FOR-A and PRF-A are C-nucleoside antibiotics known for their unusual chemical structures and remarkable biological activities. Deciphering the enzymatic mechanism for the construction of a C-nucleoside scaffold during FOR-A/PRF-A biosynthesis will not only expand the biochemical repertoire for novel enzymatic reactions but also permit target-oriented genome mining of FOR-A/PRF-A-related C-nucleoside antibiotics. Moreover, the availability of FOR-A/PRF-A biosynthetic gene clusters will pave the way for the rational generation of designer FOR-A/PRF-A derivatives with enhanced/selective bioactivity via synthetic biology strategies.
KEYWORDS: C-nucleoside antibiotics, monooxygenase, C-glycosidic bond, combinatorial biosynthesis, genome mining
ABSTRACT
Formycin A (FOR-A) and pyrazofurin A (PRF-A) are purine-related C-nucleoside antibiotics in which ribose and a pyrazole-derived base are linked by a C-glycosidic bond. However, the logic underlying the biosynthesis of these molecules has remained largely unexplored. Here, we report the discovery of the pathways for FOR-A and PRF-A biosynthesis from diverse actinobacteria and propose that their biosynthesis is likely initiated by a lysine N6-monooxygenase. Moreover, we show that forT and prfT (involved in FOR-A and PRF-A biosynthesis, respectively) mutants are correspondingly capable of accumulating the unexpected pyrazole-related intermediates 4-amino-3,5-dicarboxypyrazole and 3,5-dicarboxy-4-oxo-4,5-dihydropyrazole. We also decipher the enzymatic mechanism of ForT/PrfT for C-glycosidic bond formation in FOR-A/PRF-A biosynthesis. To our knowledge, ForT/PrfT represents an example of β-RFA-P (β-ribofuranosyl-aminobenzene 5ʹ-phosphate) synthase-like enzymes governing C-nucleoside scaffold construction in natural product biosynthesis. These data establish a foundation for combinatorial biosynthesis of related purine nucleoside antibiotics and also open the way for target-directed genome mining of PRF-A/FOR-A-related antibiotics.
IMPORTANCE FOR-A and PRF-A are C-nucleoside antibiotics known for their unusual chemical structures and remarkable biological activities. Deciphering the enzymatic mechanism for the construction of a C-nucleoside scaffold during FOR-A/PRF-A biosynthesis will not only expand the biochemical repertoire for novel enzymatic reactions but also permit target-oriented genome mining of FOR-A/PRF-A-related C-nucleoside antibiotics. Moreover, the availability of FOR-A/PRF-A biosynthetic gene clusters will pave the way for the rational generation of designer FOR-A/PRF-A derivatives with enhanced/selective bioactivity via synthetic biology strategies.
INTRODUCTION
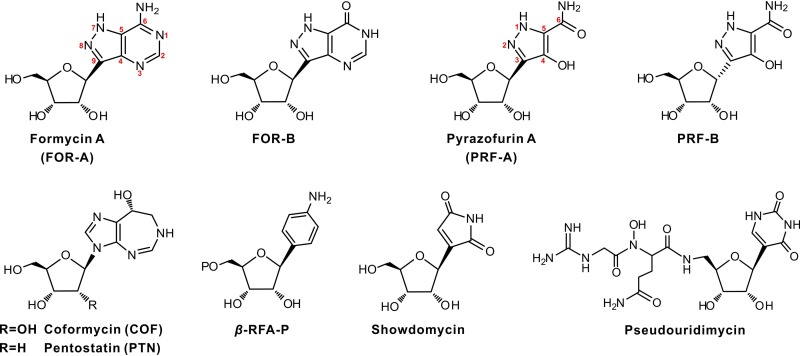
The naturally occurring C-nucleosides formycin A (FOR-A) and pyrazofurin A (PRF-A) (Fig. 1) are unusual biological molecules in which the base and ribosyl moieties are linked via a structurally conserved C-glycosidic bond (1, 2). Other important C-nucleoside antibiotics in this group include showdomycin (3) and pseudouridimycin (4) (Fig. 1). The pathway for showdomycin biosynthesis was discovered by relying on the use of the genes encoding the enzyme pair AlnA (C-glycosyltransferase) and AlnB (phosphatase) as a probe. These two enzymes catalyze the formation of the C-ribosylated aromatic polyketide alnumycin C in a two-step process (3). Recently, Sosio et al. identified the pseudouridimycin biosynthetic gene cluster by searching for the gene (contained in a gene cluster) encoding pseudouridine synthase (4).
FIG 1.
Chemical structures of FOR-A, PRF-A, and related compounds. β-RFA-P, β-ribofuranosyl-aminobenzene 5ʹ-phosphate.
FOR-A was originally isolated from the broth of Nocardia interforma in the process of screening for antitumor compounds (5), and later, this antibiotic, accompanied by FOR-B (a deaminated product of FOR-A) (Fig. 1), was discovered to be produced by Streptomyces lavendulae (6). FOR-A and FOR-B (particularly the latter) show bioactivities against Xanthomonas oryzae, a bacterial pathogen that causes rice plant disease, and influenza A virus, while only FOR-A shows antitumor and antiviral activities (7). Subsequently, FOR-A and coformycin (COF) (an adenosine deaminase inhibitor harboring a 1,3-diazepine ring) (Fig. 1) were found to be concomitantly produced by Nocardia interforma ATCC 21072 and Streptomyces kaniharaensis SF-557 (ATCC 21070) (8). This intriguing cobiosynthetic phenomenon has also been documented for other purine nucleoside antibiotic pairs, including pentostatin (PTN) and arabinofuranosyl adenine (produced by Streptomyces antibioticus NRRL 3238) (9), 2ʹ-chloropentostatin and 2ʹ-amino-2ʹ-deoxyadenosine (produced by Actinomadura sp. strain ATCC 39365) (10), and fungus-produced PTN and cordycepin (11). These antibiotic pairs all employ an unusual but general protector-protégé strategy; i.e., PTN may protect arabinofuranosyl adenine (taking this antibiotic pair as an example) from deamination by the housekeeping adenosine deaminase (9–11).
PRF-A, produced by Streptomyces candidus NRRL 3601, possesses prominent antiviral and antitumor activities, but interestingly, PRF-B (Fig. 1), the α-anomer of PRF-A coproduced by this strain in a smaller amount, shows little bioactivity (12). Previous studies indicated that PRF-A blocks the de novo biosynthesis of pyrimidine and purine by independently targeting orotidylate decarboxylase (13) and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (14).
Previous isotope feeding experiments demonstrated that glutamate and ribose play key roles in the C-glycosidic bond formation of FOR-A and indicated that the C-3-to-C-6 unit of PRF-A (C-9, C-4, C-5, and C-6 of FOR-A) is derived from C-4 to C-1 of glutamate and/or α-ketoglutarate (15). Moreover, Ochi et al. confirmed that N-3, N-7, and N-8 of FOR-A originate from the ε-amino nitrogen of lysine (16). More recently, Wang et al. identified the FOR-A biosynthetic gene cluster on the basis of previous studies (17) and deciphered the late steps to FOR-A biosynthesis, by which they established that FOR-A biosynthesis shares cross talk with the de novo purine pathway (17). Although a pathway for FOR-A biosynthesis was tentatively proposed, several important aspects still remained unknown.
In the present work, we report the identification and comparative analysis of the FOR-A/PRF-A biosynthetic gene clusters from diverse actinobacteria and further propose that the biosynthesis of FOR-A/PRF-A is likely initiated by a lysine N6-monooxygenase. We also demonstrate that a β-RFA-P (β-ribofuranosyl-aminobenzene 5ʹ-phosphate) synthase-like enzyme governs the construction of the C-glycosidic bond of FOR-A/PRF-A associated with cryptic decarboxylation.
RESULTS
Identification of FOR-A gene clusters from diverse actinobacteria.
