Abstract
Damage to neuronal tissues in mammals leads to permanent loss of tissue function that can have major health consequences. While mammals have no inherent regenerative capacity to functionally repair neuronal tissue, other species such as amphibians and teleost fish readily replace damaged tissue. The exploration of development and native regeneration can thus inform the process of inducing regeneration in non-regenerative systems, which can be used to develop new therapeutics. Increasing evidence points to an epigenetic component in the regulation of the changes in cellular gene expression necessary for regeneration. In this review, we compare evidence of epigenetic roles in development and regeneration of neuronal tissue. We have focused on three key systems of important clinical significance: the neural retina, the inner ear, and the spinal cord in regenerative and non-regenerative species. While evidence for epigenetic regulation of regeneration is still limited, changes in DNA accessibility, histone acetylation and DNA methylation have all emerged as key elements in this process. To date, most studies have used broadly acting experimental manipulations to establish a role for epigenetics in regeneration, but the advent of more targeted approaches to modify the epigenome will be critical to dissecting the relative contributions of these regulatory factors in this process and the development of methods to stimulate the regeneration in those organisms like ourselves where only limited regeneration occurs in these neural systems.
Keywords: histone, neurogenesis, retina, spinal cord, cochlea
Introduction
Loss of neurological sensory tissue through disease and damage affects the lives of many. These conditions can be lifelong disabilities, as the mammalian central nervous system (CNS) has almost no regenerative ability. A considerable effort has gone into developing new approaches towards reducing neural degeneration, but few have been clinically successful. As a result, there is an urgent need for methods to stimulate repair of neuronal tissue. The approaches being taken to repair neuronal tissue can be grouped into 1) efforts to replace lost tissue through stimulation of endogenous regeneration, or 2) through in vitro generation of neuronal structures to be transplanted to the site of damage. These regenerative efforts could help millions of patients with neuronal degeneration and damage.
In this review, we focus on the role of epigenetic regulation in the development and regeneration of neural tissue. We have focused on three key systems of important clinical significance: the neural retina, the inner ear, and the spinal cord. We begin with a short review of the phenomenon of regeneration of neurons and their axons in these systems. We next detail what is known about the role of epigenetic regulation during development of each of each of these tissues. Finally, we focus on what is known about epigenetic regulation in response to injury and during regeneration. It has become clear through studies of both regenerative and non-regenerative tissue that the epigenetic landscape is very important in proper tissue development, and in regulating cell fate transitions. By comparing the response to injury in species that regenerate robustly with those that fail to do so, we hope to gain insight into the role of the epigenome in regulating these processes.
1. Glial response to Neuronal Injury
Although most regions of the central nervous system and most sensory organs of mammals do not regenerate new neurons or sensory receptor cells, other vertebrates have a range of regenerative abilities in these same structures. Neuronal regeneration is robust in the brain and retina of teleost fish species, and new neurons are regenerated primarily from fate changes of radial and astroglial cells in these regions[1]. In fish, damage instigates a glial response which can be beneficial to regeneration through mitotic cell proliferation and initiation of neurogenesis[1]. After damage, glia upregulate common neurogenesis genes like Ascl1, Brn2, Myt1l, Sox2, and NeuroD4[1]. Regulatory factors such as Notch and Shh are also expressed in regenerating fish glia, though their roles are more complex[1,2]. By contrast, in mammals, the glial response to injury—called “reactive gliosis”—is quite different, and leads instead to the formation of a "scar" that prevents migration and potentially inhibits neurogenesis and differentiation[1].
Why is there such a difference between species in the response to injury? Gene expression differences in glia across species may partly explain the differences in response to injury. For example, radial glia are the primary progenitor for neurons in the brain[3] and gene expression analyses have shown that progenitors and some types of glia, like radial glia in the brain, or Muller glia in the retina, are highly related[4]. In regenerative species, a low-level of constitutive expression of neurogenesis-related genes may predispose these animals to regeneration throughout their lives due to continuous growth[1]. Indeed, injury to the developing nervous system in mammals can lead to much more robust regenerative responses than what is seen in adults. However, another potential answer to this question may lie in critical differences in the epigenome. In the following sections we review the evidence that epigenomic differences across species may underlie the limits to regeneration observed in mammalian neural tissue.
2. Epigenetics
The epigenomic landscape has a strong influence on cell fate determination, cell differentiation and other developmental processes that are highly relevant to regeneration[5]. The epigenome is generally characterized by chromatin states, or the interaction between DNA and histones or other packing proteins, often as facilitated by modifications to DNA or proteins[6,7]. There are two basic states of chromatin: euchromatin associated with open transcription, and heterochromatin, associated with inactive transcription[8].
Histones are a major component of chromatin, packaged together into histone octets that interact with 147bp fragments of DNA to create nucleosomes[7]. The generation of dense heterochromatin, or decompaction into euchromatin are both associated with many posttranslational modifications to the N-terminal tail structures of the core nucleosome histones. In euchromatin, lysine acetylation of histones is associated with open and accessible chromatin, due to steric interactions[7]. The best-characterized modifications are histone H3 lysine 27 acetylation (H3K27ac) and H3 lysine 4 acetylation (H3K4ac). Histone acetylation is catalyzed by Histone Acetyltransferases (HATs) and these acetyl groups are removed by Histone Deacetylases (HDACs)[6]. Euchromatin is also associated with some methyl modifications, such as histone H3K4me3 and H3K36me3[7]. Condensed heterochromatin is almost exclusively associated with histone methylation. These modifications are added to histone tails by Histone Methyltransferases (HMTases), and removed by Histone Lysine Demethylases (KDMs). The most well-characterized heterochromatic histone methyl mark is H3 lysine trimethylation (H3K27me3), which is catalyzed by Polycomb Repressive Complex 2 (PRC2)[6]. Also well studied is the repressive H3K9me2/3, though as with euchromatic histone modifications, many other modifications have been reported. However, heterochromatin and euchromatin markers can be found in combination with each other to create bivalent chromatin, which is frequently found around genes that may be rapidly activated during cell fate decisions[7,8]. The combinations of all of these histone modifications, with some redundancy, help determine the chromatin state.
In addition to epigenetic regulation through histone modifications, DNA methylation can also affect gene expression. There are many varieties of DNA methylation, but the best studied is 5-methylcytosine at C-phosphate-G (CpG) islands[8]. This type of DNA modification is most associated with heterochromatin and reduced expression. However, when found within the gene body, it is more associated with enhanced transcription. DNA methylation was previously considered to be a more permanent epigenetic modification, but has been shown to change with development and cellular activities[6]. These modifications are added by DNA methyltransferases (DNMTs) and removed by Tet-eleven translocation enzymes, APOBECs, and AID[8].
3.1. Retina: Development
The retina has proven to be an excellent system for studying neural development, due to the well-characterized cell types and its relative isolation from the rest of the CNS[9]. The retina is composed of five basic types of neurons, with many sub-types of each of these, and one intrinsic type of glia, the Müller glia (MG). In addition to the neurons and intrinsic glia, there are astrocytes that migrate into the retina from the optic nerve, microglia, and endothelial cells. The different classes of neurons and the Müller glia are all generated during development from a common progenitor, and each class of neuron arises at a specific time during development. Ganglion cells, horizontal cells and cone photoreceptors are all generated early in development, while amacrine cells, rod photoreceptors and bipolar cells are generated somewhat later in the process.
There have been several studies characterizing the epigenome of developing retina and evaluating the effects of experimentally altering histone modifications on neurogenesis in this system. DNase mapping of the developing mammalian retina demonstrated that widespread changes in chromatin accessibility correlate with changes in gene expression[10]. Early postnatal enhancer accessibility at key neurogenic genes is replaced over the course of one week by accessibility in differentiated neuronal enhancer regions, thus identifying thousands of putative regulatory regions over key stages of mouse retinal development[10]. A similar analysis of human development, identified approximately 300k DNase-accessible regions that correlate with gene expression changes[11]. This accessibility is in part informed by highly dynamic histone modifications[12]. Histone modifications, especially H3K27me3, are highly correlated with gene expression[12]. This is especially true of differentiation genes, which see many changes in the chromatin landscape during development[12].
In addition to global changes in the retinal epigenome with development, some studies have shown cell-type specific regulation, particularly for photoreceptors: rods and cones. In mice, many chromatin changes occur within the immature photoreceptor cells as they mature. For example, CCCTC-binding factor (CTCF) chromatin looping motifs show large changes with cone maturation, correlating with significant gene expression changes[13]. The epigenome of rods has a unique chromatin structure. Rather than having multiple heterochromatin foci, rod nuclei contain one extremely dense center with a unique epigenetic composition[14,15]. DNA methylation is in this case associated with inaccessible and inactive regions[16]. Cone photoreceptors have more open chromatin than rods and less methylation than rods[15]; there is evidence that the differential pattern of methylation between these cells may contribute to the unique rod chromatin architecture, since conditional deletion of three DNA methyltrasferases (DNMTs) disrupts the unique rod chromatin and thus leads to dysregulation of phototransduction genes and retinal thinning[16].
