IS26 differs from other studied ISs in the reactions that it can undertake. The differences make IS26 uniquely suited to its key role in the recruitment and spread of antibiotic resistance genes in Gram-negative bacteria. However, whether other ISs in the IS6/IS26 family can perform the same reactions is not known. IS257/IS431 and IS1216 isoforms found associated with antibiotic resistance genes in the Gram-positive bacteria staphylococci, enterococci, streptococci, and clostridia are related to IS26. However, the way that they move had not been investigated, limiting interpretation of their role in resistance gene dissemination and in the formation of cointegrates and complex resistance regions in staphylococci and enterococci. Here, they are shown to share the broad catalytic capabilities of IS26, demonstrating that it is likely that all members of the redefined IS6/IS26 family of bacterial ISs likewise are able to use both the copy-in and conservative routes.
KEYWORDS: IS1216, IS257, IS26, antibiotic resistance, insertion sequence, mobile genetic element
ABSTRACT
IS26 has been shown to form cointegrates both by a copy-in mechanism involving one insertion sequence (IS) and a target and by a targeted conservative mechanism involving two ISs. IS26 is the flagship of a group of 65 bacterial ISs in the recently redefined IS6/IS26 family. Here, whether other family members can also use two mechanisms was examined using members of the IS257/IS431 and IS1216 isoform groups, which are associated with antibiotic resistance genes in staphylococci and enterococci, respectively. Transposases Tnp257 and Tnp1216 have 39% and 47% amino acid identities, respectively, with Tnp26 and are 62% identical to one another. Using a novel transposition assay, pUC-based plasmids carrying these ISs integrated into the chromosome of a temperature-sensitive polA Escherichia coli strain grown at the restrictive temperature. In the cointegrates, the plasmid carrying IS257 was flanked by various 8-bp target site duplications, consistent with random target selection. However, in a mating-out assay, only the targeted conservative reaction was detectable at a low frequency in a recA-negative E. coli strain, indicating that IS257 is at least 100-fold less active than IS26. For IS1216, in mating-out assays, both copy-in and targeted conservative cointegrate formation were detectable at frequencies similar to those observed for IS26. Duplication of various 8-bp target sites was detected for the copy-in route. For both IS257 and IS1216, when both of the plasmids carried an IS, the targeted conservative route occurred at a significantly higher frequency than the copy-in route, and only cointegrates formed by the conservative route were detected.
IMPORTANCE IS26 differs from other studied ISs in the reactions that it can undertake. The differences make IS26 uniquely suited to its key role in the recruitment and spread of antibiotic resistance genes in Gram-negative bacteria. However, whether other ISs in the IS6/IS26 family can perform the same reactions is not known. IS257/IS431 and IS1216 isoforms found associated with antibiotic resistance genes in the Gram-positive bacteria staphylococci, enterococci, streptococci, and clostridia are related to IS26. However, the way that they move had not been investigated, limiting interpretation of their role in resistance gene dissemination and in the formation of cointegrates and complex resistance regions in staphylococci and enterococci. Here, they are shown to share the broad catalytic capabilities of IS26, demonstrating that it is likely that all members of the redefined IS6/IS26 family of bacterial ISs likewise are able to use both the copy-in and conservative routes.
INTRODUCTION
The IS6/IS26 family, hereinafter the IS26 family, includes the insertion sequences (ISs) that are most commonly found associated with antibiotic-resistance genes in resistant Gram-negative (IS26) and Gram-positive (IS257/IS431 and IS1216) bacteria (1). However, despite their importance, only a few members of the family have been examined experimentally, and only IS26 has been studied in detail.
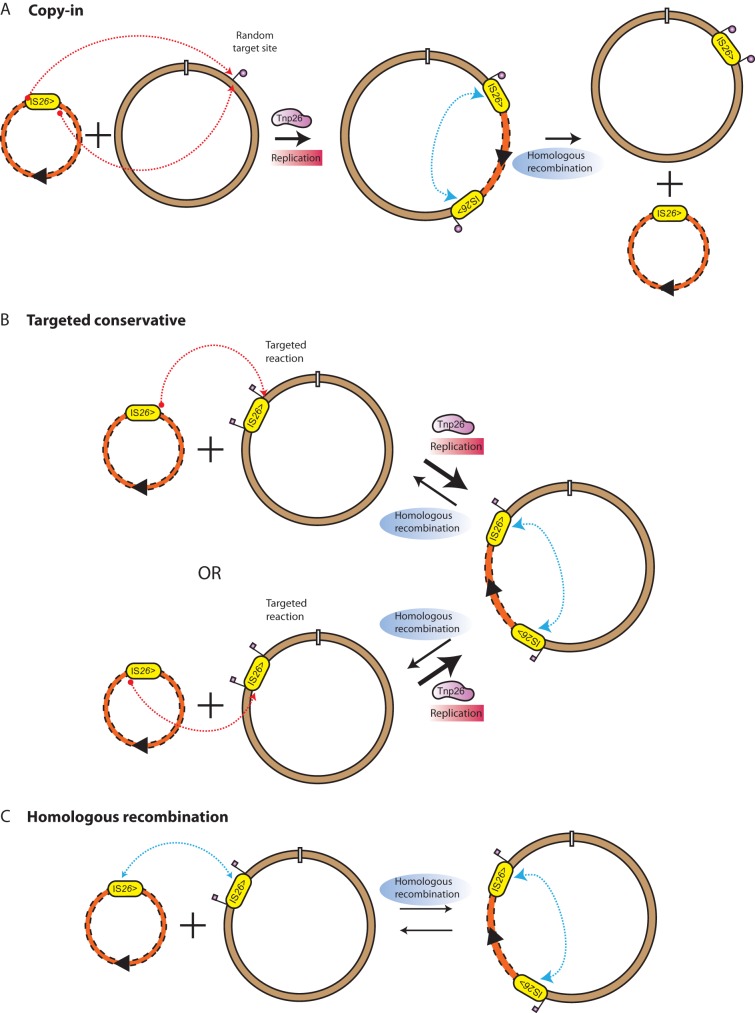
IS26 movement was first examined in the 1980s. Rather than moving alone to a new location, IS26 was shown to use a characteristic “replicative” mechanism to exclusively form cointegrates between two DNA molecules, a donor molecule containing an IS26 and a target molecule (2–4). This route duplicates the IS and the 8-bp target site (Fig. 1A). Apparent simple transposition products, namely, a single IS26 sequence surrounded by an 8-bp target site duplication (TSD) at a random site, likely arise via a cointegrate intermediate. Resolution of the cointegrate then occurs via homologous recombination between the two directly oriented IS copies in the cointegrate (Fig. 1A). The “replicative” route was later renamed “copy-in” to distinguish it from the “copy-out paste-in” mechanism used by other ISs, which was discovered subsequently and also involves a replicative step (5).
