Weak acids are widely used as food preservatives, as they are very effective at preventing the growth of most species of bacteria and fungi. However, some species of molds can survive and grow in the concentrations of weak acid employed in food and drink products, thereby causing spoilage with resultant risks for food security and health. Current knowledge of weak-acid resistance mechanisms in these fungi is limited, especially in comparison to that in yeasts. We characterized gene functions in the spoilage mold species Aspergillus niger which are important for survival and growth in the presence of weak-acid preservatives. Such identification of weak-acid resistance mechanisms in spoilage molds will help in the design of new strategies to reduce food spoilage in the future.
KEYWORDS: Aspergillus, food spoilage, fungi, transcription factors, weak-acid resistance
ABSTRACT
Propionic, sorbic, and benzoic acids are organic weak acids that are widely used as food preservatives, where they play a critical role in preventing microbial growth. In this study, we uncovered new mechanisms of weak-acid resistance in molds. By screening a library of 401 transcription factor deletion strains in Aspergillus fumigatus for sorbic acid hypersensitivity, a previously uncharacterized transcription factor was identified and named weak acid resistance A (WarA). The orthologous gene in the spoilage mold Aspergillus niger was identified and deleted. WarA was required for resistance to a range of weak acids, including sorbic, propionic, and benzoic acids. A transcriptomic analysis was performed to characterize genes regulated by WarA during sorbic acid treatment in A. niger. Several genes were significantly upregulated in the wild type compared with a ΔwarA mutant, including genes encoding putative weak-acid detoxification enzymes and transporter proteins. Among these was An14g03570, a putative ABC-type transporter which we found to be required for weak-acid resistance in A. niger. We also show that An14g03570 is a functional homologue of the Saccharomyces cerevisiae protein Pdr12p and we therefore name it PdrA. Last, resistance to sorbic acid was found to be highly heterogeneous within genetically uniform populations of ungerminated A. niger conidia, and we demonstrate that pdrA is a determinant of this heteroresistance. This study has identified novel mechanisms of weak-acid resistance in A. niger which could help inform and improve future food spoilage prevention strategies.
IMPORTANCE Weak acids are widely used as food preservatives, as they are very effective at preventing the growth of most species of bacteria and fungi. However, some species of molds can survive and grow in the concentrations of weak acid employed in food and drink products, thereby causing spoilage with resultant risks for food security and health. Current knowledge of weak-acid resistance mechanisms in these fungi is limited, especially in comparison to that in yeasts. We characterized gene functions in the spoilage mold species Aspergillus niger which are important for survival and growth in the presence of weak-acid preservatives. Such identification of weak-acid resistance mechanisms in spoilage molds will help in the design of new strategies to reduce food spoilage in the future.
INTRODUCTION
Microbiological spoilage of food and drinks is a serious threat to food security and human health. It is estimated that 25% of global food produced annually is lost due to contamination and degradation by microorganisms (1). Such spoilage also directly imperils human health, for example, due to the production of toxins by the microorganism. Preventing microbial spoilage is therefore a key to safeguarding food supply and safety. The use of chemical food preservatives to inhibit the growth of bacteria and fungi is a ubiquitous and generally effective strategy in reducing spoilage (2). Some of the most commonly used preservatives are weak organic acids such as propionic, sorbic, and benzoic acids. These are usually included in food and drink products in the form of calcium, potassium, and sodium salts, respectively.
Weak-acid preservatives are broad-spectrum antimicrobials that directly inhibit the growth of yeasts, molds, and bacteria. Although their precise mechanism of action has not yet been fully determined, it is known that weak-acid preservatives cause a reduction in cytoplasmic pH (3, 4) and inhibit nutrient uptake (5, 6). It is also known that weak acids tend to be fungistatic rather than fungicidal, especially at the levels legally permitted in food and drinks. In most cases, microbial growth is completely inhibited by the weak-acid levels used in food and drink products. However, certain species of yeasts and molds demonstrate elevated resistance to weak acids and are therefore capable of causing food spoilage (7, 8).
Weak-acid resistance can be attributed in part to the enzymatic degradation of certain acids (e.g., benzoic, sorbic, and cinnamic acids), which nullifies their antimicrobial effects. Benzoate can be catabolized through a pathway involving an initial hydroxylation step. The enzyme responsible (benzoate para-hydroxylase) has been found to be required for resistance to benzoic acid in Aspergillus niger and Aspergillus nidulans (9, 10). Sorbic and cinnamic acids (and certain other structurally related acids) are degraded by decarboxylation (11). In molds such as A. niger, a cluster of three genes is required for this process, encoding a transcription factor (SdrA), a decarboxylase (CdcA; formerly OhbA1 or FdcA) and a prenyltransferase (PadA) (12–15). The deletion of any of these genes reduces, but does not eliminate, resistance to sorbic acid. Thus, additional and as-yet-uncharacterized mechanisms of weak-acid resistance must operate in this mold species. Enzymatic decarboxylation of weak acids also occurs in numerous yeast species (16). However, contrary to the case in molds, deletion of the phenylacrylic acid decarboxylase gene (PAD1) in the yeast Saccharomyces cerevisiae does not decrease weak-acid resistance (16). Furthermore, certain spoilage yeasts do not appear to decarboxylate weak acids at all, suggesting that alternative mechanisms of resistance also operate in these species.
Mechanisms of weak-acid resistance have been best characterized in S. cerevisiae. One of the key genes required for resistance is PDR12, encoding an ATP-binding cassette (ABC) transporter (17). PDR12 is required for resistance to carboxylic acids with chain lengths between 1 and 7, proposedly by mediating the efflux of weak-acid anions from the cell in an energy-dependent manner (18). PDR12 is itself transcriptionally regulated by War1p, a Zn2Cys6 zinc finger transcription factor that binds to weak-acid response elements (WARE) in the PDR12 promoter (19). Another transcription factor, Haa1p, is also required for resistance to weak acids in S. cerevisiae by regulating the transcription of membrane multidrug transporters (Tpo2p and Tpo3p) among other, less-well-characterized genes (20). High-throughput mutant screens have helped identify many other genes which influence weak-acid resistance in S. cerevisiae. For example, Mollapour et al. (21) reported 237 genes which were required for wild-type resistance to sorbic acid and a further 34 genes which resulted in enhanced sorbic acid resistance when deleted. A similar study, also on S. cerevisiae, revealed 650 determinants of acetic acid resistance (22). Unfortunately, there is a distinct lack of equivalent data in any other fungal species, including molds. Considering the propensity of mold fungi to cause food spoilage, understanding the genetic determinants of weak-acid resistance in these species is very important.
An additional and historically overlooked determinant of antimicrobial resistance is the phenotypic heterogeneity that exists within microbial cell populations. Phenotypic heterogeneity is a phenomenon observed within isogenic cell populations, whereby individual cells can display a markedly different phenotype despite being genetically identical. This has been recognized as an important determinant of microbial cell survival in response to antimicrobial agents and other environmental stressors (23–25). Phenotypic heterogeneity in weak-acid resistance (heteroresistance) has been found in cell populations of S. cerevisiae and the spoilage yeast Zygosaccharomyces bailii (8, 26, 27). However, there has been no investigation to date of whether weak-acid resistant subpopulations exist in populations of mold spores, although heterogeneity is known to arise in A. niger spore populations as a consequence of asynchronous conidial maturation (28). The presence of weak-acid-resistant spore subpopulations could have significant implications for spoilage control strategies and is therefore worthy of investigation.
In this study, we report the identification and characterization of a novel transcription factor (weak acid resistance A) that is required for resistance to weak-acid preservatives in A. niger and A. fumigatus. Furthermore, we identify and characterize genes that are putatively regulated by WarA, including a gene encoding a putative membrane transporter protein with similarity to S. cerevisiae Pdr12p and which, we show, mediates weak-acid resistance and heteroresistance in A. niger. These data significantly enhance our understanding of weak-acid resistance in molds and highlight both similarities and differences in weak-acid resistance strategies between yeast and mold fungi.
(This article was submitted to an online preprint archive [29].)
RESULTS
A screen for transcription factor deletion strains sensitive to sorbic acid identifies warA.