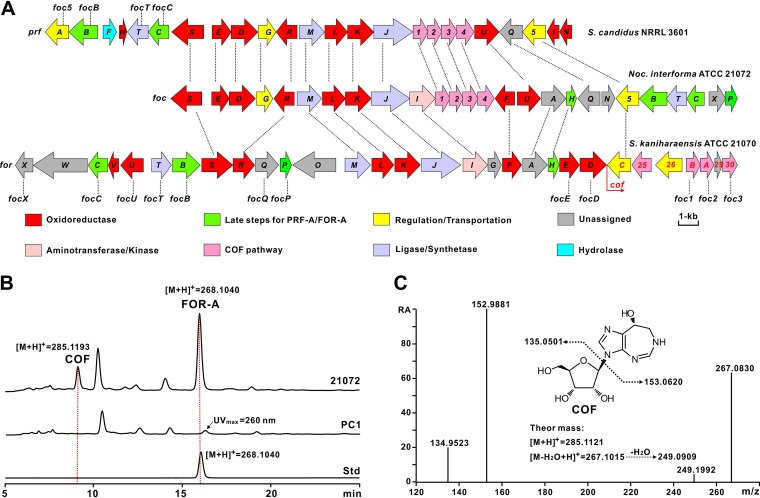
COF and PTN, which have a significant structural resemblance, were recently revealed to share highly similar biosynthetic pathways (18). To identify the target gene clusters for FOR-A biosynthesis, we sequenced the genomes of Nocardia interforma ATCC 21072 and S. kaniharaensis ATCC 21070 using the Illumina method, which rendered ca. 9.3 Mb (Nocardia interforma ATCC 21072) and ca. 8.6 Mb (S. kaniharaensis ATCC 21070) of nonredundant bases after the assembly of clean reads. We then utilized PenB (short-chain dehydrogenase) and PenC (phosphoribosylaminoimidazole-succinocarboxamide synthetase), the key enzymes for PTN biosynthesis, as query sequences to perform a BLASTP analysis (9), and the target foc and cof gene clusters, encoding the enzymes denoted Foc2/CofA (66%/49% identity to PenB) and Foc1/CofB (69%/62% identity to PenC) (Fig. 2A; see also Table S1 in the supplemental material), respectively, were detected by genome mining of Nocardia interforma ATCC 21072 and S. kaniharaensis ATCC 21070. To correlate the foc gene cluster with FOR-A biosynthesis, we deleted a 1.8-kb region (containing focI, foc1, and foc2) (Fig. S1A and Table S1). After confirmation (Fig. S1B), the resultant mutant was fermented for metabolite analysis, and high-performance liquid chromatography (HPLC) and liquid chromatography–high-resolution mass spectrometry (LC-HRMS) analyses indicated that the production of FOR-A and COF was abolished in the PC1 mutant (Fig. 2B and C and Fig. S1C), confirming the identity of the foc gene cluster for the biosynthesis of the COF–FOR-A pair.
FIG 2.
Genetic organizations and verification of the PRF-A/FOR-A and COF (and COF-related) gene clusters. (A) Genetic organizations of the PRF-A/FOR-A and COF (and COF-related) gene clusters. The PRF-A gene cluster (prf) is from S. candidus NRRL 3601, and the FOR-A and COF gene cluster (foc) is from Nocardia interforma ATCC 21072. The genes responsible for FOR-A and COF biosynthesis (for and cof) in S. kaniharaensis ATCC 21070 are also designated cof1 to cof30 (from left to right) (GenBank accession number KY705052). The designation “cof” was used for consistency with the study by Wang et al. (but cof25, cof26, cof29, and cof30 were missed or not denoted in that work) (17). For the related genes for COF (potential COF) biosynthesis in the other two strains, Arabic numerals (prf1 to prf5 and foc1 to foc5) were used for the designations. In addition, the foc genes at the top correspond to the prf genes, by which the encoded enzymes are homologous. “Late steps for PRF-A/FOR-A” indicates the enzymatic steps after C-glycosidic bond formation during PRF-A/FOR-A biosynthesis. The genes coding for homologous enzymes in these three gene clusters are correspondingly marked by a dotted line. Taking prfT (from S. candidus NRRL 3601), focT (from Nocardia interforma ATCC 21072), and forT (from S. kaniharaensis ATCC 21070) as examples, the enzymes encoded by them are interhomologous. (B) Verification of the gene cluster (foc) for the biosynthesis of FOR-A and COF in Nocardia interforma ATCC 21072. 21072 indicates the metabolites of Nocardia interforma ATCC 21072; PC1 indicates the metabolites of Nocardia interforma PC1, in which a 1.8-kb region of the foc gene cluster was deleted; and Std indicates the authentic standard of FOR-A. (C) MS/MS analysis of COF from the metabolites of Nocardia interforma ATCC 21072. “RA” indicates the relative abundance. “Theor mass” indicates the theoretical mass.
Further bioinformatic analysis led to the discovery of potential FOR-A gene clusters with organizational diversities from other actinobacterial strains, including Streptomyces resistomycificus NRRL 2290 and Salinispora arenicola CNS-205 (Fig. S1D). We then fermented these strains for metabolite analysis, and the HPLC and LC-mass spectrometry (MS) results indicated that S. resistomycificus NRRL 2290, but not Salinispora arenicola CNS-205, is capable of synthesizing FOR-A under the chosen fermentation conditions (Fig. S1E to G). These data suggested that FOR-A gene clusters highlighting organizational diversities are widely distributed among actinobacteria but that not all of these strains are conferred the ability for FOR-A production.
Discovery of the PRF-A gene cluster from S. candidus NRRL 3601.
PRF-A is structurally similar to FOR-A, which implies that both molecules should employ similar enzymatic logics for the construction of the pyrazole ring. To search for the candidate gene cluster for PRF-A biosynthesis, we sequenced the genome of S. candidus NRRL 3601, generating ca. 8.3 Mb of nonredundant bases after the assembly of preliminary data, and we then used FocJ (methionine-tRNA ligase) as the query sequence to perform a BLASTP analysis, in that the function of Spb40 (FocJ homolog) from the s56-p1 (a dipeptide natural product) pathway was recently confirmed for N-N bond formation (19). This approach leads to the location of a candidate gene cluster from the S. candidus NRRL 3601 genome, encoding PrfJ (86%/58% identities to FocJ/ForJ), PrfK (l-lysine N6-monooxygenase) (84%/62% identities to FocK/ForK), and Prf1 (phosphoribosylaminoimidazole-succinocarboxamide synthetase) (82%/60% identities to Foc1/CofB) (Fig. 2A, Table 1, and Table S1). This strongly suggested that the target gene cluster is involved in PRF-A biosynthesis.
TABLE 1.
Deduced functions of the open reading frames in the prf gene clustera
| Protein | No. of aa | Protein function | Homolog, origin | Identity (%), similarity (%) | GenBank accession no. |
|---|---|---|---|---|---|
| PrfA | 396 | MFS transporter | NbrT6, Nocardia terpenica | 72, 84 | AJO72759 |
| PrfB | 476 | Adenylosuccinate lyase | Orf67, Nocardia terpenica | 81, 89 | AJO72760 |
| PrfF | 224 | HAD family hydrolase | CLW14_2649, Streptomyces sp. strain 75 | 56, 66 | REE32254 |
| PrfH | 118 | Cupin domain-containing protein | CRH09_08295, Nocardia terpenica | 89, 94 | ATL66197 |
| PrfTb | 334 | β-RFA-P synthase-like enzyme | Orf61, Nocardia terpenica | 81, 87 | AJO72754 |
| PrfC | 343 | SAICAR synthetase | Orf62, Nocardia terpenica | 76, 84 | AJO72755 |
| PrfS | 540 | Fumarate reductase/succinate dehydrogenase | Orf63, Nocardia terpenica | 77, 83 | AJO72756 |
| PrfE | 329 | Putative monooxygenase | Sare_0593, Salinispora arenicola CNS-205 | 55, 64 | ABV96521 |
| PrfD | 444 | Putative monooxygenase | Orf75, Nocardia terpenica | 79, 86 | AJO72768 |
| PrfG | 288 | Transcriptional regulator | NbrR10, Nocardia terpenica | 59, 70 | AJO72769 |
| PrfR | 351 | Glycine/d-amino acid oxidase | EV381_1130, Actinopolyspora sp. strain DSM 45956 | 70, 79 | RZU68182 |
| PrfM | 413 | Phosphoribosylglycinamide synthetase | Orf72, Nocardia terpenica | 77, 86 | AJO72765 |
| PrfL | 390 | Saccharopine dehydrogenase | Orf71, Nocardia terpenica | 66, 77 | AJO72764 |
| PrfKb | 437 | l-Lysine N6-monooxygenase | Spb38, Streptomyces sp. strain SoC090715LN-17 | 42, 58 | BAW27702 |
| PrfJ | 668 | Methionine-tRNA ligase | Spb40, Streptomyces sp. SoC090715LN-17 | 33, 47 | BAW27704 |
| Prf1 | 250 | SAICAR synthetase | PenC, S. antibioticus | 69, 79 | AKA87338 |
| Prf2 | 233 | Short-chain dehydrogenase | PenB, S. antibioticus | 66, 79 | AKA87339 |
| Prf3 | 260 | Hydrolase | DI270_030585, Microbispora triticiradicis | 39, 54 | RGA01201 |
| Prf4 | 288 | ATP phosphoribosyltransferase | PenA, S. antibioticus | 49, 64 | AKA87340 |
| PrfU | 402 | Phthalate 4,5-dioxygenase | Orf68, Nocardia terpenica | 79, 89 | AJO72761 |
| PrfQ | 387 | Amidohydrolase | Orf65, Nocardia terpenica | 77, 84 | AJO72758 |
| Prf5 | 404 | MFS transporter | D5S19_24520, Amycolatopsis panacis | 85, 92 | RJQ80812 |
| PrfI | 156 | Flavin reductase | C8D87_103611, Lechevalieria atacamensis | 72, 79 | RAS67272 |
| PrfN | 169 | FMN reductase | C8D88_10484, Lechevalieria deserti | 71, 82 | PWK86923 |
aa, amino acids; MFS, major facilitator superfamily. SAICAR, phosphoribosylaminoimidazole-succinocarboxamide.