FIG 1.
Three routes to cointegrate formation between two molecules. (A) Copy-in route; (B) targeted conservative route; (C) homologous recombination. IS26 is indicated by yellow ovals, with orientation shown by “>.” A target site and subsequent 8-bp duplications are indicated by a vertical flag. The relative frequencies of the Tnp26-mediated and homologous-recombination-mediated reactions are indicated by the thicknesses of the arrows.
For 3 decades, copy-in cointegration was believed to be the sole movement mechanism used by IS26 and related ISs. However, IS26 was recently predicted, and then shown, to utilize a second transposase-dependent reaction to form cointegrates when both of the two DNA molecules involved carry a copy of IS26 (6). This unique reaction differs from any described for any other ISs to date and has properties akin to site-specific recombination. The reaction is targeted and occurs at one or the other end of the two ISs (7). It is also conservative, as the IS is not duplicated and a TSD is not generated (Fig. 1B) (6). Cointegration via the conservative route occurs at a frequency over 50-fold higher than with copy-in cointegrate formation (6–8), making it the preferred reaction if copies of IS26 in two different DNA molecules are available. Though the same cointegrate may be formed by homologous recombination (Fig. 1C), the transposase-catalyzed reaction has been shown to occur at a frequency over 1,000-fold higher than that of homologous recombination (8), making it the preferred reaction in a recombination-proficient host.
Recently, the relationships within a curated set of 112 ISs currently assigned to the IS6 family in ISfinder (https://isfinder.biotoul.fr/) were examined to identify the ISs that are the closest relatives of IS26 and hence most likely to also utilize the targeted conservative cointegrate formation mechanism discovered for IS26 (9). A well-differentiated group of 65 bacterial ISs was defined as the IS6/IS26 family to distinguish it from the larger IS6 family documented in ISfinder (9). The reduced family, here referred to as the IS26 family, includes six clades (clades I to VI), with IS26 belonging to clade I and members of the IS257/IS431 and IS1216 isoform groups belonging to clade II, which includes most of the ISs of Gram-positive origin (9).
Members of the IS257/IS431 group are found in Staphylococcus species. Three variants named IS257 (IS257R1, IS257R2, and IS257L) (10, 11) and three variants named IS431 (IS431L, IS431R, and IS431mec) (12) were discovered contemporaneously, and both names have been used over the years to refer to identical or closely related ISs. Here, we use IS257. IS257 isoforms range in size from 788 to 791 bp, with 18- or 20-bp terminal inverted repeats (TIRs), and share >95% nucleotide identity. The 224-amino-acid transposases share >98% amino acid identity with one another (see reference 9 for details of the variation).
Studies of available DNA sequences have inferred the ability of IS257 to form cointegrates and indicated an 8-bp TSD (13, 14). In an early study, IS257R2 was shown to be active. Cointegrates were formed between two plasmids via the untargeted copy-in route when only one plasmid contained a copy of IS257. The IS257 was duplicated and an 8-bp TSD created (15). However, the frequency of cointegrate formation was not recorded. In the same study, cointegrates were formed between two plasmids, each containing a copy of IS257, and the cointegrates appeared to have formed by recombination between the two IS257 sequences (15). As this experiment was conducted in a recombination-proficient Staphylococcus aureus strain, homologous recombination could not be excluded as the mechanism responsible, though it was claimed that the frequency of cointegration was higher than expected if homologous recombination was solely responsible (15).
IS1216 isoforms are 809 bp, with perfect 19-bp TIRs. A single study has inferred the importance of IS1216-mediated formation of cointegrate plasmids for mobilizing resistance genes from Enterococcus faecium to Enterococcus faecalis and reported the presence of an 8-bp TSD in the cointegrate (16). However, the frequency of cointegrate formation has never been quantified, and the possibility that IS1216 utilizes the targeted conservative mechanism has not been considered or examined.