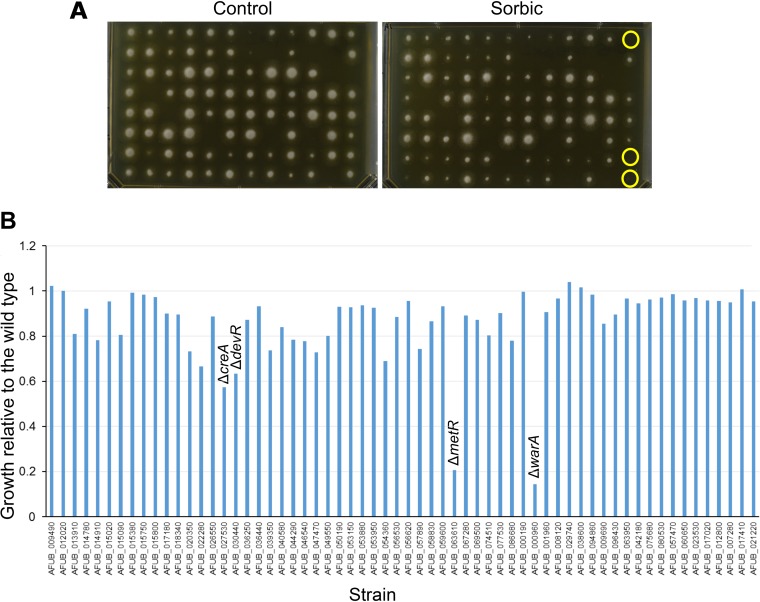
To find genes associated with weak-acid resistance, an Aspergillus fumigatus transcription factor deletant collection (30) was screened for sorbic acid sensitivity. Aspergillus fumigatus is not commonly associated with food spoilage, and it displays relatively high sensitivity to weak acids such as sorbic acid (14). However, deletant collections are available in A. fumigatus, unlike aspergilli associated with food spoilage. It was reasoned that transcription factors associated with weak-acid resistance in A. fumigatus may be conserved in related spoilage species such as A. niger. This resource comprised a library of 401 deletion strains of nonessential transcription factors. To determine sensitivity of the deletion strains, radial growth was compared on agar medium with and without sorbic acid (Fig. 1). This revealed two deletion strains which were highly sensitive to sorbic acid compared with the wild-type strain (ΔmetR and ΔAFUB_000960 mutants) (Fig. 1 and 2A). A number of other strains exhibited moderate sensitivity to sorbic acid, including the ΔcreA (ΔAFUB_027530), ΔdevA (ΔAFUB_030440) (Fig. 1B and 2A), ΔrfeD (ΔAFUB_022280), ΔAFUB_020350, and ΔAFUB_054360 mutants (Fig. 1B).
FIG 1.
Screening of A. fumigatus deletant library. (A) Example of A. fumigatus deletant library screen. Conidial suspensions of the different deletants were arrayed in 96-well plates and transferred to growth medium using a 96-pin tool. Examples of putatively sorbic acid sensitive strains are circled in yellow. (B) Sensitivity of A. fumigatus transcription factor deletion strains to sorbic acid. Sixty-two strains were identified from the initial screen in panel A as putatively sorbic acid hypersensitive and subjected to a second round of screening, as outlined in Materials and Methods. Sensitivity to sorbic acid relative to the WT strain is shown (a value of 1 indicates identical sensitivity of the deletion strain to the WT, according to radial growth). ΔwarA refers to the ΔAFUB_000960 mutant.
FIG 2.
Growth of A. fumigatus transcription factor deletion strains on medium containing weak acids. (A) Radial growth of A. fumigatus transcription factor deletion strains on agar containing 0.5 mM sorbic acid. Images were captured after 3 days of growth at 37°C. (B) Radial growth of the A. fumigatus ΔwarA mutant and wild type on agar containing weak acids. Plates were inoculated with a 10-fold dilution series of conidial suspensions; approximate numbers of conidia are indicated above the pictures. Images were captured after 2 days of growth at 28°C and are representative of 2 or 3 independent experiments. The concentrations of the acids used are given in Materials and Methods.
MetR is a basic leucine zipper domain (bZIP)-type transcription factor mediating the transcriptional regulation of genes involved in sulfur uptake and utilization (31). Because sorbic acid is known to decrease cellular uptake of some nutrients (6, 32), it was hypothesized that sulfur limitation could be a cause of sorbic acid sensitivity in the ΔmetR mutant strain. To test this, the sorbic acid sensitivity of the ΔmetR mutant strain was determined in medium supplemented with the sulfur-containing amino acid methionine. This showed that the sorbic acid sensitivity of the ΔmetR mutant strain was abolished in the presence of supplementary methionine (see Fig. S1 in the supplemental material).
Radial growth of the A. fumigatus ΔmetR mutant strain on medium containing 0.5 mM sorbic acid with or without 0.5 mM methionine. Download FIG S1, TIF file, 0.8 MB (817.4KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
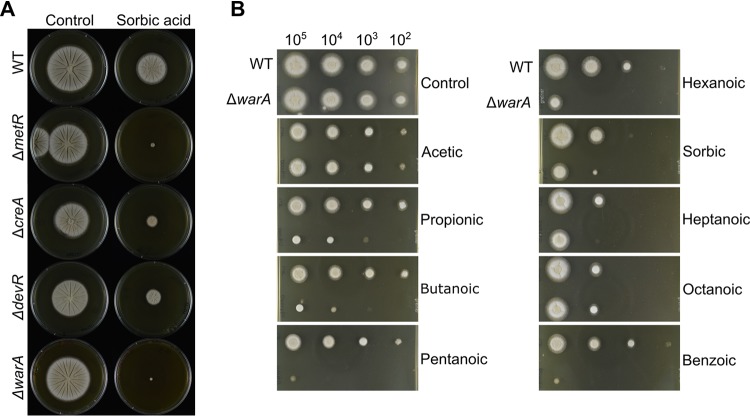
AFUB_000960 encodes a Zn2Cys6-type transcription factor which contains a fungus-specific transcription factor domain (PF11951). To further investigate the role of AFUB_000960 in weak-acid resistance, sensitivity of the deletion strain to a range of weak acids was evaluated (Fig. 2B). The ΔAFUB_000960 mutant strain was sensitive to propionic, butanoic, pentanoic, hexanoic, sorbic, and benzoic acids but not to acetic acid. Because of these phenotypes, AFUB_000960 was named weak acid resistance A (warA).
A WarA orthologue in A. niger is important for weak-acid resistance.
To determine whether orthologues of WarA are present in other species of fungi, a BLAST search using the WarA of A. fumigatus (AfuWarA) protein sequence as a query was conducted. In A. niger, this identified An08g08340, a protein with 46.2% identity and 63% similarity to the A. fumigatus WarA protein. Orthologues of AfuwarA were also found to be present in Penicillium and Botrytis spp.
Aspergillus niger is highly resistant to weak acids (especially sorbic and benzoic acids) and can readily cause food and drink spoilage. To determine whether warA is also required for weak-acid resistance in A. niger, An08g08340 was deleted by a targeted gene replacement approach, with successful deletion confirmed by PCR and Southern blotting (Fig. S2). The sensitivity of the ΔAn08g08340 mutant strain to different weak acids was evaluated (Fig. 3). In contrast to the ΔwarA mutant of A. fumigatus, the A. niger ΔAn08g08340 mutant strain demonstrated only slight sensitivity to sorbic acid, which was most apparent when individual conidia of the ΔwarA mutant strain were spread onto medium containing sorbic acid (Fig. S3A). However, the deletant was highly sensitive to propionic, butanoic, and benzoic acids, as evaluated by radial growth on agar (Fig. 3). Determination of MICs in broth also corroborated the radial growth data; the MIC in benzoic acid was 4.5 ± 0.5 mM in the wild type (WT; n = 3) compared with 3.2 ± 0.2 mM in the ΔwarA mutant (n = 3). The MIC in butanoic acid was 8.6 ± 0 mM in the WT (n = 2) and 6.9 ± 0.3 mM in the ΔwarA mutant (n = 3). Resistance was restored to WT levels when An08g08340 was reintroduced into the ΔAn08g08340 mutant strain (Fig. S4). Thus, An08g08340 has an important role in weak-acid resistance in A. niger and was named warA also in this species.
FIG 3.
Radial growth of the A. niger ΔwarA mutant growing on different weak acids. Plates were inoculated with a 10-fold dilution series of conidial suspensions; approximate numbers of conidia are indicated above the pictures. Images were captured after 2 days of growth at 28°C and are representative of 2 or 3 independent experiments. The concentrations of acids used are given in Materials and Methods.
PCR and Southern blot confirmation of warA deletion. (A) Targeted gene deletion strategy. The warA ORF was replaced with a hygromycin resistance cassette. (B) PCR confirmation of warA deletion. Primer pair 1 was used to confirm deletion of warA ORF; deletion strains are negative, and the WT is positive. Primer pair 2 was used to confirm the integration of HygR at the warA locus. (C) Southern blotting of warA and cdcA/warA deletion strains. The gDNA of strains was digested with the HindIII restriction enzyme. Membranes were hybridized with a probe consisting of digoxigenin-UTP-labeled HygR. Single bands confirm single integration of the deletion cassette into the A. niger genome. The ΔwarA_12 and ΔΔcdcA/warA_12 transformants were used for experiments. Download FIG S2, TIF file, 0.8 MB (801.3KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Growth of the ΔwarA mutant strain conidia on medium containing sorbic acid. Plates were inoculated with ∼100 conidia and incubated for 2 days (control medium) or 3 days (medium containing 1 mM sorbic acid). (B) Radial growth of the ΔΔcdcA/warA double mutant strain on medium containing 1 mM sorbic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions; approximate numbers of conidia are indicated at the top. Images were captured after 2 days of growth at 28°C and are representative of 2 or 3 independent experiments. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Radial growth of complemented ΔwarA strains on medium containing 2 mM benzoic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions. Approximate numbers of conidia are indicated above the pictures. Images were captured after 2 days of growth at 28°C. Two independent complemented lines are shown. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sorbic acid (and structurally related acids) are known to be detoxified by decarboxylation in A. niger but not in A. fumigatus (14, 15). The decarboxylation process involves three linked genes, cdcA, padA, and sdrA (12, 14). CdcA is the key enzyme involved in the decarboxylation, whereas SdrA is a transcription factor regulating the expression of CdcA, and PadA synthesizes a cofactor for CdcA. It was hypothesized that the mild sorbic acid sensitivity of the A. niger ΔwarA mutant may be due to downregulation of the cdcA, padA, or sdrA genes in the ΔwarA mutant strain. To investigate the relationship between WarA and weak-acid decarboxylation, a ΔΔcdcA/warA double mutant was constructed. The ΔΔcdcA/warA double mutant strain was more sensitive to sorbic acid than was the ΔcdcA mutant strain (Fig. S3B), suggesting a cdcA-independent role for warA in resistance of A. niger to sorbic acid.