Functional studies of chromatin modifying enzymes have shown the importance of these modifications to retinal development. For example, Lysine 27 methylation is crucial for proliferation and maintaining progenitor status in development[17-21]. Conditional deletion of Ezh2—the enzymatic component of Polycomb Repressive Complex 2 (PRC2)—in retinal progenitor cells reduces proliferation and disrupts normal development of retinal lamination and differentiation[17]. RNAseq analysis of knockout retinas shows increases in genes known to be repressed by H3K27me3 in other systems, including targets that account for changes to proliferation[17,21]. Additionally, deletion of the H3K27 demethlase Jmjd3 leads to increases in proliferation and loss of Vsx1+ bipolar cells[22]. H3K27me3 associated with bivalency is also well characterized in the mammalian retina[17,20,23]. Bivalent regions tend to have the most changes in expression during development, and K27me3 contributes significantly to amacrine cell gene bivalency[17,20]. H3K27me3 has been shown to change inversely in development with active H3K4me2[20]. Both modifications are most highly associated in the retina with gene rich areas, and genes that are highly expressed at some point in the generation of the neural retina (recently on or off), while non-retinal genes are less likely to be modified at all[17,20]. However, while loss of Ezh2 function leads to substantial changes in photoreceptor development and maturation, this does not significantly affect retinal ganglion progenitor development[13,24,25], suggesting that all cell types are not equally reliant on this form of regulation. Nevertheless, it is clear that H3K27me3 and its related enzymes are important regulators of retinal development.
In addition to histone methylation, acetylation is also important for retinal development and normal function. HDAC1 in particular plays key roles in late postnatal development and maturation, as it regulates proliferation, and later the transition in photoreceptor development from young immature neurons to mature ones in the first postnatal week[26]. In development, HDAC1 binds to Pax6 and pairs its catalytic activity to Pax6 binding sites and targets such as Ascl1, Atoh7, and NeuroD1[27]. Histone acetylation also binds Brd2 and Setd5 complexes to regulate semaphorins[28]. The role of acetylation on development becomes a common theme throughout many neuronal systems. Overall, it is clear that many different epigenetic modifications contribute to retinal development, and the combinatorial nature of this system makes it difficult to parse the relative contributions of these modifications to specific developmental processes. Even so, knowledge of the developmental epigenetic landscape provides an important context for related changes that occur after injury.
3.2. Retina: Regeneration
Although the adult mammalian retina has little ability to regenerate new neurons after their loss from injury or disease, retinal regeneration has been shown to occur in non-mammalian vertebrates[29]. Amphibians, particularly urodeles, have the capacity to completely regenerate an entirely new retina from the retinal pigmented epithelium (RPE). If the retina is removed (or even separated from the adjacent RPE cells, the cells lose their pigmentation, reduce their expression of genes characteristic of this cell fate, and express genes of embryonic retinal progenitor cells. The RPE-derived retinal progenitors undergo multiple rounds of cell division and produce a new, laminated retina that can restore functional vision to the animals. This remarkable feat is analogous to the capacity of these animals to regenerate other parts of their bodies, like their limbs, and it is very likely that the dramatic changes in gene expression and phenotypic plasticity require equally dramatic changes in the epigenome.
Teleost fish are also capable of regenerating all types of retinal neurons after injury or disease, though in this case, the source of the new neurons is not the RPE, but rather the radial-glial like Müller glia[2,30]. As noted above, the Müller glial cell is the only glial cell generated during development by the retinal progenitor, and has a gene expression profile that is in many ways quite similar to that of the embryonic progenitor cell[4,31]. A key difference between the embryonic progenitor cell and the Muller glia is the proneural transcription factor, Ascl1 This gene is expressed by retinal progenitors, but not by mature Muller glia. After injury, the Müller glia of fish express Ascl1 and this gene is necessary for the Muller glia to produce the progenitor-like cells needed for regenerating neurons. In addition to Ascl1, critical changes occur in Stat3 signaling, multiple growth factors and receptors, canonical Wnt signaling, and microRNAs, and these are all necessary for successful regeneration [2,32].
The changes in cell signaling and transcription factors leading to regeneration of new neurons in this system is accompanied by critical changes in the epigenome. The changes in DNA methylation in particular have received the most detailed analysis[33-35]. DNA methylation regulators are expressed at low levels in quiescent MG, but after injury they are upregulated within 4 days[33]. These include genes important for demethylation of existing CpGs, e.g. Gadd45, Apobec2a/b, and Tet, as well as four Dnmts. While the vast majority of DNA methylation sites in MG are not changed after injury (~98%), the small percentage of sites that do change are potentially informative. Of the 9,554 DMBs (differentially methylated bases), approximately equal numbers show increased and decreased methylation[34]. Of the 125 promoter-associated DMBs, those demethylated after injury correlate with increased gene expression. Interestingly, the promoters of pluripotency factors and regeneration-associated genes have low levels of methylation at their promoters in MG from uninjured retinas[34]. Thus, the expression of stem cell and progenitor genes appears not to be repressed via this mechanism. Nevertheless, functional studies indicate that changes in DNA methylation that occur after injury are important for the regeneration process in the fish, since knockdown of Apobec2a/b inhibits regeneration while treatment with 5-dAza increases tuba1a:gfp expression, an indicator of MG reprogramming in response to injury[33,35]. During this period of regeneration in the fish retina, regeneration associated genes are additionally regulated by HDACs[36]. HDACs are essential for regulating the regeneration process, and small molecule inhibition of their activity in the fish model reversibly blocks regeneration after damage[36]. Apart from these studies, there is little known about epigenetic changes in fish Müller glia during the regeneration process.
Changes in methylation in mammalian Müller glia have also been studied, though to a lesser extent than the fish. Similar to the zebrafish, the pluripotency factors’ promoters are hypomethylated in Muller glia from uninjured fish retina. In the immediate response period 4-12 hours after retinal damage, the transcription factor Oct4 loses nearby DNA methylation in mouse Müller glia, but it is quickly re-silenced after 24 hours[37]. The loss of methylation in mammalian Muller glia after injury is interesting, though it is not clear how widespread this phenomenon is or if it occurs at genes critical for regeneration. Future studies on DNA methylation in mammalian Müller glia could potentially shed light on this issue, given the importance of DNA methylation in developing retina.
Although fish have the capacity to regenerate neurons from MG, mammals lack this ability. Injury to the mammalian retina typically leads to MG hypertrophy and reactivity[38].Some effort has been made to stimulate the MG to generate new neurons, using growth factors and activating signaling pathways. Although some of these treatments lead to a re-entry of some MG into the mitotic cell cycle, evidence for regeneration of new neurons has been scant. Several years ago, Pollack et al demonstrated that mouse MG could be maintained in dissociated cell culture, and they used this system to test the ability of proneural transcription factors to reprogram the MG into retinal progenitors. Many progenitor genes were reactivated in the MG with Ascl1 over-expression, and a subset of the Ascl1-expressing MG generated new neurons in the cultures[39]. Analysis with ChIP demonstrated that Ascl1 induced a reduction in the repressive H3K27me3 modification and an increase in H3K27ac at key progenitor and neural genes[39]. These in vitro studies were followed up in vivo; using transgenic mice to specifically direct Ascl1 expression to MG. Ueki et al found MG from two week old mice could be reprogrammed to regenerate neurons in vivo, and the newly generated neurons had the morphology resembling bipolar cells and amacrine cells[40]. This capacity was lost as the animals matured, and analysis of the chromatin accessibility in the MG using DNase-seq showed that open chromatin near neural genes was lost in the MG, when compared with the developing retina[41]. This motivated these investigators to use HDAC inhibitors in conjunction with Ascl1-over-expression and they found that this combination promoted functional regeneration of new neurons in the adult mouse retina[41]. Comparison of the open chromatin in the Ascl1-reprogrammed MG demonstrated that new neuron and neural progenitor sites became accessible as the cells shifted to a progenitor-like state and then to neurons[41]. These studies demonstrated the feasibility of using in vivo reprogramming factors, in conjunction with epigenetic modification, to drive regeneration in a non-regenerating species.
