Abstract
Motility structures are vital in all three domains of life. In Archaea, motility is mediated by the archaellum, a rotating type IV pilus-like structure that is a unique nanomachine for swimming motility in nature. Whereas periplasmic FlaF binds the surface layer (S-layer), the structure, assembly and roles of other periplasmic components remain enigmatic, limiting our knowledge of the archaellum’s functional interactions. Here, we find that the periplasmic protein FlaG and the association with its paralogue FlaF are essential for archaellation and motility. Therefore, we determine the crystal structure of Sulfolobus acidocaldarius soluble FlaG (sFlaG), which reveals a β-sandwich fold resembling the S-layer-interacting FlaF soluble domain (sFlaF). Furthermore, we solve the sFlaG2–sFlaF2 co-crystal structure, define its heterotetrameric complex in solution by small-angle X-ray scattering and find that mutations that disrupt the complex abolish motility. Interestingly, the sFlaF and sFlaG of Pyrococcus furiosus form a globular complex, whereas sFlaG alone forms a filament, indicating that FlaF can regulate FlaG filament assembly. Strikingly, Sulfolobus cells that lack the S-layer component bound by FlaF assemble archaella but cannot swim. These collective results support a model where a FlaG filament capped by a FlaG–FlaF complex anchors the archaellum to the S-layer to allow motility.
Motility is fundamental to microbes, enabling movement to more favourable environments1. Archaea use archaella (formerly known as archaeal flagella) for motility in liquids and on surfaces2,3. Despite resembling type IV pili (T4P), responsible for grappling hook-like surface motility by extension and retraction, archaella generate thrust via a rotating filament, which is analogous, yet unrelated, to bacterial flagella4–6. Accordingly, the archaellum is termed a rotating T4P, making it a unique nanomachine for motility in nature4.
Archaellum-encoding fla operons contain 7–15 essential genes3 for archaellation and motility, as shown by studies in Sulfolobales, Halobacteriales and Methanococcales7–10. The core archaellum consists of the conserved genes flaG, flaF, flaH, flaI and flaJ3, although fla operons from the major archaeal phyla Euryarchaeota and Crenarchaeota differ in the number of archaellins and the presence of either flaX in Crenarchaea or flaCDE in Euryarchaea. The core archaellar motor complex is formed of FlaI, FlaH and FlaX11, assembling on the foundation of the inner membrane protein FlaJ (a homologue of PilC from T4P12–15. FlaI is a bifunctional ATPase essential for archaellum assembly16 and rotation17,18, in contrast to T4P where two ATPases are required for the two functions of extension and retraction in twitching motility19–21. FlaH forms a cyclic oligomer that binds ATP for its interaction with FlaI, but has never been shown to hydrolyse ATP. FlaX forms a large ring16, which may act as scaffold for the assembly of FlaH and FlaI11,22. Together, the FlaH–FlaI–FlaJ–FlaX complex generates torque, but it is unclear how this freely rotating structure anchors to the cell wall to productively rotate the archaellar filament for propulsion.
FlaF is a recently identified periplasmic archaellum component23. Its soluble domain (sFlaF) binds the major cell wall component, the S-layer (surface layer), in Sulfolobus acidocaldarius; disruption of FlaF dimerization abolishes motility23. Furthermore, the conserved archaellar monotopic membrane proteins FlaF and FlaG have been bioinformatically shown to be archaellin homologues, but lack the class III signal peptides of true archaellins24,25. However, the functions of FlaF and FlaG remain enigmatic, and the structure of FlaG is not known.
In this study, we studied S. acidocaldarius FlaG by expressing the archaellin domain of FlaG, a soluble protein (sFlaG) that is stable and flexible at pH 3, mimicking the acidic S. acidocaldarius periplasmic environment. We solved the crystal structures of sFlaG and the sFlaG–sFlaF complex, validated complex assembly in solution by small-angle X-ray scattering (SAXS), and measured complex interactions in vitro. The structures and SAXS data showed that sFlaG becomes more rigid when binding to sFlaF. Structure-based mutations that disrupt sFlaG–sFlaF tetrameric complex formation abolished motility and archaellum assembly. Surprisingly, we found that both FlaG and FlaF are released from the membrane into the periplasm in S. acidocaldarius. The mature FlaG has its N-terminal membrane domain cleaved, while mature FlaF remains in a full-length form in the periplasm. Intriguingly, the FlaG soluble domain from the euryarchaeon Pyrococcus furiosus (Pfu sFlaG) formed filaments; addition of Pfu sFlaF to Pfu sFlaG during reconstitution abolished sFlaG filament formation, yielding globular structures instead, suggesting that FlaF regulates FlaG’s filament formation. Furthermore, we found that the cells of Sulfolobus islandicus lacking the large S-layer subunit SlaA assemble archaella but cannot swim, indicating that anchoring archaella to the S-layer is required for motility. These collective structural, biochemical and functional results suggest that the FlaG–FlaF complex is critical for archaellum assembly and motility by anchoring the archaellum to the S-layer.
Results
FlaG has a β-sandwich fold that resembles FlaF and archaellins.
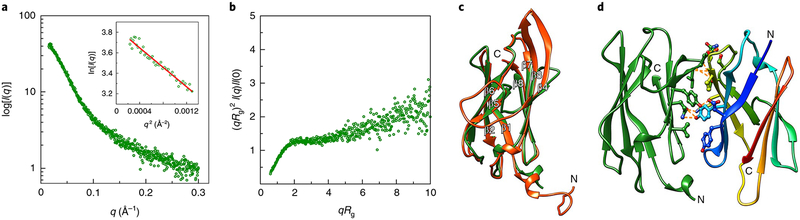
The structure and function of FlaG and how it contributes to archaellum assembly and function are unknown. To better understand FlaG, we stored sFlaG at pH 3 in citric acid buffer to mimic the optimal pH environment of S. acidocaldarius26. SAXS analysis showed that sFlaG was stable (that is, not aggregating), but highly flexible (partially folded) as shown in a dimensionless Kratky plot27–29 (Fig. 1a,b and Supplementary Table 1). sFlaG crystallized despite its flexibility and we determined its structure to 1.93 Å resolution by multi-wavelength anomalous dispersion (MAD) phasing (Protein Data Bank (PDB): 5TUH; Supplementary Table 2). The structure of sFlaG revealed a C2-symmetric dimer and a β-sandwich fold with a flexible N-terminal α-helix that resembles sFlaF23 (Fig. 1c). The N-terminal α-helices of the sFlaG dimer point in opposite directions, suggesting that alternative FlaG assembly with the membrane anchor is possible since both N termini should anchor to the membrane (Fig. 1d). Proteins, Interfaces, Structures and Assemblies (PISA) analysis30 showed that the S. acidocaldarius sFlaG dimer is unstable (ΔGassembly = −1.4 kcal mol−1 and ΔGdissociation = −4.0 kcal mol−1), which is consistent with our SAXS result showing sFlaG to be monomeric (Supplementary Table 1) and suggesting that subunit interactions via crystal packing thermodynamically stabilize sFlaG folding. These results suggest that the monomeric sFlaG is stabilized by self-assembly and folds into a β-sandwich structure mimicking sFlaF and archaellins.
