Abstract
Interleukin 11 (IL11) is a profibrotic cytokine, secreted by myofibroblasts and damaged epithelial cells. Smooth muscle cells (SMCs) also secrete IL11 under pathological conditions and express the IL11 receptor. Here we examined the effects of SMC-specific, conditional expression of murine IL11 in a transgenic mouse (Il11SMC). Within days of transgene activation, Il11SMC mice developed loose stools and progressive bleeding and rectal prolapse, which was associated with a 65% mortality by two weeks. The bowel of Il11SMC mice was inflamed, fibrotic and had a thickened wall, which was accompanied by activation of ERK and STAT3. In other organs, including the heart, lung, liver, kidney and skin there was a phenotypic spectrum of fibro-inflammation, together with consistent ERK activation. To investigate further the importance of stromal-derived IL11 in the inflammatory bowel phenotype we used a second model with fibroblast-specific expression of IL11, the Il11Fib mouse. This additional model largely phenocopied the Il11SMC bowel phenotype. These data show that IL11 secretion from the stromal niche is sufficient to drive inflammatory bowel disease in mice. Given that IL11 expression in colonic stromal cells predicts anti-TNF therapy failure in patients with ulcerative colitis or Crohn’s disease, we suggest IL11 as a therapeutic target for inflammatory bowel disease.
Introduction
Non-striated smooth muscle cells (SMCs) line the walls of hollow organs and the vasculature. In adults, SMCs are not terminally differentiated and their cellular phenotype remains plastic. A variety of extracellular cues such as humoral factors, mechanical or oxidative stress and cell-cell interactions can induce a spectrum of cellular states ranging from contractile SMCs to highly synthetic and proliferative SMCs [1]. Synthetic SMCs are associated with a wide variety of vascular pathologies such as atherosclerosis or hypertension [1] and other disorders such as asthma [2] and inflammatory bowel disease (IBD) [3]. Many fibro-inflammatory diseases have a component, or are defined by, SMC dysfunction. This is exemplified by systemic sclerosis, which presents with global organ fibrosis and specific vascular abnormalities [4] and is characterized by elevated transforming growth factor beta (TGFB) 2 and interleukin 11 (IL11) expression in dermal stromal cells [5, 6]. This co-occurrence of fibrosis and SMC dysfunction may in part be explained by molecular similarities of the fibrogenic fibroblast-to-myofibroblast conversion and the SMC contractile-to-synthetic phenotype switch. Both these cellular transitions are characterized by extracellular matrix (ECM) production, cell proliferation, invasion and migration. They can also be triggered by the same extracellular cues including TGFB family members [1, 7].
We recently identified IL11 as a critical driver of fibroblast activation in the cardiovascular system, liver and lung downstream of a variety of pro-fibrotic factors including TGFB1 [8–10]. In a study from 1999, IL11 was also found to be secreted by vascular SMCs (VSMCs) in response to pathogenic stimuli, including interleukin 1 alpha (IL1A), TGFB and tumor necrosis factor (TNF) [11]. Although IL11 is upregulated in systemic sclerosis [6], TNF-resistant ulcerative colitis [12, 13] and asthma [14] and despite SMCs being a source of IL11 [11], the effect of IL11 function in SMC biology has not been studied. To address this gap in our knowledge, we generated an inducible Il11 transgenic mouse to overexpress mouse Il11 in myosin heavy chain 11 (Myh11)-positive smooth muscle cells (Il11SMC). Here we characterized key organs that may be affected by SMC pathobiology in Il11SMC mice to better understand the role of SMC-derived IL11.
Materials and methods
Mouse models
This study was carried out in compliance with the recommendations in the Guidelines on the Care and Use of Animals for Scientific Purposes of the National Advisory Committee for Laboratory Animal Research (NACLAR). All experimental procedures were approved (2014/SHS/0925) and conducted in accordance to the SingHealth Institutional Animal Care and Use Committee (IACUC). All mice were from a C57BL/6JN genetic background and were bred and housed in individually vented cages in the same room under ABSL-1 conditions in the SingHealth Experimental Medicine Centre and provided normal chow (Specialty Feeds) and water ad libitum. All research staff involved in animal studies underwent the Responsible Care and Use of Laboratory Animal Course (RCULAC, Singapore) prior to study commencement. Animals were euthanized at endpoint by ketamine (100 mg/kg) and xylazine (10 mg/kg) given IP, followed by the removal of vital organs and tissues.
Mice were scruffed to restrict motion during tamoxifen administration IP and monitored daily for clinical signs of distress and body weights were measured thrice per week upon tamoxifen induction. When rectal inflammation/bleeding was observed, the wound was gently disinfected with 70% methylated spirits and 10% povidone-iodine. Mice that displayed behavioral abnormalities, weight loss, and/or rectal bleeding were therapeutically treated with buprenorphine (0.1 mg/kg SQ) and enrofloxacin (5 mg/kg SQ) where necessary. Animals that did not recover with treatment or presented with deteriorated symptoms including pronounced weight loss (>20% over 1 week or >10% over 24 hours) and the development of rectal prolapse were euthanized following consultation with a veterinarian prior to the study endpoint and were treated as deaths.
Smooth muscle-specific Il11 transgenic model
To direct transgene expression in smooth muscle cells, we crossed the heterozygous Rosa26-Il11 (Gt(ROSA)26Sortm1(CAG-Il11)Cook) mouse [8] to the hemizygous SMMHC-CreERT2 (B6.FVB-Tg(Myh11-cre/ERT2)1Soff/J) mouse [15] available from the Jackson Laboratory (031928 and 019079 respectively) to generate double heterozygous SMMHC-CreERT2:Rosa26-IL11 offspring (referred to here as Il11SMC mice). Only male Il11SMC mice were utilized as the Myh11-Cre/ERT2 transgene is inserted on the Y chromosome. To induce Cre-mediated Il11 transgene induction, six week old Il11SMC mice were intraperitoneal injected with 3 doses of 50 mg kg-1 tamoxifen (tam; T5648, Sigma Aldrich) or an equivalent volume of corn oil vehicle (veh; C8267, Sigma Aldrich) for a week. Single hemizygous SMMHC-CreERT2 littermates were designated as controls (referred to as CreSMC). A total of forty-seven Il11SMC mice (tam-treated n = 35; veh-treated n = 12) and twenty-seven CreSMC mice were used. Individual mice died due to bowel inflammation and bleeding (n = 7) or were humanely euthanized when mice showed signs of pronounced weight loss and rectal prolapse (n = 15).
For genotyping of mice genomic DNA, we performed polymerase chain reaction (PCR) on the tail biopsies which were obtained at the time of weaning. Genotyping was conducted in two sequential PCRs, for Myh11-Cre and Rosa26-Il11 genes separately. Agarose gel electrophoresis was subsequently conducted to confirm the respective product sizes for genotyping. Genotyping primer sequences are listed in S1 Table.
Fibroblast-specific Il11 transgenic model
To model fibroblasts-selective secretion of IL11 in vivo, we crossed the heterozygous Rosa26-IL11 mice with Col1a2-CreER mice [16] to generate double heterozygous Col1a2-CreER:Rosa26-Il11 mice (referred to as Il11Fib) [9]. For Cre-mediated Il11 transgene induction, Il11Fib mice were intraperitoneal injected with 50 mg kg-1 tamoxifen at 6 weeks of age for 10 consecutive days and the animals were sacrificed on day 21. Wildtype littermates (designated as control) were injected with an equivalent dose of tamoxifen for 10 consecutive days as controls. Both female and male mice were used.