In addition to the ability of some species to regenerate new neurons from glial or RPE-derived progenitor cells, the projection neurons of the retina—the retinal ganglion cells (RGCs)—also have the capacity to regenerate their axons after an injury to the optic nerve in some species. This type of axonal regeneration occurs naturally in fish and frogs, to a limited extent in reptiles, but not at all in birds or mammals [42]. The ganglion cells in fish and amphibians respond to injury by forming growth cones at the tips of the severed axons, and these new growth cones grow to visual centers in the brain. The growth of these axons is similar to that which occurs during development, and many of the genes required for axon growth and pathfinding are re-expressed in the ganglion cells after the injury. By contrast, in mammals, the predominant response of the RGCs to optic nerve crush (ONC) is to undergo apoptosis. Most RGCs that survive fail to form new growth cones or extend axons in the optic nerve[43].
Comparisons between the epigenome of developing RGCs, where axon growth is robust, with that of mature RGCs, where regeneration fails, might lead to a better understanding of potential limits to epigenomic reprogramming that might underlie successful regeneration. For example, the coactivator and acetyltransferase P300 is expressed during postnatal development of retinal ganglion cells[44]. However, after ONC, P300 decreases in expression in the RGCs[44]. Experimental overexpression of P300 induces axon growth and increases in acetylation of regeneration-associated genes[44]. Death of RGCs after ONC may also be due in part to epigenomic limits to reprogramming, as the addition of Trichostatin-A (TSA) after ONC improves neuronal survival independent of changes in axon regrowth or P300 activity[45]. TSA has also been reported to increase neurite outgrowth via regulation of RARb[45]. The interpretation of these results is complicated by the fact that histone acetyltransferases and HDACs have nonhistone targets and effects seen from modulating them may not be entirely due to epigenetic regulation.
4.1. Inner Ear: Development
Another peripheral sensory structure that has been extensively studied for its capacity for regeneration is the inner ear [46]. The sensory epithelia of the inner ear of vertebrates underlie the auditory and vestibular senses. The auditory sense is mediated through sensory cells in the cochlea in mammals and basilar papilla in non-mammalian vertebrates, while the vestibular sense is mediated through sensory cells in the utricular and sacullar maculae and the cristae. Fish and aquatic amphibians also have small clusters of sensory epithelia distributed along the body, called lateral line organs that are similar to vestibular sensory structures, and are used to sense water flow[47]. All of these organs contain sensory receptors, called hair cells, which transduce mechanical motion into changes in electrical activity. The sensory hair cells are typically surrounded by one or more different types of supporting cells, and particularly in the sensory epithelium of the mammalian cochlea, called the Organ of Corti, the diversity in supporting cell morphology shows many distinct types with highly specialized functions[47].
The development of the auditory and vestibular organs has been well characterized and the molecular pathways involved in hair cell and support cell fate determination is known in some detail[48]. The highly stereotypic arrangement of hair cells and support cells required for proper function in these organs has allowed investigators precision in analysis of developmental phenotypes. The inner ear develops from an invagination from the embryonic ectoderm called the otic placode, located near the developing hindbrain. The complex inner ear structures, including the sensory epithelia arise from the otic placode, though a series of morphogenic events. The sensory structures require the transcription factor Sox2 for their neural potential, and downstream of Sox2, another critical transcription factor, the proneural gene Atoh1 is necessary and sufficient for hair cell formation during development in all these structures where it has been tested, including fish, bird and mouse. The upstream regulatory pathways that direct Atoh1 to a subset of hair cell progenitor cells are known to include Notch and fibroblast growth factor (FGF) signaling, and canonical Wnt and BMP signaling molecules also interact to define the region of sensory specification[48].
There is evidence for both histone modifications and DNA methylation being important regulators of inner ear sensory development and/or lateral line development in fish. For example, the histone lysine demethylase LSD1 is required for hair cell development in zebrafish lateral line[49]. This epigenetic modulator specifically regulates Sox2 expression and other developmental processes[49]. In addition, HDACs, especially from class I, directly regulate important inner ear developmental genes, such as Atoh1, Sox2, and Pax5 in the zebrafish[50,51].
In mammalian inner ear, several studies have shown changes in histone modifications that correlate with developmental stages. Atoh1 specifically is highly regulated through epigenetics during development and is shown to have high H3K9ac near the gene in hair cells, compared to the epigenome of progenitors[52]. During development, Atoh1 loses accessibility and gains repressive H3K27me3[52]. Prosensory genes thus may be repressed in support cells via this mechanism, as the inhibition of acetylation during development reduced Atoh1 expression and hair cell production[52]. Additionally in mouse, the histone demethylase Kdm4b and the DNA methyltransferase, Dnmt3a, are necessary for early events in otic development, linking both histone and DNA methylation to the regulation of prosensory homeobox and HMG box TFs during development [53] [54]. Another histone methyltransferase important for inner ear development is Whsc1, which is responsible for trimethylation of H3K36[55-57]. Mutations of this methyltransferase lead to hair cell defects and deafness, and knockout mice have disorganized hair cell rows as well as stereociliar organization defects[57]. Epigenetic regulators have important roles in development and organization of the inner ear, and thus are important to the generation of this sensory tissue.
Prosensory genes are also regulated by larger chromatin restructuring components. In particular, chromatin domain binding 7 (Chd7) protein remodels chromatin around developmental sensory genes[58-60]. Chd7 upregulation increases expression of developmental genes such as retinoic acid-associated pathway genes and Aldh1a3[59]. This remodeler also pairs with Sox2 to regulate its targets in the inner ear[60]. The chromatin looping-related protein CTCF additionally plays major roles in proneural development in the inner ear, as loss of the protein leads to developmental defects in the inner ear[61]. CTCF appears to regulate development via key genes such as Neurog1, which acts in a neurosensory capacity[61]. The chromatin remodeler also appears to alter accessibility at promoter and enhancer regions, indicating a role in chromatin structure during inner ear development[16]. It is clear that beyond singular histone modifications, the larger chromatin structure can impact gene transcription regulating cellular fate.
4.2. Inner ear Sensory epithelia: Regeneration
Loss of hair cells in both auditory and vestibular sensory epithelia occurs normally with age and leads to age-related loss of hearing and balance. Age-related hearing impairment is widely recognized as an increasing problem in our society, but loss of balance in the elderly, and the falls that result from this instability, are one of the most common causes of bone fractures and hospitalization in this population[62]. Developing methods to stimulate hair cell regeneration in people that have lost these cells due to age, noise-damage or ototoxic drugs is a primary focus of many scientists in this field.
In the species that have the ability to regenerate new hair cells after injury, the new hair cells arise from adjacent support cells. The loss of hair cells triggers an increase in the proliferation of the nearby support cells, and many of the newly generated cells differentiate into hair cells.In the bird auditory basilar papilla, there is also evidence for direct transdifferentiation from support cells to hair cells. [63]. This regeneration of hair cells is facilitated by the activity of Atoh1, which is regulated by Notch signaling [63], in many ways mirroring the analogous process by which hair cells and support cells arise during development
The fish lateral line organs have been particularly advantageous for studies of hair cell regeneration. The process can be directly imaged in live animals, and drugs can be applied directly to the water to initiate hair cell death and to manipulate signaling pathways during regeneration. Consequently, there is a growing literature on the role of epigenetic factors in controlling the regeneration process. Some of the same epigenetic regulators that act in development appear to behave similarly in sensory regeneration. For example, pharmacological inhibition of Lsd1 inhibits supporting cell proliferation and the regeneration of new hair cells through the regulation of Wnt and Fgf signaling, as well as the cyclin dependent kinase inhibitors, p21/Cip1 and p27/Kip1 [49]. Also, treating zebrafish with the G9a methylatransferase inhibitor BIX01294 inhibits supporting cell proliferation and reduces hair cell regeneration after ototoxic damage. Inhibition of Utx/Jmjd, a H3K27demethylase, also inhibits hair cell regeneration in this system[64].
Another popular system for studying hair cell regeneration is the posthatch chick. The discovery that the chick basilar papilla regenerates hair cells in the late 1980s has stimulated a great deal of research in this area, and much of what is known at the cellular level about hair cell regeneration comes from studies in the bird[65]. Regeneration in the bird auditory basilar papilla and vestibular epithelia (e.g. the utricle) occurs in organ culture, which allows drugs that act on epigenetic modifiers to be screened in vitro. After damage to the hair cells, the support cells of the bird inner ear sensory epithelia spontaneously re-enter the mitotic cell cycle; HDAC inhibition by valproic acid (VPA) in primary cultures of chick utricle blocks the otherwise high proliferation levels; however, the HDAC inhibition does not appear to affect differentiation of those hair cells that are regenerated[66]. This is very similar to the result obtained in the fish lateral line; however, it should be noted that HDAC inhibitors can interfere with mitotic cell proliferation via non-histone actions as well.