Determination of WarA-regulated genes by transcriptomic analysis during weak-acid stress of A. niger.
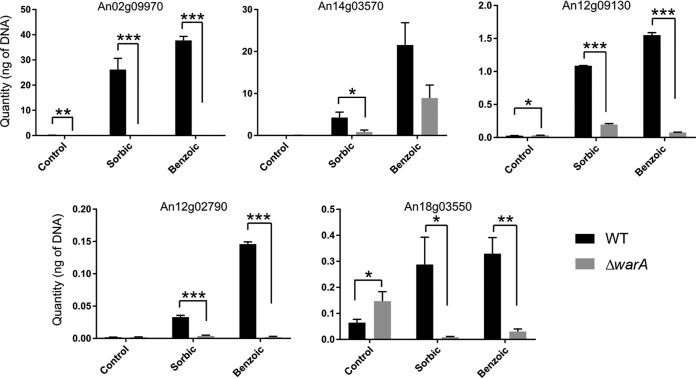
The weak-acid sensitivity of the ΔwarA mutant suggested that WarA regulates genes which are important for weak-acid resistance. Previously, genes upregulated by sorbic acid exposure in A. niger were successfully identified by exposing conidia of the wild type (WT) to sorbic acid during germination (6). In order to identify which genes are differentially regulated in the A. niger ΔwarA mutant, RNA sequencing (RNA-seq) analysis was conducted with WT and ΔwarA mutant conidia germinated in the presence or absence of sorbic acid. Germination of WT conidia for 1 h in the presence of 1 mM sorbic acid resulted in 3,274 differentially expressed genes (false-discovery rate [FDR]-adjusted P < 0.05) (1,885 upregulated and 1,467 downregulated) in comparison with conidia germinated in the absence of sorbic acid. In ΔwarA mutant conidia, 3,442 genes were differentially expressed during germination in the presence of sorbic acid (1,885 upregulated and 1,557 downregulated), in comparison with germination without sorbic acid. Importantly, a number of genes were identified that were highly upregulated in the WT during sorbic acid exposure but not in the ΔwarA mutant (Tables 1 and S1). This included a gene encoding benzoate para-hydroxylase (bphA), an enzyme known to be required for benzoate detoxification (9), which had a log2 fold change (log2FC) of 6.50 in the WT, compared with a log2FC of −0.51 in the ΔwarA mutant. A number of uncharacterized enzymes also required warA for normal upregulation by sorbic acid, e.g., An12g09130 encoding a putative dienelactone hydrolase, and An12g02790 encoding a putative isoflavone reductase (log2FC 5.95 in the WT and log2FC 0.36 in the ΔwarA mutant), as well as several genes encoding putative transporter proteins. Of particular interest among these transporters was An14g03570, an ABC-type transporter with 56% amino acid sequence similarity to S. cerevisiae Pdr12p. Pdr12p has a crucial role in weak-acid detoxification in S. cerevisiae (17, 18). To support the RNA-seq data, five of the genes showing differential expression between the WT and ΔwarA strains were selected for quantitative reverse transcription-PCR (qRT-PCR) analysis (Fig. 4). The qRT-PCR data supported the trends in gene expression seen in the RNA-seq data set; all of the selected genes had a lower transcript abundance in the ΔwarA mutant than in the WT during sorbic acid treatment. In addition, transcript abundances of the selected genes were compared by qRT-PCR during benzoic acid treatment. As expected, all of the genes upregulated by sorbic acid were also upregulated by benzoic acid and had lower transcript abundances in the ΔwarA mutant than in the WT (Fig. 4).
TABLE 1.
Transcriptomics data for selected genes upregulated in the WT during sorbic acid treatment and differentially expressed in the WT versus ΔwarA mutanta
| Gene_ID | Log2FC forb
: |
RPKM forc
: |
Function(s)d | ||
|---|---|---|---|---|---|
| WT_sorbic vs WT control | ΔwarA_sorbic vs ΔwarA control | WT_sorbic | ΔwarA_sorbic | ||
| An14g03570 | 9.35 | 7.1 | 611.3 | 146 | ABC-type transporter with similarity to S. cerevisiae Pdr12p |
| An12g09130 | 9.31 | 5.98 | 6,055.3 | 224.7 | Possible dienelactone hydrolase function |
| An02g09970 | 8.56 | 4.35 | 1,532.7 | 7.3 | Ortholog(s) have role in drug response, hexose transport, pathogenesis |
| An13g02460 | 8.24 | 3.08 | 283 | 0.3 | Protein similar to NRPS (NRPS-like) |
| An06g02170 | 7.98 | 1.51 | 80.3 | 3 | Ortholog(s) have S-adenosyl-Met-dependent methyltransferase activity |
| An13g03170 | 7.25 | 5.73 | 1,433.3 | 465.3 | Unknown |
| An12g09120 | 7.14 | 2.87 | 312.3 | 31.7 | Unknown |
| An09g03500 | 6.5 | −0.51 | 226.3 | 4 | Putative benzoate-para-hydroxylase; 3-hydroxybenzoate 4-hydroxylase |
| An12g02790 | 5.95 | 0.36 | 135.3 | 2.7 | Isoflavone reductase-phenylcoumaran benzylic ether reductase type |
| An08g07850 | 5.81 | 3.77 | 11,809.3 | 3,248.7 | Unknown |
| An01g05850 | 5.59 | 1.5 | 75.7 | 3 | Thioesterase domain protein |
| An08g01560 | 5.22 | −1.47 | 802.7 | 5.7 | Ortholog(s) have role in meiotic cell cycle, regulation of TORC1 signaling |
| An04g05240 | 5.02 | 2.84 | 833 | 259.7 | Unknown |
| An11g04385 | 4.9 | 2.02 | 74 | 5.7 | Possible ubiquitin hydrolase |
| An13g02450 | 4.21 | 0 | 492.7 | 0 | Six-hairpin glycosidase |
| An08g01980 | 3.31 | 1.71 | 309 | 74.7 | Unknown |
| An13g02290 | 3.06 | −0.95 | 73.3 | 1.7 | Possible 3-dehydroshikimate dehydratase |
| An09g05760 | 2.74 | −0.47 | 397.7 | 40 | Ortholog(s) have actomyosin contractile ring, intermediate layer localization |
| An18g03550 | 2.45 | −2.43 | 1,506.7 | 22.3 | Similar to yeast Arr3 arsenate transporter |
| An08g05750 | 2.26 | 0.76 | 1,095 | 298 | Unknown |
| An16g00700 | 0.96 | −3.07 | 345 | 20.7 | Has domain(s) with predicted 2Fe-2S cluster binding, oxidoreductase activity |
See Table S1 for a full list of genes.
FC, fold change.
RPKM, reads per kilobase per million.
NRPS, nonribosomal peptide synthase; TORC1, target of rapamycin complex 1.
FIG 4.
qRT-PCR of genes differentially regulated in WT and ΔwarA mutant strains of A. niger. Transcript abundances in WT (black bars) and ΔwarA mutant (gray bars) conidia germinated in control medium or in the presence of 1 mM sorbic acid or 1 mM benzoic acid. Error bars are standard deviation of the results from 3 technical replicates. WT and ΔwarA mutant transcript abundances were compared by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RPKM and log2FC values for A. niger genes (Excel file). Download Table S1, XLSX file, 0.9 MB (947.6KB, xlsx) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of An02g09970 and An14g03570.
