculated from the hyperpolarizing steps of the current-clamp step protocol. Input resistance is an average of three steps (2 negative and 1 positive) and is defined as the as ΔV/I. Capacitance was calculated in the current-clamp hyperpolarizing step as follows. First, the resistance was determined as voltage derivative (dV)/DI (voltage/current), and then the cell time constant (tau) was obtained, fitting the voltage changing between baseline and hyperpolarizing plateau. Capacitance was calculated as tau/resistance. Capacitance is the time constant of the voltage between the baseline and the plateau during a hyperpolarizing step. Single AP parameters were calculated as described previously.62 An event was detected as an AP when cross 0 mV and when the rising slope was more than 20 mV/ms in a range of injected current from 0 pA to 500 pA. All recordings and analyses were carried out blinded to the transduced vector.
Activity Clamp
For activity clamp experiments, current traces in voltage-clamp configuration in the presence of 4AP were recorded, holding GFP-positive interneurons (18 DIV) at −70 mV in the presence of GABAA and N-methyl-D-aspartate (NMDA) blockers. The resulting AMPA current traces were converted in conductance (G = I/V). Using Signal dynamic clamp software in conjunction with CED Power 1401-3 (CED), the conductance traces were used to inject currents into interneurons in current-clamp configuration. During recordings, the voltage of the patched neurons was read in real time and used to calculate the current to be injected from the 4AP conductance trace. To compare different cells, the conductance threshold was calculated in each neuron prior to each dynamic clamp experiment. For voltage-clamp spontaneous excitatory synaptic activity of the epileptic traces (4AP, 100 μM) and current-clamp recordings in dynamic clamp configuration, the internal and extracellular solutions were the same as described above for neuronal whole-cell patch-clamp recordings. For voltage-clamp recordings in the extracellular solution, D-AP5 (50 μM) and PTX (30 μM) were added to block GABAA and NMDA receptors, respectively. For current-clamp recordings, D-AP5 (50 μM), CNQX (10 μM), and PTX (30 μM) were added to block NMDA receptors, AMPA receptors, and GABAA receptors, respectively. Experiments were performed at room temperature (22°C–24°C). For voltage-clamp recordings, neurons with unstable resting potential and/or a leak current of more than 100 pA were discarded, and neurons were clamped at −70 mV. For current-clamp recordings, neurons with unstable resting potential and/or a bridge balance of more than 15 MΩ were discarded. Bridge balance compensation was applied, and the resting membrane potential was held at −70 mV. An AMPA conductance step protocol (Erev = 0 mV; τ = 1 ms; ΔG = 1 nS) was used to find the conductance threshold that elicited an AP, and then the epileptic conductance trace was scaled to the 15% of the conductance threshold. Neurons that were unable to generate at least one AP were therefore excluded. The sampling frequencies in voltage- and current-clamp configuration were set at 20 kHz to perfectly overlap the conductance traces with the software voltage reading. To analyze the dynamic clamp traces, an automatic MATLAB script was used41 to detect events and calculate APs parameters. An event was selected as an AP when its peak crossed 0 mV and its dV/time derivative (dt) was more than 20. Voltage threshold was calculated as the first point with a derivative of more than 20 V/s. All recordings and analyses were carried out blinded to the transduced vector. Recordings were acquired using a Multiclamp 700A amplifier (Axon Instruments, Molecular Devices, Sunnyvale, CA, USA) and Signal dynamic clamp software in conjunction with CED Power 1401-3 (CED, Cambridge Electronic Design), filtered at 10 kHz, and digitized at 50 kHz.
Ex Vivo Electrophysiology
Mice were sacrificed after deep isoflurane anesthesia, and brains were extracted. 350-μm-thick coronal sections were cut using a Leica VTS 1000 vibratome. After the cut, the slices were allowed to recover for 30 min at 32°C in modified artificial cerebrospinal fluid (ACSF) containing 92 mM sucrose, 87 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 25 mM glucose, and 10 mM MgSO4 aerated with 95% O2 and 5% CO2 (pH 7.4); slices were then allowed to recover at room temperature for at least 45 min before recording.
Current-clamp recordings were performed using a MultiClamp 700B amplifier (Molecular Devices) with pCLAMP 10 software. Pipette capacitance and resistance were always compensated. Signals were low-pass-filtered at 10 kHz and sampled at 50–100 kHz; the signal was digitized using a Digidata 1550 D/A converter (Molecular Devices).