Additionally, the demethylase Lsd1 appears to affect reactivity after damage in the cochlear ganglion neurons[67]. Inhibition of Lsd1 after cochlear damage leads to an increase in the number of ganglion cell bodies, and improved neurite density[67]. Thus, it is apparent that H3K4 methylation is protective against damage, though activity of its demethylase after damage impairs the tissue’s ability to repair itself. Unfortunately, evidence is lacking for the specific intersection of inner ear damage/regeneration and epigenetics. It is clear from developmental studies both in the inner ear and in other sensory and neuronal tissues that epigenetics plays a key role, and could be highly relevant to regeneration efforts. However, more research is needed to elucidate precise roles that epigenetic modulators could play in repairing diseased or damaged tissues.
5.1. Spinal cord: Development
The spinal cord is a critical part of the CNS, providing the hub for somatosensory input and motor output. The neurons andChxlO glia of the spinal cord arise during development in distinct domains of progenitors, patterned along the dorsal-ventral axis. Both astrocytes and oligodendroglia are generated from the progenitors located along the ventricular surface of the spinal canal. A detailed description of spinal cord development is beyond the scope of this review, though it is important to note some key findings of epigenetic regulation in the developing CNS.
As different types of neurons are generated during development they acquire unique patterns of accessible enhancers and promoters[10]. Different regions of the CNS have unique epigenomic signatures that direct region-specific expression of critical genes[68]. Similar studies to those described above for retina have shown roles for H3K27me and H3K9me during development. The activity of neurons can even change the overall chromatin accessibility of activity-related genes[69]. Synchronous activation of the dentate gyrus changes accessibility of Arc and Gabrr1 with thousands of peaks differentially associated with activity[69]. These activity changes are associated with alterations to H3K4 methylation and H3K27 acetylation around transcription start sites and active enhancers, as well as the binding activity of the chromatin looping protein CTCF[69]. This particular change to activity-related chromatin is linked to cFOS binding, and the loss of cFOS reduces chromatin restructuring[69].
5.2. Spinal cord: Regeneration
Spinal cord injury (SCI) leads to a loss of neurons at the point of impact, but more importantly frequently damages axons of passage, leading to loss in motor and sensory communication of the body with the brain distal to the lesion. This devastating injury has motivated many investigators for decades to develop methods to repair the damaged spinal cord. As for the two sensory systems described above, spinal cord regeneration occurs naturally in some species, and comparative studies among vertebrates have provided insight into the mechanisms by which successful regeneration occurs, and the points of failure where it does not.
What is known about the genes that limit successful regeneration of axons? In species where axon regeneration occurs in the spinal cord, there are key genes that change in response to injury. The changes in regulatory transcription factors are particularly interesting, since they may represent critical points of intervention. There is an intrinsic loss in the ability of neurons to grow axons in the mammalian spinal cord as the CNS matures[42]. The loss of expression of axon growth-associated genes accompanies this developmental decline in the ability of neurons to extend axons. For example, Sox11, a critical transcription factor needed for axon regeneration in peripheral nerves, is expressed in corticospinal axons that are actively extending axons in fetal and neonatal mice, but is not expressed in mature neurons or after injury[70]. Experimentally driving Sox11 in adult mouse cerebral cortical neurons promotes axon regeneration from the cortico-spinal tract. Other transcription factors, like Klf6 and Klf7, and key signaling pathways, e.g. STAT and PTEN, are also important regulators of axon growth both during development and in spinal cord regeneration[42].
What role do epigenetics play in the gene expression changes needed for axon regeneration? As noted above, developmental and neuron growth-related genes are downregulated during later development and neuronal maturation[71]. Vanketesh et al found a loss in chromatin accessibility in putative cis-regulatory regions associated with these down-regulated growth promoting genes; this downregulation is associated with changes in chromatin accessibility, while upregulated genes undergo decondensation of the chromatin[71]. For genes like Sox11 and Klf7 (another transcription factor that stimulates axon regeneration) a loss in chromatin accessibility correlates with the loss in their expression as neurons lose their intrinsic axon growth potential[71] ; however, the correlation is not perfect as not all developmentally downregulated genes lose accessibility: approximately half retain accessibility at their promoters. In the peripheral nerve, where axon regeneration is successful, chromatin remodeling genes and histone acetyltransferases show increased expression prior to the increase in axon growth-associated genes[72]. In addition, peripheral nerve injury leads to nuclear export of HDAC5, and this is required for the activation of regeneration associated genes[73]. However, axotomy specifically transports HDAC5 to distal axons, indicating the role of this protein in regeneration may be linked to non-histone acetylation targets, especially as axotomy is linked to increases in H3 acetylation[73].
This increase in acetylation after injury indicates a crucial role of acetylation in damage response and regeneration. Hypoacetylation of H4 is associated with reduced axon growth, and H4 acetylation is specifically enriched at regeneration associated gene promoters in the immediate window 6-24 hours after peripheral axotomy[74]. Histone acetyltransferase activity on regeneration-associated genes is also linked to Smad1, which may play a role in directing HATs to their targets[74]. Additionally, the lysine acetyltransferase PCAF can alter the epigenome in association with improved axon regeneration[75]. Sciatic axotomy enriches the epigenome for H3K9 acetylation, in correlation with nuclear PCAF[75]. Though these increases in acetylation may not entirely correlate with expression, the addition of PCAF to SCI damaged tissues improves neurite outgrowth[75]. These acetylation findings tie in with the evidence supporting that broad opening of chromatin surrounding cell fate conversions improves regeneration, which likely occur in a reciprocal HAT/HDAC changes in activity.
The correlation in the loss of chromatin accessibility and acetylated chromatin and the loss in the capacity for axon regeneration has prompted several investigators to test whether the barriers to axon regeneration could be overcome by HDAC inhibition (or less frequently, small molecule activation of HATs)[74-76]. The reagents used to inhibit HDACs are broad in their activity, and so a more targeted approach may ultimately be necessary; nevertheless, some positive effects have been observed in these studies. In vitro studies demonstrated that HDAC inhibitors can promote axon growth on non-permissive substrates, like myelin and CSPGs[76]. In vivo, treatment of spinal sensory neurons with either TSA or the HDAC1 inhibitor MS-275, promotes their expression of axon regeneration genes, and the degree of axon regeneration after spinal injury [74].
In addition to the evidence for histone acetylation and regeneration, DNA methylation has also been studied for a role in the regulation of regeneration-associated genes in response to damage in peripheral nerve[77]. The dorsal root ganglion sensory neurons have both a central and a peripheral branch. Axotomy of the peripheral branch, which leads to successful axon regeneration, leads to upregulation in DNA methylation; however, axotomy of the central branch, which does not stimulate regeneration, does not induce this increase in DNA methylation[77]. When Tet3 is upregulated following damage, increases in methylation of axon growth and myelination genes are seen, though differentiation and neurite outgrowth genes lose methylation[77]. In peripheral axotomy compared to central, there is also more expression of regeneration-associated genes, which are methylated within 24 hours of damage[77]. The negative regulation of regeneration-associated genes shortly after damage may be contributing to reduced tissue repair, as regeneration gene expression post-trauma is cut short.
In addition to affecting changes in axon growth-associated genes, epigenetic regulation of cell survival genes is also important for recovery after spinal cord injury. In this context, treatment with HDAC inhibitors after spinal injury has also shown some benefit. The application of HDAC inhibitors after SCI improves behavioral tests within one week of injury[78]. Neurons in the area of damage show reduced apoptosis and alleviated microglial response, plus elevated Stat1 as compared to untreated SCI[78]. HDAC inhibition of HDAC6 also promotes cell survival and neurite growth after injury[79].
What is known about the changes in gene expression that are necessary for successful regeneration of neurons in the spinal cord? In those species that regenerate new neurons in the spinal cord after injury, such as amphibians and fish, the radial glia that line the central canal are thought to be the main source of neural progenitors[1,2]. Prior to injury, these latent progenitors in the spinal cord express Olig2, Pax6, and Nkx6.1, and these genes are necessary for proper regeneration[2]. In the fish, Sox2 is additionally necessary for the mitotic cell proliferation that is necessary for production of the regenerated neurons[1]. In mammals, these genes are associated with neurogenesis during development, but are not expressed in glia after injury; therefore, much of the work in mammals has focused on developing strategies to reprogram glial cells in the spinal cord or elsewhere in the CNS to regenerate new neurons.
