Abstract
Adeno-associated viruses (AAVs) are being developed as vectors for the treatment of genetic disorders. However, pre-existing antibodies present a significant limitation to achieving optimal efficacy for the AAV gene delivery system. Efforts aimed at engineering vectors with the ability to evade the immune response include identification of residues on the virus capsid important for these interactions and changing them. Here K531 is identified as the determinant of monoclonal antibody ADK6 recognition by AAV6, and not the closely related AAV1. The AAV6-ADK6 complex structure was determined by cryo-electron microscopy and the footprint confirmed by cell-based assays. The ADK6 footprint overlaps previously identified AAV antigenic regions and neutralizes by blocking essential cell surface glycan attachment sites. This study thus expands the available repertoire of AAV-antibody information that can guide the design of host immune escaping AAV vectors able to maintain
Keywords: Adeno-associated virus, AAV, antibody, receptor, neutralization, Parvoviruses, viral vectors, epitope, antigenic footprint
INTRODUCTION
The Adeno-associated viruses (AAVs), small single stranded DNA (ssDNA) viruses with a diameter of ~260 Å, belong to the Dependoparvovirus genus of the Parvovoridae (Agbandje-McKenna and Chapman, 2006; Chapman and Agbandje-McKenna, 2006). The AAV capsid is assembled with a T=1 icosahedral architecture (Agbandje-McKenna and Chapman, 2006; Chapman and Agbandje-McKenna, 2006) from 60 copies (in total) of three overlapping capsid viral proteins (VPs), VP1, VP2, and VP3, coded from the cap open reading frame by use of alternative transcripts and start sites. The VP sequences are stochastically incorporated into the T=1 capsid in a ratio of 1:1:10 for VP1:VP2:VP3 (Buller and Rose, 1978; Rose et al., 1971; Snijder et al., 2014).
The AAVs exhibit several features which make them attractive as gene delivery vectors: they are non-pathogenic, can package non-genomic ssDNA and self-complementary DNA (scDNA) sequences, can infect dividing and non-dividing cells, and exhibit long-term gene expression (Buning et al., 2004). Currently, AAVs represent ~10% of gene therapy clinical trials worldwide (ClinicalTrials.gov) and are being developed for the treatment of several different monogenic diseases, including cystic fibrosis, hemophilia, and muscular dystrophy (Mendell et al., 2009; Nathwani et al., 2011; Wagner et al., 1999). Significantly, an AAV serotype 1 (AAV1) based vector packaging the lipoprotein lipase gene is the only approved gene therapy treatment marketed under the name Glybera™ (Gaudet et al., 2012). However, although the AAV vector system shows significant promise for gene delivery, one of the limitations is the pre-existing B-cell immunity against different AAV serotypes observed in the general population (Hurlbut et al., 2010; Li et al., 2012; Manno et al., 2006; Zadori et al., 2001). Epidemiological studies report a prevalence of 40–70% seropositivity for AAV in people depending on serotype (Boutin et al., 2010). This antibody response has the ability to neutralize AAV vectors when tested in vitro and in vivo, and eligibility for inclusion in recent clinical trials requires that patients be naïve for anti-AAV antibodies (Nathwani et al., 2011). Thus, significant effort is extended to overcoming the effects of this response. This includes the isolation of novel AAV sequences from non-human primates, other mammals, and non-mammalian species, the use of directed evolution in the presence of antibodies, and rational design of capsid variants guided by information on antigenic footprints (reviewed in (Tseng and Agbandje-McKenna, 2014)). In all these efforts, there is a need to preserve the ability of the vectors to infect and transduce the target/desired cells. Thus it is important that functionally mapped regions, including receptor binding sites and transduction determinants, which often overlap with antigenic footprints, are minimally modified in the engineered variants (reviewed in (Tseng et al., 2015).
There are currently over 150 genomic sequences of AAV isolated from humans, non-human primates, and other mammalian species. Thirteen “serotypes” have been described for that represent the range of sequence identity between these viruses (Gao et al., 2004; Gao et al., 2002; Mori et al., 2008; Schmidt et al., 2008). The AAVs are divided onto different groups (group A to F) and clonal isolates (AAV4 and AAV5) based on sequence identities that tends to trend with tissue tropism (Gao et al., 2004). The closely related AAV1 and AAV6, which have ~98% sequence identity, are the representative members of group A. These viruses differ by only 6 out of 736 amino acids in their VPI sequence, with one of the differences (residue 129) located in VP1u (the unique sequence of VP1, not present in VP 2 or VP3), and the remaining five located within the VP3 common region (Wu et al., 2006). Previously, structures determined for AAV1 and AAV6, in which only the VP3 region is ordered, showed two of the differing residues (418 and 642) were located in the interior surface of the capsid while the remaining three residues (531, 584, and 598) were located on the exterior capsid surface (Ng et al., 2010). All five VP3 residues are clustered around the icosahedral 3-fold axis of the capsid associated with receptor attachment and/or transduction determinants for several AAVs (Huang et al., 2016). Importantly, comparative analysis of single amino acid variants in AAV1 and AAV6 at the 6 positions, identified K531 as a determinant of heparan sulfate (HS) receptor binding and liver tropism, and while other studies identified K531 as a transduction determinant when combined with AAV1’s L129 (Limberis et al., 2009; Wu et al., 2006). Residues involved in sialic acid (SIA) receptor binding by AAV1 and AAV6 are identical and have been mapped to the base of the protrusions surrounding the 3-fold axis (Huang et al., 2016).