Function confirmed in vitro in this study.
Comparative analysis of the gene clusters for FOR-A and PRF-A biosynthesis.
Comparative analysis of the FOR-A gene clusters revealed that they have apparent diversities in genetic organization as well as in the total number of genes. The FOR-A gene cluster (30 genes) from S. kaniharaensis ATCC 21070 spans a 37.0-kb continuous chromosomal region, while the counterpart foc gene cluster (26 genes) from Nocardia interforma ATCC 21072 is ca. 29.4 kb in size (Fig. 2A and Table S1). As anticipated, in silico analysis indicated that the genes for FOR-A and COF biosynthesis are linked together, as reported by a recent study (17). Moreover, there are 7 additional genes (forW, forV, forO, forG, cof25, cof26, and cof29) in S. kaniharaensis ATCC 21070 (Table S1) that are proposed to execute some alternative functions during FOR-A and COF biosynthesis in this strain.
The prf gene cluster is composed of 24 genes and occupies a continuous ∼27.1-kb chromosomal region of S. candidus NRRL 3601 (Fig. 2A). The homologous prf and foc gene clusters contain several specialized genes (Fig. 2A). Of these genes, prfF (encoding a HAD family hydrolase), prfH (encoding a cupin domain-containing protein), prfI (encoding flavin reductase), and prfN (encoding flavin mononucleotide [FMN] reductase) are present only in the prf gene cluster (Table 1). Further analysis indicates that the foc gene cluster contains the specialized genes focH (5-aminoimidazole-4-carboxamide ribonucleotide transformylase) and focA (adenylosuccinate synthetase) for the late steps of FOR-A biosynthesis (Table S1). More interestingly, the candidate genes prf1, prf2, and prf4 (corresponding to foc1, foc2, and foc4) (Table 1), whose products are homologous to PenC, PenB, and PenA, respectively, from the PTN pathway, are also present in the prf gene cluster. However, more surprisingly, we were unable to identify COF-related compounds, either COF or PTN, from the culture broths of S. candidus NRRL 3601 under laboratory fermentation conditions. Moreover, two particular genes, focF (which encodes a dehydrogenase) and focI (which encodes an aminotransferase), are present only in the FOR-A gene cluster, suggesting that both genes are involved in dedicated enzymatic steps in FOR-A biosynthesis (Table S1).
ForK/PrfK functions as a lysine N6-monooxygenase.
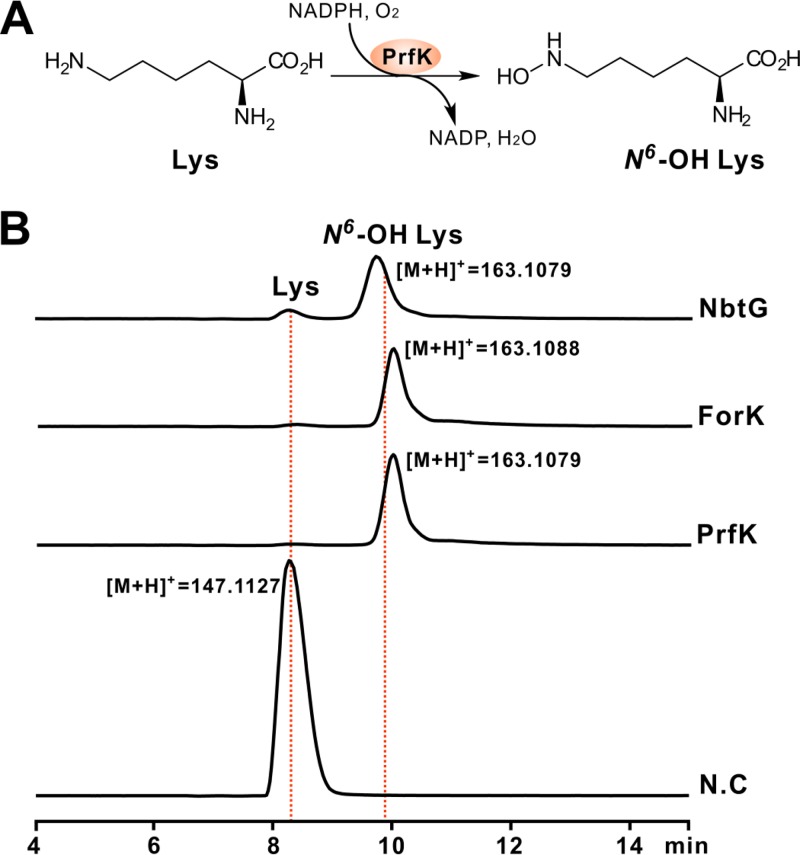
FOR-A and PRF-A feature a distinctive hydrazine moiety (N-N bond) (Fig. 1), which is also present in s56-p1 (19). A recent study revealed that N-N bond construction is governed by a cascade of two enzymes, Spb38 (lysine N6-monooxygenase) and Spb40 (methionine-tRNA ligase) (19) (Table 1 and Fig. 3A). Bioinformatic analysis of the FOR-A/PRF-A gene cluster identified the candidates ForK/PrfK (deduced lysine N6-monooxygenase) and ForJ/PrfJ (methionine-tRNA ligase) (Table S1 and Fig. S2A). To determine if ForK/PrfK fulfills the potential role of lysine N6-monooxygenase, we overexpressed and purified the protein in Escherichia coli (Fig. S2B). As expected, the ForK/PrfK protein is light yellow (Fig. S2C), a characteristic typical of flavoproteins (Fig. S2D). We then tested their enzymatic activity in vitro with l-lysine as the substrate and NADPH as a cofactor. As anticipated, LC-MS analysis indicated that the ForK/PrfK reaction mixtures are capable of generating a distinctive [M + H]+ ion of N6-OH lysine (Fig. 3B and Fig. S2E and F), the product of a control reaction with NbtG, a well-characterized lysine N6-monooxygenase (Fig. S2G and H) (20).
FIG 3.
Biochemical characterization of ForK/PrfK as a lysine (Lys) N6-monooxygenase. (A) Scheme of the ForK/PrfK-catalyzed reaction. (B) LC-MS analysis of the ForK/PrfK reaction using lysine as the substrate and NADPH as a cofactor. NbtG indicates the NbtG reaction as a positive control. NbtG (GenBank accession number BAD55606) is from Nocardia farcinica IFM 10152. ForK indicates the ForK reaction, and PrfK indicates the PrfK reaction. N.C, reaction mixture without an enzyme added as a negative control.