6. Pioneering Factors
The generation of induced neurons (iN) from somatic cells in culture mimics some of the changes that occur in in vivo regeneration, and much can be learned from this method of neurogenesis. Many of these studies have been carried out in fibroblasts, where viral over-expression of Brn2, Ascl1, and Myt1l (collectively known as the BAM factors) in fibroblasts induces the cells to generate functional neurons[80,81]. In fish and amphibians, CNS neuronal regeneration occurs via the neuronal glia, which reprogram their fate and act as progenitors in regenerative species. Several groups have shown that mammalian glia can also be reprogramed by infecting mature astrocytes or NG2+ cell with similar transcription factors, including Ascl1, Pax6, Brn2, Neurog2, and Sox2 to generate functional neurons[82]. The combination of Neurog2 and Ascl1 has been proven to generate functional neurons not only in cultured astrocytes, but additionally in the endogenous neural progenitor cells (NG2+ cells) of the injured spinal cord[83]. Furthermore, downstream targets of these factors, NeuroD4 and Insm1, can independently be used to reprogram astrocytes into mature neurons. Gotz and colleagues have monitored the chromatin state changes during the reprogramming process and find that there is an increase in the repressive histone modification H4K20me3 around the Neurod4 gene, in astrocytes as they mature in culture, and this modification appears to be downstream of binding by the transcriptional co-repressor, REST[84]. Whether this mechanism can be generalized to other neural and neural progenitor genes in astrocytes awaits additional investigation. In general, the addition of one to two transcription factors to glia can be sufficient to reprogram the cells to a neural or neural progenitor state. These factors act as "pioneering" factors, capable of restructuring the epigenome to induce widespread expression changes. As seen in fibroblast studies with Ascl1, addition of the pioneering factor does in fact cause widespread chromatin accessibility changes within 2-5 days of induced expression[81]. The addition of HDAC inhibitors to a pioneering factor-based iN system can additionally exploit these chromatin changes and have been shown to improve the genesis of new mature neurons. This pioneering activity is one key to understanding fate changes in glial cells as they are induced to regenerate neuronal tissues.
Conclusions
Increasing evidence indicates that epigenetic regulation is important for the development and function of neuronal tissue. Epigenetic regulation also plays an important role in the maintenance of cell state, and therefore is likely to have important roles in the changes in cell state that accompany the response of neural tissue to injury and subsequent changes associated with regeneration; however, at this time the field has really only just begun to uncover the mechanisms that adapt the epigenome for regeneration, to better understand the factors that limit this process in the mammalian CNS and sensory organs. Although there are relatively few studies in this area, the limited evidence to date supports some initial conclusions. Histone acetylation is a key regulator of development and regeneration in neural tissues. Acetylation is associated with open chromatin and more active transcription, and as a result, changes in chromatin acetylation should precede the changes in gene expression that are necessary for regeneration. Studies in neural systems that spontaneously regenerate after injury support the hypothesis that histone acetylation is necessary for regeneration; however, an important caveat is that inhibitors of HDACs may have effects on cell proliferation that are independent of their effects on chromatin accessibility and transcriptional activation. This may explain the seemingly contradictory results that HDAC inhibition can block regenerative proliferation of support cells in chick inner ear, while in other studies is critical for opening up new sites in the chromatin to facilitate glial reprogramming to neural progenitors and neurons in the mouse retina. In addition to the caveat that HDACs act on proteins other than histones, a second caveat is that not all HDACs are the same, and that they participate in many different complexes depending on the other proteins present in the cell. Broad methods to modify the epigenome, such as small molecule HDAC inhibitors, affect many different HDACs and many different complexes, and it is difficult to know which of these are critical in any given context. Thus, future studies focused on specific HDAC and HAT family members will undoubtedly provide a more nuanced view of the roles of acetylation. This conclusion is highlighted by the results on peripheral nerve regeneration and HDAC3 and HDAC5, where nuclear cytoplasmic shuttling provides an additional level of control. Despite these caveats, it is clear from even this limited number of examples that cellular reprogramming can be facilitated by manipulations in histone acetylation and thus will likely be important to consider in the development of regenerative therapies in sensory systems and CNS(Fig. 1).
Fig.1.
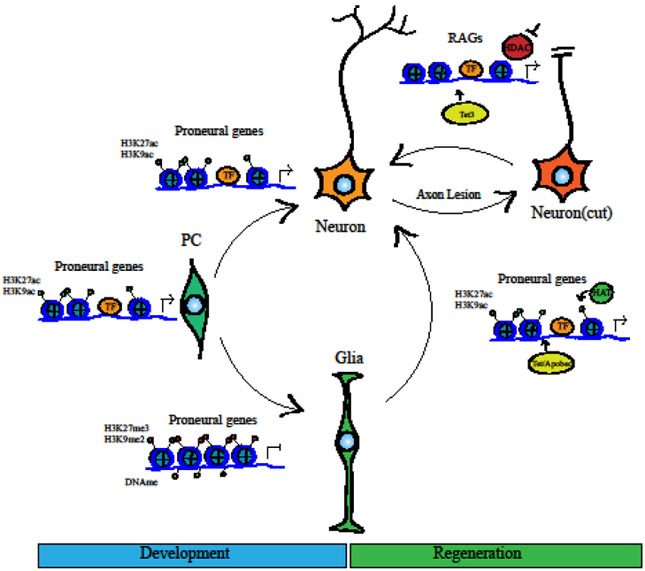
Development of neural tissue begins from a pluripotent progenitor cell that differentiates into neurons or support cells, which is in part determined by chromatin accessibility at proneural genes. These proneural genes are made accessible in neuronal progenitor cells via histone tail acetyl modifications. Such modifications leave chromatin open for gene regulators such as neurogenic transcription factors which promote proneural gene expression. Glial support cells are more frequently closed at proneural sites by histone modification and DNA methylation, which lead to inaccessibility at cis-regulatory binding motifs in the genome. This epigenetic regulation shuts down neural gene expression and promotes the glial fate. In contrast, neurons retain proneural accessibility with histone acetylation and accessibility to transcription factor binding sites, similar to that of progenitor cells. After tissue damage, in order for functional reprogramming to occur, glial support cells utilize pioneering transcription factors to open chromatin around proneural genes, which coincides with increasing acetylation and demethylation of DNA. These epigenomic landscape alterations promote proneural gene expression and fate changes to neuronal cell types. After axon lesion, regeneration-associated genes in the damaged neuron lose DNA methylation, and axon regrowth is aided by the activity of HATs and small molecule inhibition of HDACs which permit regenerative gene expression.
In addition to histone acetylation, methylation of histone tails also has clear roles in neuronal development. H3K27 trimethylation is historically the best-studied histone methyl mark and appears to broadly regulate transcription. Repressive histone modifications such as this correlate especially to the shutdown of early development and neurogenesis gene programs(Fig. 1). During inner ear development, the gene coding for the proneural transcription factor Atoh1 gains the repressive chromatin modification H3K27me3 as the genesis of new hair cells is complete, and this may relate to the lack of regeneration of new hair cells in mature mouse inner ear. In the retina, conditional deletion of Ezh2 (necessary for the repressive H3K27me3 modification) leads to a reduction in proliferation of the progenitors, likely via an increase in the cyclin dependent kinase p21cip. Re-entry of glia into the mitotic cycle after injury may require Ezh2 to repress this inhibitory factor, and supplementing Ezh2 to glia could facilitate regenerative proliferation. These types of modifications also can contribute strongly to epigenetic memory and therefore can affect fate changes in regeneration in many tissue types, thus targeted rewriting of repressive histone modifications could be beneficial in future attempts to reprogram cells in efforts to regenerate neuronal tissue.
DNA methylation acts as a much more stable and consistent repressor of gene expression as compared to histone modifications and thus may be more of a barrier to regeneration. During development, prosensory and proneural gene programs, while partially regulated by histone modifications, also appear to be repressed via DNA methylation. Importantly, DNA methylation appears to change in response to damage. Changes in DNA methylation accompany the spontaneous regeneration that occurs after injury to the fish retina, and functional studies support a role for demethylation in retinal regeneration in this species(Fig. 1). However, it is not clear which genes needed for regeneration are repressed by this method, since the pluripotency and regeneration-associated gene promoters are not among those demethylated after injury. DNA methylation changes also accompany the successful regeneration in peripheral nerves, suggesting that axon regeneration is controlled by this mechanism. Thus, in non-regenerative systems, the lack of appropriate changes in DNA methylation may prevent expression of regeneration-associated genes. To date there have been relatively few studies that test this hypothesis, though the availability of small molecule DNMT inhibitors should facilitate in vivo testing.
In addition to the general regulators of the epigenome discussed above, the role of transcription factors in regulating gene expression changes cannot be overstated. While many studies have demonstrated that repressive chromatin can prevent gene expression, transcription factors can engage the genome and under the proper circumstances reactive genes in regions of heterochromatin. During fate changes and regeneration, the most dynamic changes to the epigenome appear to be in enhancer regions where transcription factors are binding. Thus transcription factor activity regulates fate changes beyond simple expression via changes to the epigenome(Fig. 1). Some transcription factors that are particularly effective at stimulating reprogramming, such as Sox2 and Ascl1 are particularly effective in inducing neuronal fate changes because they induce major chromatin remodeling. These so-called pioneering transcription factors likely interact with chromatin remodelers to regulate the accessibility of their gene targets, and as a result the epigenetic state changes can be driven by transcription factors. While individually, many components discussed here can change cellular fate and be beneficial in attempts for regeneration, a combinatorial approach is most likely to be effective in creating the most effective regeneration program for any given neuronal tissue. By using neurogenic pioneering transcription factors, somatic cells may take advantage of native developmental pathways that act in targeted manners to change transcriptional programs and effect chromatin remodeling changes; however, the manipulation of chromatin remodelers to increase accessibility at regeneration-associated genes can alter and improve regeneration both in vitro and in vivo. Thus, a combination of neurogenic transcription factor expression with targeted epigenetic remodeling may produce the best results in generating new neurons in diseased and damaged tissue.