Fig. 1 |. The sFlaG crystal structure exhibits a β-sandwich fold that is stable in pH 3 buffer solution as shown by SAXS.
a, SAXS scattering profile of sFlaG in pH 3 buffer is monodispersed. Inset: Guinier plot showing monodispersal. b, The dimensionless Kratky plot shows that sFlaG is partially folded in solution. A dimensionless Kratky plot of a well-folded globular protein should exhibit a bell-shaped peak that converges to the q axis at high q. c, Overlay of the crystal structure of sFlaG (green) with the sFlaF structure (orange-red; PDB: 4P94) shows resembling β-sandwich folds. The main difference between the two structures are the loops between β3 and β4 and between β4 and β5. The β-strands are labelled according to the sFlaF structure. d, The dimer interface of the sFlaG structure shows hydrogen bonds (orange dashed lines) and hydrophobic interactions (shown as sticks). However, sFlaG was classified as a weak dimer by the PISA program30. sFlaG topology is coloured in rainbow from blue (N terminus) to red (C terminus) in one of the subunits.
sFlaG and sFlaF form a heterotetrameric complex.
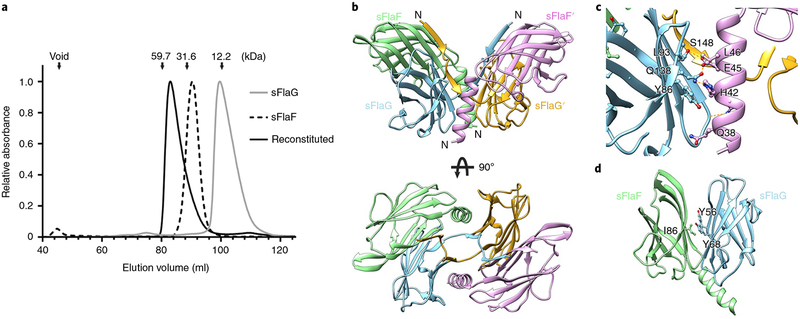
Given the need for FlaG self-interactions to stabilize its fold, we reasoned that FlaF and FlaG could form a complex. We first confirmed that monomeric and dimeric sFlaF are stable at pH 3 showing a globular peak in size-exclusion chromatography (SEC) (Supplementary Fig. 1). sFlaF and sFlaG did not form hetero-oligomers by simple mixing in pH 3 solution. Therefore, we developed a protocol to test and reconstitute a heterocomplex of sFlaG–sFlaF (see Methods). The peak of the reconstituted sample shifted towards higher molecular weight compared to the individual protein runs in SEC, indicating the formation of an sFlaG–sFlaF complex (Fig. 2a).
Fig. 2 |. sFlaG interacts with sFlaF to form a heterotetrameric complex.
a, Normalized absorbance at λ = 280 nm from SEC was plotted against elution volume. Individual sFlaG (grey line) and sFlaF (black dashed line) were eluted as monomer and dimer, respectively. The elution volume of reconstituted protein (black solid line) shifted towards a higher molecular weight, indicating the presence of a tetrameric complex. The protein molecular weight was calculated based on molecular weight standards (shown by the arrows). b, Overview of the heterotetrameric sFlaG–sFlaF complex crystal structure. Two sFlaG and two sFlaF β-sandwich fold subunits form a heterotetramer, which is stabilized by a long N-terminal α-helix of sFlaF and two short N-terminal β-strands of sFlaG. c, The interactions between the α-helix of sFlaF (bright purple) and curved β-sheet of sFlaG (light blue) consist of the interactions of hydrogen bonds (orange dashed lines) and the cation–π interactions of H42 and Y86. d, The sFlaG–sFlaF dimer was stabilized by hydrophobic residues. Residue I86 of sFlaF (shown with sticks) is located at the centre of the dimer interface.
To gain an insight into this complex, we determined the crystal structure of the sFlaG–sFlaF complex at 2.47 Å resolution (Supplementary Table 2). The solved structure exhibited four molecules per asymmetric unit that retained the β-sandwich fold. The most stable complex identified from crystal packing contained two sFlaG and two sFlaF subunits (Fig. 2b) based on PISA analysis showing the sFlaG–sFlaF heterotetrameric complex as a stable complex with a surface area approximately 20,820 Å2 and buried area approximately 9,790 Å2. Interestingly, flexible and partially unfolded sFlaG in solution was stabilized by sFlaF forming a complex, and the N termini of both sFlaG and sFlaF were stabilized between subunits, further confirming the tetrameric formation in solution by SAXS (Fig. 2b, Supplementary Figs. 2 and 3 and Table 1). Surprisingly, two sFlaG intertwined through two short N-terminal β-strands that formed antiparallel β-sheets with adjacent subunits of sFlaG (via residues 45–48) and sFlaF (via residues 38–41) instead of forming a predicted α-helix (Fig. 2b and Supplementary Fig. 4c,d). The N-terminal region of sFlaF exhibited a long α-helix interacting with the adjacent sFlaG through a curved β-sheet that further stabilized the tetramer complex by means of an hydrogen-bonding network and by cation–π interactions between sFlaFH42 and sFlaGY86 (Fig. 2c). Collectively, the crystal structure of our complex and SAXS solution measurements indicate stabilizing interactions between sFlaG and sFlaF to form a heterotetramer.
The FlaG–FlaF complex is required for archaellum assembly and motility.
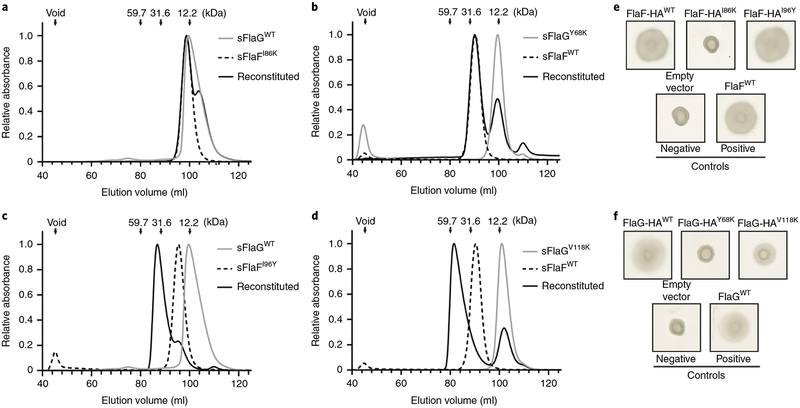
To test the function of the sFlaG–sFlaF complex, we created mutations that disrupted different binding interfaces identified in the crystal structure. Protein variants were reconstituted to evaluate complex formation by SEC and their impacts on motility using cell-based motility assays (Fig. 3 and Supplementary Fig. 5). sFlaFI86K has been shown to disrupt the sFlaF dimer23. Remarkably, residue I86 of sFlaF lays at the centre of the dimer interface of the sFlaG–sFlaF complex, opposite to Y56 and Y68 on sFlaG (Fig. 2d), resembling the sFlaF dimer (Supplementary Fig. 6a). Either the FlaFI86K or sFlaGY68K substitution disrupted sFlaG–sFlaF tetrameric complex formation (Fig. 3a,b) and did not restore motility in ΔaapFΔflaF or ΔaapFΔflaG cells, respectively (Fig. 3e,f). Furthermore, mutations (sFlaGQ138F and sFlaGS148A) designed to disrupt the interaction between the N-terminal α-helix of sFlaF and the β-sheet of sFlaG resulted in unstable proteins, preventing further analysis of this interaction for tetramer formation and motility (Fig. 2c). The sFlaFI96Y variant was generated to reduce the interaction between the heterodimers sFlaG–sFlaF and sFlaG″–sFlaF″ (Supplementary Fig. 6b); the sFlaGV118K variant was created to disrupt possible sFlaG homodimer interaction (Supplementary Fig. 6c) based on the sFlaG dimer structure. The sFlaFI96Y and sFlaGV118K variants had minor or no effect on complex formation or cell motility restoration in ΔaapFΔflaF or ΔaapFΔflaG cells (Fig. 3c–f). To further test this, we solved the structure of the FlaGV118K mutant complex with sFlaF and found that the interface was preserved (Supplementary Fig. 6c). Notably, all FlaF and FlaG variants used in the motility assays were expressed at the same level for the motility assays (Supplementary Fig. 7). Interestingly, when the FlaF variants were overexpressed in complementation assays in ΔaapFΔflaF cells, none of the cells were motile (Supplementary Fig. 8a) in contrast to the tested FlaG variant overexpression in ΔaapFΔflaG cells (Supplementary Fig. 8b). These data suggest that FlaG can be accommodated at high protein levels, but FlaF concentration must be regulated for cell motility. Furthermore, we observed that FlaFI86K and FlaGY68K variants that failed to form complexes in vitro could not assemble archaella in S. acidocaldarius cells (Supplementary Fig. 9). Taken together, these results suggest that sFlaF interacts with sFlaG to form a functional complex that is essential for both motility and archaellum assembly.