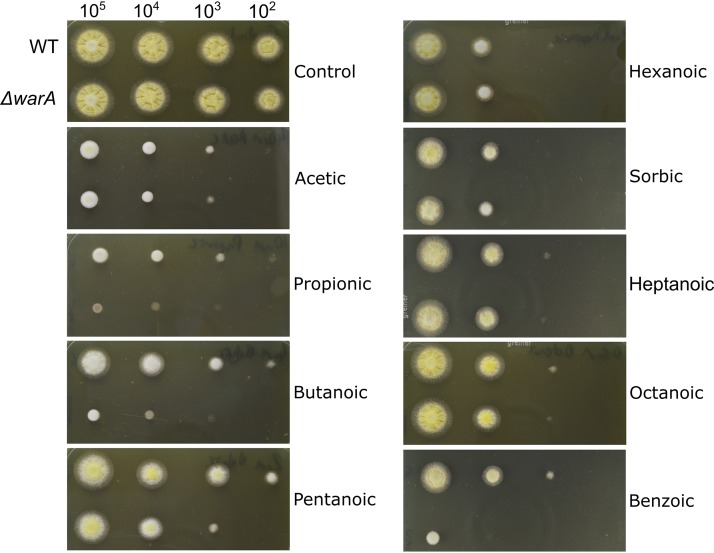
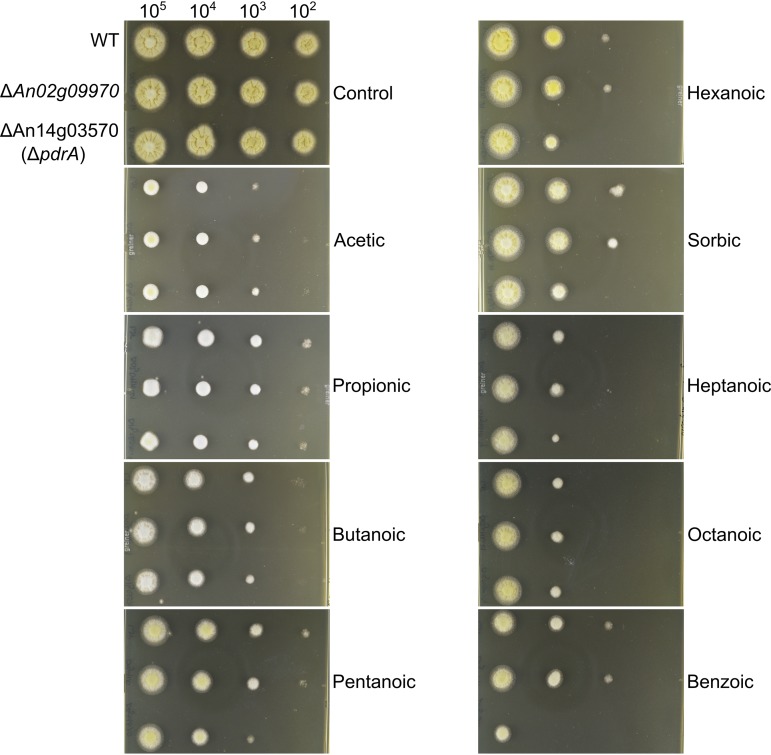
The transcriptomic analysis identified genes that are downregulated in the A. niger ΔwarA mutant, relative to the WT strain, during weak-acid stress. These genes may therefore have a role in weak-acid resistance. To investigate this, two genes of interest (An02g09970 and An14g03570) were selected for further characterization. An02g09970 encodes a putative transmembrane transporter of the major facilitator superfamily (MFS) and was selected for further investigation due to its extremely high transcript abundance during sorbic acid treatment and large disparity in transcript abundances between the WT and ΔwarA mutant strains (log2FC of 8.56 in the WT versus log2FC of 4.35 in the ΔwarA mutant) (Table 1). The protein also shares significant sequence similarity with Tpo2 and Tpo3, S. cerevisiae proteins involved in resistance to acetic, propionic, and benzoic acids (20). An14g03570 encodes an ABC-type transporter with similarity to the weak-acid detoxification protein Pdr12p in S. cerevisiae, as stated above. Both genes were deleted in A. niger by a targeted gene replacement approach, and mutant genotypes were confirmed by PCR and Southern blotting (Fig. S5). The sensitivity of the constructed deletion strains to weak acids was then evaluated. The ΔAn02g09970 mutant did not exhibit altered sensitivity to any of the weak acids tested (Fig. 5). However, the ΔAn14g03570 mutant was more sensitive to sorbic, pentanoic, and benzoic acids (Fig. 5 and Table 2), and resistance was restored to WT levels when An14g03570 was reintroduced into the ΔAn14g03570 mutant strain (Fig. S6). Because of the similarity in sequence and function between An14g03570 and Pdr12p, An14g03570 was named PdrA.
FIG 5.
Radial growth of ΔAn02g09970 and ΔAn14g03570 (ΔpdrA) mutant strains growing on weak acids. Plates were inoculated with a 10-fold dilution series of conidial suspensions.
TABLE 2.
MIC values for A. niger WT and the ΔAn14g03570 (ΔpdrA) mutant strain
| Acid | MIC (mM) fora
: |
|
|---|---|---|
| WT | ΔpdrA mutant | |
| Benzoic | 4.67 ± 0.12 | 3.40 ± 0.20 |
| Pentanoic | 3.70 ± 0.35 | 2.60 ± 0.30 |
| Sorbic | 5.00 ± 0.35 | 3.80 ± 0.35 |
Values are averages of 3 biological replicates ± the standard deviation.
PCR and Southern blotting of An02g09970 and An14g03570 (pdrA) deletion strains. (A) Targeted gene deletion strategy. The An02g09970 and An14g03570 ORFs were replaced with a hygromycin resistance cassette. (B) PCR confirmation of An02g09970 deletion. Primer pair 1 was used to confirm deletion of the An02g09970 ORF; deletion strains are negative, and the WT is positive. Primer pairs 2 and 3 were used to confirm the integration of HygR at the An02g09970 locus. (C) PCR confirmation of An14g03570 deletion. Primer pair 1 was used to confirm deletion of the An14g03570 ORF; deletion strains are negative, and the WT is positive. Primer pairs 2 and 3 were used to confirm integration of HygR at the An14g03570 locus. (D) Southern blotting of the ΔAn02g09970 and ΔAn14g03570 mutant strains. The gDNA of the strains was digested with the restriction enzymes XbaI (for the ΔAn02g09970 mutant) or EcoRV (for the ΔAn14g03570 mutant). Membranes were hybridized with a probe consisting of digoxigenin-UDP-labeled HygR. Single bands confirm single integration of the deletion cassette into the A. niger genome (multiple integrations apparent for the ΔAn02g09970_39 mutant). The ΔAn14g03570_4 and ΔAn02g09970_29 transformants were used for experiments. Download FIG S5, TIF file, 1.0 MB (1,000.4KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Radial growth of complemented ΔpdrA mutant strains on medium containing 2 mM benzoic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions. Two independent complemented lines are shown. A ΔpdrA mutant strain containing the empty pAN7.1BAR plasmid is also shown. Download FIG S6, TIF file, 0.8 MB (861.6KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
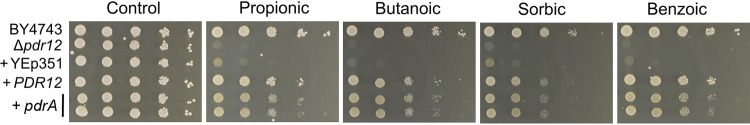
Complementation of the S. cerevisiae Δpdr12 mutant strain with PdrA (An14g03570).
The above-mentioned results showed that pdrA is required for resistance of A. niger to certain weak acids. Because PdrA has significant protein sequence similarity with S. cerevisiae Pdr12p (36% sequence identity, 56% similarity), it was hypothesized that these proteins could be functional homologues. To test this hypothesis, functional complementation of the S. cerevisiae Δpdr12 mutant strain was attempted. The cDNA sequence of A. niger pdrA was cloned between the S. cerevisiae PDR12 promoter and terminator to allow for native regulation of pdrA in response to weak-acid stress in S. cerevisiae. The resulting plasmid (Fig. S7) was transformed into a S. cerevisiae Δpdr12 mutant strain. Transformants were tested for sensitivity to a range of weak acids. As hypothesized, pdrA could indeed functionally complement PDR12, as sensitivity of the Δpdr12 mutant strain to weak acids was largely rescued in cells transformed to express pdrA (Fig. 6). The resultant level of resistance was similar to that evident in Δpdr12 mutant cells expressing PDR12 from the same vector backbone.
FIG 6.
Growth of S. cerevisiae complemented strains on weak acids. Tenfold dilution series of S. cerevisiae strains (isogenic with the BY4743 wild type) were inoculated onto medium containing weak acids. The Δpdr12 mutant strain was transformed with either empty plasmid (+YEp351), YEp351 plasmid containing PDR12 (+PDR12), or YEp351 plasmid containing the pdrA ORF and PDR12 promoter and terminator (+pdrA). Two independent transformants of the +pdrA strain are shown.
Plasmid map of YEp351 containing An14g03570 (pdrA). Download FIG S7, TIF file, 0.6 MB (659.6KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
pdrA is a determinant of heteroresistance to sorbic acid in A. niger conidia.