Table 1.
Summary of primary literature cited on the subject of epigenetics in neuron development and regeneration.
| Authors | Year | Title | System | Species | Focus | Epigenetics |
|---|---|---|---|---|---|---|
| N Fujimura et al | 2018 | Polycomb repression complex 2 is required for the maintenance of retinal progenitor cells and balanced retinal differentiation | Retina | Mouse | Development | H3K27me3 |
| R Raeisossadati | 2018 | Small Molecule GSK-J1 Affects Differentiation of Specific Neuronal Subtypes in Developing Rat Retina | Rat | H3K27me3 | ||
| L Cheng et al | 2018 | Ezh2 does not mediate retinal ganglion cell homeostasis or their susceptibility to injury | Mouse | H3K27me3 | ||
| G Villain et al | 2018 | miR-126-5p promotes retinal endothelial cell survival through SetD5 regulation in neurons. | Mouse | Histone Acetylation | ||
| A Hoshino et al. | 2017 | Molecular Anatomy of the Developing Human Retina. | Human | Chromatin Accessibility | ||
| I. Aldiri et al. | 2017 | The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. | Mouse, Human | Chromatin Accessibility | ||
| JM Daum et al. | 2017 | The formation of the light-sensing compartment of cone photoreceptors coincides with a transcriptional switch | Mouse | Chromatin Accessibility, Looping | ||
| TL Davis, I Rebay | 2017 | Antagonistic regulation of the second mitotic wave by Eyes absent-Sine oculis and Combgap coordinates proliferation and specification in the Drosophila retina. | Drosophila | Chromatin Occupancy, Polycomb | ||
| AEO Hughes et al | 2017 | Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors | Mouse | Chromatin Structure | ||
| RK Singh et al | 2017 | Dnmt1, Dnmt3a and Dnmt3b cooperate in photoreceptor and outer plexiform layer development in the mammalian retina | Mouse | DNA methylation | ||
| K Ueno et al | 2017 | Analysis of Müller glia specific genes and their histone modification using Hes1-promoter driven EGFP expressing mouse | Mouse | H3K27me3, H3K4me2 | ||
| RC Ferrira et al | 2017 | Histone Deacetylase 1 Is Essential for Rod Photoreceptor Differentiation by Regulating Acetylation at Histone H3 Lysine 9 and Histone H4 Lysine 12 in the Mouse Retina. | Mouse | Histone Acetylation | ||
| N Yan et al | 2016 | Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1 | Mouse | H3K27me3 | ||
| M.S. Wilken et al. | 2015 | DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements | Mouse | Chromatin Accessibility | ||
| J Zhang et al | 2015 | Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation. | Mouse | H3K27me3 | ||
| EY Popova et al | 2012 | Stage and Gene Specific Signatures Defined by Histones H3K4me2 and H3K27me3 Accompany Mammalian Retina Maturation In Vivo | Mouse | H3K27me3, H3K4me2 | ||
| BE Bernstein et al | 2006 | A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells | Mouse | H3K27me3, H3K4me2 | ||
| J de Melo et al | 2003 | Dlx1,Dlx2,Pax6,Brn3b, andChx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina | Mouse | Histone Acetylation | ||
| S Mitra et al | 2018 | Histone Deacetylase-Mediated Müller Glia Reprogramming through Her4.1-Lin28a Axis Is Essential for Retina Regeneration in Zebrafish. | Zebrafish | Regeneration | Histone Acetylation | |
| NL Jorstad et al | 2017 | Stimulation of functional neuronal regeneration from Müller glia in adult mice | Mouse | Chromatin Accessibility | ||
| LI Reyes-Aguirre | 2016 | Oct4 Methylation-Mediated Silencing As an Epigenetic Barrier Preventing Müller Glia Dedifferentiation in a Murine Model of Retinal Injury | Mouse | DNA methylation | ||
| Y Ueki et al | 2015 | Transgenic expression of the proneural transcription factor Ascl1 in Müller glia stimulates retinal regeneration in young mice | Mouse | Chromatin Accessibility | ||
| C Powell, E Cornblath, D Goldman | 2014 | Zinc-binding domain-dependent, deaminase-independent actions of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 2 (Apobec2), mediate its effect on zebrafish retina regeneration | Zebrafish | DNA methylation | ||
| Y Koriyama et al | 2014 | Neuritogenic Activity of Trichostatin A in Adult Rat Retinal Ganglion Cells Through Acetylation of Histone H3 Lysine 9 and RARβ Induction | Rat | Histone Acetylation | ||
| C Powell et al | 2013 | Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration. | Zebrafish | DNA methylation | ||
| J Pollak et al | 2013 | ASCL1 reprograms mouse Müller glia into neurogenic retinal progenitors | Mouse | H3K27me3/ac | ||
| C Powell, F Elsaeidi, D Goldman | 2012 | Injury-dependent Müller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b | Zebrafis h | DNA methylation | ||
| P Gaub et al | 2011 | The histone acetyltransferase p300 promotes intrinsic axonal regeneration | Rat | Histone Acetylation | ||
| Y He et al | 2016 | LSD1 is Required for Hair Cell Regeneration in Zebrafish | Inner Ear | Zebrafish | Development | H3K4/9me |
| JO Shin et al | 2018 | CTCF Regulates Otic Neurogenesis via Histone Modification in the Neurog1 Locus | Mouse | Chromatin Looping | ||
| H Yao et al | 2018 | CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development | Mouse | Chromatin Structure | ||
| D Roellig and ME Bronner | 2016 | The epigenetic modifier DNMT3A is necessary for proper otic placode formation | Chick | DNA methylation | ||
| Y He et al | 2016 | Histone deacetylase 1 is required for the development of the zebrafish inner ear | Zebrafish | Histone Acetylation | ||
| M Ahmed, K Ura, and A Streit | 2015 | Auditory hair cell defects as potential cause for sensorineural deafness in Wolf-Hirschhorn syndrome | Mouse | H3K36me3 | ||
| ZP Stojanova, T Kwan, and N Segil | 2015 | Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea | Mouse | H3K9ac | ||
| RA Uribe | 2015 | Histone demethylase KDM4B regulates otic vesicle invagination via epigenetic control of Dlx3 expression | Chick | H3K9me3, H3K936me3 | ||
| Y He et al | 2014 | Effect of histone deacetylase inhibitors trichostatin A and valproic acid on hair cell regeneration in zebrafish lateral line neuromasts | Zebrafish | Histone Acetylation | ||
| E Engelen et al | 2011 | Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes | Mouse | Chromatin Structure | ||
| EA Hurd et al | 2010 | The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear | Mouse | Chromatin Structure | ||
| K Nimura et al | 2009 | A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome | Mouse, Human | H3K36me3 | ||
| D Tang et al | 2016 | Inhibition of H3K9me2 Reduces Hair Cell Regeneration after Hair Cell Loss in the Zebrafish Lateral Line by Down-Regulating the Wnt and Fgf Signaling Pathways | Zebrafish | Regeneration | H3K9me2 | |
| A Li et al | 2015 | Lysine-specific demethylase 1 inhibitors protect cochlear spiral ganglion neurons against cisplatin-induced damage | Mouse | H3K4me2 | ||
| EL Slattery et al | 2009 | Epigenetic Influences on Sensory Regeneration: Histone Deacetylases Regulate Supporting Cell Proliferation in the Avian Utricle | Chick | Histone Acetylation | ||
| A Visel et al | 2013 | A high-resolution enhancer atlas of the developing telencephalon | Spinal Cord | Mouse, Human | Development | Transcription Regulation |
| Y Su et al | 2017 | Neuronal activity modifies the chromatin accessibility landscape in the adult brain | Mouse | Chromatin accessibility, looping | ||
| I Venkatesh et al | 2018 | Developmental chromatin restriction of pro-growth gene networks acts as an epigenetic barrier to axon regeneration in cortical neurons | Mouse | Regeneration | Chromatin Accessibility | |
| S Chen et al | 2018 | Valproic acid attenuates traumatic spinal cord injury-induced inflammation via STAT1 and NF-κB pathway dependent of HDAC3 | Rat | Histone Acetylation | ||
| YHE Loh et al | 2017 | Comprehensive mapping of 5-hydroxymethylcytosin e epigenetic dynamics in axon regeneration | Mouse | DNA methylation | ||
| R Puttagunta et al | 2014 | PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system | Mouse | H3K9ac | ||
| Y Cho et al | 2013 | Injury-Induced HDAC5 Nuclear Export Is Essential for Axon Regeneration | Mouse | Histone Acetylation | ||
| MJ Finelli et al | 2013 | Epigenetic Regulation of Sensory Axon Regeneration after Spinal Cord Injury | Mouse | Histone Acetylation | ||
| P Gaub et al | 2010 | HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation | Rat | Histone Acetylation | ||
| MA Rivieccio et al | 2009 | HDAC6 is a target for protection and regeneration following injury in the nervous system | CHO | Histone Acetylation | ||
| OL Wapinski et al | 2017 | Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons | Culture | MEF | Regeneration | Chromatin Accessibility |
| G Masserdotti et al | 2015 | Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes | Mouse | Histone Methylation |
Acknowledgements