Several capsid antibodies generated against AAV1 cross-react with AAV6 and capsid-antibody complex structures have shown similar footprints on the AAV1 and AAV6 capsids (Kuck et al., 2007); (Gurda et al., 2012). One of these antibodies, ADK1a, which neutralizes both AAV1 and AAV6, has a footprint overlapping with the SIA binding site suggesting a mechanism of neutralization. However, ADK6, generated against the AAV6 capsid, does not recognize AAV1 (Sonntag et al., 2011). In this study, the determinant of the selective reactivity of AAV6 to ADK6 was identified by cryo-electron microscopy (cryo-EM) and image reconstruction (cryo-reconstruction). This was confirmed with site-directed mutagenesis followed by native immunoblots and in vitro neutralization assays. The goal was to determine which of the three AAV1/AAV6 capsid surface amino acids (E531K, F584L, A598V; AAV/AAV6 residue types (Ng et al., 2010)) conferred this selective recognition. Residue 531 was identified as the determinant of this differential recognition despite the ADK6 footprint covering all three AAV1/6 differing residues and containing residues which overlap with the previously mapped footprints for anti-AAV1 capsid antibodies ((Tseng et al., 2015). Significantly, as mentioned above, K531 is the determinant of HS binding by AAV6, a phenotype that is absent in AAV1, and dictates liver tropism (Wu et al., 2006). The ADK6 footprint also overlapped the previously mapped common SIA binding site residues, N447, S472, V473, N500, T502, and W503, for AAV1 and AAV6. These observations indicate a block of both HS and SIA binding as the mechanism of ADK6 neutralization of AAV6. This study thus provides residue level information that can aid future efforts to engineer AAV1 and AAV6 vectors with desired tissue tropism and antibody escape properties.
MATERIALS AND METHODS
Production and purification of AAV6 virus-like particles
The production and purification of AAV6 virus-like particles (VLPs) using the Baculovirus/SF9 expression system has been previously described (DiMattia et al., 2005; Miller et al., 2006; Ng et al., 2010). Briefly, a baculovirus packaging a gene containing the DNA for expressing the AAV6 VP2 and VP3 was made using the Bac-to-Bac system according to the manufacturer’s instructions (Invitrogen) and used to infect SF9 cells. The cell pellet from the infection was resuspended in TNTM buffer (25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.2% Triton X-100, 2 mM MgCl2 ), freeze/thawed 3X with Benzonase (Promega) treatment at 37°C after the second thaw, and clarified by centrifugation at 10,000 rpm in a JA-20 rotor at 4°C for 20 min. The supernatant was loaded on a 20% sucrose cushion (w/v of sucrose in TNTM buffer) and the sample centrifuged at 45,000 rpm on a Ti70 rotor at 4°C for 3 hr. The supernatant was discarded and the pellet was resuspended in TNTM and left stirring overnight at 4°C. The resuspended pellet was loaded onto a 5–40% (w/v) sucrose step gradient and the sample centrifuged at 35,000 rpm in an SW41 rotor for at 4°C for 3 hr. The VLP containing fraction were collected by fractionation, dialyzed into Buffer A (20 mM Tris-HCl, pH 8.5, 15 mM NaCl) and the sample further purified by ion exchange chromatography before use.
A 1 ml anion exchange (Q column, GE Healthcare) was equilibrated with Buffer A and Buffer B (20 mM Tris-HCl, pH 8.5, 500 mM NaCl). The sample was loaded on the column at a flow rate of 0.5 ml/min, the column was washed with 10 column volume (CV) of Buffer A, and the sample was eluted with a 5 CV gradient from 0 – 100% Buffer B (Zolotukhin et al., 2002). Five hundred microliter fractions were collected and screened to identify the fractions containing AAV6 VP. These fractions were pooled, buffer exchanged into PBS, concentrated to 1 mg/ml, and analyzed by SDS PAGE and negative stain electron microscopy (EM) to check for purify and capsid integrity, respectively.
Purification of ADK6 IgG antibodies
The ADK6 immunoglobulin G (IgG) antibody was produced by the University of Florida Hybridoma Core Lab as previously described (Kuck et al., 2007; Tseng et al., 2016). The ADK6 hybridoma supernatant was diluted 1:5 with PBS and loaded onto a 1 ml HiTrap protein G HP column (GE Healthcare), washed with 10 CV of PBS, eluted with 0.5 ml of Glycine-HCl at pH 2.7, and neutralized with 50 μl of neutralization buffer (1 M Tris-HCl pH 10). The purified IgG was buffer exchanged into 20 mM Sodium Phosphate pH 7.0, 10 mM EDTA, and concentrated for papain cleavage.
ADK6 fragment antibody (FAb) generation and purification
Cysteine HCl was added to the papain digestion buffer (20 mM sodium phosphate pH 7.0, 10 mM EDTA) immediately prior to use. Concentrated IgGs were incubated with immobilized papain (Pierce) at an enzyme:substrate ratio of 1:160 at 37°C for 16 h. An equal volume of papain stop buffer (10 mM Tris-HCl pH 7.5) was added to stop the cleavage process and the mixture was centrifuged at 1500 × g for 2 min to separate the sample from the immobilized papain. The FAbs were separated from the undigested IgG and fragment crystallizable (Fc) fragments on a HiTrap Protein A column (GE Healthcare). The FAbs were collected in the wash and flowthrough fractions, buffer exchanged into PBS, and concentrated for use.