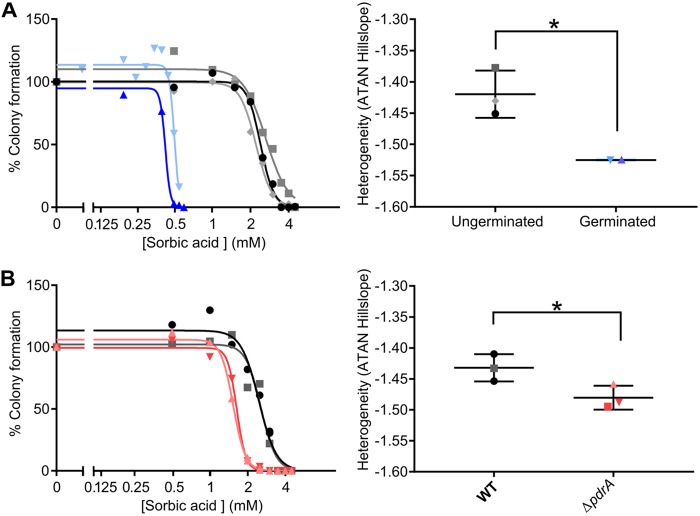
Resistance to weak-acid preservatives has been found to be heterogeneous between individual cells of genetically uniform cell populations of the yeasts Zygosaccharomyces bailii and S. cerevisiae (8, 26, 27). We observed a similar phenomenon in populations of A. niger conidia, whereby small subpopulations were capable of germinating and forming colonies at high concentrations (>4 mM) of sorbic acid (Fig. 7A). Conidia harvested from colonies that grew on these high concentrations of sorbic acid did not retain increased resistance upon direct reinoculation to sorbic acid-containing medium (data not shown), suggesting that these were transient, nonheritable phenotypes, i.e., not due to genotypic variants within the population. To test whether this heteroresistance had its origin in the ungerminated conidial state, conidia were also pregerminated for 6 h before spread plating onto sorbic acid-containing medium. This showed that germinated conidia were much more susceptible to sorbic acid (Fig. 7A). Moreover, resistance to sorbic acid among pregerminated conidia was much more homogeneous than when the resistance assay commenced with ungerminated conidia, as evidenced by the gradients of the dose inhibition curves; such dose-response curves reflect heterogeneity, with shallower curves indicating greater heterogeneity (23, 33). Thus, at least some factors determining heteroresistance to sorbic acid are specific to ungerminated conidia and are lost upon germination. Given the contributions of warA and pdrA to sorbic acid resistance in A. niger, it was tested whether these genes could be determinants of heteroresistance. The dose-response curves of ΔpdrA and ΔwarA mutant conidia demonstrated that pdrA makes a significant contribution to sorbic acid heteroresistance in A. niger conidia, whereas warA does not (Fig. 7B, data for ΔwarA mutant not shown).
FIG 7.
Sorbic acid dose response curves for A. niger conidia. (A) Dose-response curves of germinated (blue lines) and ungerminated (black/gray lines) WT conidia, and comparison of slope values. Dose-response curve slope values were compared with 2-way Welch’s t test (P = 0.0404,) n = 2 or 3. (B) Dose-response curves of WT (black/gray lines) and ΔpdrA mutant (pink/red lines) conidia and comparison of dose-response curve slope values, compared using a 2-way Welch’s t test (P = 0.0468), n = 3. Two representative independent experiments are shown in the dose-response curve. ATAN, arctangent.
DISCUSSION
This study reports the discovery of novel factors determining weak-acid resistance in molds. By screening >400 transcription factor deletion strains in A. fumigatus, we discovered a previously uncharacterized transcription factor that is required for resistance to certain weak organic acids. This transcription factor, here named weak acid resistance A (warA), was also found to be present in the food spoilage mold A. niger, where it also plays an important role in weak-acid resistance. However, WarA appears to mediate resistance to different weak acids in A. fumigatus and A. niger. In A. fumigatus, the ΔwarA mutant strain is particularly sensitive to linear-chain acids 3 to 6 carbons in length, whereas in A. niger, the ΔwarA mutant strain is most sensitive to propionic, butanoic, and benzoic acids but exhibited less sensitivity to 5- and 6-carbon acids. Such differences in acid sensitivity may reflect differences in the WarA regulon between A. niger and A. fumigatus or in the divergence in gene function within the WarA regulon.
We sought to gain insight to the WarA regulon in A. niger by conducting a comparative transcriptomics experiment between germinating WT and ΔwarA conidia treated with sorbic acid. This approach identified several genes that appear to be regulated (either directly or indirectly) by WarA. These include a number of putative enzymes and transporter proteins, offering several candidates for future studies of weak-acid resistance mechanisms in A. niger. Among these candidates, we attempted to characterize two putative transporter protein-encoding genes. The first of these functions, An02g09970, is a transporter of the major facilitator superfamily, with sequence similarity to Tpo2p and Tpo3p in S. cerevisiae. Tpo2p and Tpo3p are transporters of the DHA1 (drug:H+ antiporter-1) family and are known to be required for resistance to acetic, propionic, and benzoic acids (20). However, the deletion of An02g09970 did not sensitize A. niger to any of the acids tested, and so the role of this gene remains unknown. It is possible that this transporter is responsible for detoxification of other xenobiotics not tested here (if indeed it has a role in detoxification at all), or that An02g09970 is functionally redundant with other A. niger genes. We also attempted to characterize pdrA, encoding an ABC-type transporter. The deletion of pdrA resulted in increased sensitivity to pentanoic, hexanoic, sorbic, and benzoic acids, substantiating a role for this protein in weak-acid resistance. Importantly, we were able to demonstrate that pdrA is a functional homologue of PDR12 in S. cerevisiae. Pdr12p is a key protein involved in weak-acid resistance of S. cerevisiae (17), where it is thought to efflux weak-acid anions from the cytoplasm in an energy-dependent manner (18). The identification of PdrA as a functional homologue of Pdr12p in a mold species such as A. niger shows that a similar mechanism of weak-acid detoxification by active efflux may operate in yeasts and molds. Interestingly, the Δpdr12 mutant functional complementation experiment demonstrated that pdrA confers resistance to a broader range of weak acids than was suggested by the weak-acid sensitivity of the ΔpdrA mutant strain. For example, pdrA complemented the propionic acid sensitivity of the S. cerevisiae Δpdr12 mutant, but the A. niger ΔpdrA mutant strain was not more sensitive to propionic acid than was the WT. This may indicate the presence of multiple, redundant mechanisms for resistance to certain weak acids in A. niger which may not operate in S. cerevisiae.
pdrA, as well as several other candidate WarA-regulated genes, were all upregulated in response to both sorbic and benzoic acids. This suggests a degree of overlap between transcriptomic responses to different weak acids, as also found in S. cerevisiae (34). Thus, although we characterized the WarA regulon by comparative transcriptomics in response only to sorbic acid in the present study, it is likely that many of the differentially expressed genes would be similarly regulated in response to other weak acids. There may be relevant consensus sequences within WarA-regulated genes, although these are not apparent from promoter sequence alignments we have carried out. In S. cerevisiae, a cis-acting weak-acid response element (WARE) was discovered in the promoter of PDR12 which is required for PDR12 induction by the transcription factor War1p (19).
The regulation of WarA itself is also an outstanding question. Recent evidence in S. cerevisiae suggests that weak-acid anions bind directly to the transcription factors War1p and Haa1p, thereby regulating their DNA-binding transcriptional activation (35). However, WarA shares very little sequence homology with either War1p or Haa1p. In fact, a BLAST search of the S. cerevisiae protein database with the WarA protein sequence yields no hits at all. Nevertheless, a similar mechanism of transcription factor activation cannot be ruled out for WarA, particularly as direct ligand binding has been established for a number of Zn2Cys6 family transcription factors (of which WarA is a member), including Pdr1p, Pdr3p, Leu3p, and Put3p (36–38).
During the course of this study, experiments with sorbic acid determined that genetically uniform populations of A. niger conidia demonstrate heteroresistance to this weak acid. Phenotypic heterogeneity within microbial cell populations has been demonstrated in a number of fungi in response to environmental stresses (reviewed in reference 23); however, this is the first report of weak-acid heteroresistance in fungal conidia. Interestingly, heteroresistance was decreased within 6 h of conidial germination, suggesting that at least some factors underlying this heterogeneity are limited to ungerminated conidia and are lost upon germination. Resistance to sorbic acid was also markedly lower in germinated conidia, which has also recently found to be the case for propionic acid (39). Heteroresistance to weak acids in fungal conidia has significant implications for the food industry, because spoilage of products may occur due to contamination with just a few conidia from a highly resistant subpopulation. Thus, future spoilage control strategies may have to take into account the presence of weak-acid heteroresistance, perhaps by specifically targeting resistant subpopulations.
Conidia of the ΔpdrA mutant strain showed a significantly more homogeneous response to sorbic acid. Heteroresistance typically arises from gene expression heterogeneity (or noise) (40), so the present results suggest that pdrA could be expressed heterogeneously within conidial populations; the conidia expressing more pdrA are potentially able to withstand sorbic acid stress. It is also noted that the deletion of pdrA did not eliminate sorbic acid heteroresistance in A. niger, so it is likely that other genes also contribute.