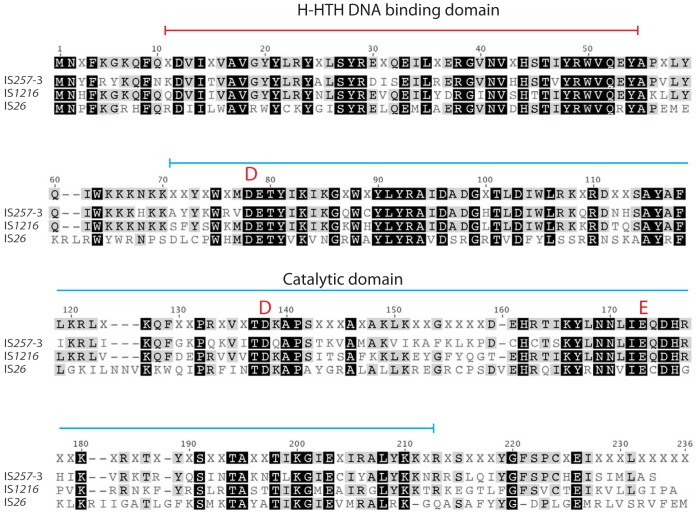
Here, to determine whether the targeted conservative cointegrate formation mechanism is unique to IS26 or whether other distantly related ISs in the IS26 family can also use this mechanism, IS257 and IS1216 were examined. The transposases of IS26, IS1216, and IS257 share between 39 and 62% amino acid identity with each other (Table 1), and an alignment of the transposases (Fig. 2) shows high conservation throughout the helix helix-turn-helix (H-HTH) DNA binding domain and the catalytic domain (see Fig. S1 in reference 9 for a full alignment of the IS26 family). To eliminate the detection of products of homologous recombination, cointegration was experimentally determined in a recA-negative Escherichia coli strain using well-established assays for detecting and quantifying cointegrate formation via both the untargeted copy-in route and the targeted conservative route.
TABLE 1.
Transposase amino acid similarity
| IS | % amino acid similarity to transposase of: |
|||
|---|---|---|---|---|
| IS26 | IS257R2 | IS257-3 | IS1216 | |
| IS26 | 100.0 | 38.7 | 39.1 | 46.7 |
| IS257R2 | 100.0 | 99.1 | 61.9 | |
| IS257-3 | 100.0 | 61.8 | ||
| IS1216 | 100.0 | |||
FIG 2.
Alignment of the amino acid sequences of the transposases of IS26, IS257-3, and IS1216. The extents of the H-HTH putative DNA binding domain and the DDE catalytic domain are marked above the sequences. The completely conserved DDE residues are marked by red letters. Amino acids are indicated as follows: black background, 100% similarity; dark-gray background, 80 to 99% similarity; unshaded letters, less than 79% similarity.
RESULTS
IS257-mediated copy-in cointegrate formation.
IS257-mediated cointegrate formation was initially examined in E. coli using a mating-out assay to detect cointegrates formed between the conjugative plasmid R388 (trimethoprim resistant [Tpr]) and pRMH1008 (ampicillin resistant [Apr]), a small, nonconjugative nonmobilizable plasmid containing IS257-3. However, cointegrate formation between pRMH1008 (IS257-3) and R388 was not detected (see below). To eliminate the possibility that the IS257-3 variant chosen was defective, a second variant (IS257R2) was cloned from pSK41 to generate plasmid pRMH1009. The transposase of IS257R2 differs from the transposase of IS257-3 at three positions: 37E→G, 75V→I, and 96D→E. Cointegrate formation was also not detected between pRMH1009 and R388, suggesting that either the reaction occurred at a frequency below the limit of detection of this assay (∼8 × 10−8 cointegrates per transconjugant) or that IS257, which is found exclusively in Staphylococcus spp., may not be active in E. coli.
In order to determine whether IS257-mediated cointegrate formation occurs at a frequency below that detectable by the standard mating-out assay, a polA mutant strain, E. coli MM383, which produces a DNA polymerase I (PolI) that is defective at 42°C, was used to detect cointegrate formation as described previously (17). Resistance to Ap (mediated by the blaTEM-1 gene in the pUC19 backbone) after growth at the nonpermissive temperature is indicative of the incorporation of the plasmid into the chromosome. The IS26-containing pUC19 derivative pRMH977, which is known to form cointegrates (6), was first tested to validate the assay. When pUC19 was used as a control, i.e., there was no IS present to mediate cointegrate formation, only 0.03% of cells retained resistance to Ap following 24 h of growth at 42°C without Ap selection (Fig. 3). In contrast, when MM383 containing pRMH977 was grown for 24 h at the nonpermissive temperature without selection, 1.67% (average from three independent experiments) of cells retained Ap resistance (Fig. 3).
FIG 3.
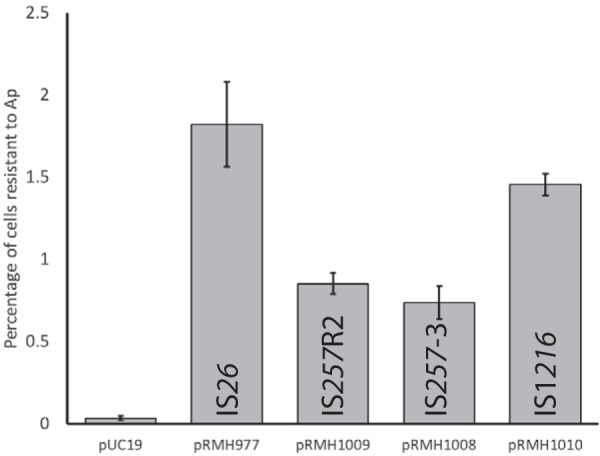
Cointegrate formation between pUC19 derivatives and the MM383 chromosome. The percentage of cells resistant to ampicillin (Ap), indicative of stable cointegrate formation between the plasmid and the MM383 chromosome, is shown as the means of results from three independent experiments. Error bars indicate the standard errors of the means.