AAV6-ADK6 FAb complex formation
AAV6 VLPs at a concentration of 1 mg/ml and ADK6 FAbs at a concentration of 1 mg/ml were mixed at a molar ratio of 1:1 and 2:1 FAb:VP binding site, and the mixture was incubated at 4°C for 1 h. The complexes were examined by negative stain EM on a Spirit microscope (FEI) to confirm capsid decoration by FAbs prior to vitrification for cryo-EM data collection.
AAV6-ADK6 FAb complex cryo-EM data collection
Three microliters of the AAV6-ADK6 FAb complex mixture were loaded onto C-Flat holey carbon grids (CF-2/2-4C-50, Protochips Inc.) that were glow discharged for 1 min prior to use, and vitrified by plunge freezing into liquefied ethane in a Vitrobot Mark IV (FEI). The frozen grids were transferred to liquid nitrogen and then into a FEI Technai TF20 transmission electron microscope operating at 200kV. Cryo micrographs were collected using a defocus range of 2.5–3.0 μm and total dosage of 20e−/Å2 per image. Thirty-six micrographs were collected with a Gatan Ultra Scan 4000 CCD camera at a step size of 1.82 Å/pixel.
Cryo-EM and image reconstruction of the AAV6-ADK6 FAb complex
The RobEM software package (http://cryoEM.ucsd.edu/programs.shtm) was used to extract decorated AAV6 VLPs (complexed) particles from each micrograph. The defocus level for each micrograph was estimated using the CTFFIND3 application (Mindell and Grigorieff, 2003) incorporated into the AUTO3DEM application (Yan et al., 2007a; Yan et al., 2007b). Preprocessing of the selected particles to remove blemishes, correct linear gradient, normalize, and apodize the images used the “autopp” subroutine within the AUTO3DEM software package and an initial model, at ~25 Å resolution, was generated for searching and initiating the refinement of each particle origin and orientation using the same application (Yan et al., 2007a). Following an initial 10 cycles of search and refinement, the data set was “re-boxed” and “re-centered” using the refined particle center and orientation information, and the images were corrected to compensate for the effect of phase reversal in the contrast transfer function (CTF) followed by additional cycles of refinement also within AUTO3DEM (Yan et al., 2007b). The final resolution was determined by the Fourier shell correlation (FSC) threshold of 0.5 (van Heel and Schatz, 2005). The images of the reconstructed map were illustrated using the Chimera software package (Pettersen et al., 2004).
Pseudo-atomic model fitting and identification of the ADK6 antibody footprint
The AAV6 60mer VP3 capsid coordinates were generated by icosahedral matrix multiplication using the Oligomer generator subroutine using the Viperdb online server (http://viperdb.scripps.edu/) (Carrillo-Tripp et al., 2009) from the AAV6 crystal structure (RCSB PDB ID no: 3OAH). The coordinates were fitted into the cryo-EM reconstructed complex density map by rigid body rotations and translations using the Chimera program (Pettersen et al., 2004). This 60mer docked with a correlation coefficient (CC) of 0.94. To enable model building into the FAb density, a difference map, subtracting a scaled density map generated for the docked 60mer model from the AAV6-ADK6 complex map, was generated. A generic FAb structure (PDB ID no: 2FBJ) was fitted into the resulting positive difference density map representing the FAb interacting with the reference AAV6 VP3 monomer (chain A) using the Chimera program (Pettersen et al., 2004). The coordinates for the reference monomer was extracted from the docked 60mer and together with the docked Fab model was used to generate a 60mer using Viperdb (http://viperdb.scripps.edu/) (Carrillo-Tripp et al., 2009). This complex 60mer was then re-docked into the reconstructed complex density map and the CC was similar at 0.93. To determine the interacting residues between the AAV6 capsid and ADK6 FAbs the PDBePISA (http://www.ebi.ac.uk/msd-srv/prot_int/) (Krissinel and Henrick, 2007) and COOT (Emsley et al., 2010) software packages were used. Images for the co-ordinates of the fitted complex were generated using the PyMol program (http://www.pymol.org/) (Schrödinger, 2017).
Recombinant wild-type and variant AAV1 and AAV6 vector production and purification
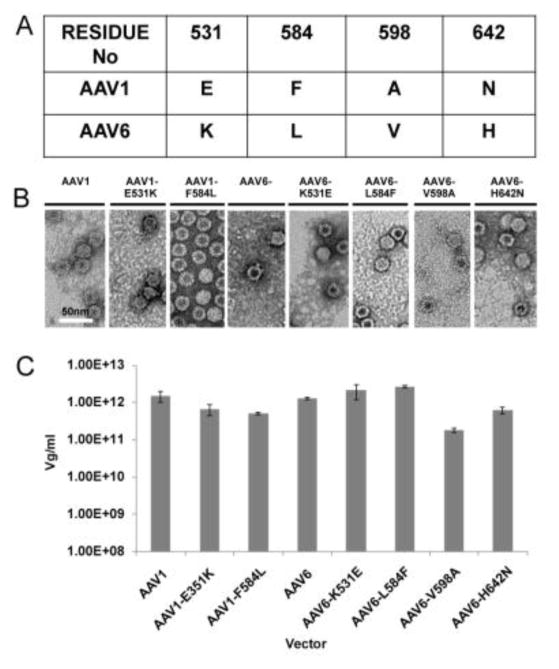
To identify the critical residue determinant for ADK6 recognition by AAV6 (and not AAV1) recombinant wild-type (WT) AAV1 (rAAV1) and AAV6 (rAAV6), and reciprocal single site-directed variants, at the equivalent capsid surface amino acid positions 531 and 584, K and F in AAV6 and E and L in AAV1, respectively, located within the ADK6 footprint, and AAV6-V598A were made for testing by native immunoblot and infectivity in the presence of ADK6. As a negative control of a differing non-footprint residue (interior capsid surface) and control for AAV6 capsid assembly in the presence of the amino acid substitution, the reciprocal AAV6-H642N variant was also created for testing. These variants were made as previously described in the pXR1 and pXR6 backgrounds, for AAV1 and AAV6, respectively (Wu et al., 2006).