Cells were held at 30°C–32°C. The extracellular solution contained 125 mM NaCl, 25 mM NaHCO3, 2 mM CaCl2, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgSO4, and 10 mM D-glucose aerated with 95% O2 and 5% CO2 (pH 7.4). The patch pipette contained 124 mM KH2PO4, 5 mM KCl, 2 mM MgCl2, 10 mM NaCl, 10 mM HEPES, 0.5 mM EGTA, 2 mM Na-ATP, and 0.2 mM Na-GTP (pH 7.25, adjusted with KOH).
Ctrl-and Scn1a-dCas9A PV+ interneurons were identified via tdTomato and GFP expression visualized with epifluorescence microscopy. The input/output relationship was determined by plotting AP frequency in response to progressive 500-ms, 50-pA current step injection. An AP was defined as spikes having a rising slope of more than 20 V/s and an amplitude exceeding −15 mV. Maximal steady-state firing frequency was defined as the maximal mean firing frequency in response to a current injection. Input resistance (Rm) was calculated from a −50 pA step from the resting membrane potential. AP amplitude was calculated from the AP threshold, defined as the voltage at which the first derivative (dV/dt) of the AP waveform reached 10 mV/ms, to the absolute value of the AP peak for the first spike obtained at the rheobase (defined as the minimal current injection able to elicit neuronal firing, determined through 10-pA current steps). Spike width was determined at half-amplitude (half-width) between the AP threshold and peak. Spike frequency adaptation (SFA) was calculated as the ratio of the first to the 10th inter-spike interval (ISI1/ISI10). Maximal rise and decay slope were defined, respectively, as the maximal and minimal value of the first derivative of the AP waveform.
Intracerebroventricular Injections
Neonatal mice were anesthetized in ice for 3 min. 5 μl of viral suspension containing two AAVs (titer, 1013 viral genomes [vg]/mL) (TRE-dCas9-VP64 and pU6-sgCrtl/sg1P-mDlx5enh-rtTa-T2A-Tomato/GFP, 1:1), and 0.05% Fast Green FCF (Sigma Aldrich) was injected into lateral ventricles using a Hamilton syringe with a 33G needle. After injections, pups were placed on a warming pad until they regained normal color and movement. Subsequently, they were rubbed with bedding to prevent rejection before reintroducing the mother into the cage. Dox was administered immediately in drinking water or food. One week after the injections, mice were genotyped.
Immunostaining
Cells and neurons were fixed in ice-cold 4% paraformaldehyde (PFA) in phosphate buffer (PB) for 20 min. Mice were anesthetized with ketamine/xylazine and t perfused with 0.1 M PB at room temperature at pH 7.4 with freshly prepared PFA in PB. Tissues were post-fixed in 4% PFA overnight and then soaked in cryoprotective solution (30% sucrose in PBS). Tissues were sectioned using a cryostat after optimal cutting temperature (OCT) compound embedding in dry ice. For immunofluorescence, free-floating, 30-μm-thick coronal sections or plated cells were rinsed in PBS and incubated for 20 min with 2% Triton X-100, and 3% BSA for 1 h was used to saturate the nonspecific binding site before overnight incubation with the primary antibody (diluted in a solution containing 1% BSA and Triton X-100 at room temperature). Following incubation, sections were rinsed three times in PBS and incubated for 1 h with the secondary antibody.
Primary antibodies for the following epitopes were used: red fluorescent protein (RFP) (rabbit, 1:500, MBL International), GFP (chicken, 1:500, Molecular Probes), Calbindin (mouse, 1:200, Swant), PV (mouse, 1:500, Swant), somatostatin (SST) (rat, 1:200, Millipore), NPY (rabbit, 1:500, Immunostar), VIP (rabbit, 1:500, Immunostar), Map2 (mouse, 1:250, Immunological Science), GABA (rabbit, 1:1,000, Sigma-Aldrich). Slices and cell coverslips were mounted with fluorescent mounting medium (Dako). Images were captured with a Nikon Eclipse 600 fluorescence microscope.