The authors acknowledge Grant #TA-RM-0614-0650-UWA from the Foundation Fighting Blindness to T.A.R., NIH NEI 1R01EY021482 to T.A.R., Allen Distinguished Investigator Award (Paul G. Allen Family Foundation) to T.A.R. We thank members of the Reh and Bermingham-McDonogh laboratories for their review and valuable discussion regarding the manuscript. We are grateful to Dr. Levi Todd and Wes Jenkins for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Alunni A, Bally-Cuif L, A comparative view of regenerative neurogenesis in vertebrates., Development. 143 (2016) 741–53. doi: 10.1242/dev.122796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gemberling M et al. , The zebrafish as a model for complex tissue regeneration, Trends Genet. 29 (2013) 611–620. doi: 10.1016/J.TIG.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Noctor SC et al. , Neurons derived from radial glial cells establish radial units in neocortex, Nature. 409 (2001) 714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- [4].Nelson BR et al. , Genome-wide analysis of Müller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate., PLoS One. 6 (2011) e22817. doi: 10.1371/journal.pone.0022817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ronan JL, Wu W, Crabtree GR, From neural development to cognition: unexpected roles for chromatin, Nat Rev Genet. 14 (2013) 347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jenuwein T, Allis CD, Translating the histone code., Science. 293 (2001) 1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- [7].Kouzarides T, Chromatin Modifications and Their Function, Cell. 128 (2007) 693–705. doi: 10.1016/J.CELL.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [8].Corso-Dïaz X et al. , Epigenetic control of gene regulation during development and disease: A view from the retina, Prog. Retin. Eye Res 65 (2018) 1–27. doi: 10.1016/J.PRETEYERES.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brzezinski JA, Reh TA, Photoreceptor cell fate specification in vertebrates., Development. 142 (2015) 3263–73. doi: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wilken MS et al. , DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements, Epigenetics Chromatin. 8 (2015) 8. doi: 10.1186/1756-8935-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hoshino A et al. , Molecular Anatomy of the Developing Human Retina., Dev. Cell 43 (2017) 763–779.e4. doi: 10.1016/j.devcel.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aldiri I et al. , The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis., Neuron. 94 (2017) 550–568.e10. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Daum JM et al. , The formation of the light-sensing compartment of cone photoreceptors coincides with a transcriptional switch, Elife. 6 (2017). doi: 10.7554/eLife.31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hughes AEO et al. , Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors, Sci. Rep 7 (2017) 43184. doi: 10.1038/srep43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mo A et al. , Epigenomic landscapes of retinal rods and cones, Elife. 5 (2016). doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Singh RK et al. , Dnmt1, Dnmt3a and Dnmt3b cooperate in photoreceptor and outer plexiform layer development in the mammalian retina, Exp. Eye Res 159 (2017) 132–146. doi: 10.1016/J.EXER.2016.11.014. [DOI] [PubMed] [Google Scholar]
- [17].Fujimura N et al. , Polycomb repression complex 2 is required for the maintenance of retinal progenitor cells and balanced retinal differentiation, Dev. Biol 433 (2018) 47–60. doi: 10.1016/J.YDBIO.2017.11.004. [DOI] [PubMed] [Google Scholar]
- [18].Ueno K et al. , Analysis of Müller glia specific genes and their histone modification using Hes1-promoter driven EGFP expressing mouse, Sci. Rep 7 (2017) 3578. doi: 10.1038/s41598-017-03874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yan N et al. , Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1, Sci. Rep 6 (2016) 33887. doi: 10.1038/srep33887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Popova EY et al. , Stage and Gene Specific Signatures Defined by Histones H3K4me2 and H3K27me3 Accompany Mammalian Retina Maturation In Vivo, PLoS One. 7 (2012) e46867. doi: 10.1371/journal.pone.0046867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang J et al. , Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation., Dev. Biol 403 (2015) 128–38. doi: 10.1016/j.ydbio.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Raeisossadati R et al. , Small Molecule GSK-J1 Affects Differentiation of Specific Neuronal Subtypes in Developing Rat Retina, Mol. Neurobiol (2018) 1–12. doi: 10.1007/s12035-018-1197-3. [DOI] [PubMed] [Google Scholar]
- [23].Bernstein BE et al. , A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells, Cell. 125 (2006) 315–326. doi: 10.1016/J.CELL.2006.02.041. [DOI] [PubMed] [Google Scholar]
- [24].Cheng L et al. , Ezh2 does not mediate retinal ganglion cell homeostasis or their susceptibility to injury, PLoS One. 13 (2018) e0191853. doi: 10.1371/journal.pone.0191853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Davis TL, Rebay I, Antagonistic regulation of the second mitotic wave by Eyes absent-Sine oculis and Combgap coordinates proliferation and specification in the Drosophila retina., Development. 144 (2017) 2640–2651. doi: 10.1242/dev.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ferreira RC et al. , Histone Deacetylase 1 Is Essential for Rod Photoreceptor Differentiation by Regulating Acetylation at Histone H3 Lysine 9 and Histone H4 Lysine 12 in the Mouse Retina., J. Biol. Chem 292 (2017) 2422–2440. doi: 10.1074/jbc.M116.756643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].de Melo J et al. , Dlx1,Dlx2,Pax6,Brn3b, andChx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina, J. Comp. Neurol 461 (2003) 187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- [28].Villain G et al. , miR-126-5p promotes retinal endothelial cell survival through SetD5 regulation in neurons., Development. 145 (2018) dev156232. doi: 10.1242/dev.156232. [DOI] [PubMed] [Google Scholar]
- [29].Lamba D, Karl M, Reh T, Neural regeneration and cell replacement: a view from the eye., Cell Stem Cell. 2 (2008) 538–49. doi: 10.1016/j.stem.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fausett BV, Goldman D, A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina., J. Neurosci 26 (2006) 6303–13. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Blackshaw S et al. , Genomic analysis of mouse retinal development., PLoS Biol. 2 (2004) E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goldman D, Müller glial cell reprogramming and retina regeneration., Nat. Rev. Neurosci 15 (2014) 431–42. doi: 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Powell C, Elsaeidi F, Goldman D, Injury-dependent Muller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b, J Neurosci. 32 (2012) 1096–1109. doi: 10.1523/JNEUROSCI.5603-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Powell C et al. , Analysis of DNA methylation reveals a partial reprogramming of the Müller glia genome during retina regeneration., Proc. Natl. Acad. Sci. U. S. A 110 (2013) 19814–9. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Powell C, Cornblath E, Goldman D, Zinc-binding domain-dependent, deaminase-independent actions of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 2 (Apobec2), mediate its effect on zebrafish retina regeneration, J Biol Chem. 289 (2014) 28924–28941. doi: 10.1074/jbc.M114.603043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mitra S et al. , Histone Deacetylase-Mediated Müller Glia Reprogramming through Her4.1-Lin28a Axis Is Essential for Retina Regeneration in Zebrafish., IScience. 7 (2018) 68–84. doi: 10.1016/j.isci.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reyes-Aguirre LI, Lamas M, Oct4 Methylation-Mediated Silencing As an Epigenetic Barrier Preventing Müller Glia Dedifferentiation in a Murine Model of Retinal Injury, Front. Neurosci 10 (2016) 523. doi: 10.3389/fnins.2016.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bringmann A et al. , Cellular signaling and factors involved in Muller cell gliosis: neuroprotective and detrimental effects, Prog Retin Eye Res. 28 (2009) 423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- [39].Poliak J et al. , ASCL1 reprograms mouse Müller glia into neurogenic retinal progenitors, Development. 140 (2013) 2619–2631. doi: 10.1242/DEV.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ueki Y et al. , Transgenic expression of the proneural transcription factor Ascl1 in Müller glia stimulates retinal regeneration in young mice, Proc. Natl. Acad. Sci 112 (2015) 13717–13722. doi: 10.1073/PNAS.1510595112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jorstad NL et al. , Stimulation of functional neuronal regeneration from Müller glia in adult mice, Nature. 548 (2017) 103–107. doi: 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu K et al. , Neuronal intrinsic mechanisms of axon regeneration, Annu Rev Neurosci. 34 (2011) 131–152. doi: 10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- [43].Lim J-HA et al. , Neural activity promotes long-distance, target-specific regeneration of adult retinal axons., Nat. Neurosci 19 (2016) 1073–84. doi: 10.1038/nn.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gaub P et al. , The histone acetyltransferase p300 promotes intrinsic axonal regeneration, Brain. 134 (2011) 2134–2148. doi: 10.1093/brain/awr142. [DOI] [PubMed] [Google Scholar]