To produce WT and variant rAAV1 and rAAV6 vectors, monolayers of HEK293 cells, at 70% confluency, were triply transfected with 18μg of WT and mutant pXRAAV1 and pXRAAV6 plasmids, 18μg of pTR-UF3-Luciferase (luciferase gene between AAV2 inverted terminal repeats), and 54μg of the helper plasmid pXX6, with 190μl of Polyethyleneimine (1 mg/ml) per 15 cm plate. Ten 15 cm plates were transfected for each vector followed by incubation at 37°C for 72 h in the presence of 5% CO2. Post transfection, the cells were harvested and centrifuged at 1100 × g for 15 min. The supernatant for each vector was precipitated with 10% PEG 8000 (Fisher) and the cell pellets were resuspended in 10 ml TD buffer (1X PBS, 5 mM MgCl2 and 2.5 mM KCl, pH7.4) and freeze/thawed 3X to release virus from the cells. Both the PEG precipitated virus resuspended in TD buffer and the resuspended cell lysate were Benzonase (Novagen) treated at 37°C for 1 h followed by clarification by centrifugation at 10,000 rpm in a JA-20 rotor at 4°C for 20 min. The genome containing vectors were separated from the empty capsids by a step Iodixanol gradient (Zolotukhin et al., 2002). In brief, the clarified supernatants were loaded onto a 15–60% step iodixanol gradient. The 40/60% interface or vector containing fractions was collected and diluted with 10×TD buffer. The genome containing vectors were further purified by AVB (Thermo Fisher) affinity column chromatography (Mietzsch et al., 2014). A 1ml AVB column was equilibrated with 10ml TD buffer, and the diluted vector containing fractions were loaded at 1ml/min. The purified vector was eluted with 10 ml or 10 CV elution buffer (0.1M Na Acetate pH=2.5 and 0.75M NaCl at 0.5ml fractions). The elution fractions were diluted with 50μl neutralization buffer (1M Tris-HCl pH=10). The purified vectors were buffer exchanged into PBS buffer and quantified by UV spectrometry analysis and qPCR.
Native immunoblots
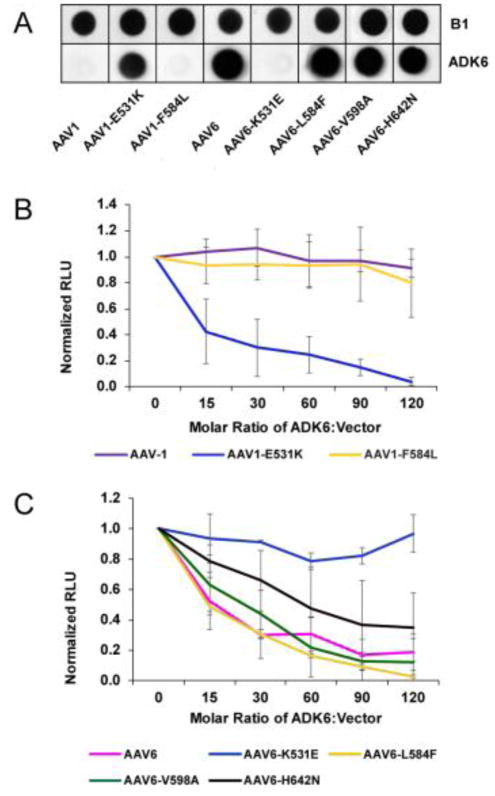
To confirm the interaction of AAV6 with ADK6 and the lack of recognition of AAV1 by the antibody, and to further delineate the specific residue(s) important for the capsid antibody binding, the purified rAAV1 and rAAV6 vectors and the single residue variants, rAAV1-E531K, rAAV1-F584L, rAAV6-K531E, rAAV6-L584F, rAAV6-H642N vectors (Fig. 2A) were probed by native immunoblot with ADK6. One hundred ng of purified intact vectors were loaded on a nitrocellulose membrane. The membrane was blocked with 5% milk (w/v) in PBS and 0.05%Tween (1 h), followed by 1 h incubation with ADK6 (0.5 mg/ml) diluted 1:3000. The rAAV-ADK6 complexes were probed with the Horse Radish Peroxidase (HRP) anti-mouse secondary antibody diluted 1:5000. The membrane was finally probed with chemiluminescent and detected on a Kodak film. The film image was documented with a Gel Doc (Biorad). As a positive control 100ng of rAAV vectors were boiled at 100°C for 5 min and the sample loaded on a nitrocellulose membrane. The sample was then probed with the B1 antibody that recognizes the C-terminus of the rAAV1 and rAAV6 VP. This C-terminus epitope is only available when the capsid is denatured (Wobus et al., 2000).
Figure 2.
Production and purification of WT and variant AAV1 and AAV6 vectors. A) Residue positions and type for surface amino acids plus internal residue 642 which differ between AAV1 and AAV6. B) Negative stained EM of WT and variant AAV1 and AAV6 vectors. The scale bar is shown in white on the first EM image. C) Quantitation of purified vector genome titer determined by qPCR for WT and variant AAV1 and AAV6 vectors.