In summary, this study markedly advances our understanding of weak-acid resistance mechanisms in A. niger. The identification of WarA as a key transcription factor involved in weak-acid resistance allowed us in turn to identify many more genes which may also be important. Further work is required to determine how all these genes may contribute to weak-acid resistance. Moreover, we demonstrated here that a key weak-acid resistance mechanism operates in both S. cerevisiae and A. niger in the form of the functionally homologous ABC transporters Pdr12p and PdrA, respectively.
MATERIALS AND METHODS
Strains and media.
The Aspergillus fumigatus transcription factor deletant library, derived from wild-type strain MFIG001, was constructed by homologous recombination using gene replacement cassettes and transformation methodologies described previously (30, 41). Studies in Aspergillus niger were performed in the A. niger N402 background (referred to as the A. niger WT throughout) and an A. niger ΔcdcA mutant strain (14). Aspergillus strains were cultivated on slopes of potato dextrose agar (PDA) (Sigma) for 7 days at 28°C. Conidia were harvested using 0.1% (vol/vol) Tween 80 and filtered through a 40-μm cell strainer (Fisher) before counting on a hemocytometer. Studies in S. cerevisiae used the BY4743 background and isogenic Δpdr12 mutant strain cultivated on YEPD agar (2% glucose, 2% Bacto peptone [Oxoid], 1% yeast extract [Oxoid], 1.5% agar) at 30°C. The S. cerevisiae strains were obtained from Euroscarf (Frankfurt). Growth assays with weak acids (see below) were performed on YEPD agar (pH 4).
Deletant library screening and growth assays.
The first round of A. fumigatus transcription factor deletion library screening was performed in a 96-well array format. Conidial suspensions of the A. fumigatus strains were initially supplied in a 40% glycerol–0.01% phosphate-buffered saline (PBS) solution at a concentration of 4 × 107 ml−1. These were subsequently arrayed in 96-well plates at a concentration of 4 × 105 conidia ml−1 in 0.01% Tween 20, transferred using a 96-pin tool to Nunc OmniTray single-well plates containing YEPD agar (pH 4), and then incubated at 28°C for 2 to 3 days. Radial growth was measured using ImageJ and compared between the control medium and medium containing sorbic acid. The second round of screening was performed on 90-mm petri dishes. Plates were inoculated with 105 conidia and incubated at 37°C for 3 days. Radial growth was compared between the control medium (YEPD) and the same medium containing sorbic acid.
Subsequent growth assays with weak acids on solid medium were performed in Aspergillus spp. by inoculating YEPD agar (pH 4) supplemented with weak acids with 5 μl of conidial suspension, containing 105 to 102 conidia, and subsequent incubation at 28°C for 2 to 3 days. The weak-acid concentrations used for A. fumigatus were 15 mM acetic acid, 4 mM propionic acid, 1.5 mM butanoic acid, 0.75 mM pentanoic acid, 0.2 mM sorbic acid, 0.25 mM hexanoic acid, 0.5 mM benzoic acid, 0.08 mM heptanoic acid, and 0.05 mM octanoic acid. The weak-acid concentrations used for A. niger were 40 mM acetic acid, 10 mM propionic acid, 4 mM butanoic acid, 2 mM pentanoic acid, 1.5 mM sorbic acid, 2 mM hexanoic acid, 2 mM benzoic acid, 1 mM heptanoic acid, and 0.75 mM octanoic acid.
The MICs of weak acids were determined by placing 10 ml of YEPD broth (pH 4) into 30-ml McCartney bottles and inoculating with 104 conidia. The bottles were incubated statically at 28°C for 28 days, and the concentration of acids required to completely inhibit visible growth was recorded. The concentrations of acids used were at 0.2 mM increments for benzoic and sorbic acids, 0.3 mM increments for pentanoic and butanoic acids, and 2 mM increments for propionic acid.
Dose-response curves were generated by harvesting conidia as stated above, diluting to 500 spores ml−1, and spreading 200 μl of this onto YEPD agar (pH 4) containing sorbic acid. Plates were incubated at 28°C for up to 28 days and the colonies counted. For pregerminated conidia, conidia were first inoculated into 10 ml of YEPD-Tween 80 (6.66 ml YEPD and 3.33 ml of 0.1% Tween 80) to a final concentration of 500 spores ml−1 and incubated statically at 28°C for 6 h before spread plating and incubation as described above. For quantitative comparison of heteroresistance, Hill slopes were fitted to plots (% viability versus log10[sorbic acid]) using the Prism software, and arctangent values for the slopes were calculated with Excel to estimate relative heterogeneity (a shallower slope indicating higher heterogeneity) (33, 42).
RNA-seq and qRT-PCR.
For preparation of RNA, conidia of A. niger N402 were inoculated into 1 liter of YEPD broth (pH 4) to a final concentration of 106 conidia ml−1 and incubated at 28°C for 1 h, with shaking at 150 rpm. For sorbic acid or benzoic acid treatments, the medium was supplemented with 1 mM sorbic or 1 mM benzoic acid for the 1-h incubation, as these concentrations inhibit conidial germination over the course of the experiment but are not lethal (∼25% of the MIC values for these acids). Conidia were harvested by filtration through a Corning vacuum filtration unit and immediately used for RNA extraction. RNA was extracted using a Norgen Biotek plant/fungi total RNA extraction kit, as per the manufacturer’s instructions.
RNA-seq analysis was performed by the University of Liverpool Centre for Genomic Research. Three biological replicates were performed for each time point under each condition. Between 463 and 1,000 ng of total RNA (depending on available material) was poly(A) treated using the NEBNext poly(A) mRNA magnetic isolation module and subsequently purified using AMPure RNA XP beads. Successful depletion of rRNA was confirmed using Qubit fluorometric quantification (Thermo Fisher) and an Agilent 2100 Bioanalyzer. All of the depleted RNA was used as input material for the NEBNext Ultra directional RNA library prep kit for Illumina. Following 15 cycles of amplification, the libraries were purified using AMPure XP beads. Each library was quantified using a Qubit fluorometer and the size distribution assessed using the Bioanalyzer. These final libraries were pooled in equimolar amounts using the Qubit and Bioanalyzer data. The quantity was assessed using a Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit, while the quality and average fragment size were assessed using the high-sensitivity DNA kit on the Agilent Bioanalyzer. The RNA libraries were sequenced on an Illumina HiSeq 4000 platform with version 1 chemistry using sequencing by synthesis (SBS) technology to generate 2 × 150-bp paired-end reads. Initial processing and quality assessment of the sequence data were performed as follows. Briefly, base calling and demultiplexing of indexed reads were performed using CASAVA version 1.8.2 (Illumina). The raw FASTQ files were trimmed to remove Illumina adapter sequences using Cutadapt version 1.2.1 (43). The option “-O 3” was set, so the 3′ end of any reads which matched the adapter sequence over a stretch of at least 3 bp was trimmed off. The reads were further trimmed to remove low-quality bases, using Sickle version 1.200, with a minimum window quality score of 20. After trimming, reads shorter than 20 bp were removed. Reads were aligned to the A. niger CBS 588.13 genome sequence (http://www.aspergillusgenome.org/download/sequence/A_niger_CBS_513_88/current/A_niger_CBS_513_88_current_chromosomes.fasta.gz) using Tophat version 2.1.0 (44). The expression of each gene was calculated from the alignment files using HTseq-count (45). The raw count data were also converted into fragments per kilobase per million (FPKM) read values. The count numbers per gene were used during the subsequent differential expression analysis. All of the differential gene expression (DGE) analyses were performed in the R (version 3.3.3) environment using the DESeq2 package (46). Significantly differentially expressed genes were defined as those with an FDR-adjusted P value of <0.05.
For qRT-PCR analysis of gene expression, RNA was extracted as stated above. Genomic DNA was removed using the Turbo DNase-free kit (Invitrogen). cDNA was synthesized using SuperScript IV reverse transcriptase (Invitrogen) and oligo d(T)20 primer (Invitrogen), according to the manufacturer’s instructions. Transcripts were amplified using SYBR green master mix on an Applied Biosystems 7500 real-time PCR instrument and quantified against a standard curve of A. niger genomic DNA. The primer pairs used are listed in Table S2.
List of primers used in this study (Excel file). Download Table S2, XLSX file, 0.02 MB (20.7KB, xlsx) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene deletion studies and complementation in A. niger.