To examine IS257-mediated cointegrate formation, MM383 containing either pRMH1008 (IS257-3) or pRMH1009 (IS257R2) was grown for 24 h without selection at the nonpermissive temperature. After being plated on selective media, 0.85% or 0.74% of cells (means from three independent experiments) from the MM383/pRMH1008 or MM383/pRMH1009 culture, respectively, retained Ap resistance (Fig. 3). To determine whether the Ap resistance was due to cointegrate formation between pRMH1008 or pRMH1009 and the MM383 chromosome or was due to residual free plasmid, 30 presumptive cointegrates (15 each from pRMH1008/MM383 and pRMH1009/MM383) were regrown for a second 24-h cycle at 42°C without Ap selection. In all 30 cases, 100% of cells in the culture retained resistance to Ap, confirming that IS257-3 and IS257R2 had formed stable cointegrates with the MM383 chromosome.
Fifteen cointegrates formed between pRMH1009 (IS257R2) and the MM383 chromosome were subjected to inverse PCR and sequenced using outward-facing primers in IS257R2 (RH2736 and RH2737) to determine the location of pRMH1009 and to determine whether an 8-bp target site duplication was generated. pRMH1009 was integrated at 15 different locations and in both possible orientations within the MM383 chromosome (Table 2). In each instance, the two copies of IS257R2 were in the same orientation and an 8-bp TSD had been generated, as expected for an IS26 family member.
TABLE 2.
Locations of cointegrates formed between pRMH1009 (IS257R2) and E. coli MM383 chromosome
| Cointegrate | TSD locationa | TSD sequence | Orientationb |
|---|---|---|---|
| 1 | 1056978–1056985 | ATGGGGGA | 1 |
| 2 | 2168492–2168499 | TGCCACTG | 1 |
| 3 | 1504489–1504496 | CAGTGGGT | 2 |
| 4 | 4325421–4325428 | GGCAAGAT | 1 |
| 5 | 3569123–3569130 | ATACGACG | 2 |
| 6 | 3421503–3421510 | CTAACTGG | 2 |
| 7 | 2966437–2966444 | ACGGAGAT | 2 |
| 8 | 37426–37433 | TTGAGTGG | 1 |
| 9 | 4412738–4412745 | TGCTACTA | 2 |
| 10 | 203519–203526 | GAAGAACT | 1 |
| 11 | 3308058–3308065 | TCGGATTT | 1 |
| 12 | 549300–549307 | TGATCGCA | 2 |
| 13 | 1852645–1852652 | CAACGACA | 2 |
| 14 | 3931278–3931285 | CTAAGCAC | 1 |
| 15 | 4002153–4002160 | CGCCAATG | 2 |
Location in the E. coli K-12 reference sequence (GenBank accession number U00096.3).
Orientation 1 is defined as that of the tnp257 of IS257R2, which is in the same orientation as the positive strand of the K-12 chromosome. Orientation 2 is defined as that of the tnp257 of IS257R2, which is in the orientation opposite to that of the positive strand of the K-12 chromosome.
IS257-mediated targeted conservative cointegrate formation.
The ability of IS257 to perform targeted conservative cointegrate formation was tested in a recA-negative background to ensure that all events detected were catalyzed by the transposase. Cointegration was tested using pRMH1008 (IS257-3 Apr) and R388::IS257-3 Tpr or pRMH1009 (IS257R2 Apr) and R388::IS257R2 (Tpr). IS257-3 in pRMH1008 formed Apr Tpr cointegrates with R388::IS257-3 at an average frequency of 5.11 × 10−6 cointegrates per transconjugant (Table 3). When pRMH1009 was tested, Apr Tpr cointegrates were formed at a similar average frequency of 3.59 × 10−6 cointegrates per transconjugant (Table 3). These frequencies are 40- to 60-fold lower than the values obtained here (Table 3) and our previously reported values (2.1 × 10−4 [6] and 2.9 × 10−4 [7]) for the reaction between two wild-type IS26 sequences under the same conditions.
TABLE 3.
Cointegrate formation frequencies
| IS type and IS | Target | Cointegration frequencya
|
|
|---|---|---|---|
| Range | Mean (SD) | ||
| Untargeted replicative | |||
| IS257-3 | R388 | <4.32 × 10–8 to <9.45 × 10–8 | <7.47 × 10–8 |
| IS257R2 | R388 | <5.55 × 10–8 to <8.99 × 10–8 | <7.84 × 10–8 |
| IS1216 | R388 | 9.00 × 10–8 to 4.14 × 10–7 | 4.47 × 10–7 (3.74 × 10–7) |
| IS26 | R388 | 2.1 × 10–7 to 7.03 × 10–7 | 5.14 × 10–7 (2.66 × 10–7) |
| Targeted conservative | |||
| IS257-3 | R388::IS257-3 | 3.10 × 10–6 to 6.67 × 10–6 | 5.11 × 10–6 (1.82 × 10–6) |
| IS257R2 | R388::IS257R2 | 2.30 × 10–6 to 5.91 × 10–6 | 3.59 × 10–6 (2.02 × 10–6) |
| IS1216 | R388::IS1216 | 4.33 × 10–5 to 9.38 × 10–5 | 6.99 × 10–5 (2.53 × 10–5) |
| IS26 | R388::IS26 | 3.29 × 10–4 to 6.09 × 10–4 | 4.88 × 10–4 (1.44 × 10–4) |
Frequency was measured as the number of cointegrates per transconjugant. The number of replicates was 3 in every case.