Fig. 3 |. Mutations that disrupt heterotetrameric complex formation by sFlaG and sFlaF also abolish swimming motility in S. acidocaldarius.
a–d, SEC was used to detect the complex formation by the following sFlaG and sFlaF variants on reconstitution: sFlaGWT + sFlaFI86K (a), sFlaGY68K + sFlaFWT (b), sFlaGWT + sFlaFI96Y (c) and sFlaGV118K + sFlaFWT (d). A swimming assay was applied to assess the effect of FlaG and FlaF variants on motility in S. acidocaldarius. e, Motility assays of in trans complementation of ΔaapFΔflaF mutants with expression vectors carrying different FlaF species under the control of an arabinose-inducible promoter. No arabinose was added to overexpress FlaF since basal expression of the promoter was sufficient to restore motility. Complementation with untagged FlaFWT and empty vector (pSVAaraFX-HA) served as positive and negative controls, respectively. f, Motility assays of in trans complementation of ΔaapFΔflaG mutants with expression vectors carrying different FlaG species under the control of an arabinose-inducible promoter. Expression was induced by adding 0.4% l-arabinose. Complementation with untagged FlaGWT and empty vector (pSVAaraFX-HA) served as positive and negative controls, respectively.
FlaG and FlaF are post-translationally processed and released from the membrane into the periplasm.
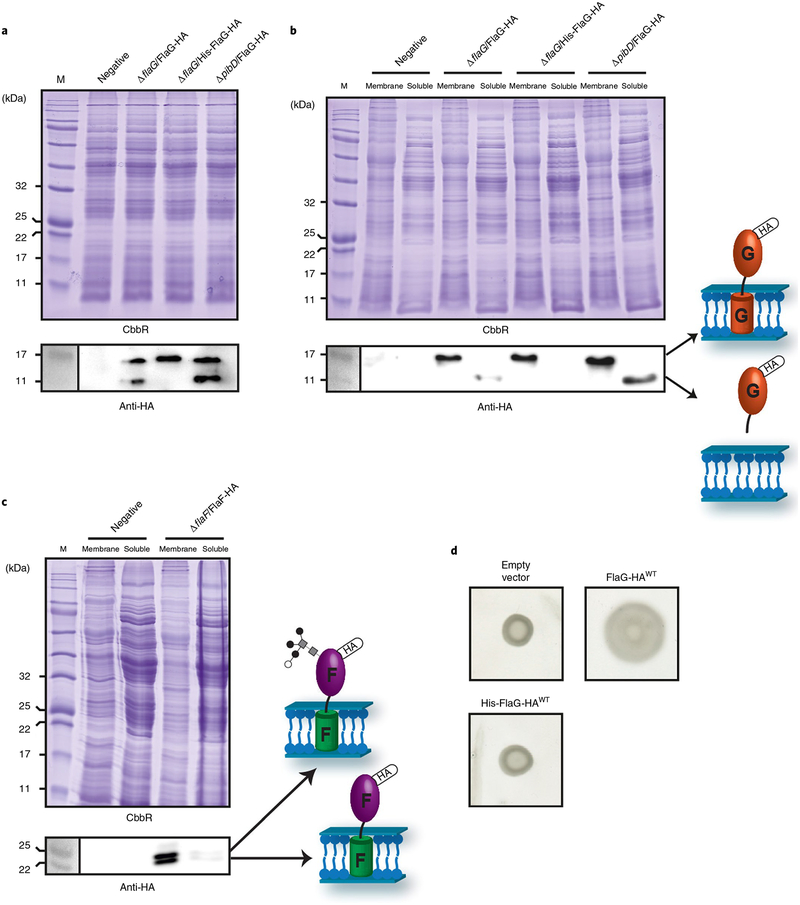
The results showing that the sFlaG–sFlaF complex is critical for motility and that FlaF can interact with the S-layer suggest that FlaG–FlaF do not reside in the membrane only, but must emerge into the periplasm to reach the S-layer to function. To test this idea, C-terminal haemagglutinin (HA)-tagged FlaG and FlaF were ectopically expressed in S. acidocaldarius and checked for their localization in the membrane and soluble fractions including the periplasm. Two specific bands for FlaG-HA, corresponding to the full-length FlaG (upper band) and a cleaved FlaG species (lower band), were detected by anti-HA immunoblotting using whole-cell lysate (Fig. 4a and Supplementary Fig. 7b). Any possible inclusion bodies were removed by low-spin centrifugation. On ultracentrifugation, both FlaG protein species could be separated into two fractions: the full-length FlaG localized in the membrane fraction; and the cleaved FlaG visible in the soluble fraction. This finding indicates that the N-terminal transmembrane domain of FlaG is processed to release it into the periplasm (Fig. 4b). In contrast, anti-HA immunoblotting with the whole-cell lysate did not reveal a cleaved species for FlaF-HA (Supplementary Fig. 7a). Instead, a higher molecular weight species due to N-linked glycosylation at N92 was detected in the membrane fraction; loss of the glycosylation did not impact cell motility (Fig. 4c and Supplementary Fig. 10). Interestingly, both glycosylated and non-glycosylated full-length FlaF were detected in the soluble fraction, although band intensity was not as intense as in the membrane fraction (Fig. 4c). Taken together, these results indicate that both FlaF and FlaG are processed and released into the periplasm.
Fig. 4 |. Both FlaF and FlaG are found in the periplasm, and maturation of FlaG N-terminal cleavage is essential for cell motility.
a, Whole-cell fractions of ΔaapFΔflaG or ΔaapFΔpibD cells expressing FlaG-HA or His-FlaG-HA were analysed by Coomassie Brilliant stain and anti-HA immunoblot: ΔaapFΔflaG pSVAaraFX-HA (negative), ΔaapFΔflaG pSVA4060 (FlaG-HA), ΔaapFΔflaG pSVA5730 (His-FlaG-HA) and ΔaapFΔpibD pSVA4060 (FlaG-HA). b, Membrane and soluble fractions of ΔaapFΔflaG or ΔaapFΔpibD cells expressing FlaG-HA or His-FlaG-HA were analysed by Coomassie Brilliant stain and anti-HA immunoblot. c, Membrane and soluble fractions of ΔaapFΔflaF cells expressing HA-tagged FlaF analysed by Coomassie Brilliant stain and anti-HA immunoblot. F, FlaF. d, Effect of N-terminal His-tagged FlaG on the swimming behaviour of S. acidocaldarius.
Processing of the FlaG N terminus is essential for motility.