Colon length was measured from the caecum to the anus. The most distal half was taken for histology and the adjacent part was portioned and immediately snap frozen in liquid nitrogen for downstream molecular work (hydroxyproline assay, western blot analysis and quantitative polymerase chain reaction assessment). The excised heart was halved from the base to mid ventricle for histology and the remainder separated into 3 portions for molecular work. The left lung was isolated for histology and the right lung separated into 3 portions for molecular work. The right lobe of the liver was excised for histology and the left lobe separated into 3 portions for molecular work. The left kidney was fixed for histology and the right kidney separated in thirds for molecular work. The dorsal skin was harvested and halved for histology and molecular work.
Hydroxyproline assay
The amount of total tissue collagen was quantified using colorimetric detection of hydroxyproline using the Quickzyme Total Collagen assay kit (Quickzyme Biosciences) performed according to the manufacturer’s protocol. All samples were run in duplicate and absorbance at 570 nm was detected on a SpectraMax M3 fluorescence microplate reader using SoftMax Pro version 6.2.1 software (Molecular Devices).
Fecal calprotectin (S100A8/A9) levels
To characterize inflammation in the gut, we investigated levels of fecal calprotectin in the Il11SMC and Il11Fib mice using the mouse S100A8/A9 heterodimer duoset ELISA kit (DY8596-05, R&D systems). Calprotectin is a biomarker for inflammatory activity and has been clinically applied as a diagnostic tool for inflammatory bowel diseases [17, 18]. Stool samples were collected in a 1.5 ml tube and diluted with 50x (weight per volume) of extraction buffer (0.1 M Tris, 0.15 M NaCl, 1.0 M urea, 10 mM CaCl2, 0.1 M citric acid monohydrate, 5 g/l BSA (pH 8.0)) with the assumption of fecal density to be 1 g/ml. Samples were homogenized until no large particles were present. Homogenate was transferred into a fresh tube and further centrifuged at 10,000 g at 4 °C for 20 minutes. The supernatant was assessed for S100A8/A9 levels by ELISA as per the manufacturer’s instructions.
RT-qPCR
Total RNA was extracted from snap-frozen tissues using RNAzol RT (R4533, Sigma-Aldrich) followed by Purelink RNA mini kit (12183025, Invitrogen) purification. The cDNA was prepared using iScript cDNA synthesis kit (1708891, Bio-Rad) as per the manufacturer’s instructions. Quantitative RT-PCR gene expression analysis was performed on duplicate samples using fast SYBR green (Qiagen) technology using the ViiA 7 Real-Time PCR System (Applied Biosystem). RT-qPCR primers are listed in S2 Table. Expression data were normalized to Gapdh mRNA expression levels and the 2-ΔΔCT method was used to calculate the fold change.
Immunoblotting
Western blots were carried out on total protein extracts from mouse tissues. Frozen tissues were homogenized and lyzed in radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (Roche) followed by centrifugation. Equal amounts of protein lysates were separated by SDS-PAGE, transferred onto PVDF membrane and immunoblotted for pERK1/2 (4370, CST), ERK1/2 (4695, CST), pSTAT3 (4113, CST), STAT3 (4904, CST), GAPDH (2118, CST) and IL11 (X203, Aldevron). Proteins were visualized with appropriate secondary antibodies anti-rabbit HRP (7074, CST) and anti-mouse HRP (7076, CST).
Histology
Tissues from Il11SMC and Il11Fib mice were fixed in 10% neutral-buffered formalin for 24–48 hours, tissue processed and paraffin-embedded. Sections were obtained at 5 μm and stained with Masson’s trichrome staining for collagen. Brightfield photomicrographs of the sections were randomly captured by a researcher blinded to the treatment groups using the Olympus BX51 microscope and Image-Pro Premier 9.2 (Media Cybernetics).
Photomicrographs of the colon taken at 200X magnification were used to calculate muscle wall thickness. The distance between the inner and outer circumference of the muscularis propria was measured using the incremental distance tool at a calibrated step size of 25 μm on Image-Pro Premier 9.2 (Media Cybernetics). A total of 75 to 250 measurements across three to five photomicrographs per section were taken and averages reported per photomicrograph. Muscle thickness was reported as an average across 3 cross-sections of the colon per animal.
Photomicrographs of the dorsal skin were captured in 3 fields per section at 100X magnification and used to calculate epidermal and dermal thickness. The epidermis was measured from the stratum basale to the stratum granulosum using hand-drawn line segments on Image-Pro Premier 9.2 (Media Cybernetics). The dermis was measured from the dermal-epidermal junction to the hypodermis. Measurements were recorded using the incremental distance tool at a calibrated step size of 50 μm on Image-Pro Premier 9.2 (Media Cybernetics). A total of 75 to 200 measurements across three photomicrographs per section were taken and averages reported per photomicrograph. Overall epidermal and dermal thickness was reported as an average across the 3 fields per animal.
Fibrosis quantification was conducted as referenced [19]. Color deconvolution version 1.5 plugin using the Masson Trichrome vector on ImageJ (version 1.52a, NIH) and thresholding was applied for area quantification. Perivascular fibrosis was measured as a ratio of the fibrosis area to the vessel area. Vascular hypertrophy was quantified as the ratio of media wall area to the lumen area.
Immunohistochemistry
Paraffin-embedded colon tissue were sectioned at 5 μm, deparaffinized and permeabilized with Triton X-100 (Sigma-Aldrich) and heat antigen retrieved with Bull’s Eye Decloaker (Biocare Medical). Slides were then blocked for endogenous peroxidase with Bloxall™ blocking solution (Vector Lab) followed by blocking with either 3% bovine serum albumin, or mouse on mouse blocking reagent (Vector Lab). Anti-IL11 (ab10558; PA5-36544, Invitrogen), anti-CD45 (1:100; ab10558, Abcam), anti-LGALS3 (1 μg/ml; CL8942AP, Cedarlane) amd anti-LAMP2 (1 μg/ml; 550292, BD Bioscience) were added and incubated overnight at 4°C. Anti-rabbit (1:100; ab27478, Abcam) and anti-rat IgG (1 μg/ml; sc-2026, Santa Cruz) isotype controls were added as respective negative controls. Slides were incubated with anti-rabbit IgG peroxidase (1:500, A0545, Sigma-Aldrich) and anti-rat IgG peroxidase (MP-7404, Vector Lab) followed by chromogen development with ImmPACT® DAB peroxidase substrate kit (SK-4105, Vector Lab) according to manufacturer’s instructions. Lastly, Gill’s haematoxylin (H-3401, Vector Lab) was added for nuclear counterstain. To control for unspecific binding, primary antibody isotype controls were included and images are presented in S3 Fig.
Statistical analysis
Data are presented as mean ± standard deviation or median ± range as stated in the figure legends. Statistical analyses were performed on GraphPad Prism 8 software (version 8.1.2). Outliers (ROUT 2%, GraphPad Prism software) were removed prior to analyses. Comparison of survival curves was analyzed with the log-rank Mantel-Cox test. Bodyweight progression was analyzed with two-way ANOVA with Sidak multiple comparisons. A comparison of mice strains for all other parameters was analyzed with a two-tailed unpaired t-test. The criterion for statistical significance was established at P < 0.05.