PCR screening of 10 Apr Tpr cointegrates from each of the three experiments confirmed that in all instances, pRMH1008 or pRMH1009 had been incorporated adjacent to the existing IS in R388::IS257-3 or R388::IS257R2, respectively. Hence, the IS257 variants IS257-3 and IS257R2 are able to perform the targeted conservative cointegrate formation reaction previously described for IS26.
Cointegrate formation mediated by IS1216.
The ability of IS1216 to perform untargeted copy-in cointegrate formation had never been tested previously. With the standard mating-out assay, cointegrates formed between R388 (Tpr) and pRMH1010 (IS1216 Apr) were detected. The reaction between pRMH1010 and R388 generated Apr Tpr cointegrates at a frequency of 4.47 × 10−7 cointegrates per transconjugant, averaged from three independent experiments (Table 2). This is comparable to the frequency of cointegrate formation demonstrated here (Table 2) and reported previously for IS26 via this route (7, 18, 19). Fifteen Apr Tpr cointegrates (five from each of three independent experiments) were subjected to inverse PCR and sequencing to determine the location of the integrated pUC-based plasmid in the R388 backbone. Cointegrates had formed at 15 different positions in R388 (Fig. 4) in both possible orientations. In each instance, IS1216 had been duplicated, the two copies of IS1216 were in direct orientation to each other, and an 8-bp TSD was generated.
FIG 4.
Cointegrate formation between pRMH1010 (IS1216) and R388. The R388 backbone is drawn to scale from GenBank accession no. BR000038 with key resistance genes, genes involved in replication (repA), and genes involved in conjugative transfer (tra) shown as arrows inside the circular backbone. Arrows pointing toward the circular backbone indicate the location of 15 mapped R388::pRMH1010 cointegrates, and the sequence of the 8-bp duplication of the target is shown. Blue lettering indicates that the cointegrate was in orientation 1 (tnp1216 is in the same orientation as the R388 repA gene), and red lettering indicates that the cointegrate was in orientation 2 (tnp1216 is in the orientation opposite to that of R388 repA).
Untargeted cointegration was also demonstrated using the temperature-sensitive MM383 assay. After 24 h of growth without selection at the nonpermissive temperature, 1.45% of colonies retained Ap resistance (Fig. 3), indicative of cointegrate formation. This is comparable to the frequency of the presence of IS26 (1.82%) and approximately 2-fold higher than the frequency of the two IS257 isoforms tested, consistent with the frequency demonstrated in the standard mating-out assay.
Targeted conservative cointegrate formation was also measured using pRMH1010 (Apr) and R388::IS1216 (Tpr). IS1216 in pRMH1010 formed Apr Tpr cointegrates with R388::IS1216 at an average frequency of 6.99 × 10−5 cointegrates per transconjugant (Table 2). This frequency is 150-fold higher than the frequency of the untargeted copy-in reaction reported above. It is similar to the frequency obtained for the reaction between two IS26 sequences here (Table 2) and previously (6, 7, 19). PCR screening of 10 streptomycin-resistant (Smr) and Apr Tpr colonies from each of the three independent targeted conservative experiments confirmed that in all instances, pRMH1010 had incorporated adjacent to IS1216 in R388::IS1216 via the targeted conservative cointegration mechanism.
The tnp26, tnp257, and tnp1216 genes are expressed equally in pUC19.
We considered the possibility that differences between the levels of tnp257, tnp26, and tnp1216 transcription in the pUC19-based constructs are responsible for differences in the cointegration frequencies of IS257 versus those of IS26 and IS1216. The level of tnp expression was quantified relative to that of the constitutively expressed blaTEM-1 gene via reverse transcription-quantitative PCR (RT-qPCR) analysis of RNA isolated from constructs containing IS26 (pRMH977), IS257R2 (pRMH1009), and IS1216 (pRMH1010). No significant differences between tnp transcript levels were observed in three independent experiments (Table 4), indicating that the level of tnp expression is unlikely to be the cause of the lower cointegrate formation frequency of IS257.
TABLE 4.
Expression of transposase genes in pUC19
| Plasmid | IS (transposase gene) | Expressiona |
|---|---|---|
| pRMH977 | IS26 (tnp26) | 1 |
| pRMH1008 | IS257-3 (tnp257-3) | 0.93 (0.88–1.13) |
| pRMH1009 | IS257R2 (tnp257R2) | 1.08 (0.79–1.33) |
| pRMH1010 | IS1216 (tnp1216) | 0.84 (0.67–1.21) |
Expression relative to tnp26 expression in pRMH977. tnp expression was determined in three independent experiments; the mean is reported and the range shown in parentheses.
DISCUSSION
We predicted that the shared characteristics of members of the IS26 family, namely, their related transposases and conserved TIRs, may indicate an ability to perform the copy-in and targeted conservative cointegration reactions (9). Here, we have experimentally shown for the first time that the IS26 family members IS257 and IS1216 found in Gram-positive species form cointegrates by both the copy-in and conservative routes and hence share the dual-mechanistic cointegrate formation capability previously demonstrated only for IS26. However, the low frequency of cointegrate formation exhibited by IS257 via both the copy-in and the targeted conservative route is surprising given the prevalence of IS257 in many staphylococcal chromosomes and plasmids. This is an important step forward, as it extends this mechanism beyond the Gram-negative species in which IS26 is found. Given the extent of the differences between Tnp26, Tnp257, and Tnp1216, it seems reasonable to conclude that all IS26 family members can perform this reaction. Like IS26, the ability to function in two different modes has likely contributed to the success that IS257 and IS1216 have had in mobilizing antibiotic resistance genes and shaping the genomes of the species in which they reside.