Although neither FlaG nor FlaF harbours a positively charged class III signal peptide at its N terminus24, we tested if FlaG processing is specific and relevant for swimming motility in S. acidocaldarius. To see whether FlaG processing depends on PibD peptidase, which cuts prearchaellins, we expressed FlaG-HA in a PibD deletion strain (ΔaapFΔpibD) and found that FlaG cleavage is PibD-independent (Fig. 4a,b). To test whether cleavage is required for motility, we created an insertion variant with a six-histidine (His6)-tag inserted after the Met1 residue of FlaG-HA. This insertion inhibited FlaG processing; cleaved FlaG was not detected in the soluble fraction and only full-length FlaG remained in the membrane fraction (Fig. 4a,b). Importantly, this FlaG insertion variant abolished archaellum assembly and did not restore motility in ΔaapFΔflaG cells (Supplementary Fig. 9a and Fig. 4d). Although how FlaG is cleaved remains unclear, these data suggest that the FlaG transmembrane domain is specifically cleaved in a PibD-independent manner that allows cleaved FlaG to escape into the periplasm to play an essential role in archaellum assembly and motility.
Interaction of FlaG–FlaF with the S-layer is essential for motility but not assembly.
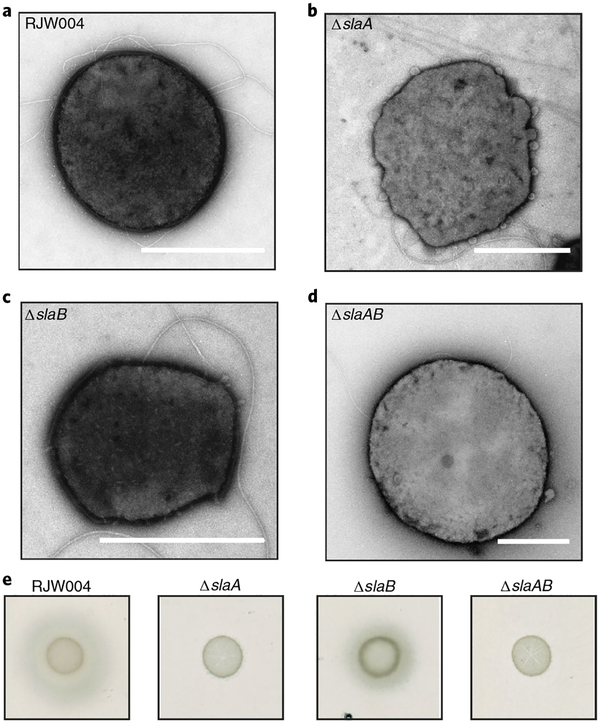
Based on these findings, we reasoned that the FlaG–FlaF complex may span the periplasm to anchor the archaellum to the S-layer. To test this idea, we examined the impact of S-layer mutations of SlaA, a large protein forming the crystalline cell surface and SlaB, a smaller protein anchoring SlaA to the membrane. Notably, a recent transposon screen in S. islandicus using transmission electron microscopy (TEM) and scanning electron microscopy revealed that S-layer proteins are not essential for cell viability31, with deletion mutants showing impaired cell morphology32. As observed previously, we found that cells lacking SlaB assembled a SlaA layer around the cell, but in an irregular manner compared to the wild type (WT; RJW004)32. ΔslaA and ΔslaAB showed notable growth defects without the S-layer31; ΔslaA did not have an S-layer, but its membrane had a deformed shape with numerous extrusions (Fig. 5b). Strikingly, all of the mutant cells still assembled archaella despite lacking S-layer proteins (Fig. 5a–d). Compared to WT, ΔslaB exhibited a smaller halo around the cell spot on motility plates, indicating a small defect in motility. In contrast, both ΔslaA and ΔslaAB were non-motile (Fig. 5e) although they showed slight growth on the motility plates. These results support the notion that the S-layer is not required to assemble archaella, but an S-layer that interacts with the FlaG–FlaF complex is required for rotation of the archaellar filament.
Fig. 5 |. S-layer protein SlaA deletion cells impair cell motility while maintaining archaella in S. islandicus.
a–d, Cell morphology of each S. islandicus mutant was visualized under TEM: WT (RJW004) cells (a), slaA deletion knockout cells (b), slaB deletion knockout cells (c) and slaAB deletion knockout cells (d). e, Effect of sla deletion mutants on motility in S. islandicus. Scale bars, 1 μm.
Pfu sFlaG forms filaments in vitro regulated by sFlaF.
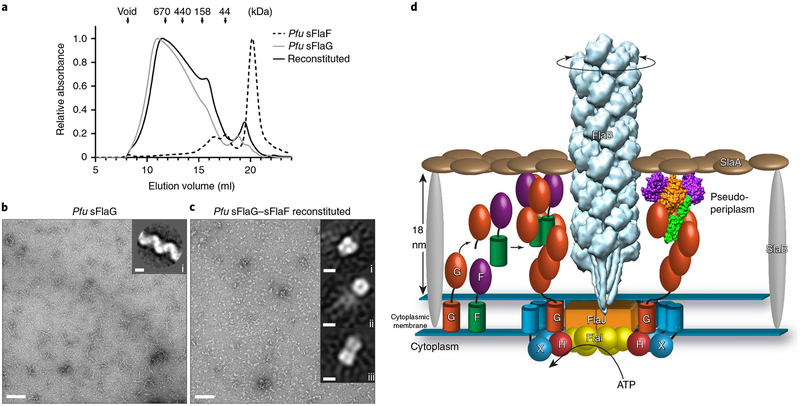
Our observations that FlaG and FlaF have a structural resemblance to true archaellins, are extruded from the membrane and interact with the S-layer, led us to ask if they may form filaments. Since the S. acidocaldarius soluble domains do not form stable oligomers larger than tetramers (Fig. 2a), we tested for filament formation of Pfu sFlaG and sFlaF33. Despite their low sequence identities (approximately 28%) to S. acidocaldarius, FlaF and FlaG are predicted to have similar secondary structures (Supplementary Fig. 11a). While Pfu sFlaF eluted as a monomer (<20 kDa) from the size-exclusion column, Pfu sFlaG formed a broad, high-molecular-weight peak (>150 kDa; Fig. 6a and Supplementary Fig. 11b–d). On reconstitution, elution entirely shifted the monomeric Pfu sFlaF peak to the Pfu sFlaG fractions, indicating interaction. Interestingly, negatively stained TEM images from the SEC fractions of Pfu sFlaG revealed 5–7 nm wide helical filaments (Fig. 6b), while the TEM images of corresponding fractions of coeluted Pfu sFlaG and sFlaF instead showed smaller globular knobs and no filaments (Fig. 6c). These combined data show that FlaG alone can form filaments, while addition of FlaF limits filament formation.
Fig. 6 |. Pfu sFlaF limits Pfu sFlaG filament formation that results in our working model.
a, Normalized absorbance at λ = 280 nm from SEC was plotted against elution volume. Individual Pfu sFlaF (black dashed line) eluted at low molecular weight, probably as a monomer. Individual Pfu sFlaG (grey line) elutes as a broad peak at higher molecular weight (> 150 kDa). On reconstitution (black solid line), Pfu sFlaG elution behaviour is only slightly altered, since Pfu sFlaF completely shifts towards the sFlaG fraction, representing FlaG–FlaF complex formation. The molecular weight standards are indicatedby the arrows. b,c, Negatively stained micrographs (scale bars, 90 nm) from Pfu sFlaG and Pfu sFlaG–sFlaF SEC fraction at 15 ml elution volume, analysed with TEM. Insets: two-dimensional class averages are shown (scale bars, 5 nm). While Pfu sFlaG alone forms spiral-shaped filaments (b and inset i in b), the Pfu sFlaG–sFlaF complex forms globular complexes (c) that could be averaged into three different classes (insets i–iii in c). d, Integrated testable model consistent with structural and mutational data for the archaellum assembly machinery in S. acidocaldarius. Pre-archaellin FlaB is processed byPibD peptidase (peptidase A24) and assembled to the archaellum filament. The proposed stator protein complex FlaG–FlaF is located in the periplasmic space, approximately 18 nm to the S-layer. Cleaved FlaG (matured form) forms the filament and is capped by the FlaG–FlaF complex, which interactswith the S-layer (SlaA). The motor complex is assembled by the scaffold protein FlaX, which forms a ring structure approximately 30 nm in diameter and interacts with cytoplasmic hexameric ATPase FlaI and the ATP-binding protein FlaH, where they interact with the inner membrane protein FlaJ. Finally, the archaellum filament rotates on the ATP consumption by FlaI. F, FlaF; G, FlaG; H, FlaH; X, FlaX.