Results
Expression of Il11 in smooth muscle cells results in ill health and early mortality
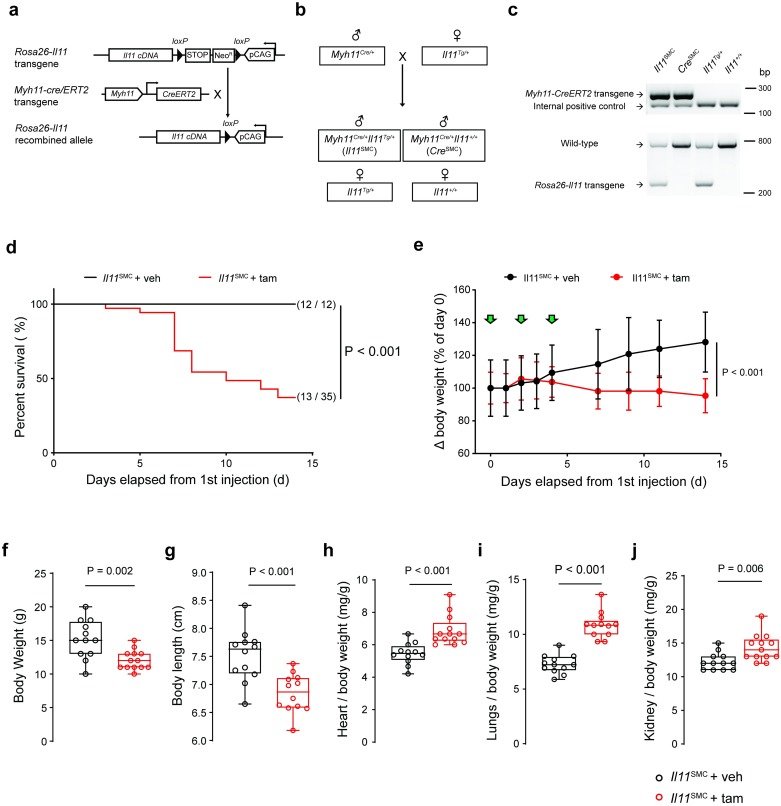
We generated an Il11SMC mouse model that overexpresses IL11 specifically in Myh11+ve SMCs: Conditional transgenic mice with mouse Il11 inserted into the Rosa26 locus (Rosa26-Il11-Tg) [8] were crossed with smooth muscle-specific Myh11-cre/ERT2 mice [15] (Fig 1a and 1b). We then injected tamoxifen (tam) three times at day 0, 3 and 5 into 6-week old Il11SMC mice to induce recombination in Myh11+ve cells and monitored the survival and body weight for 14 days. Following tam-induced Il11 expression in SMCs, mice started dying from day three onwards, with only 37% of Il11SMC mice surviving to day 14. This was significantly different from the survival of either vehicle (veh)-treated Il11SMC animals or tam-treated CreSMC control mice, which were unaffected and both had 100% survival (both P < 0.001; Fig 1d and S1b Fig). Starting from day four onwards, tam-treated Il11SMC mice progressively lost weight as compared to veh -treated and tam-treated CreSMC controls (both P < 0.001; Fig 1e and S1d Fig). Following two weeks of tam-induced Il11 expression, Il11SMC mice were significantly smaller in body weight and length as compared to tam-treated CreSMC controls (both P < 0.001; S1f and S1g Fig) and veh-treated Il11SMC mice (P = 0.002 and P < 0.001 respectively; Fig 1f and 1g). In contrast, the indexed weight of the heart, lung and kidney in tam-treated Il11SMC animals was significantly elevated (PHeart < 0.001; PLung < 0.001; PKidney = 0.006) when compared to veh-treated mice (Fig 1h). We did not observe differences in liver weight or colon length in veh or tam treated Il11SMC animals (data not shown).
Fig 1. Expression of Il11 in smooth muscle cells is associated with body weight loss, elevated organ weights and spontaneous death.
(a) Schematic diagram of the targeted expression of Il11 in Myh11+ve SMC. In Rosa26-Il11 mice, a floxed cassette containing both the neomycin (neo) resistance and stop elements is positioned before the murine Il11 transgene cassette, which undergoes tamoxifen (tam) initiated Cre-mediated recombination when crossed to the Myh11-Cre/ERT2 mouse. (b) Breeding scheme to generate Myh11Cre/+Rosa26Il11/+ (Il11SMC) and Myh11Cre/+Rosa26+/+ (CreSMC) offspring mice. Note that the Myh11-Cre gene is expressed on the Y chromosome and therefore only male offspring carry the transgene. (c) Genotyping of tail biopsy DNA. A 287 bp band indicates the presence of the Cre transgene whereas the 180 bp band determines the presence of the internal positive control (top gel). Polymerase chain reaction with the Rosa26-Il11 primer set detects a 270 bp band indicative of the Rosa26-Il11 transgene whereas the 727 bp band indicates the presence of the wild-type transgene (bottom gel). Uncropped blots are presented in S2 Fig. (d) Survival curve of Il11SMC mice treated with tam (n = 35) and corn oil vehicle (veh; n = 12) mice following tamoxifen initiation at day 0 and followed until day 14. Survival curves were compared using the log-rank Mantel-Cox test. (e) Body weight changes (expressed as percentage of day 0 body weight) in Il11SMC mice treated with tam or veh (n = 8 per group). Green arrows denote individual injections. Statistical analyses by two-way ANOVA with Sidak multiple comparisons; data expressed as mean ± standard deviation. (f) Collated body weights (left) and (g) body lengths (right) of Il11SMC mice treated with tam or veh measured at d14 post initial tamoxifen dose (n = 12–13 per group). (h) Organ weights of the heart, (i) lung and (j) kidney normalized to body weight in Il11SMC mice treated with tam or veh (n = 12–13 per group). All comparisons were conducted in mice 14 days post-veh and tam treatment. Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
IL11 expression causes severe inflammatory bowel disease associated with fibrosis
The most obvious and striking feature of Il11SMC mice treated with tam was progressive rectal prolapse and pale loose stool formation from as early as day three after gene induction (Fig 2a and S1c Fig). Gross anatomical inspection of the gastrointestinal tract revealed inflammation and swelling of the small and large intestines of tam-treated Il11SMC mice when compared to veh-treated controls (Fig 2b). Intestinal inflammation was specifically indicated by an increase in fecal calprotectin, a biomarker used to monitor disease activity in human colitis, in tam-treated Il11SMC mice when compared to veh treatment (P < 0.001; Fig 2c). Masson’s trichrome staining of the colon indicated a very large increase in collagen deposition (P < 0.001; Fig 2d and 2e). Histology also showed a significant increase in the thickness of the smooth muscle-dominant muscularis propria (P = 0.040; Fig 2f). Quantitative hydroxyproline assessments revealed an increase in colonic collagen content in Il11SMC mice after tam treatment (P < 0.001; Fig 2g), confirming the histological data.
Fig 2. Il11 expression results in fibro-inflammatory disease of the colon.