Neutralization assays
To determine if binding of ADK6 to the WT and variant rAAV vectors is neutralizing in vitro, purified vectors were used to infect HEK293T cells in the presence of the antibody as previously described (Tseng et al., 2015). Briefly, HEK293T cells were seeded in 96 well plates at 1.25×104 cells/well for 24 h to reach 70% confluency. The purified WT and mutant vectors (2.5×109 viral genome (vg)) were incubated at virus particle:ADK6 IgG molecular ratios of 1:0, 1:15, 1:30, 1:60, 1:90 and 1:120 in PBS in a final volume of 30 μl in unsupplemented DMEM (Gibco) at 37°C for 1 h. After this incubation period, the media was aspirated from the cells, the complex sample was added to 70 μl of DMEM supplemented with 10% FBS and 1% Antibiotic and antimytotic (ABAM), and the mixture was added to the cells. The cells were incubated at 37°C for 48 h in the presence of 5% CO2. The cells were harvested, washed with PBS, lysed, and transduction level determined by the Luciferase Assay System (Promega) according to the manufacturer’s instruction.
RESULTS and DISCUSSION
Cryo-reconstruction of the AAV6-ADK6 complex
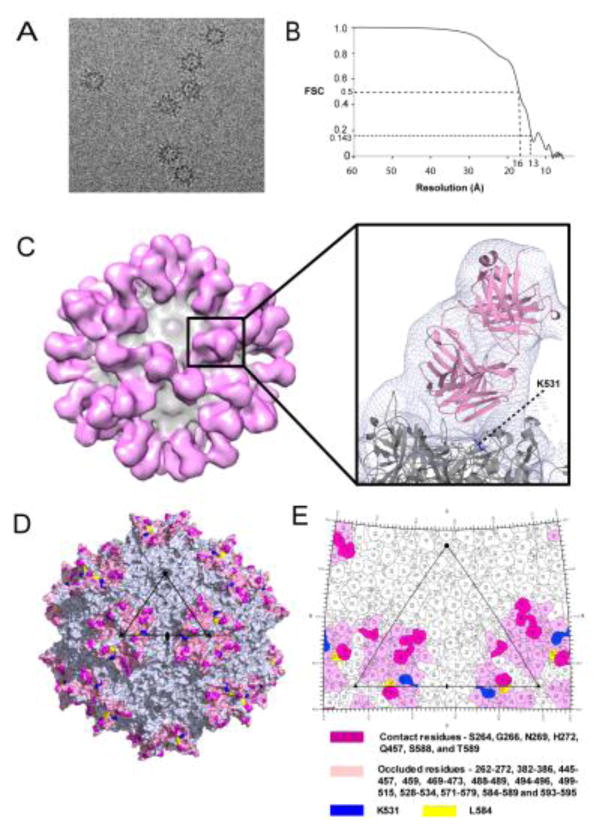
The cryo-EM reconstruction of the AAV6:ADK6 FAb complex, determined to 16 Å resolution (FSC 0.5), shows ADK6 FAbs bound to the side of the 3-fold protrusion across the 2-fold axis of the AAV6 capsid surface suggesting bivalent binding (Fig. 1A–C). Consistent with this observation, a complex with an intact ADK6 IgG (un-cleaved) showed a similar structure to the FAb alone with no ordered density for the Fc (data not shown). The ADK6 binding site on the AAV6 capsid is within a common antigenic site utilized by the other AAVs, the 3-fold protrusions and the 2/5-fold wall (Tseng et al., 2015). These epitopes are regions of high sequence and structural diversity and shown to be important for receptor recognition and transduction by the AAVs. The predicted contact residues between AAV6 and the ADK6 FAb in the pseudo-atomic model fitted into the reconstructed density map, with CC of 0.93, are shown in (Fig. 1C–E).
Figure 1.
Cryo-EM and image reconstruction of the AAV6-ADK6 FAb complex. A) Example cryo-electron micrograph of AAV6-ADK6 capsid-FAb complex. Projections from the capsid surface indicate FAb decoration. B) Fourier Shell Correlation plotted against resolution (FSC plot) for the AAV6-ADK6 structure. The estimated resolution is 16 and 13 Å at FSC 0.5 and 0.143, respectively. C) Surface density representation of the cryo-reconstructed AAAV6-ADK6 FAb complex structure viewed along the icosahedral 2-fold axis. The capsid density is colored gray and the FAb colored light pink. Right: Zoomed in image of the complex density map and docked pseudo-atomic model of the capsid (grey) and FAb (pink). Residue K531 is shown in blue and labeled. D) AAV6 capsid surface image (in gray) with FAb predicted contact residues in hot pink, occluded residues (as defined in the text) in light pink, and K531 and L584, within the occluded region, in blue (basic) and yellow (hydrophobic), respectively. The viral asymmetric unit, bounded by a 5-fold and two 3-fold axes intercepted by a 2-fold axis, is depicted by a large black triangle. The 2-, 3-, and 5-fold axes are represented by an oval, triangle, and pentagon, respectively. E) 2D “roadmap” projection of the residues within a viral asymmetric unit. The residues are colored as in (D). The images were generated by the Chimera (Pettersen et al., 2004), PyMol (Schrödinger, 2017), and RIVEM programs (Xiao and Rossmann, 2007) for panels C, D, and E, respectively.