Discussion
Limited information on the structure and roles of the periplasmic proteins has made it unclear how the archaellum is anchored within the cell envelope and how this facilitates motility. Cryogenic electron microscopy imaging has only resolved parts of the archaellum motor complex in the cytoplasm, while potential periplasmic components could not be assessed34,35. FlaF is the only subunit of the archaellum known to interact with the S-layer23. In this study, we find that FlaG forms a complex with FlaF via its C-terminal, archaellin-like domain, as shown in our crystal structures.
We infer that FlaF most probably interacts with the large subunit of the S-layer, SlaA, for motility, since motility is abolished in mutants lacking SlaA but not SlaB. Furthermore, S. islandicus cells lacking SlaA assemble archaella, yet they are non-motile. These results support the idea that FlaF interaction with SlaA is required for motility (not assembly), but raises the question of how the FlaG–FlaF complex may interact with SlaA while their N-terminal transmembrane regions are embedded in the membrane 18 nm away. We discovered that the FlaG N terminus must be cleaved in a PibD-independent manner for motility, which would enable FlaG complexes in the periplasmic space. The mechanism and determining sequence for FlaG cleavage merits further investigation. Full-length FlaF, on the other hand, localizes in the membrane and soluble periplasm fractions. How full-length FlaF enters the periplasm is unknown, but this would be enabled if the FlaF membrane domain is buried in a filament similar to true archaellins and to T4P36. Whereas a membrane-bound FlaG–FlaF tetramer is only 4.5 nm in height, a FlaG-containing filament may bridge the 18 nm gap between the membrane and the S-layer.
We more rigorously tested this idea in the conserved fla operon of the experimentally more tractable Pfu system33. We found that Pfu sFlaG formed apparently unbounded filaments, whereas the reconstituted Pfu sFlaG–sFlaF complex formed size-limited knob-like structures, suggesting that FlaF can cap and regulate FlaG filament formation. Therefore, we propose a model where FlaG–FlaF complexes cap FlaG filaments, which are sufficiently long to attach to the S-layer (Fig. 6d). This model would explain why overexpression of FlaG restores ΔaapFΔflaG motility whereas FlaF overexpression leads to non-motile cells in S. acidocaldarius, since increased FlaF concentration could prematurely halt FlaG filament formation. Yet, how FlaG forms the filament identified in this study and interacts with the FlaG–FlaF complex merits future investigation and high-resolution cryogenic electron microscopy studies.
For S. acidocaldarius sFlaG, we may not have observed the filament seen with the Pfu homologue due to the truncation construct or the absence of a cofactor or ligand as recently seen in the FlaB1 filament of Methanococcus maripaludis37. Alternatively, specific coexpression interdependencies may exist, as seen in the analogous bacterial flagellar stator complexes that must be coexpressed38. Furthermore, lack of density for a putative FlaG–FlaF structure in the in situ subtomogram average structure of the Pyrococcus archaellum motor35 may reflect archaellar filament flexibility precluding alignment of extracytoplasmic components. Indeed, stator complexes in Escherichia coli and Salmonella flagellar motors are similarly absent in subtomogram average structures due to their dynamic and potentially discontinuous positioning39.
Our characterization of the sFlaG–sFlaF tetrameric complex, together with the filaments formed by Pfu sFlaG, contributes fundamental insights into archaellum component structure, assembly and function. In the current integrated model (Fig. 6d), mature FlaG forms a periplasmic filament that is capped by the heterotetrameric FlaG–FlaF complex. The FlaG–FlaF complex binds to the S-layer protein SlaA and may therefore act as an archaellum stator. In S. acidocaldarius, the cytoplasmic motor complex, which consists of FlaI, FlaH and FlaX, forms a ring structure that interacts with the inner membrane protein FlaJ. The proposed periplasmic stator complex is currently best understood as a filament composed of FlaG and FlaG–FlaF complex with dual roles in FlaB assembly and motility implied by the combined structural and functional data presented in this study.
By analogy to the bacterial system, a potential torque-generating interface between rotor and stator is probably localized to the inner membrane. Therefore, we reason that, at the bottom of the stator filament, the transmembrane domains of FlaG may interact with the transmembrane region of the only conserved membrane protein in the motor, that is, FlaJ. In turn, some FlaG subunits undergo maturation losing their membrane anchor region, allowing filament assembly with the proposed FlaG–FlaF cap binding to the SlaA subunit of the S-layer to enable torque generation. How the FlaG–FlaF complex is involved in torque generation will merit future investigations.
Methods
Plasmid construction and site-directed mutagenesis.
Full-length flaG (Saci_1176) and full-length flaF (Saci_1175) were amplified using the 8236/8215 and 8237/8215 primers, respectively, with S. acidocaldarius MW001 genomic DNA serving as the template. The products were cloned into pSVAaraFX-stop and pSVAaraFX-HA to yield tag-less and HA-tagged versions of both proteins. An N- and C-terminal truncated His6-tagged construct of Saci FlaG (NΔ31/CΔ3) was generated for heterologous expression in E. coli because full-length FlaG produced in E. coli was not soluble. NΔ31/CΔ3 FlaG (sFlaG) construct was amplified using the 3749 and 2143 primers and incorporated into a pET-Duet1 backbone using EcoRI/HindIII restriction sites, resulting in pSVA2826. N-terminal truncated Pfu FlaF (NΔ40, PF0334) and Pfu FlaG (NΔ31, PF0333) were amplified using the primer pairs 3741/3742 and 6657/6658, respectively. Both inserts were incorporated into the pET-Duet 1 backbone using EcoRI/HindIII restriction sites, yielding pSVA 2822 and pSVA 4014, respectively. Site-directed mutagenesis was performed using PCR on pSVA4060, pSVA2826 for flaG and on pSVA4061, pSVA1921 for flaF, respectively. Plasmids and oligonucleotides used are listed in Supplementary Table 3.
Recombinant protein production and purification.
Chemically competent E. coli Rosetta (DE3) cells carrying the pLysS plasmid were freshly transformed with the pET-based vector and cultivated overnight in Luria–Bertani medium supplemented with 50 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol. Luria–Bertani medium (1 l) containing ampicillin and chloramphenicol was inoculated using 10 ml of the pre-culture. Cells were grown at 37 °C until reaching an optical density OD600 of 0.4 for Saci sFlaF (and variants), 0.75 for Saci sFlaG (and variants), 0.7 for Pfu sFlaF and 0.9 for Pfu sFlaG. Protein overproduction was induced using 0.5 mM isopropyl β-d-1-thiogalactopyranoside and the induction was carried out for 3 h at 37 °C. Cells were collected by centrifugation and cell pellets were flash-frozen in liquid nitrogen and kept at −80 °C until further use.