(a) Representative images of the Il11SMC mice before (d0) and up to 7 days (d7) treatment with either corn oil vehicle (veh) or tamoxifen (tam). Presence of rectal prolapse are indicated with white arrows. Images represent the same animal across time points not taken to the same scale. (b) Excised gastrointestinal tract of representative Il11SMC mice at day 14 post-treatment with veh or tam. Scale bar represents 5 cm. (c) Fecal calprotectin in representative Il11SMC mice treated with veh or tam assessed by ELISA (n = 6–8 per group). (d) Representative cross-section and longitudinal section of the colon stained with Masson’s trichrome (left) and at 200X magnification (right). Scale bar of cross and longitudinal sections represents 500 μm and at 200X magnification represents 200 μm. (e) Colon fibrosis determined as a percentage of collagen positive area (blue) from histological images taken at 200X magnification (n = 6 per group). (f) Tunica muscularis (smooth muscle) thickness of the colon (n = 6 per group). (g) Total collagen content assessed by hydroxyproline assay and expressed as fold change (FC) of veh-treated Il11SMC mice (n = 10–11 per group). (h) Representative images of the colonic smooth muscle and crypts taken at 400X magnification for Masson’s trichrome and immunohistochemistry staining for IL11, cluster of differentiation 45 (CD45), lysosome-associated membrane protein 2 (LAMP2), and galectin-3 (LGALS3) (n = 3 per group). Black arrows denote focal staining of positive cells, white arrows denote myenteric plexus which are positive for IL11 and CD45 expression, and white arrowheads denotes leukocyte aggregation. Scale bars represent 100 μm. All comparisons were conducted in organs harvested from mice 14 days post-veh and tam treatment. Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
We then performed an immunohistochemical staining for IL11, CD45, lysosomal-associated membrane protein 2 (LAMP2) and lectin, galactose binding, soluble 3 (LGALS3) in the smooth muscle and crypt compartment of the colon in veh and tam-treated Il11SMC mice (Fig 2h). In Il11SMC mice, IL11 staining was diffuse in the smooth muscle, perhaps with a higher background staining, and also localized more strongly to other stromal cells that are likely fibroblasts, which express the IL11 receptor [8]. Furthermore, CD45+ve leukocytes were increased in the fibrotic regions and crypts of the tam-treated Il11SMC mouse colon. LAMP2 and LGALS3 are markers for epithelial cells and activated macrophages contributing to intestinal inflammation. Tam-treated Il11SMC colon demonstrated increased expression of LAMP2 and LGALS3 in the epithelial cells and leukocytes of the crypts, consistent with inflammation in these regions [20–23]. In tam-treated Il11SMC treated with Tam as compared to veh-treated mice, there was also activation of leukocytes in the Peyer’s patches of the colon, which are a primary site of mucosal immune response (Fig 3a), as well as in localized areas of disrupted villi architecture (Fig 3b). Interestingly, the myenteric plexus of the colon demonstrated ganglionic hyperplasia and fibrosis (Fig 3c), typical of neuroinflammation associated with inflammatory bowel disease.
Fig 3. Il11 expression in smooth muscle cells leads to lymphoid cell aggregates, villi distortion and ganglionic hyperplasia.
(a) Peyer’s patch of tam-treated Il11SMC mice showed increased expression of IL11, CD45, LAMP2 and LGALS3 compared to veh-treated controls. (b) Cross- and longitudinal sections of the mucosa region of the colon in tam-treated Il11SMC mice have inflammatory cell infiltrates that extend from the submucosa to the mucosa region resulting in distortion of villi architecture. (c) Ganglionic hyperplasia and fibrosis in the myenteric plexus of tam-treated Il11SMC mice compared to vehicle controls. White arrowheads denote ganglionic cells. Black boxes were imaged at 400X magnification. All scale bars represent 200 μm.
IL11 expression in smooth muscle cells activates non-canonical IL11 signaling pathways
Given that smooth muscle cells are expressed in the walls of most organs, including the vasculature, bronchi, gastrointestinal and abdominal organs, we sought to confirm the expression of Il11 in Il11SMC mice across tissues and performed western blotting at 14 days after tamoxifen administration. This confirmed that IL11 protein was significantly upregulated at the protein level across all tissues tested (Pcolon = 0.034; Pheart = 0.002; Plung = 0.039; Pliver < 0.001; Pkidney = 0.004; and Pskin = 0.004; Fig 4).
Fig 4. Il11SMC mice exhibit activated ERK1/2 signaling across organs.
Immunoblots of IL11 expression, phospho- (p) and total ERK1/2 and STAT3 protein in (a) colon, (b) heart, (c) liver, (d) lung, (e) kidney, (f) skin tissue of Il11SMC mice treated with vehicle (veh) or tamoxifen (tam) (n = 3 per group). Dotted boxes in the immunoblots represent the veh-treated (black) and tam-treated (red) groups for all organs. All comparisons were conducted in organs harvested from mice 14 days post-veh or tam treatment.
IL11 is a member of the IL6 family of cytokines, which are considered to signal via the Janus Kinase (JAK)/Signal Transducer and Activator of Transcription (STAT) pathway [24]. However, we recently showed that the IL11 effect, both in vitro in fibroblasts and in vivo at the tissue level, is also dependent on non-canonical signaling via extracellular signal-regulated kinase (ERK) [8–10]. To investigate both canonical and non-canonical signaling pathways after Il11 expression, we performed western blotting of phosphorylated (p) STAT3 or ERK1/2 and total protein levels and derived indices of kinase activation by normalizing phosphorylation amounts to total protein levels (Fig 4). At baseline, ERK was phosphorylated at low levels in most tissues except for the skin. Upon IL11 expression, we detected a strong and significant activation of ERK in all tissues (Pcolon = 0.002; Pheart = 0.004; Plung = 0.049; Pliver = 0.056; Pkidney = 0.001; and Pskin < 0.001; Fig 4). STAT3 phosphorylation was unchanged in the heart, lung and liver but was elevated in the colon and skin (P = 0.05 and 0.001 respectively; Fig 4). In contrast, total levels of STAT3 appeared to be increased in the liver and kidney of tam-treated Il11SMC animals (Fig 4d and 4e). Overall, while both pathways were affected, ERK signaling was consistently activated across tissues tested whereas STAT3 was not.
IL11 destroys tissue integrity and promotes collagen deposition
To investigate the effect of Il11 expression in SMCs on tissue composition beyond the colon, we performed histological analyses of the heart, lung, liver, kidney and skin. Masson’s trichrome staining was used to visualize collagen and quantify extracellular matrix deposition. In the heart, we observed collagen deposition in the perivascular region (P = 0.002; Fig 5a and 5b). We also observed vascular hypertrophy (P = 0.019; Fig 5c) and mild ventricular hypertrophy in the absence of dilatation (data not shown). Hydroxyproline assay of the whole heart confirmed cardiac fibrosis (P = 0.026; Fig 5d). In the lung, Ashcroft scores of pulmonary histological images showed lung damage after tam-induced Il11 expression (P < 0.001; Fig 5e and 5f). Masson’s trichrome staining indicated elevated collagen expression throughout the lung in Il11SMC mice and pulmonary fibrosis was confirmed by the hydroxyproline assay (P = 0.001; Fig 5g).
Fig 5. Il11 expression in smooth muscle cells causes fibrosis across organs.