Surgery for Electrode Implantation, Seizure Induction, and EEG
At least 5 days before recordings, epidural stainless steel screw electrodes (0.9 mm in diameter and 3 mm long) were surgically implanted under intraperitoneal anesthesia (100 mg/kg ketamine, 10 mg/kg xylazine) and secured using dental cement (Ketac Cem, ESPE Dental, Seefeld, Germany). Two active electrodes were placed on the right and left parietal areas (2 mm lateral to the midline, 1 mm posterior to the bregma) and one over the occipital area (1 mm posterior to lambda) as a common reference.
For video EEG recording, the implanted electrodes were connected via flexible cables to an amplifier, and the EEG signal was sampled at 256 Hz, coded with 16 bits, and digitally saved using a System Plus device (Micromed, Mogliano Veneto, Italy). To obtain a good signal-to-noise ratio for seizure display, after acquisition, EEG traces were bandpass-filtered between 0.3 and 10 Hz. Video EEG recordings were inspected to detect seizures, defined as high-amplitude (at least twice the baseline) rhythmic discharges lasting at least 5 s. We defined the beginning of the seizure as the first EEG change; the end of the seizure was defined as the end of ictal EEG activity.
To induce seizures, we adopted a protocol modified from Oakley et al.16 Mice were placed in a glass beaker and heated with an infrared heat lamp (HL-1, Phisitemp, Clifton, New Jersey) controlled by a TCAT-2DF thermocontroller (Phisitemp, Clifton, New Jersey). Mouse rectal temperature was continuously monitored with a RET-4 probe (Phisitemp, Clifton, New Jersey). Seizures were identified by EEG recording and video analysis. First, mice were recorded at baseline for 15 min, and then seizures were evoked by progressively increasing the body temperature by 0.5°C every 30 s. The heating bulb was then promptly switched off to allow recovery; the mice were then monitored until the EEG and temperature returned to the baseline or until death occurred.
For EEG analysis, Neuroscore (Data Sciences International, St. Paul, MN) was used. For spike detection, EEG traces were first bandpass-filtered between 5 and 70 Hz. Threshold temperature, seizure duration, number of spikes during the attack, and spike frequency were considered. Spikes were detected by threshold analysis and then visually inspected to reject artifacts. All recordings and analysis were carried out blinded to the transduced viruses. Seizure severity was scored using a modification of the Racine scale.66
KA-Induced Febrile Seizures in Pups
A rectal temperature probe was used in P14–P17 pups to measure the basal body temperature before each injection. A single LPS injection (2 μg/mouse, Sigma, L4516) was administrated intraperitoneally (i.p.) 2 h before the experiment to induce fever and increase the temperature of the pups. Low-dose i.p. KA injections (5 mg/kg, Tocris, 0222) were performed every hour, and seizures were scored using the Racine scale every 10 min. Mice were culled after grade 5 was reached, and the time taken to reach grade 5 was used as a readout of seizure susceptibility. Seizure scoring was performed blinded to the identity of the injected virus.
Statistical Analysis
The results were analyzed with GraphPad Prism. Mean comparisons among different groups were performed with Student’s t test or two-way ANOVA followed by Bonferroni’s multiple comparisons test. In the case of non-normally distributed data, median comparisons between two groups were performed with a Mann-Whitney U test. The normality in the data distribution was assessed using the D’Agostino and Pearson omnibus test. For seizure score comparison, the chi-square test was employed. Individual statistical analyses and details regarding experimental design are described in detail alongside each experiment in Results and in the figure legends.
Author Contributions
G.C. and G.L. performed the experiments and analyzed data. C.D.B. designed and tested the sgRNAs and Scn1a transcriptional activation. J.C. performed in vivo WT experiments. N.V., G.M., and S. Bido performed viral injections in mice. L.M. developed the computational analysis. S. Brusco, V.C., S.M., and L.L. performed and analyzed the in vivo EEG recordings. S.G. produced AAVs. T.C. contributed to in vitro electrophysiology. F.B. contributed to analysis of the electrophysiological recordings. S.S. and D.M.K. contributed to the design of the experiments. V.B. supervised, coordinated, and supported the project and wrote the paper with G.C. and G.L.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We are thankful to Dr. K. Yamakawa for Scn1a mutant mice; L. Muzio, S. Levi, and D. Zacchetti for providing valuable reagents; S. Comai and M. Simonato for sharing the in vivo EEG recording instrumentation; C. Butti and E. Fraviga for technical help; and D. Bonanomi and all members of the Broccoli lab for helpful discussions. We acknowledge the FRACTAL core facility for expert supervision of flow cytometry. This work was supported by the Associazione Gruppo Famiglie Dravet (to V.B.), European Union FP7 Integrating Project “Desire” (602531 to F.B. and V.B.), the Cariplo Foundation (2016-0532 to G.C.), the Italian Ministry of Health (GR-2016-02363972 to G.C.), the Telethon Foundation (GGP19249 to G.C.), a Marie Curie individual fellowship (Marie Skłodowska-Curie grant agreement no. 658418 to G.L.), and an MRC gene therapy grant (MR/L01095X/1 to D.M.K. and S.S.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.08.018.