IS257 had previously been examined in more detail than IS1216, largely due to the association between IS257 and determinants conferring resistance to antibiotics (aminoglycosides, bleomycin, mupirocin, tetracycline, trimethoprim, and virginiamycin), heavy metals (cadmium and mercury), antiseptics, and disinfectants in Staphylococcus aureus (1, 20). However, whether IS257 is responsible for mobilizing these determinants is not always clear. There are numerous examples of small plasmids carrying resistance determinants [e.g., tet(K), aadD, and erm(C)] being integrated into a chromosome or other plasmids via IS257-mediated cointegration and generating an 8-bp TSD (14, 21). However, we could find only one example of the apparent movement of a transposon, Tn4003, resulting in the creation of an adjacent target site duplication (13). Further analysis of the now-extensive sequence data available will be needed to determine the true role of IS257 in moving resistance genes in a transposon-like structure.
IS1216 is associated with an increasing number of resistance genes in Enterococcus species (1, 18, 22), Clostridium perfringens (23), and S. aureus (24), including genes conferring resistance to penicillin, vancomycin, streptomycin, tetracycline-minocycline, gentamicin, kanamycin, and tobramycin, among others. However, like that of IS257, the role of IS1216 in mobilizing these resistance genes is not always clear, and there are only a very limited number of cases where a TSD has been documented (16, 24).
Whereas IS26 variants differ at only a few positions (19) and the IS1216 isoforms (IS1216, IS1216V, and IS1216E) also differ at only a few positions, sharing at least 98.8% nucleotide identity to one another, the sequence divergence among the IS257 isoforms is much greater (9). The degree of divergence of the IS257 isoforms likely indicates a significantly longer evolutionary history of IS257 in staphylococci, and it is possible that over time, IS257 may have acquired mutations that have regulated the transposase activity to mitigate potentially deleterious effects in the host.
The findings reported here shed light on how IS257 and IS1216 form cointegrates and hence how they may mobilize antibiotic resistance genes. Clearly, further work on these key players in the modern resistance story in Gram-positive bacteria is warranted.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli DH5α (supE44 ΔlacU169 [ϕ80 lacZΔM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used to propagate plasmids. E. coli UB5201 (pro met recA, nalidixic acid resistance [Nxr]) was used as a donor in mating-out experiments, and E. coli UB1637 (lys his trp lac recA Smr) was used as a recipient. MM383 [F– lacZ53 λ– thyA36 IN(rrnD-rrnE)1 rpsL151(Smr) polA12(ts) rha-5 deoC2] (25), a temperature-sensitive polA E. coli K-12 mutant, was used in temperature-sensitive cointegration assays, and an isogenic strain without the polA mutation, MM384 [F– lacZ53 λ– thyA36 IN(rrnD-rrnE)1 rpsL151(Smr) rha-5 deoC2), was included as a control. Antibiotics (Sigma) were added at the following concentrations to either Mueller-Hinton broth or Mueller-Hinton agar: ampicillin, 100 μg/ml; nalidixic acid, 25 μg/ml; streptomycin, 25 μg/ml; and trimethoprim, 25 μg/ml.
Plasmid construction.
The plasmids used in this study are listed in Table 5. Gibson assembly (New England Biolabs, USA) was used to generate pRMH1008, pRMH1009, pRMH1010, R388::IS257-3, R388::IS257R2, and R388:IS1216 using the primers listed in Table S1 in the supplemental material under standard manufacturer conditions. Inserts were cloned into the BamHI site of pUC19 or into the HindIII site of R388. pSK41 (26) DNA was used as the template for IS257-3 and IS257R2, and pJEG040 (27) DNA was used as the template for IS1216. The pUC19 universal primers were used to confirm the presence of the insert in pUC19, and primers RH2735 and RH2563 were used to confirm the presence of the insert in R388. PCR and routine sequencing of PCR products were performed as previously described (6) using primers listed in Table S1. Plasmid DNA was isolated by alkaline lysis as previously described (6).
TABLE 5.
Plasmids used in this study
| Plasmid | Description | Inserta | Resistance phenotypeb | Reference |
|---|---|---|---|---|
| pRMH1008 | IS257-3 in pUC19c | Bases 45556–100 from pSK41 | Ap | This study |
| pRMH1009 | IS257R2 in pUC19c | Bases 22809–23851 from pSK41 | Ap | This study |
| pRMH1010 | IS1216 in pUC19c | Bases 25151–26035 from pJEG040 | Ap | This study |
| R388 | IncW plasmid | Su Tp | 33 | |
| R388::IS257-3 | IS257-3 in R388d | Bases 45556–100 from pSK41 | Su Tp | This study |
| R388::IS257R2 | IS257R2 in R388d | Bases 22809–23851 from pSK41 | Su Tp | This study |
| R388::IS1216 | IS1216 in R388d | Bases 25151–26035 from pJEG040 | Su Tp | This study |
Primers used in this study. Download Table S1, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2020 Harmer and Hall.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mating-out cointegration assays.