(a) Representative Masson’s trichrome stained mid-ventricle sections of the heart harvested at 14 days post-vehicle (veh) or tamoxifen (tam) initiation (left) and 200X magnification images demonstrating perivascular fibrosis (right). Scale bars for mid-ventricle sections and 200X magnification denote 500 μm and 200 μm respectively. (b) Perivascular fibrosis quantification of histological images from veh- and tam-treated Il11SMC mice at 200X magnification (n = 6 per group). (c) Vascular hypertrophy quantification of veh- and tam-treated Il11SMC mice (n = 6 per group). (d) Total collagen content in the heart assessed by hydroxyproline assay and shown as fold change (FC) of veh- and tam-treated Il11SMC mice (n = 7–8 per group). (e) Representative Masson’s trichrome stained whole lung sections (left) and 200X magnification images (right). Scale bars for whole lung sections and 200X magnification denote 500 μm and 200 μm respectively. (f) Pulmonary fibrosis quantification as assessed by the Ashcroft score (n = 6–8 per group). (g) Total collagen content in the lung assessed by hydroxyproline assay as above (n = 7–8 per group). (h) Representative Masson’s trichrome stained liver sections taken at 400X magnification demonstrating perisinusoidal fibrosis. Scale bar at 400X magnification indicates 100 μm. (i) Fibrosis quantification of liver sections (400X magnification) from veh- and tam-treated Il11SMC mice (n = 6 per group). (j) Total collagen content in the liver assessed by hydroxyproline assay as above (n = 10–11 per group). (k) Representative Masson’s trichrome stained cross-section of the kidney (left) and 200X magnification images (right). Scale bars for the cross-section of the kidney and 200X magnification denote 500 μm and 200 μm respectively. (l) Fibrosis quantification of kidney sections (200X magnification) from veh- and tam-treated Il11SMC mice (n = 6 per group). (m) Total collagen content in the kidney assessed by hydroxyproline assay as above (n = 10–11 per group). (n) Representative Masson’s trichrome stained section of the dorsal skin at 100X magnification (left) and at 400X magnification (right). Scale bar at 100X and 400X magnification represents 200 μm and 100 μm respectively. (o) Dermal and (p) epidermal thickness of the dorsal skin. (q) Total collagen content in the skin assessed by hydroxyproline assay as above (n = 10–11 per group). All comparisons were conducted in organs harvested from mice 14 days post-veh and tam treatment. Statistical analyses by two-tailed unpaired t-test; data shown as median ± IQR, whiskers represent the minimum and maximum values.
The effect of Il11 expression on the liver was overall mild and characterized by perisinusoidal fibrosis (Fig 5h to 5j). Renal tissue structure was also affected only mildly, with limited fibrosis occurring around the blood vessels (Fig 5k to 5m). The effect of IL11 on the skin of tam-treated Il11SMC animals was more profound and both the dermal and epidermal thickness was significantly increased (Fig 5n to 5p; P = 0.041 and P = 0.001 respectively). Dorsal skin sections showed that epidermal and dermal cell infiltrates were increased and the adipose tissue layer in the hypodermis was largely depleted. Confirming Masson’s trichrome staining of skin sections, we observed increased collagen deposition in the skin of tam-treated Il11SMC mice using the hydroxyproline assay (P < 0.001; Fig 5q).
IL11 secretion from smooth muscle cells drives fibrogenic gene expression
We assessed the RNA expression of fibrogenic genes to complement our histology studies, which further substantiated the presence of multi-organ fibrosis. Reverse transcription-polymerase chain reaction (RT-PCR) was performed using RNA from colonic, ventricular, pulmonary, hepatic, renal and skin tissue of veh- or tam-treated Il11SMC mice. Collagen, type I, alpha 1 (Col1a1) RNA was significantly upregulated in all tissues (Pcolon = 0.005; Pheart = 0.005; Plung < 0.001; Pliver = 0.016; Pkidney = 0.022; Pskin = 0.016; Fig 6), confirming the effect of Il11 expression on global organ fibrosis that we observed on the protein level (Fig 5). Additional markers for fibrosis such as collagen, type I, alpha 2 (Cola1a2), collagen, type III, alpha 1 (Col3a1), fibronectin 1 (Fn1), tissue inhibitor of metalloproteinase 1 (Timp1) and matrix metallopeptidase 2 (Mmp2) were also assessed via RT-PCR (Fig 6a to 6f). These genes were elevated in most tissues of tam-treated Il11SMC mice. Timp1 transcripts were significantly upregulated in the heart (P < 0.001), lung (P = 0.004), liver (P = 0.003), kidney (P = 0.003) and skin (P = 0.017), which is a recognized feature of pathological ECM remodeling [25].
Fig 6. Relative gene expression of fibrogenic genes in organs from tam-treated Il11SMC mice.
Relative mRNA expression of collagen type 1a1 (Col1a1), type 1a2 (Col1a2), type 3a1 (Col3a1), fibronectin-1 (Fn1), tissue inhibitor of metalloproteinase 1 (Timp1) and matrix metalloproteinase 2 (Mmp2) normalized to Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression in the (a) colon, (b) heart, (c) lung, (d) liver, (e) kidney and (f) skin. All comparisons were conducted 14 days post-veh (black) and tam (red) initiated mice. Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
IL11 secreted from smooth muscle cells causes inflammation across tissues
In addition to fibrosis, SMC-driven diseases are often characterized by tissue inflammation. To better understand whether IL11 secretion from SMCs can contribute to this pathology, we performed RT-PCR experiments of inflammatory marker genes across multiple tissues. Interleukin 6 (IL6) also signals via gp130, similar to IL11, but its specific IL6 receptor subunit is expressed on a different subset of cells, most of which belong to the immune system [8]. IL6 is also a well-established therapeutic target for inflammatory diseases such as rheumatoid arthritis [26]. Upon tam-induced Il11 expression in Il11SMC mice, we found Il6 mRNA to be significantly upregulated across all tissues tested (Pcolon = 0.001; Pheart < 0.001; Plung = 0.015; Pliver = 0.007; Pkidney < 0.001; and Pskin = 0.003; Fig 7).
Fig 7. Relative gene expression of inflammatory genes in organs from tam-treated Il11SMC mice.
Relative mRNA expression of interleukin 6 (Il6), C-C motif chemokine ligand 2 (Ccl2), C-C motif chemokine ligand 5 (Ccl5) normalized to Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression in the (a) colon, (b) heart, (c) lung, (d) liver, (e) kidney and (f) skin respectively. All comparisons were conducted in 14 days post-veh (black) and tam (red) initiated mice. Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
In the colon, we also detected increased RNA expression of the inflammatory chemokine C-C motif chemokine ligand 2 (Ccl2) (P = 0.017), whereas C-C motif chemokine ligand 5 (Ccl5) was not significantly elevated but trended upwards (P = 0.141). Interestingly, these inflammatory chemokines are upregulated in the colonic mucosa of IBD patients [27, 28]. However, CCL2 transcripts, and not CCL5 transcripts, were found to be expressed in vessel-associated cells such as SMCs in IBD [27]. Given that Il11SMC mice express Il11 in SMCs, it is consistent that the transcript expression of the chemokine expressed in this particular cellular niche in the colon is most affected. In the skin, all three inflammatory markers tested were highly upregulated. This points to an inflammatory gene expression signature in the skin that is reminiscent of that seen in systemic sclerosis, since IL6, CCL2, and CCL5 are elevated in the serum of patients [29, 30]. Of note, CCL2 levels were correlated with the extent of skin fibrosis in systemic sclerosis, a pathogenic feature also triggered by IL11 expression in SMCs (Fig 7) [29].