Gene deletion studies were performed in A. niger N402, the open reading frames (ORFs) of the target genes being replaced by a hygromycin resistance cassette (Fig. S2 and S6). Gene deletion cassettes were constructed by gap repair cloning in S. cerevisiae (47). Briefly, the hygromycin resistance cassette and the regions approximately 1 kb upstream and downstream of each target gene were amplified from genomic DNA by PCR (the primers used listed in Table S2). The hygromycin resistance cassette had a 20- to 30-bp homology with the 1-kb flanking regions, and each flanking region also had 20- to 30-bp homology with the multiple-cloning site of the YEp351 plasmid. The PCR products and HindIII-linearized YEp351 plasmid were transformed into S. cerevisiae BY4743, and transformants were selected by leucine prototrophy. Successful construction of the gene deletion cassettes was confirmed by PCR. The resulting gene deletion cassettes were amplified by PCR and purified using PCR purification columns (Macherey-Nagel) to produce a final linear gene deletion cassette. All PCRs were performed using Phusion high-fidelity DNA polymerase (New England BioLabs). Production of protoplasts and their transformation were performed using standard methods (48). Transformants were selected using 200 μg ml−1 hygromycin (Roche) and confirmed by PCR and Southern blotting (Fig. S2 and S6), using standard methods (49).
For complementation of A. niger gene deletion strains, the genes in question (warA and pdrA [An14g03570]) were amplified by PCR (primers listed in Table S2) and cloned into the SbfI site of the pAN7.1BAR plasmid (Fig. S8), which contains the BAR gene as a selectable marker (replacing the original hygromycin resistance cassette [50] and imparting resistance to phosphinothricin). PCR amplification included ∼1 kb upstream and ∼300 bp downstream of the ORF. Transformation of the resulting plasmids was performed as described above, except that transformants were selected using 5 mg ml−1 dl-phosphinothricin (Carbosynth) in YDA agar (yeast nitrogen base without amino acids, including 1.7 g liter−1 ammonium sulfate, 10 g liter−1 glucose, 2.25 g liter−1 ammonium nitrate, and 1 M sucrose; pH was adjusted to 7.0 using Na2HPO4, solidified with 1.2% [wt/vol] agar) (51). Transformants were subjected to an additional round of selection by growth on YDA agar containing 5 mg ml−1 dl-phosphinothricin.
Plasmid map of pAN7.1BAR. Download FIG S8, TIF file, 0.6 MB (646.7KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cloning and complementation in S. cerevisiae.
Complementation studies were performed in the S. cerevisiae Δpdr12 mutant strain. The complementation plasmid (Fig. S7) was constructed by yeast gap repair cloning (47). Briefly, the PDR12 promoter and terminator and PdrA ORF were amplified by PCR (primers listed in Table S2). Each amplified fragment included a 20- to 30-bp region of homology either with the YEp351 plasmid or with a neighboring fragment. The PCR products and HindIII-linearized YEp351 plasmid were transformed into the S. cerevisiae Δpdr12 mutant, and transformants were selected by leucine prototrophy (47). Complementation was also performed with the PDR12 ORF as a positive control for successful complementation. The S. cerevisiae Δpdr12 mutant strain was also transformed with the empty YEp351 plasmid as a negative control.
Data availability.
RNA-seq data have been deposited in GenBank under BioProject no. PRJNA594492.
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council (grant BB/N017129/1). This is an Industry Partnering Award in conjunction with Lucozade Ribena Suntory and Mologic Ltd. This work was also supported by the Wellcome Trust (grant 208396/Z/17/Z).
REFERENCES
- 1.Bondi M, Messi P, Halami PM, Papadopoulou C, de Niederhausern S. 2014. Emerging microbial concerns in food safety and new control measures. Biomed Res Int 2014:251512. doi: 10.1155/2014/251512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaczmarek M, Avery SV, Singleton I. 2019. Microbes associated with fresh produce: sources, types and methods to reduce spoilage and contamination. Adv Appl Microbiol 107:29–82. doi: 10.1016/bs.aambs.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Salmond CV, Kroll RG, Booth IR. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J Gen Microbiol 130:2845–2850. doi: 10.1099/00221287-130-11-2845. [DOI] [PubMed] [Google Scholar]
- 4.Plumridge A, Hesse SJA, Watson AJ, Lowe KC, Stratford M, Archer DB. 2004. The weak acid preservative sorbic acid inhibits conidial germination and mycelial growth of Aspergillus niger through intracellular acidification. Appl Environ Microbiol 70:3506–3511. doi: 10.1128/AEM.70.6.3506-3511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freese E, Sheu CW, Galliers E. 1973. Function of lipophilic acids as antimicrobial food additives. Nature 241:321–325. doi: 10.1038/241321a0. [DOI] [PubMed] [Google Scholar]
- 6.Novodvorska M, Stratford M, Blythe MJ, Wilson R, Beniston RG, Archer DB. 2016. Metabolic activity in dormant conidia of Aspergillus niger and developmental changes during conidial outgrowth. Fungal Genet Biol 94:23–31. doi: 10.1016/j.fgb.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warth AD. 1989. Relationships between the resistance of yeasts to acetic, propanoic and benzoic acids and to methyl paraben and pH. Int J Food Microbiol 8:343–349. doi: 10.1016/0168-1605(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 8.Stratford M, Steels H, Nebe-von-Caron G, Novodvorska M, Hayer K, Archer DB. 2013. Extreme resistance to weak-acid preservatives in the spoilage yeast Zygosaccharomyces bailii. Int J Food Microbiol 166:126–134. doi: 10.1016/j.ijfoodmicro.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boschloo JG, Moonen E, van Gorcom RFM, Hermes HFM, Bos CJ. 1991. Genetic analysis of Aspergillus niger mutants defective in benzoate-4-hydroxylase function. Curr Genet 19:261–264. doi: 10.1007/bf00355052. [DOI] [PubMed] [Google Scholar]
- 10.Fraser JA, Davis MA, Hynes MJ. 2002. The genes gmdA, encoding an amidase, and bzuA, encoding a cytochrome P450, are required for benzamide utilization in Aspergillus nidulans. Fungal Genet Biol 35:135–146. doi: 10.1006/fgbi.2001.1307. [DOI] [PubMed] [Google Scholar]
- 11.Stratford M, Plumridge A, Pleasants MW, Novodvorska M, Baker-Glenn CAG, Pattenden G, Archer DB. 2012. Mapping the structural requirements of inducers and substrates for decarboxylation of weak acid preservatives by the food spoilage mould Aspergillus niger. Int J Food Microbiol 157:375–383. doi: 10.1016/j.ijfoodmicro.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Lubbers RJM, Dilokpimol A, Navarro J, Peng M, Wang M, Lipzen A, Ng V, Grigoriev IV, Visser J, Hilden KS, de Vries RP. 2019. Cinnamic acid and sorbic acid conversion are mediated by the same transcriptional regulator in Aspergillus niger. Front Bioeng Biotechnol 7:249. doi: 10.3389/fbioe.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payne KAP, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJW, Trivedi DK, Goodacre R, Beveridge R, Barran P, Rigby SEJ, Scrutton NS, Hay S, Leys D. 2015. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 522:497–501. doi: 10.1038/nature14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plumridge A, Melin P, Stratford M, Novodvorska M, Shunburne L, Dyer PS, Roubos JA, Menke H, Stark J, Stam H, Archer DB. 2010. The decarboxylation of the weak-acid preservative, sorbic acid, is encoded by linked genes in Aspergillus spp. Fungal Genet Biol 47:683–692. doi: 10.1016/j.fgb.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Plumridge A, Stratford M, Lowe KC, Archer DB. 2008. The weak-acid preservative sorbic acid is decarboxylated and detoxified by a phenylacrylic acid decarboxylase, PadA1, in the spoilage mold Aspergillus niger. Appl Environ Microbiol 74:550–552. doi: 10.1128/AEM.02105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stratford M, Plumridge A, Archer DB. 2007. Decarboxylation of sorbic acid by spoilage yeasts is associated with the PAD1 gene. Appl Environ Microbiol 73:6534–6542. doi: 10.1128/AEM.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piper P, Mahé Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Mühlbauer M, Coote P, Kuchler K. 1998. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J 17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holyoak CD, Bracey D, Piper PW, Kuchler K, Coote PJ. 1999. The Saccharomyces cerevisiae weak-acid-inducible ABC transporter Pdr12 transports fluorescein and preservative anions from the cytosol by an energy-dependent mechanism. J Bacteriol 181:4644–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kren A, Mamnun YM, Bauer BE, Schuller C, Wolfger H, Hatzixanthis K, Mollapour M, Gregori C, Piper P, Kuchler K. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol Cell Biol 23:1775–1785. doi: 10.1128/mcb.23.5.1775-1785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sá-Correia I. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun 337:95–103. doi: 10.1016/j.bbrc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Mollapour M, Fong D, Balakrishnan K, Harris N, Thompson S, Schüller C, Kuchler K, Piper PW. 2004. Screening the yeast deletant mutant collection for hypersensitivity and hyper‐resistance to sorbate, a weak organic acid food preservative. Yeast 21:927–946. doi: 10.1002/yea.1141. [DOI] [PubMed] [Google Scholar]
- 22.Mira NP, Palma M, Guerreiro JF, Sá-Correia I. 2010. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewitt SK, Foster DS, Dyer PS, Avery SV. 2016. Phenotypic heterogeneity in fungi: importance and methodology. Fungal Biol Rev 30:176–184. doi: 10.1016/j.fbr.2016.09.002. [DOI] [Google Scholar]