Contributor Information
Gaia Colasante, Email: colasante.gaia@hsr.it.
Vania Broccoli, Email: broccoli.vania@hsr.it.
Supplemental Information
References
- 1.Kullmann D.M. Neurological channelopathies. Annu. Rev. Neurosci. 2010;33:151–172. doi: 10.1146/annurev-neuro-060909-153122. [DOI] [PubMed] [Google Scholar]
- 2.Dravet C. Dravet syndrome history. Dev. Med. Child Neurol. 2011;53:1–6. doi: 10.1111/j.1469-8749.2011.03964.x. [DOI] [PubMed] [Google Scholar]
- 3.Meisler M.H., Kearney J.A. Sodium channel mutations in epilepsy and other neurological disorders. J. Clin. Invest. 2005;115:2010-7. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nickels K.C., Wirrell E.C. Cognitive and Social Outcomes of Epileptic Encephalopathies. Semin. Pediatr. Neurol. 2017;24:264–275. doi: 10.1016/j.spen.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Kasperaviciute D., Catarino C.B., Matarin M., Leu C., Novy J., Tostevin A., Leal B., Hessel E.V., Hallmann K., Hildebrand M.S., UK Brain Expression Consortium Epilepsy, hippocampal sclerosis and febrile seizures linked by common genetic variation around SCN1A. Brain. 2013;136:3140–3150. doi: 10.1093/brain/awt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cetica V., Chiari S., Mei D., Parrini E., Grisotto L., Marini C., Pucatti D., Ferrari A., Sicca F., Specchio N. Clinical and genetic factors predicting Dravet syndrome in infants with SCN1A mutations. Neurology. 2017;88:1037–1044. doi: 10.1212/WNL.0000000000003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marini C., Scheffer I.E., Nabbout R., Suls A., De Jonghe P., Zara F., Guerrini R. The genetics of Dravet syndrome. Epilepsia. 2011;52(Suppl 2):24–29. doi: 10.1111/j.1528-1167.2011.02997.x. [DOI] [PubMed] [Google Scholar]
- 8.Yu F.H., Mantegazza M., Westenbroek R.E., Robbins C.A., Kalume F., Burton K.A., Spain W.J., McKnight G.S., Scheuer T., Catterall W.A. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat. Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]
- 9.Han S., Tai C., Westenbroek R.E., Yu F.H., Cheah C.S., Potter G.B., Rubenstein J.L., Scheuer T., de la Iglesia H.O., Catterall W.A. Autistic-like behaviour in Scn1a+/- mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogiwara I., Miyamoto H., Morita N., Atapour N., Mazaki E., Inoue I., Takeuchi T., Itohara S., Yanagawa Y., Obata K. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J. Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito S., Ogiwara I., Yamada K., Miyamoto H., Hensch T.K., Osawa M., Yamakawa K. Mouse with Nav1.1 haploinsufficiency, a model for Dravet syndrome, exhibits lowered sociability and learning impairment. Neurobiol. Dis. 2013;49:29–40. doi: 10.1016/j.nbd.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Hedrich U.B.S., Liautard C., Kirschenbaum D., Pofahl M., Lavigne J., Liu Y., Theiss S., Slotta J., Escayg A., Dihné M. Impaired action potential initiation in GABAergic interneurons causes hyperexcitable networks in an epileptic mouse model carrying a human Na(V)1.1 mutation. J. Neurosci. 2014;34:14874–14889. doi: 10.1523/JNEUROSCI.0721-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogiwara I., Iwasato T., Miyamoto H., Iwata R., Yamagata T., Mazaki E., Yanagawa Y., Tamamaki N., Hensch T.K., Itohara S., Yamakawa K. Nav1.1 haploinsufficiency in excitatory neurons ameliorates seizure-associated sudden death in a mouse model of Dravet syndrome. Hum. Mol. Genet. 2013;22:4784–4804. doi: 10.1093/hmg/ddt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai C., Abe Y., Westenbroek R.E., Scheuer T., Catterall W.A. Impaired excitability of somatostatin- and parvalbumin-expressing cortical interneurons in a mouse model of Dravet syndrome. Proc. Natl. Acad. Sci. USA. 2014;111:E3139–E3148. doi: 10.1073/pnas.1411131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatsukawa T., Ogiwara I., Mazaki E., Shimohata A., Yamakawa K. Impairments in social novelty recognition and spatial memory in mice with conditional deletion of Scn1a in parvalbumin-expressing cells. Neurobiol. Dis. 2018;112:24–34. doi: 10.1016/j.nbd.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Oakley J.C., Kalume F., Yu F.H., Scheuer T., Catterall W.A. Temperature- and age-dependent seizures in a mouse model of severe myoclonic epilepsy in infancy. Proc. Natl. Acad. Sci. USA. 2009;106:3994–3999. doi: 10.1073/pnas.0813330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirrell E.C. Treatment of Dravet Syndrome. Can. J. Neurol. Sci. 2016;43:S13–S18. doi: 10.1017/cjn.2016.249. [DOI] [PubMed] [Google Scholar]