Fibroblast-selective expression of Il11 recapitulates the features of colonic inflammatory phenotype seen in Il11SMC mice
We have previously described a model of Il11 expression in fibroblasts (Il11Fib) that drives fibrosis in the heart, kidney, and lung [8, 9]. To examine further the effect of Il11 expression in stromal cells on the colon, we studied colonic phenotypes in this second model of Il11 expression from the stromal niche (Fig 8a). Gross examination of the gastrointestinal tract of Il11Fib mice revealed macroscopic appearances consistent with inflammation of the colon to a similar extent as in Il11SMC mice (Fig 8b). The total gastrointestinal gut length of Il11Fib mice was unchanged overall but the colon length alone was reduced (P = 0.030; Fig 8b to 8d), which is a feature of experimental colitis in mice [31]. In this model, as compared to Il11SMC mice, we detected Il6 but not Ccl2 or Ccl5, upregulation in the colon (Fig 8d and 8e). Inflammation of the gut was apparent in the Il11Fib model as fecal calprotectin was significantly elevated (P = 0.003; Fig 8f). Histological examination revealed marked colonic dilation and increased SMC thickness (Fig 8g and 8h). In contrast to the Il11SMC model of Il11 expression, colonic fibrosis as determined by histology, hydroxyproline assay or ECM gene expression, was not significantly different between tam-treated Il11Fib and controls (data not shown). Taken together, fibroblast-driven Il11 expression recapitulates primarily the SMC-driven inflammatory, but not the fibrotic, phenotype in the mouse.
Fig 8. Mice with fibroblast-specific Il11 expression develop inflammatory bowel disease.
(a) Schematic diagram demonstrating tamoxifen (tam) injection procedure in 6-week-old Il11Fib and wildtype (control) littermates. (b) Excised gastrointestinal tract of representative Il11Fib mice at day 21 compared to controls. Scale bar represents 5 cm. (c) Indexed GIT length in reference to body length (BL) was unchanged in Il11Fib mice but (d) indexed colon length was markedly reduced as compared to controls (n = 4 per group). (e) Expression of inflammatory genes (Il6, Ccl2 and Ccl5) in the colon tissue of Il11Fib mice as compared to controls (n = 4–5 per group). (f) Fecal calprotectin in stool samples collected from Il11Fib and control mice (n = 4–5 per group) as assessed by ELISA. (g) Representative cross-sections of the colon of Il11Fib and control mice stained with Masson’s Trichrome (left) and at 200X magnification (right) (n = 6 biological replicates). Scale bars indicate 500 μm and 200 μm respectively. (h) Thickness of the smooth muscle layer (muscularis propria) in tam-treated Il11Fib mice compared to controls (n = 6 per group). All comparisons were conducted in 21 days post-tam initiation in control (black) and Il11Fib (green) mice. Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
Discussion
In humans, IL11 is highly upregulated in the colonic mucosa of patients with either ulcerative colitis or Crohn’s disease who do not respond to anti-TNF therapy, with recent single cell RNA-seq studies localizing IL11 to inflammatory mucosal stromal cells [32–34]. To better understand the effect of IL11 in the colon, recombinant human IL11 has been used in rodent models of IBD [34–38] and it was suggested that IL11 may have a protective role in the bowel. However, a caveat with these studies is that human IL11 was administered to rodents despite the fact that human IL11 does not activate mouse stromal cells [8]. More recently, we have found that human IL11 unexpectedly acts as an inhibitor of endogenous mouse IL11 activity in the liver [39]. Thus, previous studies in IBD that showed that when human IL11 is injected into mice it protects them from IBD may paradoxically support the opposite conclusion: IL11 is not protective at all, but a driver of IBD. In light of this, there is a great need to assess the effects of species-specific IL11 in the mouse, which we undertook in this study by expressing murine Il11 in SMCs or fibroblasts in adult mice.
To enable our studies, we developed the Il11SMC mouse as a tool to study the effect of murine IL11 secreted from SMCs, an established source of IL11 in the vasculature, airway, and colon [11, 40, 41]. Surprisingly, expression of Il11 in SMCs was sufficient to induce severe colonic inflammation and rectal prolapse within 3 days, which was followed by early mortality in Il11SMC animals. We also documented increased colonic muscle thickness, which is a characteristic of the dextran sulphate sodium-induced colitis model [42]. In humans, histological features of clinical colitis include architectural distortion, shortening and size variation of crypts, immune cell infiltration, and granuloma formation [43]. Occurrences of architectural distortion of the glands and crypts in these mice were rare but present, although this may be reflective of the very short duration of IL11 expression. In contrast, these mice demonstrated thicker muscularis mucosa, increased immune cell infiltration, increased pro-inflammatory markers LAMP2 and LGALS3 in epithelial cells and the stroma and increased fibrosis in the mucosa sharing close similarities to intestinal fibrosis as observed in patients with ulcerative colitis [44]. Interestingly, IL11 expression in smooth muscle cells demonstrated signs of neuroinflammation in the myenteric plexus, which has been observed in inflammatory bowel disease [45].
We explored further the IL11 effect in the bowel using an additional model that expresses mouse Il11 in a second stromal cell type: the fibroblast. This complementary model also develops severe diarrhea and inflammation of the small intestines and colon, reinforcing the data generated in the Il11SMC mice. In this model, the colon becomes distended with thicker muscularis mucosa, suggesting that IL11 secreted from fibroblasts acts in a paracrine fashion to cause smooth muscle hypertrophy. A lack of grossly detectable intestinal fibrosis in the colon in this model, which is very different to findings in the heart, kidney and lung [8,9], may reflect differing cellular composition of fibroblasts and smooth muscle cells in the intestinal wall, where smooth muscle cells appear to play a larger role. This would be consistent with the suggestion that smooth muscle hyperplasia and hypertrophy contributes mostly to the fibrostenosis and inflammation in IBD [46] and underlies the colonic contractile dysfunction [47].
Taken together these data show that Il11 expression in stromal cells is sufficient to cause an IBD phenotype and challenges the earlier data, based on the use of recombinant human IL11 in the mouse, that IL11 is protective in the bowel. Considered along with patient studies that show IL11 to be highly upregulated in the colonic mucosa of patients with ulcerative colitis or Crohn’s disease and that IL11 predicts treatment failure [32–34], our results highlight IL11 as a promising therapeutic target for IBD, particularly in the context of anti-TNF therapy resistance.