- 24.Band VI, Weiss DS. 2019. Heteroresistance: a cause of unexplained antibiotic treatment failure? PLoS Pathog 15:e1007726. doi: 10.1371/journal.ppat.1007726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson DI, Nicoloff H, Hjort K. 2019. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 17:479–496. doi: 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 26.Stratford M, Steels H, Nebe-von-Caron G, Avery SV, Novodvorska M, Archer DB. 2014. Population heterogeneity and dynamics in starter culture and lag phase adaptation of the spoilage yeast Zygosaccharomyces bailii to weak acid preservatives. Int J Food Microbiol 181:40–47. doi: 10.1016/j.ijfoodmicro.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Niño M, Marquina M, Swinnen S, Rodríguez-Porrata B, Nevoigt E, Ariño J. 2015. The cytosolic pH of individual Saccharomyces cerevisiae cells is a key factor in acetic acid tolerance. Appl Environ Microbiol 81:7813–7821. doi: 10.1128/AEM.02313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teertstra WR, Tegelaar M, Dijksterhuis J, Golovina EA, Ohm RA, Wösten H. 2017. Maturation of conidia on conidiophores of Aspergillus niger. Fungal Genet Biol 98:61–70. doi: 10.1016/j.fgb.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Geoghegan IA, Stratford M, Bromley M, Archer DB, Avery SV. 2019. Weak acid resistance A (WarA), a novel transcription factor required for regulation of weak-acid resistance and spore-spore heterogeneity in Aspergillus niger. bioRxiv doi: 10.1101/788141. [DOI] [PMC free article] [PubMed]
- 30.Furukawa T, van Rhijn N, Fraczek M, Gsaller F, Davies E, Carr P, Gago S, Fortune-Grant R, Rahman S, Mabey Glisenan J, Houlder E, Kowalski CH, Raj S, Paul S, Parker JE, Kelly S, Cramer RA, Latge J-P, Cook P, Moye-Rowley S, Bignell E, Bowyer P, Bromley MJ. The negative cofactor 2 complex is a master regulator of drug resistance in Aspergillus fumigatus. Nat Commun, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natorff R, Sieńko M, Brzywczy J, Paszewski A. 2003. The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol Microbiol 49:1081–1094. doi: 10.1046/j.1365-2958.2003.03617.x. [DOI] [PubMed] [Google Scholar]
- 32.Melin P, Stratford M, Plumridge A, Archer DB. 2008. Auxotrophy for uridine increases the sensitivity of Aspergillus niger to weak-acid preservatives. Microbiology 154:1251–1257. doi: 10.1099/mic.0.2007/014332-0. [DOI] [PubMed] [Google Scholar]
- 33.Holland SL, Reader T, Dyer PS, Avery SV. 2014. Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environ Microbiol 16:1729–1740. doi: 10.1111/1462-2920.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legras JL, Erny C, Le Jeune C, Lollier M, Adolphe Y, Demuyter C, Delobel P, Blondin B, Karst F. 2010. Activation of two different resistance mechanisms in Saccharomyces cerevisiae upon exposure to octanoic and decanoic acids. Appl Environ Microbiol 76:7526–7535. doi: 10.1128/AEM.01280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MS, Cho KH, Park KH, Jang J, Hahn JS. 2018. Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res 47:1211–1224. doi: 10.1093/nar/gky1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur JK, Arthanari H, Yang F, Pan S-J, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Näär AM. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 37.Sze J-Y, Woontner M, Jaehning JA, Kohlhaw GB. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258:1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- 38.Sellick CA, Reece RJ. 2003. Modulation of transcription factor function by an amino acid: activation of Put3p by proline. EMBO J 22:5147–5153. doi: 10.1093/emboj/cdg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dijksterhuis J, Meijer M, van Doorn T, Houbraken J, Bruinenberg P. 2019. The preservative propionic acid differentially affects survival of conidia and germ tubes of feed spoilage fungi. Int J Food Microbiol 306:108258. doi: 10.1016/j.ijfoodmicro.2019.108258. [DOI] [PubMed] [Google Scholar]
- 40.Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C, Fraczek MG, Dineen L, Lebedinec R, Macheleidt J, Heinekamp T, Delneri D, Bowyer P, Brakhage AA, Bromley M. 2019. High‐throughput gene replacement in Aspergillus fumigatus. Curr Protoc Microbiol 54:e88. doi: 10.1002/cpmc.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stratford M, Steels H, Novodvorska M, Archer DB, Avery SV. 2018. Extreme osmotolerance and halotolerance in food-relevant yeasts and the role of glycerol-dependent cell individuality. Front Microbiol 9:3238. doi: 10.3389/fmicb.2018.03238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 44.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballance DJ, Turner G. 1985. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36:321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- 49.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 50.Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel C. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 51.Ahuja M, Punekar NS. 2008. Phosphinothricin resistance in Aspergillus niger and its utility as a selectable transformation marker. Fungal Genet Biol 45:1103–1110. doi: 10.1016/j.fgb.2008.04.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Radial growth of the A. fumigatus ΔmetR mutant strain on medium containing 0.5 mM sorbic acid with or without 0.5 mM methionine. Download FIG S1, TIF file, 0.8 MB (817.4KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCR and Southern blot confirmation of warA deletion. (A) Targeted gene deletion strategy. The warA ORF was replaced with a hygromycin resistance cassette. (B) PCR confirmation of warA deletion. Primer pair 1 was used to confirm deletion of warA ORF; deletion strains are negative, and the WT is positive. Primer pair 2 was used to confirm the integration of HygR at the warA locus. (C) Southern blotting of warA and cdcA/warA deletion strains. The gDNA of strains was digested with the HindIII restriction enzyme. Membranes were hybridized with a probe consisting of digoxigenin-UTP-labeled HygR. Single bands confirm single integration of the deletion cassette into the A. niger genome. The ΔwarA_12 and ΔΔcdcA/warA_12 transformants were used for experiments. Download FIG S2, TIF file, 0.8 MB (801.3KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Growth of the ΔwarA mutant strain conidia on medium containing sorbic acid. Plates were inoculated with ∼100 conidia and incubated for 2 days (control medium) or 3 days (medium containing 1 mM sorbic acid). (B) Radial growth of the ΔΔcdcA/warA double mutant strain on medium containing 1 mM sorbic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions; approximate numbers of conidia are indicated at the top. Images were captured after 2 days of growth at 28°C and are representative of 2 or 3 independent experiments. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Radial growth of complemented ΔwarA strains on medium containing 2 mM benzoic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions. Approximate numbers of conidia are indicated above the pictures. Images were captured after 2 days of growth at 28°C. Two independent complemented lines are shown. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RPKM and log2FC values for A. niger genes (Excel file). Download Table S1, XLSX file, 0.9 MB (947.6KB, xlsx) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PCR and Southern blotting of An02g09970 and An14g03570 (pdrA) deletion strains. (A) Targeted gene deletion strategy. The An02g09970 and An14g03570 ORFs were replaced with a hygromycin resistance cassette. (B) PCR confirmation of An02g09970 deletion. Primer pair 1 was used to confirm deletion of the An02g09970 ORF; deletion strains are negative, and the WT is positive. Primer pairs 2 and 3 were used to confirm the integration of HygR at the An02g09970 locus. (C) PCR confirmation of An14g03570 deletion. Primer pair 1 was used to confirm deletion of the An14g03570 ORF; deletion strains are negative, and the WT is positive. Primer pairs 2 and 3 were used to confirm integration of HygR at the An14g03570 locus. (D) Southern blotting of the ΔAn02g09970 and ΔAn14g03570 mutant strains. The gDNA of the strains was digested with the restriction enzymes XbaI (for the ΔAn02g09970 mutant) or EcoRV (for the ΔAn14g03570 mutant). Membranes were hybridized with a probe consisting of digoxigenin-UDP-labeled HygR. Single bands confirm single integration of the deletion cassette into the A. niger genome (multiple integrations apparent for the ΔAn02g09970_39 mutant). The ΔAn14g03570_4 and ΔAn02g09970_29 transformants were used for experiments. Download FIG S5, TIF file, 1.0 MB (1,000.4KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Radial growth of complemented ΔpdrA mutant strains on medium containing 2 mM benzoic acid. Plates were inoculated with a 10-fold dilution series of conidial suspensions. Two independent complemented lines are shown. A ΔpdrA mutant strain containing the empty pAN7.1BAR plasmid is also shown. Download FIG S6, TIF file, 0.8 MB (861.6KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid map of YEp351 containing An14g03570 (pdrA). Download FIG S7, TIF file, 0.6 MB (659.6KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of primers used in this study (Excel file). Download Table S2, XLSX file, 0.02 MB (20.7KB, xlsx) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid map of pAN7.1BAR. Download FIG S8, TIF file, 0.6 MB (646.7KB, tif) .
Copyright © 2020 Geoghegan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq data have been deposited in GenBank under BioProject no. PRJNA594492.