The negative-control reaction mixtures, without an enzyme added, were unable to produce the expected MS peaks (Fig. 3B). Moreover, we tested the cofactor specificity of the two enzymes, and the LC-MS results indicated that the reaction mixtures with NADH as a cofactor are also able to give the anticipated [M + H]+ ion of N6-OH lysine, confirming that NADH could also support the activity of PrfK/ForK (Fig. S2I). Taken together, these enzymatic data established that PrfK/ForK is a lysine N6-monooxygenase likely responsible for the initiation of PRF-A/FOR-A biosynthesis.
The ΔforT and ΔprfT mutants accumulate the corresponding intermediates ADCP and DCOP.
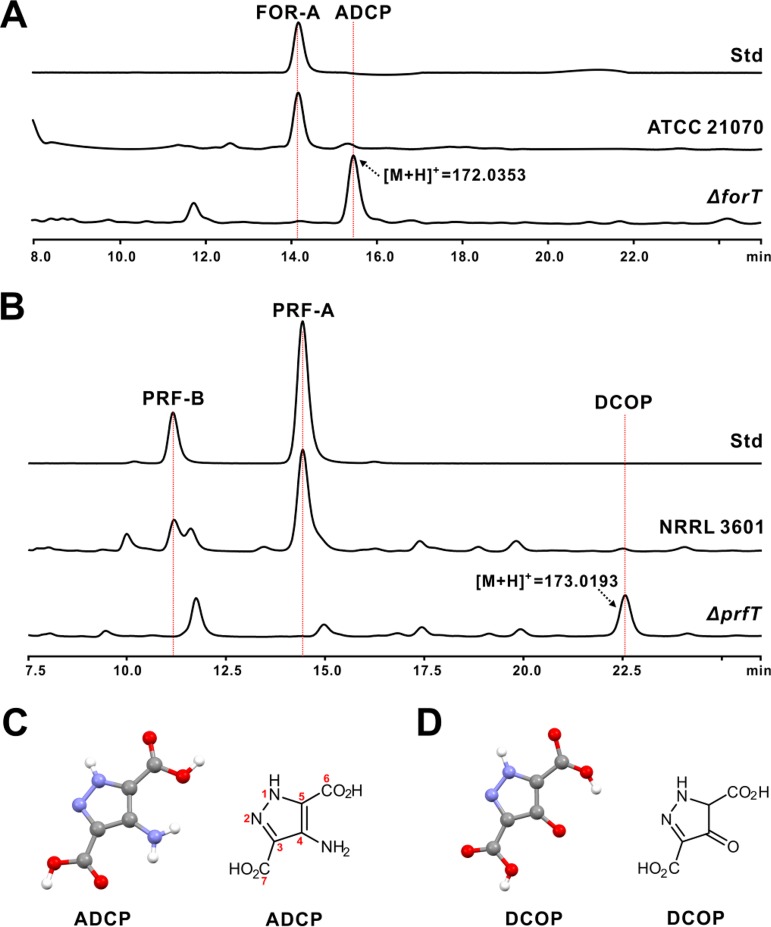
FOR-A/PRF-A features an unusual C-nucleoside scaffold, but the biosynthetic construction of the C-C bond has long remained mysterious. In silico investigation of the FOR-A/PRF-A gene cluster revealed that forT and prfT encode a deduced β-RFA-P synthase-like enzyme (Table 1 and Table S1). To investigate the in vivo functional role of forT and prfT, we deleted them in frame using a CRISPR-Cas9 strategy (Fig. S3A to D) (21). As S. kaniharaensis ATCC 21070 proved more amenable to in-frame deletion manipulation than Nocardia interforma ATCC 21072, it was used in this study for the following genetic experiments. After confirmation by a combined PCR-and-sequencing analysis (Fig. S3A and B), the ΔforT mutant was fermented for metabolite analysis. HPLC analysis indicated that this mutant could not generate the distinctive FOR-A peak but was capable of yielding a new peak (4-amino-3,5-dicarboxypyrazole [ADCP]) (Fig. 4A and Fig. S3E to G). LC-MS analysis of ADCP indicates that it shows an apparent [M + H]+ ion at m/z 172.0353 (Fig. 4A) and a specific fragment ion at m/z 153.8110 (Fig. S3H). Likewise, the confirmed ΔprfT mutant (Fig. S3C and D) lacks the capacity for PRF-A and PRF-B production (Fig. S3I and J) but could also generate a new peak (3,5-dicarboxy-4-oxo-4,5-dihydropyrazole [DCOP]) (Fig. 4B and Fig. S3K). LC-MS analysis of the DCOP peak indicates that it shows an apparent [M + H]+ ion at m/z 173.0193 (Fig. 4B and Fig. S3L).
FIG 4.
HPLC analysis of the metabolites produced by the ΔforT and ΔprfT mutants. (A) HPLC analysis of the metabolites produced by the S. kaniharaensis ΔforT mutant. Std indicates the authentic standard of FOR-A, ATCC 21070 indicates the metabolites produced by wild-type S. kaniharaensis ATCC 21070, and ΔforT indicates the metabolites of the ΔforT mutant. (B) HPLC traces of the metabolites produced by the S. candidus ΔprfT mutant. Std indicates the authentic standard of PRF-A, NRRL 3601 indicates the metabolites produced by wild-type S. candidus NRRL 3601, and ΔprfT indicates the metabolites of the ΔprfT mutant. (C) Crystal and chemical structures of ADCP. (D) Crystal and chemical structures of DCOP. The numbering for the atoms of DCOP corresponds to those of ADCP.
To determine the chemical structures of ADCP and DCOP, we initially attempted to purify them via HPLC for nuclear magnetic resonance (NMR) analysis. However, this approach failed to give rise to any apparently detectable 1H NMR signals for structure elucidation, and we were therefore prompted to optimize the conditions for ADCP/DCOP crystallization for X-ray crystallography determination. This approach showed that ADCP/DCOP harbors a highly conjugated five-membered structural system, in which two N atoms (N-1 and N-2) are linked together to form an unusual N-N bond (Fig. 4C and D and Table S2). It is this unusual structure of ADCP/DCOP that results in the lack of detectable 1H signals during NMR analysis. Taken together, the combined genetic and crystallography data suggested that prfT and forT are most likely the candidate genes for C-C bond formation during PRF-A/FOR-A biosynthesis.
PrfT/ForT is a β-RFA-P synthase-like enzyme for C-glycosidic bond formation during PRF-A/FOR-A biosynthesis.
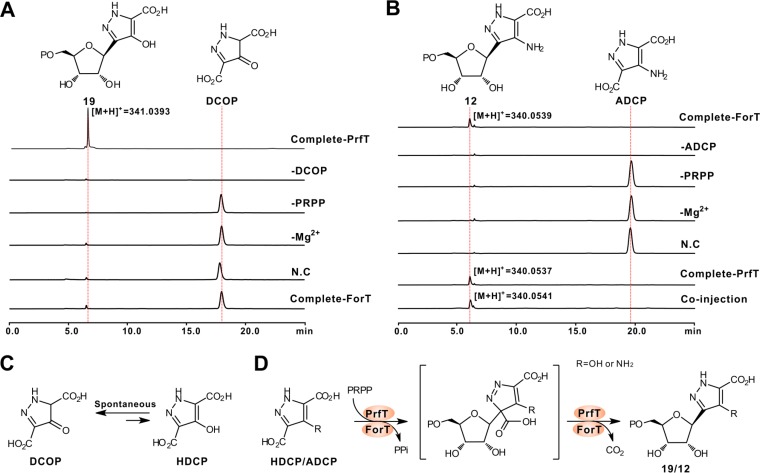
Bioinformatic analysis indicated that PrfT/ForT (Table 1 and Table S1) exhibits 81%/64% identities to Orf61 (hypothetic β-RFA-P synthase) from Nocardia terpenica (Fig. S4A). β-RFA-P synthase is responsible for the first committed step of methanopterin biosynthesis, catalyzing the conversion of phosphoribosyl pyrophosphate (PRPP) and p-aminobenzoate to β-RFA-P (Fig. 1) and CO2 (22). To obtain direct biochemical evidence that PrfT/ForT governs C-glycosidic bond construction during PRF-A/FOR-A biosynthesis, we overexpressed and purified these proteins from E. coli to near homogeneity and assayed them in vitro using PRPP and DCOP/ADCP as the substrates (Fig. S4B and C). The HPLC traces indicated that the product of the PrfT reaction is capable of producing a new peak, which is absent from that of the negative control (the reaction mixture without PrfT added) (Fig. 5A, C, and D and Fig. S4D). Further LC-MS analysis indicated that the product of the PrfT reaction exhibits an obvious [M + H]+ ion at m/z 341.0393 (Fig. 5A and Fig. S4E and F). Moreover, the reaction mixture without the divalent metal Mg2+ is not able to generate the characteristic peak of the product. We therefore purified the target compound for further 1H, 13C, and two-dimensional (2D) NMR analyses (Fig. S5 and Table S3), and its chemical structure, on the basis of the combined NMR and tandem MS (MS/MS) analyses, was finally determined as compound 19.