Donors for cointegration assays were generated via conjugation of either R388 (Sur Tpr) or an R388 derivative containing the ISs of interest into E. coli UB5201 (recA mutant, Nxr) cells containing nonconjugative pUC19-derived plasmids containing IS257-3 (pRMH1008 Apr), IS257R2 (pRMH1009 Apr), or IS1216 (pRMH1010 Apr). Cointegrate formation was assessed by mating these strains with UB1637 (recA mutant, Smr) and selecting for Apr Smr Tpr colonies. pRMH977 (IS26) and R388::IS26, as previously tested (6, 7, 19), were included as a comparison. The transposition frequency was calculated as the number of Apr Smr Tpr transconjugants (cointegrates) per Tpr Smr transconjugant (R388 or R388 derivative). Targeted conservative cointegrate formation in R388::IS257-3, R388::IS257R2, or R388::IS1216 was detected by PCR mapping across each IS into the R388 backbone using primers RH2563 and RH2735 (Table S1), flanking the R388 HindIII site, in combination with primers internal to IS1216 (RH2738 and RH2739) or internal to IS257R2 (RH2736 and RH2737) (Table S1).
Temperature-sensitive cointegration assay.
When cointegrate formation was below the limit of detection using the standard cointegration assay, a temperature-sensitive polA mutant strain, MM383, was used to detect cointegrate formation between the IS-containing pUC19-derived plasmid and the chromosome. ColE1-derived plasmids, such as pUC19, require DNA polymerase I (PolI) to initiate replication (28), and MM383 is PolI defective at 42°C, resulting in the loss of a pUC19-derived plasmid when grown at the nonpermissive temperature unless it is incorporated into the chromosome, e.g., via IS-mediated cointegrate formation.
pUC19 (Apr), pRMH1008 (pUC19::IS257-3 Apr), or pRMH1009 (pUC19::IS257R2 Apr) was transformed into MM383 (Smr) by electroporation as described previously (8). The resulting Apr Smr transformant was purified and grown at 32°C overnight (∼16 h) in 5 ml LB supplemented with Ap and Sm. One milliliter of overnight culture was inoculated into 100 ml of prewarmed LB without ampicillin selection for the plasmid and grown at 42°C for 24 h. At the end of the growth period, the culture was serially diluted in 0.9% (wt/vol) saline and plated onto LB agar supplemented with Sm to select for all MM383 cells or supplemented with Ap and Sm to select for MM383 with the plasmid integrated and incubated overnight at 32°C. Resistance to Ap was indicative of cointegrate formation between the pUC19-derived construct and the chromosome. Fifteen Apr Smr colonies were subjected to a second round of growth at the nonpermissive temperature to ensure that Apr was stably maintained, verifying that Apr was indicative of cointegrate formation, rather than the presence of residual free plasmid.
Inverse PCR and sequencing.
Inverse PCR (29) and sequencing were used to map the junctions of the pUC plasmid with the chromosome or R388 formed via the untargeted copy-in reaction. Whole-cell DNA was prepared by alkaline lysis (30). NEBcutter version 2.0 (http://nc2.neb.com/NEBcutter2) (31) was used to identify restriction enzymes that would digest the backbone frequently but would not digest the IS or pUC19-derived fragment (i.e., the internal cointegrate fragment). Two micrograms of whole-cell DNA was digested with 5 units of BtgI (for cointegrates formed by IS257) or 5 units of BsmI (for cointegrates formed by IS1216) at 37°C for 2 h. Ten nanograms of digested DNA was added to a 10-μl ligation reaction mixture (200 U T4 DNA ligase, 1.0 μl T4 DNA ligase buffer) and incubated at room temperature for 8 h. Three microliters of the ligation reaction mixture was used as the template in an inverse PCR performed using primers internal to the ISs: primers RH2736 and RH2737 (Table S1) for cointegrates formed by pRMH1008 and pRMH1009 and primers RH2738 and RH2739 (Table S1) for cointegrates formed by pRMH1010. To determine the cointegrate boundaries, products from the inverse PCR were visualized on a 1% Tris-acetate-EDTA (TAE) gel, followed by gel extraction and sequencing with primers RH2736/RH2737 or RH2738/RH2739 as described previously (6).
qRT-PCR.
Quantitative real-time PCR was performed as described previously (32), using primers (Table S1) RH1464 and RH1465 to detect the expression of tnp26, RH2740 and RH2741 to detect tnp257R2 or tnp257-3, and RH2742 and RH2743 to detect tnp1216. Constitutively expressed blaTEM-1 from the plasmid backbone was used as an endogenous control (primers RH1466 and RH1467). Real-time PCR was performed in triplicate on independent biological-replicate samples.
ACKNOWLEDGMENTS
We thank Neville Firth (University of Sydney) for providing pSK41 and for stimulating discussions and Slade Jensen (Western Sydney University) for providing pJEG040.
This work was supported by the National Health and Medical Research Council of Australia (grant 1141540).