E. coli cell pellets containing heterologously produced Saci sFlaG variants were thawed on ice and resuspended in 50 mM MES buffer (pH 6.5), 200 mM NaCl, 1× protease inhibitor cocktail (Roche) and traces of DNase I (Roche), before disruption in an LM-10 Microfluidizer (Microfluidics). His6-tagged Saci sFlaG variants were purified using nickel affinity chromatography. After applying the cell-free lysate on the column, the resin was washed with 50 mM MES (pH 6.5) and 200 mM NaCl, using an imidazole gradient of 10 to 50 mM on a gravity column. Saci sFlaG was eluted with 25 mM glycine-HCl, 300 mM NaCl, 5% glycerol, 1 M l-arginine and 100 mM imidazole (pH 10.5). The elution fraction was immediately precipitated using 80% ammonium sulfate. The resulting precipitate was resuspended in 25 mM citric acid/sodium citrate (pH 3) and 150 mM NaCl and dialysed overnight against the latter buffer to remove residual ammonium sulfate. E. coli cell pellets containing heterologously produced Saci sFlaF variants were thawed on ice and resuspended in 50 mM Tris (pH 8), 150 mM NaCl, 1× protease inhibitor cocktail and traces of DNase I before disruption in an LM-10 Microfluidizer. His6-tagged sFlaF variants were purified using nickel affinity chromatography. After loading the cell-free lysate on the column, the resin was washed with 50 mM Tris-HCl (pH 8) and 150 mM NaCl and Saci sFlaF was eluted using an imidazole gradient of 10–250 mM. Fractions containing the highest Saci sFlaF concentrations were immediately precipitated using 80% ammonium sulfate and shifted to 25 mM citric acid/sodium citrate (pH 3) and 150 mM NaCl exactly as described for Saci sFlaG. E. coli cell pellets containing heterologously produced Pfu sFlaF and sFlaG were thawed and resuspended in 150 mM NaCl, 50 mM Tris-HCl (pH 8), 0.5% (v/v) Triton X-100 and 10 mM imidazole. Additionally, 10 mM dithiothreitol were added to the thawed cell pellet comprising heterologously expressed Pfu His6-sFlaF. Traces of DNase I were added before cell lysis using an MS 73 probe sonicator (SONOPLUS; Bandelin) with 40% amplitude for 30 min.
For Pfu sFlaF, the lysate was incubated at 60 °C for 10 min. The heat-treated lysate was cleared by centrifugation at 7,400g for 25 min and the supernatant was applied on a nickel affinity gravity column (Sigma-Aldrich). The column was washed with 50 mM Tris-HCl (pH 8), 150 mM NaCl and 10 mM dithiothreitol, and protein was eluted with a stepwise imidazole gradient from 10 to 500 mM.
For FlaG, the lysate was centrifuged at 186,000g for 40 min. The supernatant was applied to a nickel affinity gravity column. The column was washed with 50 mM Tris-HCl (pH 8) and 150 mM NaCl before applying a stepwise imidazole gradient from 20 to 600 mM.
Protein complex reconstitution and SEC.
To reconstitute the Saci sFlaG/sFlaF protein complex, Saci sFlaG was precipitated using 80% saturated ammonium sulfate and the precipitated protein was spun down by centrifugation in a tabletop centrifuge at maximum speed. The sFlaG pellet was resuspended in an equal volume of sFlaF with equal concentration in 25 mM citric acid/sodium citrate buffer (pH 3) and 150 mM NaCl. The reconstituted complex was dialysed overnight against the same buffer. The dialysed sample (2 ml) was further loaded onto a Superdex 200 (16/600) size-exclusion column (GE Healthcare Life Sciences), which was pre-equilibrated with the same buffer to separate the reconstituted complex from excess individual proteins. Fractions containing the sFlaG–sFlaF protein complex were pooled, flash-frozen in liquid nitrogen and stored at −80 °C until further use. Pfu sFlaF was precipitated using 80% saturated ammonium sulfate; the precipitated protein was pelleted at 7,400g for 20 min at room temperature. The resulting pellet was resuspended in an equal volume of Pfu sFlaG. The mixture was dialysed against 50 mM Tris-HCl (pH 8) and 150 mM NaCl overnight in a Slide-A-Lyzer dialysis chamber (molecular weight cut-off = 2 kDa; Thermo Fisher Scientific). The dialysed sample was centrifuged at 7,400g for10 min at room temperature and the supernatant was used for SEC on a Superose 6 (10/300) increase column (GE Healthcare Life Sciences).
Motility assays.
Motility assays were performed to monitor the swimming ability of S. acidocaldarius and S. islandicus cells according to a modified protocol developed by Lassak et al.7. Briefly, S. acidocaldarius strains, transformed with the expression plasmids of the FlaG or FlaF variants, were cultivated aerobically at 75 °C in Brock medium (pH 3) supplemented with 0.1% N-Z-Amine (Sigma Aldrich) and 0.2% dextrin under shaking conditions. Cell culture (5 μl) in logarithmic growth phase was spotted on 0.15% GelRite plates containing Brock medium, supplemented with 0.005% N-Z-Amine and 0.4% l-arabinose or no sugar. Plates were incubated for 5–6 d in a humid chamber at 75 °C. S. islandicus strains were cultivated in standing flasks aerobically at 75 °C in Brock medium (pH 3), 0.1% tryptone, 0.2% sucrose, 50 μg ml−1 agmatine and 10 μg ml−1 uracil. Cell culture (5 μl) in logarithmic growth phase was spotted on 0.15% GelRite plates containing Brock medium, supplemented with 0.005% tryptone, 50 μg ml−1 agmatine and 10 μg ml−1 uracil. Plates were incubated for 4–5 d in a humid chamber at 75 °C.
Preparation of immunoblot samples.
To assay the production and localization of variant proteins in S. acidocaldarius, α-HA immunoblotting was performed on whole-cell, soluble or membrane fractions. To induce plasmid-based protein production under control of the ara promoter and simultaneously stress the cells to induce archaella production, 50 ml of S. acidocaldarius cultures were grown to stationary phase and collected by centrifugation; the resulting pellets were resuspended in Brock medium (pH 3) supplemented with only 0.4% l-arabinose. Induction/starvation was carried out for 4 h. Whole-cell samples were taken, centrifuged in a tabletop centrifuge at maximum speed for 2 min and the resulting pellets were resuspended in 1× SDS loading dye to reach a theoretical optical density of 10. The remaining culture was pelleted by centrifugation at 5,000g for 10 min. The cell pellet was resuspended in 5 ml of 50 mM Tris-HCl, 150 mM NaCl (pH 8) buffer and cells were lysed by sonication at 40% amplitude for 5 min with 15 s pulse and 5 s break. Cell debris was removed by centrifugation at 5,000g for15 min. The resulting supernatant was applied to ultracentrifugation at 175,000g for 45 min to separate membrane and soluble fractions. To obtain protein samples of the soluble fraction, 1 ml of the supernatant was removed from the tube directly after centrifugation and precipitated using 40% (w/v) trichloroacetic acid. After incubation on ice for 15 min, precipitated proteins were pelleted by centrifugation in a tabletop centrifuge at maximum speed for 20 min. Protein pellets were resuspended in 90 μl of 1× SDS loading dye (Sigma Aldrich) and pH was neutralized by adding 10 μl of saturated Tris solution. The membrane pellet was resuspended in 500 μl of 50 mM Tris-HCl, 150 mM NaCl and 0.05% (w/v) n-dodecyl β-d-maltoside (Carl Roth). Next, 80 μl of membranes were mixed with 20 μl of 5× SDS loading dye to yield the membrane fraction samples for SDS–polyacrylamide gel electrophoresis and immunoblot. The final membrane fraction was about 1.25× dilution compared to the soluble fraction.
SAXS.
