Summary
Ubiquinone (UQ) is a conserved polyprenylated lipid essential to cellular respiration. Two papers, one in this issue of Cell Chemical Biology (Chehade et al., 2019) and another in Molecular Cell (Lohman et al., 2019), identify lipid-binding proteins that play crucial roles in chaperoning UQ-intermediates.
Ubiquinone, also known as Coenzyme Q or UQ, is an essential redox-active lipid that functions in cellular energy metabolism. The reversible reduction and oxidation of the quinone ring to the hydroquinone (UQ + 2 e− + 2 H+ ⇔ UQH2) allows UQ to function as an acceptor and donor of electrons and protons in respiratory electron transport chains. Such transport establishes the H+ gradient used to produce ATP in prokaryotes and eukaryotes. The redox activity of UQ also enables it to serve as an electron acceptor in the synthesis of pyrimidines, and in the oxidation of sulfide, choline, dimethylglycine, sarcosine, glycerol-3-phosphate, proline, and fatty acyl-CoA substrates in beta-oxidation (Alcazar-Fabra et al., 2018).
The redox active ring of UQ is decorated with a long and extremely hydrophobic polyisoprenoid tail (Figure 1). The number of isoprene units in the tail of UQn is species dependent (UQ6 in Saccharomyces cerevisiae, UQ8 in Escherichia coli, and UQ10 in humans). Membrane biophysical and modeling studies indicate that the polyisoprenoid tails of UQn isoforms of UQ6 or greater are positioned at the mid-plane of the membrane bilayer (Quinn, 2012). This location enables reduced UQH2 to function as a lipid-soluble antioxidant that chain-terminates lipid peroxidation reactions and preserves membrane fluidity and function (Bentinger et al., 2010). Two papers, one in this issue of Cell Chemical Biology (Chehade et al., 2019) and another in Molecular Cell (Lohman et al., 2019), show that the extremely hydrophobic UQ-intermediates are ligands for lipid binding domains of proteins in UQ biosynthetic complexes.
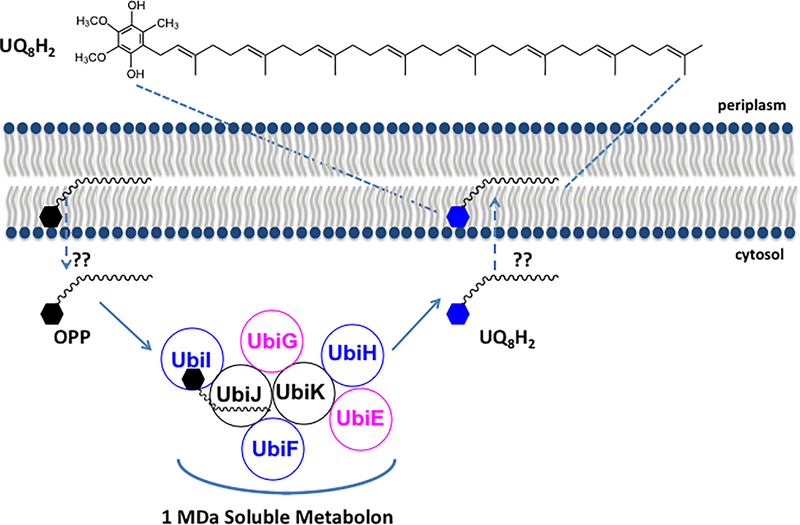
Figure 1.
A 1 MDa soluble metabolon synthesizes UQ8 in E. coli. The long octaprenyl tail of UQ8 and UQ8-intermediates in E. coli are thought to reside at the midplane of the membrane bilayer. Chehade et al. (2019) show that octaprenyl phenol (OPP), an early intermediate in E. coli UQ8 biosynthesis, is converted in six steps to the final product UQ8H2, by a high molecular mass soluble metabolon located in the cytosol. Five Ubi enzymes (shown in blue and pink) are organized around the accessory proteins UbiJ and UbiK. UbiJ contains a sterol carrier protein 2 (SCP2) domain, responsible for binding the UQ8-intermediates. The steps whereby OPP is extracted from the membrane and the UQ8H2 product is delivered back to the membrane are designated by the dashed arrows and represent intriguing and unknown trafficking steps (??).
In E. coli the length of the polyisoprenoid tail (UQ8) is determined by a polyisoprenyl-diphosphate synthase, IspB (Figure 2). The tail is then attached to 4-hydroxybenzoic acid, the aromatic ring precursor of UQ, by an integral membrane protein UbiA, to form the first polyprenylated-aromatic ring intermediate. Subsequent ring modification steps include a decarboxylation step (mediated by UbiD and UbiX), followed by a series of hydroxylation and methylation steps to produce the fully substituted hydroquinone product, UQH2.