FIG 5.
Biochemical characterization of PrfT and ForT as β-RFA-P synthase-like enzymes for C-glycosidic bond construction. (A) HPLC traces of the PrfT-catalyzed reaction. Complete-PrfT, complete reaction of PrfT with DCOP and PRPP as the substrates; −DCOP, PrfT reaction without DCOP; −PRPP, PrfT reaction without PRPP; -Mg2+, PrfT reaction without Mg2+; N.C, PrfT reaction lacking an enzyme as the negative control; Complete-ForT, complete reaction of ForT with DCOP and PRPP as the substrate. (B) HPLC traces of the ForT-catalyzed reaction. Complete-ForT, complete reaction of ForT using ADCP and PRPP as the substrates; −ADCP, ForT reaction without ADCP; −PRPP, ForT reaction without PRPP; −Mg2+, ForT reaction without Mg2+; N.C, ForT reaction without an enzyme as a negative control; Complete-PrfT, complete reaction of PrfT with ADCP and PRPP as the substrates; Coinjection, coinjection of the ForT reaction mixture and the PrfT reaction mixture. (C) Scheme of the tautomerization reaction between DCOP and HDCP (4-hydroxy-3,5-dicarboxypyrazole). (D) Proposed mechanism for the PrfT/ForT-catalyzed reaction. When the R group is OH (NH2), the reaction is from HDCP to compound 19 (ADCP to compound 12).
Similarly, the ForT reaction mixtures, as analyzed by HPLC analysis, are also capable of producing a novel characteristic peak (compound 12) (Fig. 5B and Fig. S4G), which is distinctly absent from the reaction mixtures without ForT/Mg2+. Additional MS/MS analyses showed that the target peak could produce an apparent [M + H]+ ion at m/z 340.0539 (Fig. 5B), with major fragment ions completely consistent with the theoretical fragmentation pattern of compound 12 (Fig. S4H and I). We then tested the substrate flexibility of PrfT for these compounds, and the results indicated that this enzyme could accept ADCP to form compound 12 (Fig. 5B and Fig. S4J to M) but that, conversely, ForT is not capable of recognizing DCOP as the substrate (Fig. 5A).
Subsequently, we examined the specificities of divalent metal ions for PrfT/ForT activity. Of the 6 divalent metal ions selected, Mn2+ and Co2+ are able to maintain maximal activity for PrfT (100%), but this indicates either dramatically decreased activity in the presence of Ni2+/Zn2+ or negative activity with Ca2+/Cu2+ added instead (Fig. S4N). For the ForT reaction, Co2+ could also support its maximal activity, while Mn2+ (61%), Ni2+ (52%), Zn2+ (84%), or Ca2+/Cu2+ (0%) is just partially able or unable to replace Mg2+ to support the enzymatic activity of ForT (Fig. S4O). All of these biochemical data demonstrate that PrfT/ForT functions as an unusual β-RFA-P synthase-like enzyme for C-glycosidic bond formation during PRF-A/FOR-A biosynthesis.
DISCUSSION
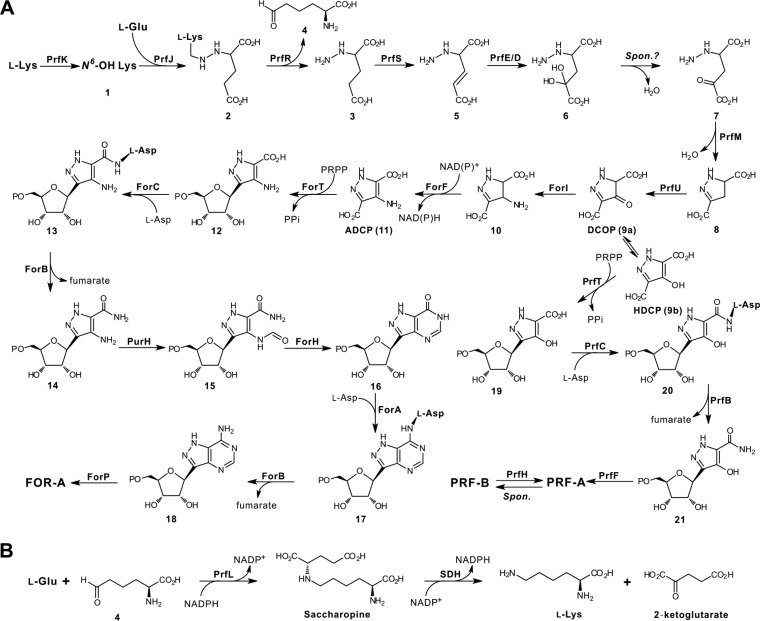
Previous metabolic labeling experiments determined that glutamate and lysine were the precursors for FOR-A and PRF-A biosynthesis and that the ribosyl moiety of both antibiotics was obtained directly from primary metabolism (15, 16). In the present study, this assignment is shown to be essentially correct. Insight into the biosynthetic gene cluster of PRF-A/FOR-A (only the PRF enzymes for common steps of both antibiotics are listed in Fig. 6) resulted in the identification of the enzymes for the construction of the pyrazole ring system (Fig. 6A). PRF-A biosynthesis is proposed to be initiated by PrfK hydroxylating l-lysine to form N6-OH lysine (compound 1), which, in common with most lysine N6-monooxygenases, prefers NADPH as a cofactor (20). Subsequently, compound 1 and l-glutamate are utilized by PrfJ (methionine-tRNA ligase) to construct the N-N bond of compound 2. PrfR (amino acid oxidase) then catalyzes the reaction to afford compound 3 (with leaving of compound 4), which is subsequently dehydrogenated by PrfS to form compound 5. From this, we tentatively propose that compound 5 is hydroxylated by PrfE/D in a sequential manner at the C-4 position to generate compound 6, which would be successively dehydrated either spontaneously or via an unknown enzymatic strategy to give rise to compound 7. Further investigation of the PRF-A gene cluster resulted in the identification of a candidate enzyme, PrfM (phosphoribosyl-glycinamide synthetase-like protein), which usually catalyzes the second step in the de novo purine pathway (23). We therefore postulate that compound 7, under the catalysis of PrfM, is subsequently converted to compound 8 for the completion of pyrazole ring construction. Thereafter, we propose that compound 8, after oxidation to DCOP (compound 9a), will undergo further sequential reactions to complete PRF-A biosynthesis (Fig. 6A).
FIG 6.
Proposed biosynthetic pathways to FOR-A and PRF-A. (A) Proposed pathways to FOR-A and PRF-A. We propose that they share identical steps at the early stage prior to the construction of C-nucleoside scaffolds. HDCP, 4-hydroxy-3,5-dicarboxypyrazole. (B) Proposed salvage pathway for the regeneration of lysine during FOR-A/PRF-A biosynthesis. SDH, short-chain dehydrogenase, which is from the primary metabolic pathway for lysine regeneration. Spon., spontaneous.
As for FOR-A biosynthesis, we deduce that DCOP acts as a potential amino group acceptor and is modified by the aminotransferase ForI to compound 10, which is then dehydrogenated to give ADCP (compound 11). Actually, these particular enzymes, the aminotransferase (ForI) and dehydrogenase (ForF), are absent from the PRF-A pathway, implying that they should perform these definite functions to accomplish ADCP biosynthesis (Fig. 6A), and related biochemical studies in our laboratory are now ongoing. Once the C-glycosidic bond is constructed, the late enzymatic steps are highly similar to those in the de novo purine pathway, which has already been illustrated in two recent studies (17, 24).