The sample sFlaG or sFlaG–sFlaF complex variants that had been run through the size-exclusion column were concentrated in a concentrator to prepare the three concentration series of SAXS samples. SAXS data were collected at the SIBYLS Beamline (BL12.3.1) of the Advanced Light Source (ALS) facility40,41. Data were processed and analysed using the SCÅTTER program (v.3.1r; http://www.bioisis.net/tutorial)27,42. SAXS data were processed according to the guidelines described by Trewhella et al.43; the parameters and statistics are listed in Supplementary Table 1. The model of sFlaG–sFlaF tetramer with missing N-terminal region and His tag from crystal structure was generated by MODELLER (v.9.17)44 in Chimera (v.1.11.2)42, followed by multistate modelling using the MultiFoXS web server (v.master.61ca85a; https://modbase.compbio.ucsf.edu/multifoxs/)45. The flexible residues of sFlaF (1–17) and sFlaG (1–34) from the constructs were chosen to generate 10,000 conformations. The best-fit model was fitted to the SAXS experimental data of the sFlaG–sFlaF complex using the FoXS web server (v.1.0; https://modbase.compbio.ucsf.edu/foxs/)46. The 0.01–0.30 and 0.01–0.36 q ranges were plotted for sFlaG (Fig. 1a) and sFlaG–sFlaF (Supplementary Fig. 3a) preparation, respectively due to the higher noise at high q range. SAXS data were deposited with the Small Angle Scattering Biological Data Bank (SASBDB; https://www.sasbdb.org/) and are summarized in Supplementary Table 1.
Crystallization and structure determination.
sFlaG native crystals were grownby mixing 2 ul of protein (3.5 mg ml−1 in 25 mM citric acid/sodium citrate (pH 3) and 150 mM NaCl), 2 μl of water and 2 μl of crystallization solution (13% PEG3350, 0.2 M sodium malonate (pH 5) and 3% 1,4-dioxane) at 15 °C. Many thin rod crystals were grown initially in 2 d. After 1 week, plate crystals were also formed. A single plate crystal was then picked and transferred to the reservoir solution with 20% ethylene glycol added as cryoprotectant and flash-frozen in liquid nitrogen. sFlaG heavy atom derivatives were obtained by soaking an sFlaG plate crystal with 10 mM K2PtCl4 for 45 min. The soaked crystal was back-soaked with the reservoir solution for 2 min and then transferred to the cryoprotectant solution with 20% ethylene glycol followed by 25% ethylene glycol and flash-frozen in liquid nitrogen for data collection. The X-ray diffraction data of the sFlaG crystals were collected at the SIBYLS Beamline (BL12.3.1) at Advance Light Source, Lawrence Berkeley National Laboratory41. The X-ray data of the native sFlaG crystal was collected at 1.0714 Å and the derivative crystal of sFlaG with heavy atom was collected using MAD data collection with three wavelengths of platinum experimental edges: peak; remote; and inflection (Supplementary Table 2). The X-ray data at each wavelength were collected entirely in 30° wedge for 180° with inverse beam before collecting the next wavelength. The X-ray data of native and derivative sFlaG was integrated by the XDS program (v.0.52)47. The integration file was then merged and scaled using the aimless program (v.0.5.25) in the CCP4 suite48. The MAD phasing solution was found using autosol (v.1.10) in the Phenix suite (v.1.10_2155)49. Four platinum sites were found and the phase information was used to generate the sFlaG model. This model was further built in phenix.autobuild (v.1.10) and the resolution was extended to 1.93 Å using the native sFlaG dataset. The resulting model was further fitted in the Coot program (v.0.8.9) and refined by phenix.refine (v.1.10) (Supplementary Table 2). The deposited sFlaG structure (PDB: 5TUH) contains residues in the regions of 97.6% favoured, 2.4% allowed and 0% outliers in a Ramachandran plot.
The sFlaG–sFlaF (WT and V118K) protein complexes were crystallized by mixing an equal volume of protein complex (3 mg ml−1 in 25 mM citric acid/sodium citrate (pH 3) and 150 mM NaCl) with crystallization solution (18% polyethylene glycol 6000 (Fluka), 0.1 M Tris, pH 7.5 and 0.2 M NaBr) and incubated at 15 °C. The hexagonal bipyramid crystals were formed after 3 d. A single crystal was transferred into the cryoprotectant solution containing reservoir and 10% ethylene glycol followed by 20% ethylene glycol and flash-frozen in liquid nitrogen. The sFlaG–sFlaF protein crystal data were collected at the SIBYLS Beamline (BL12.3.1) at ALS. The sFlaG–sFlaF complex structure was solved by molecular replacement using Phaser (v.2.5.6) in the Phenix suite, with sFlaF (PDB: 4P94) and sFlaG (PDB: 5TUH) as the search models. The twin fraction of the X-ray diffraction data detected was approximately 0.240 based on l-test. Twin law (h,-h-k,-l) was applied during refinement using phenix.refine. The sFlaG–sFlaF structure was further fitted by Coot and refined by phenix.refine (Supplementary Table 2). The deposited sFlaG–sFlaF structure (PDB: 5TUG) and sFlaGV118K–sFlaF structure (PDB: 6PBK) contains residues in the regions of 96.9% favoured, 2.7% allowed and 0.4% outliers and 96.3% favoured, 3.3% allowed and 0.4% outliers, respectively in a Ramachandran plot.
TEM of cells from Sulfolobus species.
S. islandicus WT and S-layer mutant cultures were adsorbed onto 200-mesh Carbon Type-B grids (Ted Pella) and stained with 2% (w/v) uranyl acetate. The grids were observed under a Philips CM200 transmission electron microscope operated at 120 kV. Images were taken with a Peltier-cooled Tietz 2k × 2k charge-coupled device (CCD) camera (TVIPS).
S. acidocaldarius strains were adsorbed either onto either 400-mesh Ultrathin Carbon Type-A grids (Ted Pella) or 300-mesh Carbon grids (catalogue no. AGS162–3; Plano GmbH) and stained with 2% (w/v) uranyl acetate. The grids were observed under a Philips CM10 transmission electron microscope operated at 80 kV. Images were taken with an air-cooled BioScan 792 1024 × 1024 CCD camera (Gatan).
TEM of Pfu sFlaG–sFlaF.
Five-microlitre samples were applied on glow-discharged, carbon-coated 300-mesh copper grids (Agar Scientific) and incubated for 1 min at room temperature. The liquid was blotted away using What man paper and samples were stained with 2% (w/v) uranyl acetate. Imaging was performed using FEI Tecnai T12 twin-lens microscope (LAB6) equipped with a F126 CCD camera (TVIPS). Images were visualized with 3DMOD (v.4.9.0)50,51.
Negative stain electron microscopy data acquisition and image processing of Pfu sFlaG and sFlaG–sFlaF complex.
The images of the negatively stained sFlaG filaments were taken with a Tecnai T12 twin-lens microscope using a F216 CCD camera with a pixel size of 2.586 Å. Images of the negatively stained sFlaG–sFlaF complex were taken with a FEI Tecnai G2-F20 microscope using a Falcon II detector with a pixel size of 2.538 Å. Scipion v.1.2 (ref.52) was used for processing. Particles were picked using Xmipp3 (ref.53) and contrast transfer function-corrected54. Classification was done with RELION (v.3.0)55.
Statistics and reproducibility.
FlaG–FlaF complex (WT) reconstitution and SEC were performed three times independently (Fig. 2a). FlaG–FlaF variant complex reconstitutions and SEC assays were performed twice independently (Fig. 3a–d and Supplementary Fig. 5). Motility assays and immunoblots showing protein levels were performed three times independently with similar results (Fig. 3e,f and Supplementary Figs. 7 and 8). Gels and immunoblots showing FlaG cleavage and FlaF glycosylation, as well as the respective motility assays (Fig. 4 and Supplementary Fig. 10), were performed three times independently with similar results. Motility assays of the S. islandicus S-layer mutants (Fig. 5e) were performed twice independently with similar results. The associated electron micrographs (Fig. 5a–d) show representative cells from imaging sets of at least five biological replicates per strain. Archaella were frequently observed for all strains. Electron microscopy to visualize the archaellation of S. acidocaldarius (SupplementaryFig. 9) cells was performed for at least two grids per strain originating from two independent transformations. One representative micrograph per strain is depicted. Filaments were regularly observed for motile cells, whereas no filaments were observed for non-motile cells.