- 18.Chiron C., Dulac O. The pharmacologic treatment of Dravet syndrome. Epilepsia. 2011;52(Suppl 2):72–75. doi: 10.1111/j.1528-1167.2011.03007.x. [DOI] [PubMed] [Google Scholar]
- 19.Griffin A., Hamling K.R., Knupp K., Hong S., Lee L.P., Baraban S.C. Clemizole and modulators of serotonin signalling suppress seizures in Dravet syndrome. Brain. 2017;140:669–683. doi: 10.1093/brain/aww342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sourbron J., Schneider H., Kecskés A., Liu Y., Buening E.M., Lagae L., Smolders I., de Witte P. Serotonergic Modulation as Effective Treatment for Dravet Syndrome in a Zebrafish Mutant Model. ACS Chem. Neurosci. 2016;7:588–598. doi: 10.1021/acschemneuro.5b00342. [DOI] [PubMed] [Google Scholar]
- 21.Devinsky O., Cross J.H., Wright S. Trial of Cannabidiol for Drug-Resistant Seizures in the Dravet Syndrome. N. Engl. J. Med. 2017;377:699–700. doi: 10.1056/NEJMc1708349. [DOI] [PubMed] [Google Scholar]
- 22.Murlidharan G., Samulski R.J., Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 2014;7:76. doi: 10.3389/fnmol.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kay M.A., Glorioso J.C., Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat. Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 24.Wright A.V., Nuñez J.K., Doudna J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Gao C., Chen W., Ma W., Li X., Shi Y., Zhang H., Zhang L., Long Y., Xu H. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry. 2016;6:e703. doi: 10.1038/tp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao H.-K., Hatanaka F., Araoka T., Reddy P., Wu M.Z., Sui Y., Yamauchi T., Sakurai M., O’Keefe D.D., Núñez-Delicado E. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell. 2017;171:1495–1507.e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., Shu J., Dadon D., Young R.A., Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell. 2016;167:233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H., Liu J., Zhou C., Gao N., Rao Z., Li H., Hu X., Li C., Yao X., Shen X. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci. 2018;21:440–446. doi: 10.1038/s41593-017-0060-6. [DOI] [PubMed] [Google Scholar]
- 34.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simeonov D.R., Gowen B.G., Boontanrart M., Roth T.L., Gagnon J.D., Mumbach M.R., Satpathy A.T., Lee Y., Bray N.L., Chan A.Y. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearns N.A., Genga R.M., Enuameh M.S., Garber M., Wolfe S.A., Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matharu N., Rattanasopha S., Tamura S., Maliskova L., Wang Y., Bernard A., Hardin A., Eckalbar W.L., Vaisse C., Ahituv N. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science. 2019;363 doi: 10.1126/science.aau0629. eaau0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama T., Ogiwara I., Ito K., Kaneda M., Mazaki E., Osaka H., Ohtani H., Inoue Y., Fujiwara T., Uematsu M. Deletions of SCN1A 5′ genomic region with promoter activity in Dravet syndrome. Hum. Mutat. 2010;31:820–829. doi: 10.1002/humu.21275. [DOI] [PubMed] [Google Scholar]