There are several other genes (prfHLIN) whose functional roles are still ambiguous. In particular, the product of prfH (cupin domain-containing protein) is probably responsible for the conversion of PRF-B to PRF-A, although it is also possible for this reaction to occur spontaneously. Hence, PrfH is tentatively proposed to be an isomerase during PRF-A biosynthesis, and PrfL is a potential saccharopine dehydrogenase, which normally plays a key role in lysine biosynthesis (25). Accordingly, we propose that PrfL could be responsible for a salvage pathway for lysine regeneration during PRF-A biosynthesis (Fig. 6B). The two remaining proteins, PrfI and PrfN, are apparently homologous to flavin/FMN reductase and could be involved in the reductive recycling of the FMN/flavin adenine dinucleotide (FAD) cofactor in PRF-A biosynthesis.
C-Nucleoside antibiotics have increasingly gained interest for clinical uses in the past decades (2), but their biosynthetic logics have long been underappreciated due to their difficult-to-access pathways. Indeed, nature has developed several diverse strategies for C-glycosidic bond construction during the biosynthesis of this group of antibiotics. Showdomycin biosynthesis uses a YeiN-like C-glycosyltransferase to build the C-C bond (3), while the biosynthesis of pseudouridimycin and malayamycin exploits a tRNA pseudouridylate synthase (TruD-like) for C-glycosidic bond construction (4, 26). In the present study, FOR-A and PRF-A are shown to harness a totally different β-RFA-P synthase-like enzyme (ForT/PrfT) to catalyze the formation of C-glycosides (Fig. 6A). The usual role for β-RFA-P synthase has exclusively been in the biosynthesis of the modified folate methanopterin (22). In this respect, ForT/PrfT represents an example of this kind of enzyme responsible for C-glycosidic bond formation in natural product biosynthesis. Hence, similar to β-RFA-P synthase (22), we predict that ForT/PrfT should employ an SN1-like enzymatic strategy associated with cryptic decarboxylation for the construction of the C-glycosidic bond. Notably, the single β-RFA-P synthase-like enzyme (ForT/PrfT) could be used as a promising probe for the rational mining of related C-nucleoside antibiotics, and we have already discovered several potential PRF-A/FOR-A group antibiotic pathways from the currently available reservoir of microbial genomes (see Fig. S6 in the supplemental material).
In summary, we report the pathways for FOR-A and PRF-A biosynthesis using a genomics-led approach and have delineated the associated FOR-A gene cluster diversity in actinobacteria. We also propose that the biosynthesis of FOR-A and PRF-A is likely initiated by a lysine N6-monooxygenase and show that PrfT/ForT adopts a unique strategy for C-glycosidic bond formation in nucleoside antibiotic biosynthesis. We anticipate that the C-glycosyltransferases shown in this study may serve as an inspiration for future catalyst design to generate analogs of potential therapeutic value.
MATERIALS AND METHODS
General materials and methods.
Strains and plasmids used in this study are listed in Table 2, and primers are listed in Table 3. General methods employed in this work were performed according to standard protocols described previously by Green and Sambrook (27) or Kieser et al. (28).
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Nocardia interforma ATCC 21072 | Wild-type FOR-A producer | ATCC |
| S. kaniharaensis ATCC 21070 | Wild-type FOR-A producer | ATCC |
| S. resistomycificus NRRL 2290 | New FOR-A producer | NRRL |
| Salinispora arenicola CNS-205 | Potential FOR-A producer | 29 |
| S. candidus NRRL 3601 | Wild-type PRF-A producer | NRRL |
| Nocardia interforma PC1 | ATCC 21072 derivative with deletion of a 1.8-kb region (containing focI, foc1, and foc2) | This study |
| S. kaniharaensis ΔforT | ATCC 21070 derivative with in-frame deletion of forT | This study |
| S. candidus ΔprfT | NRRL 3601 derivative with in-frame deletion of prfT | This study |
| E. coli DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG | Gibco-BRL |
| E. coli Rosetta(DE3)/pLysS | F− ompT hsdSB(rB− mB−) gal dcm λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) pLysS(Cmlr) | Novagen |
| E. coli ET12567(pUZ8002) | ET12567 containing helper plasmid pUZ8002 | 28 |
| Plasmids | ||
| pEASY-Blunt | pUCori lacZ f1 ori; Kan Amp | TransGen Biotech |
| pET28a | neo reppMB1; T7 promoter | Novagen |
| pET28a/forT | pET28a derivative carrying an NdeI-EcoRI fragment containing forT | This study |
| pET28a/forK | pET28a derivative carrying an NdeI-EcoRI fragment containing forK | This study |
| pET28a/prfT | pET28a derivative carrying an NdeI-EcoRI fragment containing prfT | This study |
| pET28a/prfK | pET28a derivative carrying an NdeI-EcoRI fragment containing prfK | This study |
| pET28a/nbtG | pET28a derivative carrying an NdeI-EcoRI fragment containing nbtG | This study |
| pCHW351 | pOJ446 derivative carrying a 2.8-kb XbaI-BglII left arm, a 2.9-kb BglII blunt right arm, and tsr for the replacement of focI, foc1, and foc2 | This study |
| pCHW352 | pCRISPR-Cas9 derivative carrying an XbaI-NcoI gRNA, a 2.0-kb left arm, and a 1.8-kb right arm for in-frame deletion of forT | This study |
| pCHW353 | pCRISPR-Cas9 derivative carrying an XbaI-NcoI gRNA, a 1.8-kb left arm, and a 2.2-kb right arm for in-frame deletion of prfT | This study |
TABLE 3.
PCR primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Ni-lfdsaicar-LF | GGTGGCGGTCAACAACGA |
| Ni-lfdsaicar-LR | GGTGGCGACGAAGAACATCA |
| Ni-lfdsaicar-RF | GTCCATATGGCACTTGACCGACCC |
| Ni-lfdsaicar-RR | GGAATTCAGGAGACGACGGTGGT |
| LfdsaicarF | GTCCATATGCCGGAGACTTTCGAT |
| LfdsaicarR | GGAATTCAGCGCCCCGTGTTCAG |
| forT-cas9-gRNA-F | GATGGGTCAACGAGATCACCTTCACGTTTTAGAG |
| forT-cas9-gRNA-R | CTAGCTCTAAAACGTGAAGGTGATCTCGTTGACC |
| forT-cas9-LF | TCGTCGAAGGCACTAGAAGGCCAGCGGCAGCACGACCTTT |
| forT cas9-LR | TCAGGAGACGACGGTGGTGGCGTCGGGTCGGTCAAGTGCCAT |
| forT-cas9-RF | GCCACCACCGTCGTCTCCTGA |
| forT-cas9-RR | GGTCGATCCCCGCATATAGGCGGTGACGAGTTCCAGGTAGCG |
| forT-cas9-idF | AACTTCCAGCCGTGGTAGATGC |
| forT-cas9-idR | CGGTCTTCATTGCCCTGCTTT |
| prfT-cas9-gRNA-F | GATGGGTCAACGAGCTGACCTTCACGTTTTAGAG |
| prfT-cas9-gRNA-R | CTAGCTCTAAAACGTGAAGGTCAGCTCGTTGACC |
| prfT-cas9-LF | TCGTCGAAGGCACTAGAAGGACGAGAAGTTCCGCACCAG |
| prfT-cas9-LR | TGTTGCGGACCGTGGTGAAGGATTCGCCGTCGAGGCT |
| prfT-cas9-RF | TTCACCACGGTCCGCAACA |
| prfT-cas9-RR | GGTCGATCCCCGCATATAGGCAACGGGTATCGGTTCAGC |
| prfT-cas9-idF | AAGGCGGAGTTCGGGTTC |
| prfT-cas9-idR | CGTGTTCATCGACTGGGAGA |
| forT-28a-exp-F | GCTCTAGAACCCGACTACGGACAGGACC |
| forT-28a-exp-R | GAAGATCTGTAGCCGATGCGGAACGAG |
| forK-28a-exp-F | GAAGATCTGGAGGAGCGGAACAACG |
| forK-28a-exp-R | GGAATTCACGCGGAAGACCACATCG |
| prfT-28a-exp-F | GTCCATATGCAGGGAACCGCGGTG |
| prfT-28a-exp-R | GGAATTCCTACAGCCAGGTCGCGGC |
| prfK-28a-exp-F | GTCCATATGAGCACCGGACACGAC |
| prfK-28a-exp-R | GGAATTCTCAGAACGTCAGCGCGAC |
Enzymes, chemicals, and reagents.