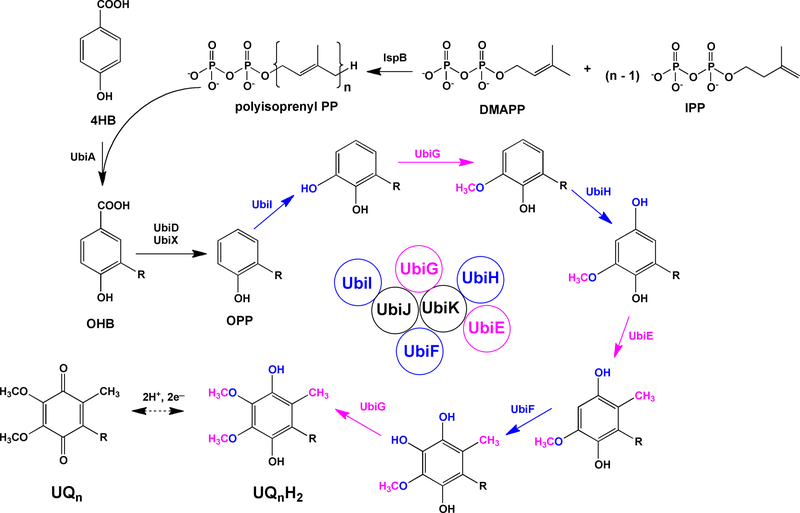
Figure 2.
The steps of UQ8 biosynthesis in E. coli. IspB produces octaprenyl-diphosphate. The octaprenyl group (designated by R) is transferred to 4-hydroxybenzoic acid (4HB) by UbiA, to form 3-octaprenyl-4-hydroxybenzoic acid (OHB). UbiD and UbiX work together to mediate decarboxylation and produce octaprenyl phenol (OPP). OPP is then subjected to alternating steps of hydroxylation (shown in blue) and methylation (shown in pink) in the order of UbiI, UbiG, UbiH, UbiE, UbiF and UbiG to form the fully substituted product, UQ8H2. UbiJ and UbiK are required accessory proteins, and UbiJ binds OPP and the other UQ8-intermediates. The order of steps in eukaryotic UQ biosynthesis is slightly different. The first hydroxylation and methylation steps in yeast and human cells are thought to precede the decarboxylation step.
Pierrel and colleagues (Chehade et al. 2019) show that five Ubi enzymes (UbiE, UbiF, UbiG, UbiH and UbiI), and two Ubi accessory polypeptides (UbiJ and UbiK), form a soluble cytosolic super-complex that carry out the final six steps of UQH2 synthesis. The authors use two-dimensional Blue Native SDS-PAGE to show that these seven Ubi polypeptides migrate together at a high molecular mass (1MDa). Bacterial two-hybrid experiments, protein mass spectrometry, and biophysical separations provide evidence for these interactions and establish the remarkable stability of this soluble metabolon. The authors note that an outstanding challenge that remains to be addressed is to determine the stoichiometry of the Ubi partner proteins that comprise the 1MDa Ubi metabolon.
Chehade et al. 2019 also demonstrate that the soluble Ubi metabolon co-purifies with UQ-intermediates. UbiJ is shown to be essential for complex formation, and the crystal structure of the sterol carrier protein 2 (SCP2) domain of UbiJ reveals a hydrophobic binding pocket that can accommodate UQ or UQ-intermediates. It will be important to determine the nature and specificity of the interaction of UbiJ with UQ and UQ-intermediates. This study not only discovers the Ubi metabolon, but it is the first to suggest that hydrophobic UQ lipid may be synthesized in the cytosol and then trafficked back to the membrane by an unknown mechanism. Thus, the work by Chehade et al. (2019) sets the stage for determining how polyisoprenoid lipids may be trafficked between membranes.