- 40.Grubb M.S., Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–1074. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris G., Leite M., Kullmann D.M., Pavlov I., Schorge S., Lignani G. Activity Clamp Provides Insights into Paradoxical Effects of the Anti-Seizure Drug Carbamazepine. J. Neurosci. 2017;37:5484–5495. doi: 10.1523/JNEUROSCI.3697-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheah C.S., Westenbroek R.E., Roden W.H., Kalume F., Oakley J.C., Jansen L.A., Catterall W.A. Correlations in timing of sodium channel expression, epilepsy, and sudden death in Dravet syndrome. Channels (Austin) 2013;7:468–472. doi: 10.4161/chan.26023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammond S.L., Leek A.N., Richman E.H., Tjalkens R.B. Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS ONE. 2017;12:e0188830. doi: 10.1371/journal.pone.0188830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.A., Guo L., Xu Q., Liu R., Lu C., Chu J. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stühmer T., Puelles L., Ekker M., Rubenstein J.L.R. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb. Cortex. 2002;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- 46.Eun B.-L., Abraham J., Mlsna L., Kim M.J., Koh S. Lipopolysaccharide potentiates hyperthermia-induced seizures. Brain Behav. 2015;5:e00348. doi: 10.1002/brb3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heida J.G., Teskey G.C., Pittman Q.J. Febrile convulsions induced by the combination of lipopolysaccharide and low-dose kainic acid enhance seizure susceptibility, not epileptogenesis, in rats. Epilepsia. 2005;46:1898–1905. doi: 10.1111/j.1528-1167.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt J.W., Catterall W.A. Biosynthesis and processing of the α subunit of the voltage-sensitive sodium channel in rat brain neurons. Cell. 1986;46:437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- 49.Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staahl B.T., Benekareddy M., Coulon-Bainier C., Banfal A.A., Floor S.N., Sabo J.K., Urnes C., Munares G.A., Ghosh A., Doudna J.A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhodes T.H., Lossin C., Vanoye C.G., Wang D.W., George A.L., Jr. Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc. Natl. Acad. Sci. USA. 2004;101:11147–11152. doi: 10.1073/pnas.0402482101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catarino C.B., Liu J.Y., Liagkouras I., Gibbons V.S., Labrum R.W., Ellis R., Woodward C., Davis M.B., Smith S.J., Cross J.H. Dravet syndrome as epileptic encephalopathy: evidence from long-term course and neuropathology. Brain. 2011;134:2982–3010. doi: 10.1093/brain/awr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 55.Chen B., Hu J., Almeida R., Liu H., Balakrishnan S., Covill-Cooke C., Lim W.A., Huang B. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016;44:e75. doi: 10.1093/nar/gkv1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrington L.B., Paez-Espino D., Staahl B.T., Chen J.S., Ma E., Kyrpides N.C., Doudna J.A. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 2017;8:1424. doi: 10.1038/s41467-017-01408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim E., Koo T., Park S.W., Kim D., Kim K., Cho H.Y., Song D.W., Lee K.J., Jung M.H., Kim S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engström P.G., Frith M.C. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 60.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colasante G., Lignani G., Rubio A., Medrihan L., Yekhlef L., Sessa A., Massimino L., Giannelli S.G., Sacchetti S., Caiazzo M. Rapid Conversion of Fibroblasts into Functional Forebrain GABAergic Interneurons by Direct Genetic Reprogramming. Cell Stem Cell. 2015;17:719–734. doi: 10.1016/j.stem.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 63.Tamamaki N., Yanagawa Y., Tomioka R., Miyazaki J., Obata K., Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J. Comp. Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 64.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Velíšková J., Velíšek L. Behavioral characterization and scoring of seizures in rodents. In: Pitkänen A., Buckmaster P.S., Galanopoulou A.S., Moshé S.L., editors. Models of Seizures and Epilepsy. Academic Press; 2017. pp. 111–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.