All of the enzymes used in this study were purchased from New England Biolabs. The chemicals and reagents were products of Sigma-Aldrich, Thermo Scientific, or J&K Scientific. The standards of FOR-A and PRF were individual products of Biorbyt and Sigma-Aldrich.
Sequencing analysis of S. kaniharaensis ATCC 21070, Nocardia interforma ATCC 21072, and S. candidus NRRL 3601.
Sequencing of the genomes of S. kaniharaensis ATCC 21070, Nocardia interforma ATCC 21072, and S. candidus NRRL 3601 was performed on an Illumina HiSeq 2500 system machine. The raw data were processed to render the resulting clean reads, which were assembled by using Velvet software (v1.2.07) to obtain the scaffold. Detailed methods and programs used for genome annotations and accurate bioinformatic analysis were performed according to protocols described previously by Xu et al. (18).
Fermentation and detection of related nucleoside antibiotics.
S. kaniharaensis ATCC 21070, Nocardia interforma ATCC 21072, and S. candidus NRRL 3601 were cultivated on YS agar (2 g yeast extract, 10 g soluble starch, and 15 g agar per liter [pH 7.3]). For fermentation, a single clone was inoculated in tryptic soy broth (TSB) and cultivated for 2 days; after that, the cultures (2%, vol/vol) were transferred to fermentation medium (16) and fermented (180 rpm at 28°C) for 7 days. For HPLC and LC-MS analyses, the fermentation beer was processed by adding oxalic acid until pH 3.0 was reached. HPLC analysis was performed using a Shimadzu LC-20AT instrument equipped with a C18 column (5 μm, 4.6 by 250 mm; Shimadzu), with an elution gradient of 10% to 30% methanol–0.15% aqueous TFA (trifluoroacetic acid) over 30 min at a flow rate of 0.5 ml/min. LC-MS analysis was carried out on a Thermo Fisher Scientific ESI-LTQ Orbitrap instrument controlled by Xcalibur in the positive mode. The parameters for MS analysis are as follows: a capillary temperature of 275°C and a capillary voltage of 35 V.
Construction of the Nocardia interforma PC1, S. kaniharaensis ΔforT, and S. candidus ΔprfT mutants.
For the construction of the mutant strain Nocardia interforma PC1, a strategy similar to the one described previously by Wu et al. (9), with Ni-lfdsaicar-LF/R (left arm) and Ni-lfdsaicar-RF/R (right arm) as primers and pOJ446 as the starter vector, was also used in this study. For the construction of the S. kaniharaensis ΔforT mutant (taking it as an example), pCHW352 (Table 2) was generated, using methods similar to the ones described previously by Mo et al. (21), and then introduced into S. kaniharaensis via conjugation. After confirmation, the conjugant, induced by thiostrepton, was screened and verified on the basis of standard methods (28). Likewise, the S. candidus ΔprfT mutant was constructed according to the protocols described above.
Overexpression and purification of the target proteins.
Synthesis of nbtG (whose sequence was optimized based on E. coli codon preference; GenBank accession number MN370059) was conducted by Synbio Tech, using pET28a as the expression vector. Expression and purification of NbtG, ForK, ForT, PrfK, and PrfT were carried out in a way similar to the one described previously by Wu et al. (9). After concentration, the purified proteins were buffer exchanged into the stock buffer (25 mM Tris, 150 mM NaCl, and 10% glycerol [pH 7.5 for ForK, ForT, PrfK, and PrfT and pH 8.0 for NbtG]) using Amicon Ultra filters. Finally, the purified proteins were flash-frozen in liquid nitrogen and stored at −80°C.
In vitro assays of NbtG, ForK, and PrfK.
Reaction mixtures (100 μl each) consisted of a solution containing 50 mM potassium phosphate buffer (pH 7.5), 1 mM l-lysine, 1 mM NAD(P)H, and 20 μg NbtG, ForK, or PrfK at 30°C. Reactions were terminated after 4 h by the addition of an equivalent volume (100 μl) of methanol. Following centrifugation to remove the protein, the reactions were analyzed by using an LC-HRMS system equipped with a reverse-phase C18 column with 0.15% TFA (95%)–methanol (5%) at a flow rate of 0.4 ml/min for 20 min. The parameters for MS analysis are the same as the ones described above.
In vitro assays of ForT and PrfT.
Reaction mixtures (100 μl each) consisting of 50 mM Tris-Cl buffer (pH 8.0), 1.5 mM PRPP, 20 mM Mg2+ (or other related divalent metal ions), 1 mM ADCP/DCOP, and 20 μg ForT/PrfT were incubated at 30°C for 4 h. After that, reactions were terminated by the addition of an equivalent volume (100 μl) of methanol, and protein was removed by centrifugation. The HPLC analysis was performed on a reverse-phase C18 column (5 μm, 4.6 by 250 mm; Shimadzu) with an elution gradient of 10% to 30% methanol–0.1% aqueous TFA (HPLC grade) over 15 min at 0.5 ml/min, the methanol system was then returned to the initial 10%, and the ratio was maintained for another 15 min. Elution was monitored at 290 nm (for the ForT reaction) or 270 nm (for the PrfT reaction) with a diode array detector (DAD), and the data were analyzed offline with Shimadzu data software.
Isolation and purification of the target metabolites ADCP and DCOP.
For the isolation of ADCP and DCOP, the fermentation beers (7 day) of mutant strains (S. kaniharaensis ΔforT and S. candidus ΔprfT) were processed by adding oxalic acid until pH 3.0 was reached and centrifuged at 6,000 rpm for 15 min. The supernatant was collected and then extracted three times with an equal volume of n-butanol. The collected extract (upper layer) was condensed and further dried by rotary evaporation and lyophilization, and the dried residues were then redissolved in 50% methanol and passed through a C18 solid-phase extraction cartridge. Finally, approximately 100 mg of the compound ADCP/DCOP could be isolated and purified from 10 liters of fermentation beers by preparative HPLC (the HPLC conditions are identical to those described above but with a flow rate of 5 ml/min).
For the purification of compound 19, the PrfT reaction (4 h) was terminated, and the reaction mixtures (100 ml) were then collected and passed through Dowex 50WX8 (H+) resin (the ratio of the reaction mixtures to resin is 3:1). The flowthrough liquid was collected and condensed. After that, about 4 mg of compound 19 was prepared by HPLC with a flow rate of 0.4 ml/min (5% methanol–0.2% aqueous TFA) over 15 min.
Single-crystal X-ray diffraction for ADCP and DCOP.
For growth of crystals of ADCP and DCOP, the solvent system (the ratio of water to methanol is 1:1) was selected to dissolve adequate ADCP and DCOP to a near-saturated state. The bottle of the sample vial was sealed by Parafilm with sporadically distributed pores. After leaving it at 4°C for about 2 weeks, needle-like crystals of ADCP and square-like crystals of DCOP appeared due to the easier evaporation of methanol and lower solubility of the compound in water. The crystals of the target compound were determined using a Bruker Apex Duo single-crystal X-ray diffractometer, and the related data, including space groups, crystal molecular structures, and intermolecular hydrogen bonds, etc., are listed in Table S2 in the supplemental material. The individual CCDC numbers for DCOP and ADCP are 1947450 and 1947811.
NMR analysis of compound 19.
NMR spectra of compound 19 were recorded on an Agilent DD2 600-MHz NMR spectrometer, using D2O as the solvent.
Accession number(s).
The DNA sequences have been deposited in the GenBank database under accession numbers KY705052 (for the FOR-A and COF gene clusters from S. kaniharaensis ATCC 21070), MH493900 (for the PRF gene cluster from S. candidus NRRL 3601), and KY682079 (for the FOR-A and COF gene clusters from Nocardia interforma ATCC 21072), as well as MN370059.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Paul R. Jensen from UCSD (CA, USA) for kindly providing us with Salinispora arenicola strain CNS-205. We also sincerely thank Pablo Sobrado from Virginia Tech (VA, USA) for the generous gift of the NbtG expression plasmid.