The AAV6 capsid surface residues which make contact with the docked ADK6 FAb model residues identified by the PDBePISA program are S264, G266, N269, H272, Q457, S588, and T589. Residues S264, G266, N269, and H272 are located on one 3-fold symmetry related monomer, Q457 on a second 3-fold related monomer, and residues S588 and T589 were located on the third 3-fold related monomer. Additionally, the ADK6 Fab footprint occludes other residues surrounding the model predicted contact residues: 262–272, 382–386, 445–457, 459, 469–473, 488–489, 494–496, 499–515, 528–534, 571–579, 584–589 and 593–595. Significantly, two of the residues that differ between AAV1 and AAV6, 531 and 584, are located in the occluded region (Fig. 1D and E). Only AAV6, but not AAV1, binds to ADK6 (Sonntag et al., 2011). Thus, the footprint implied that the specificity of the AAV6:ADK6 interaction is dictated by 531, 584 or both.
Reciprocal AAV1 and AAV6 vectors are comparable to wild–type in capsid assembly, genome titer, and transduction efficiency
The AAV1 and AAV6 variants purified by AVB column chromatography showed assembled capsids when visualized by negative stain EM (Fig. 2B), and had packaged genome titers in the 1010 – 1013 vg/ml range, which is comparable to WT virus (Fig. 2C). The AAV6-H642N variant, with a change on the interior surface of the capsid, also assembled capsids and packaged genome at levels comparable to WT. The transduction phenotype of the WT and variants were compared in the absence of antibody. The AAV1 variants had transduction efficiencies of ~160% compared to the WT virus while AAV6 variants were in the range ~95 (for AAV6-V598A) to 160% (for AAV6-L584F). The transduction efficiency of AAV6-K531E was ~120% compared to WT AAV6. These observations confirmed that the mutations made had no significant effect on capsid assembly, genome packaging or transduction efficiency.
AAV6 K531 is responsible for ADK6 recognition
Immunoblots of AAV1 and AAV6 with ADK6 confirmed the recognition of AAV6 by ADK6 and the escape by AAV1 (Fig. 3A). Importantly, ADK6 recognizes variant AAV1-E531K, with an E to K change switching the AAV1 residue type to AAV6, but not AAV1-F584 with an L to F switch at the second occluded position. This observation identified AAV6-K531 as being the determinant of ADK6 recognition. Consistent with this conclusion, ADK6 recognizes AAV6-L584F and AAV6-H642N and not AAV6-K531E (Fig. 3A). AAV1 and AAV1-F584L escape from ADK6 while AAV1-E531K is neutralized by ADK6 at a molar ratio of ~15 FAb molecules per capsid (Fig. 3B). This is only 25% binding site occupancy. ADK6 results in 50% inhibition of transduction for AAV6 and AAV6-L584F, also at a molar ratio of ~15 FAb molecules per capsid, AAV6-V598A with ~20 FAb molecules, and AAV6-H642N with ~60 FAb molecules (Fig. 3C). On the other hand, AAV6-K531E escapes antibody recognition at up to a molar ratio of 120 FAb molecules per capsid, a saturation of 2 FAb molecules per VP binding site (Fig. 3C). These observations confirm the role of K531 as the determinant of the specificity of ADK6 for AAV6 and highlights the important contributions of capsid residues that may not make direct contact with FAb residues but are part of the footprint in virus-antibody interactions.
Figure 3.
ADK6 binding and neutralization assays. A) Immunoblot of denatured WT and variant AAV1 and AAV6 vectors detected by B1 (which recognizes a linear epitope of the C-terminus of the VP1/2/3 common region) (top) and by ADK6 (which recognizes native capsids). ADK6 interacts specifically with AAV6, and this interaction is lost in mutant AAV6-K531E and restored in AAV1-E531K. B and C) Neutralization assays for WT and variants of AAV1 and AAV6, respectively, in the presence of ADK6. Transduction is normalized to the luciferase signal for WT virus at zero antibody concentration.
ADK6 binding is predicted to sterically hinder AAV6 glycan binding and has a footprint that overlaps previously defined epitopes
AAV6 is a dual glycan receptor binding serotype that utilizes both HS and SIA for cellular infection (Huang et al., 2016; Wu et al., 2006). The ADK6 footprint covers a large area of the AAV6 capsid surface including the previously structurally mapped SIA glycan receptor binding site in addition to K531 reported to be responsible for its HS binding (Fig. 4A and 4B) (Huang et al., 2013; Wu et al., 2006). The ability of ADK6 to block transduction by AAV6 indicates that ADK6 neutralizes AAV6 infection at the entry step, likely by steric hindrance of both glycan interactions. The block of SIA interaction is similar to the mechanism proposed for ADK1a neutralization of AAV1 and AAV6 which shares regions of the ADK6 footprint at the top of the 3-fold region (Huang et al., 2016; Tseng and Agbandje-McKenna, 2014). This mechanism is different to those reported for the A20 neutralization of AAV2 and ADK8 neutralization of AAV8 (Gurda et al., 2012; Huttner et al., 2003; McCraw et al., 2012; Tseng et al., 2015). These antibodies are proposed to neutralize at a post-entry step, with A20 acting in the nucleus and ADK8 blocking nuclear entry and resulting in perinuclear accumulation. Importantly, while an AAV8 cell surface glycan receptor is unknown, AAV2 binds its HS receptor at the 3-fold axis, a region disparate from the A20 footprint located at the 2/-5-fold wall (McCraw et al., 2012).
Figure 4.