- [45].Koriyama Y et al. , Neuritogenic Activity of Trichostatin A in Adult Rat Retinal Ganglion Cells Through Acetylation of Histone H3 Lysine 9 and RARβ Induction, J. Pharmacol. Sci 124 (2014) 112–116. doi: 10.1254/jphs.13171SC. [DOI] [PubMed] [Google Scholar]
- [46].Bermingham-McDonogh O, Reh TA, Regulated reprogramming in the regeneration of sensory receptor cells., Neuron. 71 (2011) 389–405. doi: 10.1016/j.neuron.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Thomas ED et al. , There and back again: development and regeneration of the zebrafish lateral line system, Wiley Interdiscip Rev Dev Biol. 4 (2015) 1–16. doi: 10.1002/wdev.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu DK, Kelley MW, Molecular mechanisms of inner ear development., Cold Spring Harb. Perspect. Biol 4 (2012) a008409. doi: 10.1101/cshperspect.a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].He Y et al. , LSD1 is Required for Hair Cell Regeneration in Zebrafish, Mol. Neurobiol 53 (2016) 2421–2434. doi: 10.1007/s12035-015-9206-2. [DOI] [PubMed] [Google Scholar]
- [50].He Y et al. , Effect of histone deacetylase inhibitors trichostatin A and valproic acid on hair cell regeneration in zebrafish lateral line neuromasts, Front. Cell. Neurosci 8 (2014) 382. doi: 10.3389/fncel.2014.00382. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [51].He Y et al. , Histone deacetylase 1 is required for the development of the zebrafish inner ear, Sci. Rep 6 (2016) 16535. doi: 10.1038/srep16535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Stojanova ZP, Kwan T, Segil N, Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea, Development. 142 (2015) 3529–3536. doi: 10.1242/DEV.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Uribe RA et al. , Histone demethylase KDM4B regulates otic vesicle invagination via epigenetic control of Dlx3 expression, J Cell Biol. 211 (2015) 815–827. doi: 10.1083/JCB.201503071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Roellig D, Bronner ME, The epigenetic modifier DNMT3A is necessary for proper otic placode formation, Dev. Biol 411 (2016) 294–300. doi: 10.1016/J.YDBIO.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Doetzlhofer A, Avraham KB, Insights into inner ear-specific gene regulation: Epigenetics and non-coding RNAs in inner ear development and regeneration, Semin. Cell Dev. Biol 65 (2017) 69–79. doi: 10.1016/J.SEMCDB.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nimura K et al. , A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome, Nature. 460 (2009) 287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- [57].Ahmed M, Ura K, Streit A, Auditory hair cell defects as potential cause for sensorineural deafness in Wolf-Hirschhorn syndrome, Dis. Model. Mech 8 (2015) 1027–1035. doi: 10.1242/DMM.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hurd DA et al. , The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear, Development. 137 (2010) 3139–3150. doi: 10.1242/DEV.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yao H et al. , CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development, JCI Insight. 3 (2018). doi: 10.1172/JCI.INSIGHT.97440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Engelen E et al. , Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes, Nat. Genet 43 (2011) 607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- [61].Shin J-O et al. , CTCF Regulates Otic Neurogenesis via Histone Modification in the Neurog1 Locus, Moleucles and Cells. 41 (n.d.) 695–702. doi: 10.14348/MOLCELLS.2018.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Neuhauser HK et al. , Burden of Dizziness and Vertigo in the Community, Arch. Intern. Med 168 (2008) 2118. doi: 10.1001/archinte.168.19.2118. [DOI] [PubMed] [Google Scholar]
- [63].Zheng F, Cochlear hair cell regeneration after noise-induced hearing loss: Does regeneration follow development?, Hear. Res 349 (2017) 182–196. doi: 10.1016/J.HEARES.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tang D et al. , Inhibition of H3K9me2 Reduces Hair Cell Regeneration after Hair Cell Loss in the Zebrafish Lateral Line by Down-Regulating the Wnt and Fgf Signaling Pathways, Front Mol Neurosci. 9 (2016) 39. doi: 10.3389/fnmol.2016.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rubel EW, Stone JS, Stimulating hair cell regeneration: on a wing and a prayer, Nat Med. 2 (1996) 1082–1083. https://www.ncbi.nlm.nih.gov/pubmed/8837604. [DOI] [PubMed] [Google Scholar]
- [66].Slattery EL et al. , Epigenetic Influences on Sensory Regeneration: Histone Deacetylases Regulate Supporting Cell Proliferation in the Avian Utricle, J. Assoc. Res. Otolaryngol 10 (2009) 341–353. doi: 10.1007/s10162-009-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li A et al. , Lysine-specific demethylase 1 inhibitors protect cochlear spiral ganglion neurons against cisplatin-induced damage, Neuroreport. 26 (2015) 539–547. doi: 10.1097/WNR.0000000000000386. [DOI] [PubMed] [Google Scholar]
- [68].Visel A et al. , A high-resolution enhancer atlas of the developing telencephalon, Cell. 152 (2013) 895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Su Y et al. , Neuronal activity modifies the chromatin accessibility landscape in the adult brain, Nat. Neurosci 20 (2017) 476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang Z et al. , Overexpression of Sox11 Promotes Corticospinal Tract Regeneration after Spinal Injury While Interfering with Functional Recovery, J. Neurosci 35 (2015) 3139–3145. doi: 10.1523/JNEUROSCI.2832-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Venkatesh I et al. , Developmental chromatin restriction of pro-growth gene networks acts as an epigenetic barrier to axon regeneration in cortical neurons, Dev. Neurobiol (2018). doi: 10.1002/dneu.22605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Li S et al. , The transcriptional landscape of dorsal root ganglia after sciatic nerve transection., Sci. Rep 5 (2015) 16888. doi: 10.1038/srep16888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cho Y et al. , Injury-Induced HDAC5 Nuclear Export Is Essential for Axon Regeneration, Cell. 155 (2013) 894–908. doi: 10.1016/J.CELL.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Finelli MJ et al. , Epigenetic Regulation of Sensory Axon Regeneration after Spinal Cord Injury, J. Neurosci 33 (2013) 19664–19676. doi: 10.1523/JNEUROSCI.0589-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Puttagunta R et al. , PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system, Nat. Commun 5 (2014) 3527. doi: 10.1038/ncomms4527. [DOI] [PubMed] [Google Scholar]
- [76].Gaub P et al. , HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation, Cell Death Differ. 17(2010) 1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- [77].Loh Y-HE et al. Comprehensive mapping of 5-hydroxymethylcytosine epigenetic dynamics in axon regeneration, Epigenetics. 12 (2017) 77–92. doi: 10.1080/15592294.2016.1264560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chen S et al. , Valproic acid attenuates traumatic spinal cord injury-induced inflammation via STAT1 and NF-κB pathway dependent of HDAC3, J. Neuroinflammation 15 (2018) 150. doi: 10.1186/s12974-018-1193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Rivieccio MA et al. , HDAC6 is a target for protection and regeneration following injury in the nervous system, Proc. Natl. Acad. Sci 106 (2009) 19599–19604. doi: 10.1073/PNAS.0907935106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Vierbuchen T et al. , Direct conversion of fibroblasts to functional neurons by defined factors, Nature. 463 (2010) 1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wapinski OL et al. , Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons, Cell Rep. 20 (2017) 3236–3247. doi: 10.1016/J.CELREP.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gascón S et al. , Direct Neuronal Reprogramming: Achievements, Hurdles, and New Roads to Success., Cell Stem Cell. 21 (2017) 18–34. doi: 10.1016/j.stem.2017.06.011. [DOI] [PubMed] [Google Scholar]
- [83].Ohori Y et al. , Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord, J Neurosci. 26 (2006) 11948–11960. doi: 10.1523/JNEUROSCI.3127-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Masserdotti G et al. , Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes, Cell Stem Cell. (2015). doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