Multi-functional MDa biosynthetic complexes (termed the CoQ synthome or Complex Q) are also involved in the synthesis of UQ in yeast and human cells (Awad et al., 2018; Stefely and Pagliarini, 2017). The Coq polypeptides that perform similar ring modification steps in UQ biosynthesis are associated peripherally with the matrix side of the inner mitochondrial membrane. In eukaryotes, the Coq4 and Coq9 polypeptides are the likely players that bind polyisoprenoid tails of UQ (Lohman et al., 2019; Rea et al., 2010). The study by Lohman et al., (2019) shows that the COQ9 polypeptide binds UQ-intermediates and facilitates the COQ7-mediated hydroxylase step. Their structural studies and molecular dynamics simulations describe a sequential five-step process of how COQ9 enables access to the UQ intermediates in the membrane, mediated by its C-terminal amphipathic helix (Lohman et al., 2019). As COQ9 approaches the mitochondrial inner membrane, a local deformation in the membrane displaces membrane embedded UQ-intermediates from the membrane. COQ9 picks up UQ-intermediates by the hydrophobic tail, and a conserved surface patch of COQ9 allows its subsequent contact with COQ7 in a specific orientation such that the UQ intermediate is presented to the COQ7 active site. There are many interesting analogies between these two studies. Hajj Chehade et al. (2019) posit that the C-terminal domain of UbiJ forms an extended alpha helix and combines with UbiK to serve as the docking platform for the Ubi metabolon. Hence, UbiJ may chaperone UQ intermediates similar to the case described for COQ4 and COQ9.
Why involve a multi-enzyme complex in UQ biosynthesis? The UbiE-UbiK polypeptides form an exceptionally large and stable soluble metabolon that acts both to enhance catalytic efficiency and to sequester reactive and hydrophobic UQ-intermediates. UQ-intermediates contain catechol moieties and oxidized forms of UQ-intermediates form unsubstituted quinones. Such compounds are notorious for their reactivity and participation in oxidative damage and electrophilic stress (Waite, 2017).
In addition to binding the polyisoprenoid tails of the UQ-intermediates, the UbiJ, COQ4, and COQ9 polypeptides play essential roles as structural elements of the multi-subunit UQ biosynthetic complexes. In their absence, the steady-state levels of the other partner proteins are decreased. Thus, the formation of the UQ complexes may rely on the UQ-intermediates themselves as essential lipid components. Once UQ is formed, it must leave the Ubi metabolon, CoQ synthome, or Complex Q, and find its way to the membrane imbedded respiratory complexes. How this is achieved is another outstanding question. It is tempting to speculate that Coq10, a START domain-containing protein that binds and recognizes the ring moiety of UQ (Allan et al., 2013) may play an important role in mediating this process.
In summary, the findings of Chehade et al. (2019) indicate that a soluble metabolon comprised of seven polypeptides can chaperone hydrophobic UQ-intermediates and synthesize UQ in six sequential steps in the cytosol. Lohman et al. (2019) show that the COQ9 polypeptide accesses and binds analogous UQ-intermediates in the inner mitochondrial membrane and chaperones or presents them to the COQ7 partner protein in a eukaryotic biosynthetic UQ complex. Both studies will stimulate much new work assessing how these exceptionally hydrophobic UQ-intermediates and UQ itself is trafficked within and between cellular membranes.
Footnotes
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- Alcázar-Fabra M, Trevisson E, Brea-Calvo G (2018) Essays Biochem. 62, 377–398. [DOI] [PubMed] [Google Scholar]
- Allan CM, Hill S, Morvaridi S, Saiki R, Johnson JS, Liau WS, Hirano K, Kawashima T, Ji Z, Loo JA, Shepherd JN, Clarke CF (2013) Biochim. Biophys. Acta 1831, 776–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad AM, Bradley MC, Fernández-Del-Río L, Nag A, Tsui HS, Clarke CF (2018) Essays Biochem. 62, 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentinger M, Tekle M, Dallner G, (2010) Biochem. Biophys. Res. Commun 396, 74–79. [DOI] [PubMed] [Google Scholar]
- Hajj Chehade M, Pelosi L, Fyfe CD, Loiseau L, Rascalou B, Brugière S, Kazemzadeh K, Vo CD, Ciccone L, Aussel L, Couté Y, Fontecave M, Barras F, Lombard M, Pierrel F (2019) Chem. Biol 26, this issue 1–11. [DOI] [PubMed] [Google Scholar]
- Lohman DC, Aydin D, Von Bank HC, Smith RW, Linke V, Weisenhorn E, McDevitt MT, Hutchins P, Wilkerson EM, Wancewicz B, Russell J, Stefely MS, Beebe ET, Jochem A, Coon JJ, Bingman CA, Dal Peraro M, Pagliarini DJ (2019) Mol. Cell 73, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn PJ (2012) Prog. Lipid Res 51, 179–198. [DOI] [PubMed] [Google Scholar]
- Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, and Falk MJ (2010) Dev. Disabil. Res. Rev 16, 200–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Pagliarini DJ (2017) Trends Biochem. Sci 42, 824–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite JH (2017) J. Exp Biol 220, 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]