Reconstitution and SEC analysis of Pfu proteins (Fig. 6a and Supplementary Fig. 11b–d), as well as the microscopy analysis (Fig. 6b,c) were performed independently three times with similar results. Overview micrographs depict representative images of the analysed SEC fractions. The SEC profiles depicted in Supplementary Fig. 1 originate from the same runs shown in Figs. 2a and 3a for the individual sFlaFWT (dimeric) and sFlaFI86K (monomeric) to have a direct comparison of the elution behaviour.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
P.T. received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 686647. S.-V.A. was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) under project no. 403222702-SFB 1381. The work was conducted at ALS, a national user facility operated by the Lawrence Berkeley National Laboratory on behalf of the US Department of Energy’s Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by US Department of Energy Office of Biological and Environmental Research. Additional support comes from the National Institutes of Health (NIH) grant no. P30GM124169. J.A.T. acknowledges start-up funds from the University of Texas Safety Tracking and Reporting System (NIH grant no. R35CA220430) and the Robert A. Welch Chair in Chemistry (grant no. CFS127800-80-101399-50). C.Z., R.L.W. and R.J.W. are supported by a grant from NASA (National Aeronautics and Space Administration) through the NASA Astrobiology Institute under cooperative agreement no. NNA13AA91A, issued through the Science Mission Directorate. We thank S. Robinson from the Microscopy Suite at the Beckman Institute for Advance Science and Technology, University of Illinois at Urbana-Champaign, for providing TEM assistance. We especially thank K. Burnett for her assistance on the SAXS data collection of the sFlaG and sFlaG–sFlaF complexes, G. Hura for the SAXS similarity calculation suggestion and S. Classen for his assistance on the X-ray crystallography data collection at the SIBYLS Beamline (BL12.3.1). We thank A. Ghosh for technical help; B. Tutt at the Department of Scientific Publications at the MD Anderson Cancer Center for proofreading and editing our manuscript; and we thank P. Simpson for technical assistance with imaging the P. furiosus FlaF and FlaG assemblies. The TEM is operated by the University of Freiburg, Faculty of Biology, as a partner unit within the Microscopy and Image Analysis Platform, Freiburg.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-019-0622-3.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The X-ray diffraction data and coordinates of the sFlaG and sFlaG–sFlaF complex structures have been deposited with the PDB: 5TUH (sFlaG); 5TUG (sFlaGWT/sFlaFWT); and 6PBK (sFlaGV118K/sFlaFWT). The SAXS data have been deposited with the SASBDB: SASDEU7 (sFlaG); SASDES7 (sFlaGWT/sFlaFWT); SASDEV7 (sFlaGWT/sFlaFI96Y); and SASDET7 (sFlaGV118K/sFlaFWT). Source data for Fig. 4 and Supplementary Figs. 5, 7, 10 and 11 are provided with the paper.
References
- 1.Jarrell KF & McBride MJ The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol 6, 466–476 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Albers SV & Jarrell KF The archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 26, 351–362 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Jarrell KF & Albers SV The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Shahapure R, Driessen RPC, Haurat MF, Albers SV & Dame RT The archaellum: a rotating type IV pilus. Mol. Microbiol 91, 716–723 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Kinosita Y, Uchida N, Nakane D & Nishizaka T Direct observation of rotation and steps of the archaellum in the swimming halophilic archaeon Halobacterium salinarum. Nat. Microbiol 1, 16148 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Marwan W, Alam M & Oesterhelt D Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J. Bacteriol 173, 1971–1977 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassak K et al. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol 83, 110–124 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Chaban B et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol 66, 596–609 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Tripepi M, Imam S & Pohlschröder M Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J. Bacteriol 192, 3093–3102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas NA, Pawson CT & Jarrell KF Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non-flagellated cells. Mol. Genet. Genomics 265, 596–603 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Banerjee A, Neiner T, Tripp P & Albers S-V Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J. 280, 6141–6149 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Ghosh A & Albers S-V Assembly and function of the archaeal flagellum. Biochem. Soc. Trans 39, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Albers S-V, Szabó Z & Driessen AJM Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol 4, 537–547 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Ng SYM, Chaban B & Jarrell KF Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J. Mol. Microbiol. Biotechnol 11, 167–191 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Takhar HK, Kemp K, Kim M, Howell PL & Burrows LL The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem 288, 9721–9728 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhury P et al. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Mol. Microbiol 99, 674–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reindl S et al. Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol. Cell 49, 1069–1082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh A, Hartung S, van der Does C, Tainer JA & Albers S-V Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem. J 437, 43–52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakovljevic V, Leonardy S, Hoppert M & Søgaard-Andersen L PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol 190, 2411–2421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mancl JM, Black WP, Robinson H, Yang Z & Schubot FD Crystal structure of a type IV pilus assembly ATPase: insights into the molecular mechanism of PilB from Thermus thermophilus. Structure 24, 1886–1897 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Tsai CL & Tainer JA The ATPase motor turns for type IV pilus assembly. Structure 24, 1857–1859 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Banerjee A et al. FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI. J. Biol. Chem 287, 43322–43330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee A et al. FlaF is a β-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure 23, 863–872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabó Z et al. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol 189, 772–778 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albers SV, Szabó Z & Driessen AJM Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. in. J. Bacteriol 185, 3918–3925 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brock TD, Brock KM, Belly RT & Weiss RL Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol 84, 54–68 (1972). [DOI] [PubMed] [Google Scholar]
- 27.Rambo RP & Tainer JA Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikhney AG & Svergun DI A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 589, 2570–2577 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Burger VM, Arenas DJ & Stultz CM A structure-free method for quantifying conformational flexibility in proteins. Sci. Rep 6, 29040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Phillips APR, Wipfler RL, Olsen GJ & Whitaker RJ The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun 9, 4908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C et al. Cell structure changes in the hyperthermophilic crenarchaeon Sulfolobus islandicus lacking the S-Layer. mBio 10, e01589–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarrell KF, Ding Y, Nair DB & Siu S Surface appendages of Archaea: structure, function, genetics and assembly. Life 3, 86–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briegel A et al. Morphology of the archaellar motor and associated cytoplasmic cone in Thermococcus kodakaraensis. EMBO Rep. 18, 1660–1670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daum B et al. Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. eLife 6, e27470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craig L et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Meshcheryakov VA et al. High-resolution archaellum structure reveals a conserved metal-binding site. EMBO Rep. 20, e46340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Way SM, Hosking ER, Braun TF & Manson MD Mot protein assembly into the bacterial flagellum: a model based on mutational analysis of the motB gene. J. Mol. Biol 297, 7–24 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Chaban B, Coleman I & Beeby M Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci. Rep 8, 97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hura GL et al. Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nat. Methods 10, 453–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Classen S et al. Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr 46, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Trewhella J et al. 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallogr. D 73, 710–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Webb B & Sali A Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneidman-Duhovny D, Hammel M, Tainer JA & Sali A FoXS, FoXSDock and MultiFoXS: single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44, W424–W429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneidman-Duhovny D, Hammel M, Tainer JA & Sali A Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J 105, 962–974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Mastronarde DN Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol 120, 343–352 (1997). [DOI] [PubMed] [Google Scholar]
- 52.de la Rosa-Trevín JM et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 53.de la Rosa-Trevín JM et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol 184, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Mindell JA & Grigorieff N Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol 142, 334–347 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Scheres SHW RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.