REFERENCES
- 1.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mollet B, Iida S, Arber W. 1985. Gene organization and target specificity of the prokaryotic mobile genetic element IS26. Mol Gen Genet 201:198–203. doi: 10.1007/bf00425660. [DOI] [PubMed] [Google Scholar]
- 3.Mollet B, Iida S, Shepherd J, Arber W. 1983. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res 11:6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iida S, Mollet B, Meyer J, Arber W. 1984. Functional characterization of the prokaryotic mobile genetic element IS26. Mol Gen Genet 198:84–89. doi: 10.1007/bf00328705. [DOI] [PubMed] [Google Scholar]
- 5.Curcio MJ, Derbyshire KM. 2003. The outs and ins of transposition: from mu to kangaroo. Nat Rev Mol Cell Biol 4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 6.Harmer CJ, Moran RA, Hall RM. 2014. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 5:e01801-14. doi: 10.1128/mBio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmer CJ, Hall RM. 2017. Targeted conservative formation of cointegrates between two DNA molecules containing IS26 occurs via strand exchange at either IS end. Mol Microbiol 106:409–418. doi: 10.1111/mmi.13774. [DOI] [PubMed] [Google Scholar]
- 8.Harmer CJ, Hall RM, Harmer CJ, Hall RM. 2016. IS26-mediated formation of transposons carrying antibiotic resistance genes. mSphere 1:e00038-16. doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmer CJ, Hall RM. 2019. An analysis of the IS6/IS26 family of insertion sequences: is it a single family? Microb Genom 5:e000291. doi: 10.1099/mgen.0.000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouch DA, Skurray RA. 1989. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene 76:195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- 11.Rouch DA, Messerotti LJ, Loo LS, Jackson CA, Skurray RA. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol 3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 12.Barberis-Maino L, Berger-Bächi B, Weber H, Beck WD, Kayser FH. 1987. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene 59:107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- 13.Leelaporn A, Firth N, Paulsen IT, Skurray RA. 1996. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J Bacteriol 178:6070–6073. doi: 10.1128/jb.178.20.6070-6073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart PR, Dubin DT, Chikramane SG, Inglis B, Matthews PR, Poston SM. 1994. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid 31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 15.Needham C, Noble WC, Dyke KG. 1995. The Staphylococcal insertion sequence IS257 is active. Plasmid 34:198–205. doi: 10.1006/plas.1995.0005. [DOI] [PubMed] [Google Scholar]
- 16.Di Sante L, Morroni G, Brenciani A, Vignaroli C, Antonelli A, D'Andrea MM, Di Cesare A, Giovanetti E, Varaldo PE, Rossolini GM, Biavasco F. 2017. pHTbeta-promoted mobilization of non-conjugative resistance plasmids from Enterococcus faecium to Enterococcus faecalis. J Antimicrob Chemother 72:2447–2453. doi: 10.1093/jac/dkx197. [DOI] [PubMed] [Google Scholar]
- 17.Brandsma JA, van Sluis CA, van de Putte P. 1981. Use of transposons in cloning poorly selectable genes of Escherichia coli: cloning of uvrA and adjacent genes. J Bacteriol 147:682–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciric L, Brouwer MS, Mullany P, Roberts AP. 2014. Minocycline resistance in an oral Streptococcus infantis isolate is encoded by tet(S) on a novel small, low copy number plasmid. FEMS Microbiol Lett 353:106–115. doi: 10.1111/1574-6968.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pong CH, Harmer CJ, Ataide SF, Hall RM. 2019. An IS26 variant with enhanced activity. FEMS Microbiol Lett 366:fnz031. doi: 10.1093/femsle/fnz031. [DOI] [PubMed] [Google Scholar]
- 20.Firth N, Skurray RA. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist Updat 1:49–58. doi: 10.1016/s1368-7646(98)80214-8. [DOI] [PubMed] [Google Scholar]
- 21.Yui Eto K, Firth N, Davis AM, Kwong SM, Krysiak M, Lee YT, O’Brien FG, Grubb WB, Coombs GW, Bond CS, Ramsay JP. 2019. Evolution of a 72-kilobase cointegrant, conjugative multiresistance plasmid in community-associated methicillin-resistant Staphylococcus aureus isolates from the early 1990s. Antimicrob Agents Chemother 63:e01569-19. doi: 10.1128/AAC.01560-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barile S, Devirgiliis C, Perozzi G. 2012. Molecular characterization of a novel mosaic tet(S/M) gene encoding tetracycline resistance in foodborne strains of Streptococcus bovis. Microbiology 158:2353–2362. doi: 10.1099/mic.0.058206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlebois A, Jalbert LA, Harel J, Masson L, Archambault M. 2012. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One 7:e44449. doi: 10.1371/journal.pone.0044449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Andrea MM, Antonelli A, Brenciani A, Di Pilato V, Morroni G, Pollini S, Fioriti S, Giovanetti E, Rossolini GM. 2019. Characterization of Tn6349, a novel mosaic transposon carrying poxtA, cfr and other resistance determinants, inserted in the chromosome of an ST5-MRSA-II strain of clinical origin. J Antimicrob Chemother 74:2870–2875. doi: 10.1093/jac/dkz278. [DOI] [PubMed] [Google Scholar]
- 25.Monk M, Kinross J. 1972. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol 109:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180:4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hal SJ, Espedido BA, Coombs GW, Howden BP, Korman TM, Nimmo GR, Gosbell IB, Jensen SO. 2017. Polyclonal emergence of vanA vancomycin-resistant Enterococcus faecium in Australia. J Antimicrob Chemother 72:998–1001. doi: 10.1093/jac/dkw539. [DOI] [PubMed] [Google Scholar]
- 28.Dasgupta S, Masukata H, Tomizawa J. 1987. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell 51:1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Russell DW. 2006. Inverse PCR. CSH Protoc 2006:pdb.prot3487. doi: 10.1101/pdb.prot3487. [DOI] [PubMed] [Google Scholar]
- 30.Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincze T, Posfai J, Roberts RJ. 2003. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res 31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmer CJ, Hall RM. 2015. IS26-mediated precise excision of the IS26-aphA1a translocatable unit. mBio 6:e01866-15. doi: 10.1128/mBio.01866-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revilla C, Garcillan-Barcia MP, Fernandez-Lopez R, Thomson NR, Sanders M, Cheung M, Thomas CM, de la Cruz F. 2008. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob Agents Chemother 52:1472–1480. doi: 10.1128/AAC.00982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study. Download Table S1, DOCX file, 0.02 MB (17KB, docx) .
Copyright © 2020 Harmer and Hall.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.