This work was supported by grants from the National Key R&D Program of China (2018YFA0903203) and the National Natural Science Foundation of China (31770041 and 31970052).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Isono K. 1988. Nucleoside antibiotics: structure, biological activity, and biosynthesis. J Antibiot (Tokyo) 41:1711–1739. doi: 10.7164/antibiotics.41.1711. [DOI] [PubMed] [Google Scholar]
- 2.De Clercq E. 2016. C-nucleosides to be revisited. J Med Chem 59:2301–2311. doi: 10.1021/acs.jmedchem.5b01157. [DOI] [PubMed] [Google Scholar]
- 3.Palmu K, Rosenqvist P, Thapa K, Ilina Y, Siitonen V, Baral B, Mäkinen J, Belogurov G, Virta P, Niemi J, Metsä-Ketelä M. 2017. Discovery of the showdomycin gene cluster from Streptomyces showdoensis ATCC 15227 yields insight into the biosynthetic logic of C-nucleoside antibiotics. ACS Chem Biol 12:1472–1477. doi: 10.1021/acschembio.7b00078. [DOI] [PubMed] [Google Scholar]
- 4.Sosio M, Gaspari E, Iorio M, Pessina S, Medema MH, Bernasconi A, Simone M, Maffioli SI, Ebright RH, Donadio S. 2018. Analysis of the pseudouridimycin biosynthetic pathway provides insights into the formation of C-nucleoside antibiotics. Cell Chem Biol 25:540–549. doi: 10.1016/j.chembiol.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hori M, Takita T, Koyama G, Tadeuchi T, Umezawa H. 1964. A new antibiotic, formycin. J Antibiot (Tokyo) 17:96–99. [PubMed] [Google Scholar]
- 6.Aizawa S, Hidaka T, Ōtake N, Yonehara H, Isono K, Igarashi N, Suzuki S. 1965. Studies on a new antibiotic, Laurusin. Agr Biol Chem 29:375–376. doi: 10.1080/00021369.1965.10858402. [DOI] [Google Scholar]
- 7.Ishizuka M, Sawa T, Hori S, Takayama H, Takeuchi T. 1968. Biological studies on formycin and formycin B. J Antibiot (Tokyo) 21:5–12. doi: 10.7164/antibiotics.21.5. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura H, Koyama G, Iitaka Y, Ono M, Yagiawa N. 1974. Structure of coformycin, an unusual nucleoside of microbial origin. J Am Chem Soc 96:4327–4378. doi: 10.1021/ja00820a049. [DOI] [PubMed] [Google Scholar]
- 9.Wu P, Wan D, Xu G, Wang G, Ma H, Wang T, Gao Y, Qi J, Chen X, Zhu J, Li YQ, Deng Z, Chen W. 2017. An unusual protector-protege strategy for the biosynthesis of purine nucleoside antibiotics. Cell Chem Biol 24:171–181. doi: 10.1016/j.chembiol.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Xu G, Wu P, Liu J, Cai YS, Deng Z, Chen W. 2017. Biosynthesis of 2ʹ-chloropentostatin and 2ʹ-amino-2ʹ-deoxyadenosine highlights a single gene cluster responsible for two independent pathways in Actinomadura sp. strain ATCC 39365. Appl Environ Microbiol 83:e00078-17. doi: 10.1128/AEM.00078-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Y, Luo F, Shang Y, Chen P, Lu Y, Wang C. 2017. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem Biol 24:1479–1489. doi: 10.1016/j.chembiol.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Gutowski GE, Sweeney MJ, DeLong DC, Hamill RL, Gerzon K, Dyke RW. 1975. Biochemistry and biological effects of the pyrazofurins (pyrazomycins): initial clinical trial. Ann N Y Acad Sci 255:544–551. doi: 10.1111/j.1749-6632.1975.tb29257.x. [DOI] [PubMed] [Google Scholar]
- 13.Cadman E, Benz C. 1980. Uridine and cytidine metabolism following inhibition of de novo pyrimidine synthesis by pyrazofurin. Biochim Biophys Acta 609:372–382. doi: 10.1016/0005-2787(80)90111-2. [DOI] [PubMed] [Google Scholar]
- 14.Worzalla JF, Sweeney MJ. 1980. Pyrazofurin inhibition of purine biosynthesis via 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5ʹ-monophosphate formyltransferase. Cancer Res 40:1482–1485. [PubMed] [Google Scholar]
- 15.Buchanan JG, Hamblin MR, Sood GR, Wightman RH. 1980. The biosynthesis of pyrazofurin and formycin. J Chem Soc Chem Commun 19:917–918. doi: 10.1039/c39800000917. [DOI] [Google Scholar]
- 16.Ochi K, Yashima S, Eguchi Y, Matsushita K. 1979. Biosynthesis of formycin-incorporation and distribution of C-13, C-14, and N-15-labeled compounds into formycin. J Biol Chem 254:8819–8824. [PubMed] [Google Scholar]
- 17.Wang SA, Ko Y, Zeng J, Geng Y, Ren D, Ogasawara Y, Irani S, Zhang Y, Liu HW. 2019. Identification of the formycin A biosynthetic gene cluster from Streptomyces kaniharaensis illustrates the interplay between biological pyrazolopyrimidine formation and de novo purine biosynthesis. J Am Chem Soc 141:6127–6131. doi: 10.1021/jacs.9b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu G, Kong L, Gong R, Xu L, Gao Y, Jiang M, Cai YS, Hong K, Hu Y, Liu P, Deng Z, Price NPJ, Chen W. 2018. Coordinated biosynthesis of the purine nucleoside antibiotics aristeromycin and coformycin in actinomycetes. Appl Environ Microbiol 84:e01860-18. doi: 10.1128/AEM.01860-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuda K, Tomita T, Shin-Ya K, Wakimoto T, Kuzuyama T, Nishiyama M. 2018. Discovery of unprecedented hydrazine-forming machinery in bacteria. J Am Chem Soc 140:9083–9086. doi: 10.1021/jacs.8b05354. [DOI] [PubMed] [Google Scholar]
- 20.Binda C, Robinson RM, Martin Del Campo JS, Keul ND, Rodriguez PJ, Robinson HH, Mattevi A, Sobrado P. 2015. An unprecedented NADPH domain conformation in lysine monooxygenase NbtG provides insights into uncoupling of oxygen consumption from substrate hydroxylation. J Biol Chem 290:12676–12688. doi: 10.1074/jbc.M114.629485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo J, Wang S, Zhang W, Li C, Deng Z, Zhang L, Qu X. 2019. Efficient editing DNA regions with high sequence identity in actinomycetal genomes by a CRISPR-Cas9 system. Synth Syst Biotechnol 4:86–91. doi: 10.1016/j.synbio.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasche ME, White RH. 1998. Mechanism for the enzymatic formation of 4-(beta-D-ribofuranosyl)aminobenzene 5′-phosphate during the biosynthesis of methanopterin. Biochemistry 37:11343–11351. doi: 10.1021/bi973086q. [DOI] [PubMed] [Google Scholar]
- 23.Sampei G, Baba S, Kanagawa M, Yanai H, Ishii T, Kawai H, Fukai Y, Ebihara A, Nakagawa N, Kawai G. 2010. Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria. J Biochem 148:429–438. doi: 10.1093/jb/mvq088. [DOI] [PubMed] [Google Scholar]
- 24.Ko Y, Wang SA, Ogasawara Y, Ruszczycky MW, Liu HW. 2017. Identification and characterization of enzymes catalyzing pyrazolopyrimidine formation in the biosynthesis of formycin A. Org Lett 19:1426–1429. doi: 10.1021/acs.orglett.7b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burk DL, Hwang J, Kwok E, Marrone L, Goodfellow V, Dmitrienko GI, Berghuis AM. 2007. Structural studies of the final enzyme in the alpha-aminoadipate pathway-saccharopine dehydrogenase from Saccharomyces cerevisiae. J Mol Biol 373:745–754. doi: 10.1016/j.jmb.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 26.Hong H, Samborskyy M, Zhou Y, Leadlay PF. 2019. C-nucleoside formation in the biosynthesis of the antifungal malayamycin A. Cell Chem Biol 26:493–501. doi: 10.1016/j.chembiol.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA. 2000. Practical Streptomyces genetics, 2nd ed John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 29.Gontang EA, Gaudencio SP, Fenical W, Jensen PR. 2010. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol 76:2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.