Supporting information
(DOCX)
(DOCX)
(a) Schematic diagram demonstrating the tamoxifen (tam) injection procedure in 6-week-old Il11SMC and CreSMC mice. (b) Survival curve of tam-treated Il11SMC (n = 35) compared to CreSMC mice (n = 27) mice from 1st injection starting at 6 weeks of age. Survival curves were compared with the log-rank Mantel-Cox test. (c) Representative images of the CreSMC and Il11SMC mice before (d0) and up to 14 days (d14) post-tam initiation (left). Note the presence of pale and loose stools in Il11SMC mice (right). The presence of rectal prolapse is indicated with white arrows. Tam-treated Il11SMC images presented here are different from Fig 1c. Images were not taken to scale. (d) Baseline body weight of 6-week-old CreSMC and Il11SMC mice before induction (n = 16 per group). Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values. (e) Representative images of CreSMC and Il11SMC mice at d14 post-Tam initiation. (f) Collated body weights and (g) body lengths of tam-treated CreSMC and Il11SMC mice measured at d14 post-Tam initiation. (n = 12–17 per group). Statistical analyses by two-tailed unpaired t-test; data expressed as median ± IQR, whiskers represent the minimum and maximum values.
(TIF)
PCR products of DNA extracted from tail biopsies of 21-day-old mice by use of set primers for Myh11-Cre (left) and Rosa26-Il11 (right) (primers as listed in S1 Table) and analyzed by agarose gel electrophoresis. Dashed boxes indicate cropped blots used in Fig 1c.
(TIF)
Smooth muscle and crypt staining with rat and rabbit IgG isotype controls demonstrate no positive staining in both veh- and tam-treated Il11-Tg colon. Scale bar represents 100 μm.
(TIF)
Acknowledgments
The authors would like to acknowledge B.L. George, E. Khin, M. Wang, J. Tan for their technical expertise and support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research was supported by the National Medical Research Council (NMRC; https://www.nmrc.gov.sg/) Singapore STaR awards to S.A.C. (NMRC/STaR/0029/2017), the NMRC Central Grant to the NHCS, Goh Foundation, Tanoto Foundation and a grant from the Fondation Leducq. S.S. is supported by the Goh Foundation and Charles Toh Cardiovascular Fellowship and by the National Medical Research Council Young Individual Research Grant (MOH-OFYIRG18nov-0003). A.A.W. is supported by the NMRC YIRG (NMRC/OFYIRG/0053/2017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiological Reviews. 2004. pp. 767–801. 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- 2.Johnson PR, Roth M, Tamm M, Hughes M, Ge Q, King G, et al. Airway smooth muscle cell proliferation is increased in asthma. Am J Respir Crit Care Med. 2001;164: 474–477. 10.1164/ajrccm.164.3.2010109 [DOI] [PubMed] [Google Scholar]
- 3.Shea-Donohue T, Notari L, Sun R, Zhao A. Mechanisms of smooth muscle responses to inflammation. Neurogastroenterology & Motility. 2012. pp. 802–811. 10.1111/j.1365-2982.2012.01986.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrick AL. Vascular function in systemic sclerosis. Current Opinion in Rheumatology. 2000. pp. 527–533. 10.1097/00002281-200011000-00009 [DOI] [PubMed] [Google Scholar]
- 5.Shin JY, Beckett JD, Bagirzadeh R, Creamer TJ, Shah AA, McMahan Z, et al. Epigenetic activation and memory at a TGFB2 enhancer in systemic sclerosis. Science Translational Medicine. 2019. p. eaaw0790. 10.1126/scitranslmed.aaw0790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denton CP, Ong VH, Xu S, Chen-Harris H, Modrusan Z, Lafyatis R, et al. Therapeutic interleukin-6 blockade reverses transforming growth factor-beta pathway activation in dermal fibroblasts: insights from the faSScinate clinical trial in systemic sclerosis. Ann Rheum Dis. 2018;77: 1362–1371. 10.1136/annrheumdis-2018-213031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leask A, Abraham DJ. TGF-β signaling and the fibrotic response. The FASEB Journal. 2004. pp. 816–827. 10.1096/fj.03-1273rev [DOI] [PubMed] [Google Scholar]
- 8.Schafer S, Viswanathan S, Widjaja AA, Lim W-W, Moreno-Moral A, DeLaughter DM, et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552: 110–115. 10.1038/nature24676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng B, Dong J, Viswanathan S, D’Agostino G, Widjaja AA, Lim W-W, et al. IL-11 is a therapeutic target in idiopathic pulmonary fibrosis. 10.1101/336537 [DOI] [PubMed] [Google Scholar]
- 10.Widjaja AA, Singh BK, Adami E, Viswanathan S, Dong J, D’Agostino GA, et al. Inhibiting Interleukin 11 Signaling Reduces Hepatocyte Death and Liver Fibrosis, Inflammation, and Steatosis in Mouse Models of Non-Alcoholic Steatohepatitis. Gastroenterology. 2019. [DOI] [PubMed] [Google Scholar]
- 11.Taki H, Sakai T, Sugiyama E, Mino T, Kuroda A, Taki K, et al. Monokine stimulation of interleukin-11 production by human vascular smooth muscle cells in vitro. Atherosclerosis. 1999;144: 375–380. 10.1016/s0021-9150(99)00009-x [DOI] [PubMed] [Google Scholar]
- 12.Arijs I, Li K, Toedter G, Quintens R, Van Lommel L, Van Steen K, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58: 1612–1619. 10.1136/gut.2009.178665 [DOI] [PubMed] [Google Scholar]
- 13.Smillie CS, Biton M, Ordovas-Montañes J, Sullivan KM, Burgin G, Graham DB, et al. Rewiring of the cellular and inter-cellular landscape of the human colon during ulcerative colitis. 10.1101/455451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minshall E, Chakir J, Laviolette M, Molet S, Zhu Z, Olivenstein R, et al. IL-11 expression is increased in severe asthma: association with epithelial cells and eosinophils. J Allergy Clin Immunol. 2000;105: 232–238. 10.1016/s0091-6749(00)90070-8 [DOI] [PubMed] [Google Scholar]
- 15.Wirth A, Benyó Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14: 64–68. 10.1038/nm1666 [DOI] [PubMed] [Google Scholar]
- 16.Zheng B, Zhang Z, Black CM, de Crombrugghe B, Denton CP. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002;160: 1609–1617. 10.1016/S0002-9440(10)61108-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in Inflammation. Frontiers in Immunology. 2018. 10.3389/fimmu.2018.01298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konikoff MR, Denson LA. Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflammatory Bowel Diseases. 2006. pp. 524–534. 10.1097/00054725-200606000-00013 [DOI] [PubMed] [Google Scholar]
- 19.Dahab GM, Kheriza MM, El-Beltagi HM, Fouda A-MM, Sharaf El-Din OA. Digital quantification of fibrosis in liver biopsy sections: Description of a new method by Photoshop software. Journal of Gastroenterology and Hepatology. 2004. pp. 78–85. 10.1111/j.1440-1746.2004.03183.x [DOI] [PubMed] [Google Scholar]