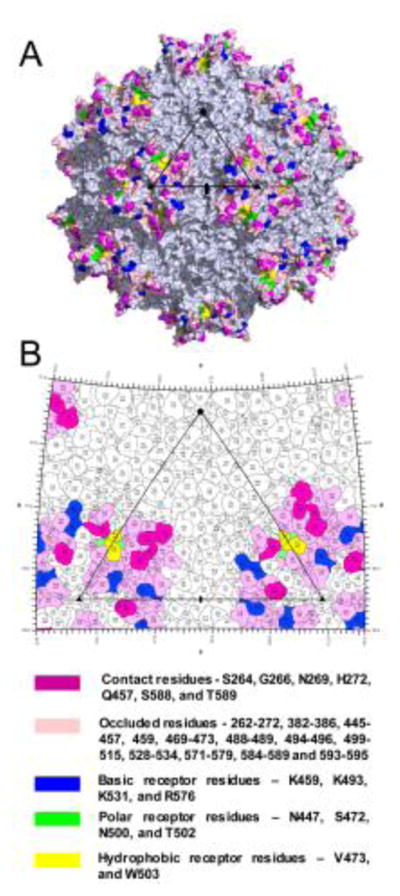
The functional surface of AAV6. A and B) Capsid surface and 2D “roadmap” projection of AAV6, respectively. The SIA and HS receptor binding residues are colored blue, green, and yellow for basic, polar, and hydrophobic residues, respectively. The remaining residue colors are as indicated below B). The viral asymmetric unit are depicted as in figure 1. The images for panels A and B were generated in the PyMol (Schrödinger, 2017) and RIVEM programs (Xiao and Rossmann, 2007), respectively.
The ADK6 footprint on AAV6 overlaps that previously mapped for other AAV-antibody interactions, including AAV8-ADK8, AAV1-ADK1a, and regions of AAV1-5H7, AAV6-5H7, and AAV2-A20 on the 3-fold protrusions and the 2/5-fold walls (Gurda et al., 2012; Tseng and Agbandje-McKenna, 2014; Tseng et al., 2015; Tseng et al., 2016). This structure thus adds to the library of antigenic footprint information being accumulated for the AAVs and will inform the engineering of a generation of AAV vectors with the ability to evade pre-existing host immune responses during the vector re-administration.
Summary
This study, using a combination of structure, site directed mutagenesis, and cell binding assays, identified a single residue, K531, as conferring antigenic selectivity between the closely related AAV1 and AAV6. Significantly, repeat administration of the approved AAV1 based lipoprotein lipase gene vector treatment will require the use of an antigenic variant with similar transduction properties. The observation that position 531 of AAV1/6 is able to provide immune escape properties is information that can inform AAV1 engineering for future use in these patients as well as the development of additional AAV1 and AAV6 vectors.
Highlights.
Reciprocal vectors generated for AAV1 and AAV6, which differ by 6 residues, are comparable to the wild-type vectors in capsid assembly, genome titer, and transduction efficiency.
The ADK6 footprint on the AAV6 capsid, revealed by cryo-EM and confirmed by mutagenesis and cellular transduction assays, identified a single residue, K531, as the determinant of ADK6 recognition.
ADK6 binding is predicted to sterically hinder AAV6 glycan binding and has a footprint that overlaps previously defined antibody epitopes.
Acknowledgments
We thank the University of Florida (UF) Interdisciplinary Center for Biotechnology Research (ICBR) electron microscopy lab for providing negative-stain EM services. This work was supported by NIH R01 GM082946 to R.M and MAM.
Footnotes
Conflict of interest statement.
M. Agbandje-McKenna (MAM) is a SAB member for Voyager Therapeutics, Inc., and AGTC, has a sponsored research agreement with AGTC and Voyager Therapeutics, and is a consultant for Intima Biociences, Inc. MAM is a co-founder of StrideBio, Inc. This is a biopharmaceutical company with interest in developing AAV vectors for gene delivery application. Jude Samulski is the scientific founder of Bamboo Therapeutics, Asklepios Biopharmaceutics, Chatham Therapeutics, and Merlin. These companies also have interest in the development of AAV for gene delivery applications.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agbandje-McKenna M, Chapman MS. Structure-function relationships. In: Bloom ME, SFC, Linden RM, Parrish CR, Kerr JR, editors. Parvoviruses. Edward Arnold, Ltd; London: 2006. pp. 125–139. [Google Scholar]
- Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- Buller RM, Rose JA. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buning H, Braun-Falco M, Hallek M. Progress in the use of adeno-associated viral vectors for gene therapy. Cells Tissues Organs. 2004;177:139–150. doi: 10.1159/000079988. [DOI] [PubMed] [Google Scholar]
- Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL, 3rd, Reddy VS. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MS, Agbandje-McKenna M. Atomic structure of viral particles. In: Bloom ME, SFC, Linden RM, Parrish CR, Kerr JR, editors. Parvoviruses. Edward Arnold, Ltd; London: 2006. pp. 109–123. [Google Scholar]
- DiMattia M, Govindasamy L, Levy HC, Gurda-Whitaker B, Kalina A, Kohlbrenner E, Chiorini JA, McKenna R, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. Production, purification, crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 5. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:917–921. doi: 10.1107/S1744309105028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet D, Methot J, Kastelein J. Gene therapy for lipoprotein lipase deficiency. Curr Opin Lipidol. 2012;23:310–320. doi: 10.1097/MOL.0b013e3283555a7e. [DOI] [PubMed] [Google Scholar]
- Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LY, Patel A, Ng R, Miller EB, Halder S, McKenna R, Asokan A, Agbandje-McKenna M. Characterization of the Adeno-Associated Virus 1 and 6 Sialic Acid Binding Site. J Virol. 2016;90:5219–5230. doi: 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hartley AV, Yin Y, Herskowitz JH, Lah JJ, Ressler KJ. AAV2 production with optimized N/P ratio and PEI-mediated transfection results in low toxicity and high titer for in vitro and in vivo applications. J Virol Methods. 2013;193:270–277. doi: 10.1016/j.jviromet.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut GD, Ziegler RJ, Nietupski JB, Foley JW, Woodworth LA, Meyers E, Bercury SD, Pande NN, Souza DW, Bree MP, Lukason MJ, Marshall J, Cheng SH, Scheule RK. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol Ther. 2010;18:1983–1994. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner NA, Girod A, Perabo L, Edbauer D, Kleinschmidt JA, Buning H, Hallek M. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 2003;10:2139–2147. doi: 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kuck D, Kern A, Kleinschmidt JA. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J Virol Methods. 2007;140:17–24. doi: 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Li C, Narkbunnam N, Samulski RJ, Asokan A, Hu G, Jacobson LJ, Manco-Johnson MJ, Monahan PE. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther. 2012;19:288–294. doi: 10.1038/gt.2011.90. [DOI] [PubMed] [Google Scholar]