- 20.Sundblad V, Quintar AA, Morosi LG, Niveloni SI, Cabanne A, Smecuol E, et al. Galectins in Intestinal Inflammation: Galectin-1 Expression Delineates Response to Treatment in Celiac Disease Patients. Front Immunol. 2018;9: 379 10.3389/fimmu.2018.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong S, Hughes RC. Macrophage surface glycoproteins binding to galectin-3 (Mac-2-antigen). Glycoconj J. 1997;14: 267–274. 10.1023/a:1018554124545 [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Lee J, Hyun J, Park J, Joo H, Shin T. Expression and immunohistochemical localization of galectin-3 in various mouse tissues. Cell Biology International. 2007. pp. 655–662. 10.1016/j.cellbi.2006.11.036 [DOI] [PubMed] [Google Scholar]
- 23.Díaz-Alvarez L, Ortega E. The Many Roles of Galectin-3, a Multifaceted Molecule, in Innate Immune Responses against Pathogens. Mediators Inflamm. 2017;2017: 9247574 10.1155/2017/9247574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taga T, Kishimoto T. gp130 AND THE INTERLEUKIN-6 FAMILY OF CYTOKINES. Annual Review of Immunology. 1997. pp. 797–819. 10.1146/annurev.immunol.15.1.797 [DOI] [PubMed] [Google Scholar]
- 25.Birkedal-Hansen H. Proteolytic remodeling of extracellular matrix. Current Opinion in Cell Biology. 1995. pp. 728–735. 10.1016/0955-0674(95)80116-2 [DOI] [PubMed] [Google Scholar]
- 26.Rossi J-F, Lu Z-Y, Jourdan M, Klein B. Interleukin-6 as a therapeutic target. Clin Cancer Res. 2015;21: 1248–1257. 10.1158/1078-0432.CCR-14-2291 [DOI] [PubMed] [Google Scholar]
- 27.Mazzucchelli L, Hauser C, Zgraggen K, Wagner HE, Hess MW, Laissue JA, et al. Differential in situ expression of the genes encoding the chemokines MCP-1 and RANTES in human inflammatory bowel disease. J Pathol. 1996;178: 201–206. [DOI] [PubMed] [Google Scholar]
- 28.Grimm MC, Elsbury SK, Pavli P, Doe WF. Enhanced expression and production of monocyte chemoattractant protein-1 in inflammatory bowel disease mucosa. J Leukoc Biol. 1996;59: 804–812. 10.1002/jlb.59.6.804 [DOI] [PubMed] [Google Scholar]
- 29.Bandinelli F, Del Rosso A, Gabrielli A, Giacomelli R, Bartoli F, Guiducci S, et al. CCL2, CCL3 and CCL5 chemokines in systemic sclerosis: the correlation with SSc clinical features and the effect of prostaglandin E1 treatment. Clin Exp Rheumatol. 2012;30: S44–9. [PubMed] [Google Scholar]
- 30.Hasegawa M, Sato S, Fujimoto M, Ihn H, Kikuchi K, Takehara K. Serum levels of interleukin 6 (IL-6), oncostatin M, soluble IL-6 receptor, and soluble gp130 in patients with systemic sclerosis. J Rheumatol. 1998;25: 308–313. [PubMed] [Google Scholar]
- 31.Diaz-Granados N, Howe K, Lu J, McKay DM. Dextran Sulfate Sodium-Induced Colonic Histopathology, but not Altered Epithelial Ion Transport, Is Reduced by Inhibition of Phosphodiesterase Activity. The American Journal of Pathology. 2000. pp. 2169–2177. 10.1016/S0002-9440(10)65087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arijs I, Li K, Toedter G, Quintens R, Van Lommel L, Van Steen K, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58: 1612–1619. 10.1136/gut.2009.178665 [DOI] [PubMed] [Google Scholar]
- 33.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019;178: 714–730.e22. 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arijs I, Quintens R, Van Lommel L, Van Steen K, De Hertogh G, Lemaire K, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis. 2010;16: 2090–2098. 10.1002/ibd.21301 [DOI] [PubMed] [Google Scholar]
- 35.Qiu BS, Pfeiffer CJ, Keith JC Jr. Protection by recombinant human interleukin-11 against experimental TNB-induced colitis in rats. Dig Dis Sci. 1996;41: 1625–1630. 10.1007/bf02087911 [DOI] [PubMed] [Google Scholar]
- 36.Gibson DL, Montero M, Ropeleski MJ, Bergstrom KSB, Ma C, Ghosh S, et al. Interleukin-11 reduces TLR4-induced colitis in TLR2-deficient mice and restores intestinal STAT3 signaling. Gastroenterology. 2010;139: 1277–1288. 10.1053/j.gastro.2010.06.057 [DOI] [PubMed] [Google Scholar]
- 37.Orazi A, Du X, Yang Z, Kashai M, Williams DA. Interleukin-11 prevents apoptosis and accelerates recovery of small intestinal mucosa in mice treated with combined chemotherapy and radiation. Lab Invest. 1996;75: 33–42. [PubMed] [Google Scholar]
- 38.Boerma M, Wang J, Burnett AF, Santin AD, Roman JJ, Hauer-Jensen M. Local administration of interleukin-11 ameliorates intestinal radiation injury in rats. Cancer Res. 2007;67: 9501–9506. 10.1158/0008-5472.CAN-07-0810 [DOI] [PubMed] [Google Scholar]
- 39.Widjaja AA, Dong J, Adami E, Viswanathan S, Ng B, Singh BK, et al. Redefining Interleukin 11 as a regeneration-limiting hepatotoxin. 10.1101/830018 [DOI] [PubMed] [Google Scholar]
- 40.Elias JA, Wu Y, Zheng T, Panettieri R. Cytokine- and virus-stimulated airway smooth muscle cells produce IL-11 and other IL-6-type cytokines. Am J Physiol. 1997;273: L648–55. 10.1152/ajplung.1997.273.3.L648 [DOI] [PubMed] [Google Scholar]
- 41.Shi X-Z, Sarna SK. Transcriptional regulation of inflammatory mediators secreted by human colonic circular smooth muscle cells. Am J Physiol Gastrointest Liver Physiol. 2005;289: G274–84. 10.1152/ajpgi.00512.2004 [DOI] [PubMed] [Google Scholar]
- 42.Jin B-R, Chung K-S, Cheon S-Y, Lee M, Hwang S, Noh Hwang S, et al. Rosmarinic acid suppresses colonic inflammation in dextran sulphate sodium (DSS)-induced mice via dual inhibition of NF-κB and STAT3 activation. Sci Rep. 2017;7: 46252 10.1038/srep46252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeRoche TC, Xiao S-Y, Liu X. Histological evaluation in ulcerative colitis. Gastroenterology Report. 2014. pp. 178–192. 10.1093/gastro/gou031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Bruyn JR, Meijer SL, Wildenberg ME, Bemelman WA, van den Brink GR, D’Haens GR. Development of Fibrosis in Acute and Longstanding Ulcerative Colitis. J Crohns Colitis. 2015;9: 966–972. 10.1093/ecco-jcc/jjv133 [DOI] [PubMed] [Google Scholar]
- 45.Lakhan SE, Kirchgessner A. Neuroinflammation in inflammatory bowel disease. Journal of Neuroinflammation. 2010. p. 37 10.1186/1742-2094-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W, Lu C, Hirota C, Iacucci M, Ghosh S, Gui X. Smooth Muscle Hyperplasia/Hypertrophy is the Most Prominent Histological Change in Crohn’s Fibrostenosing Bowel Strictures: A Semiquantitative Analysis by Using a Novel Histological Grading Scheme. Journal of Crohn’s and Colitis. 2017. pp. 92–104. 10.1093/ecco-jcc/jjw126 [DOI] [PubMed] [Google Scholar]
- 47.Collins SM, Khan I, Vallance B, Hogaboam C. The role of smooth muscle in intestinal inflammation. Inflammatory Bowel Disease. 1994. pp. 162–169. 10.1007/978-94-009-0371-5_16 [DOI] [Google Scholar]