- Limberis MP, Vandenberghe LH, Zhang L, Pickles RJ, Wilson JM. Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol Ther. 2009;17:294–301. doi: 10.1038/mt.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, Ozelo MC, Hoots K, Blatt P, Konkle B, Dake M, Kaye R, Razavi M, Zajko A, Zehnder J, Rustagi PK, Nakai H, Chew A, Leonard D, Wright JF, Lessard RR, Sommer JM, Tigges M, Sabatino D, Luk A, Jiang H, Mingozzi F, Couto L, Ertl HC, High KA, Kay MA. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- McCraw DM, O’Donnell JK, Taylor KA, Stagg SM, Chapman MS. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology. 2012;431:40–49. doi: 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Rodino-Klapac LR, Rosales-Quintero X, Kota J, Coley BD, Galloway G, Craenen JM, Lewis S, Malik V, Shilling C, Byrne BJ, Conlon T, Campbell KJ, Bremer WG, Viollet L, Walker CM, Sahenk Z, Clark KR. Limb-girdle muscular dystrophy type 2D gene therapy restores alpha-sarcoglycan and associated proteins. Ann Neurol. 2009;66:290–297. doi: 10.1002/ana.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietzsch M, Grasse S, Zurawski C, Weger S, Bennett A, Agbandje-McKenna M, Muzyczka N, Zolotukhin S, Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EB, Gurda-Whitaker B, Govindasamy L, McKenna R, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:1271–1274. doi: 10.1107/S1744309106048184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mori S, Takeuchi T, Enomoto Y, Kondo K, Sato K, Ono F, Sata T, Kanda T. Tissue distribution of cynomolgus adeno-associated viruses AAV10, AAV11, and AAVcy.7 in naturally infected monkeys. Arch Virol. 2008;153:375–380. doi: 10.1007/s00705-007-1097-8. [DOI] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O’Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol. 2010;84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rose JA, Maizel JV, Jr, Inman JK, Shatkin AJ. Structural proteins of adenovirus-associated viruses. J Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Voutetakis A, Afione S, Zheng C, Mandikian D, Chiorini JA. Adeno-associated virus type 12 (AAV12): a novel AAV serotype with sialic acid- and heparan sulfate proteoglycan-independent transduction activity. J Virol. 2008;82:1399–1406. doi: 10.1128/JVI.02012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System, Version 2.0. 2017. [Google Scholar]
- Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJ. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J Am Chem Soc. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol. 2011;85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YS, Agbandje-McKenna M. Mapping the AAV Capsid Host Antibody Response toward the Development of Second Generation Gene Delivery Vectors. Front Immunol. 2014;5:9. doi: 10.3389/fimmu.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YS, Gurda BL, Chipman P, McKenna R, Afione S, Chiorini JA, Muzyczka N, Olson NH, Baker TS, Kleinschmidt J, Agbandje-McKenna M. Adeno-associated virus serotype 1 (AAV1)- and AAV5-antibody complex structures reveal evolutionary commonalities in parvovirus antigenic reactivity. J Virol. 2015;89:1794–1808. doi: 10.1128/JVI.02710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YS, Vliet KV, Rao L, McKenna R, Byrne BJ, Asokan A, Agbandje-McKenna M. Generation and characterization of anti-Adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies. J Virol Methods. 2016;236:105–110. doi: 10.1016/j.jviromet.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wagner JA, Messner AH, Moran ML, Daifuku R, Kouyama K, Desch JK, Manley S, Norbash AM, Conrad CK, Friborg S, Reynolds T, Guggino WB, Moss RB, Carter BJ, Wine JJ, Flotte TR, Gardner P. Safety and biological efficacy of an adeno-associated virus vector-cystic fibrosis transmembrane regulator (AAV-CFTR) in the cystic fibrosis maxillary sinus. Laryngoscope. 1999;109:266–274. doi: 10.1097/00005537-199902000-00017. [DOI] [PubMed] [Google Scholar]
- Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol. 2000;74:9281–9293. doi: 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M, Samulski RJ. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J Struct Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007a;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM--an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol. 2007b;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P. A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell. 2001;1:291–302. doi: 10.1016/s1534-5807(01)00031-4. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, Flotte TR, Byrne BJ, Snyder RO. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]