SUMMARY
Dosage compensation, which corrects for the imbalance in X-linked gene expression between XX females and XY males, represents a model for how genes are targeted for coordinated regulation. However, the mechanism by which dosage compensation complexes identify the X chromosome during early development remains unknown because of the difficulty of sexing embryos before zygotic transcription using X- or Y-linked reporter transgenes. We used meiotic drive to sex Drosophila embryos before zygotic transcription and ChIP-seq to measure the dynamics of dosage compensation factor targeting. The Drosophila male-specific lethal dosage compensation complex (MSLc) requires the ubiquitous zinc-finger protein chromatin-linked adaptor for MSL proteins (CLAMP) to identify the X chromosome. We observe a multi-stage process in which MSLc first identifies CLAMP binding sites throughout the genome, followed by concentration at the strongest X-linked MSLc sites. We provide insight into the dynamics of binding site recognition by a large transcription complex during early development.
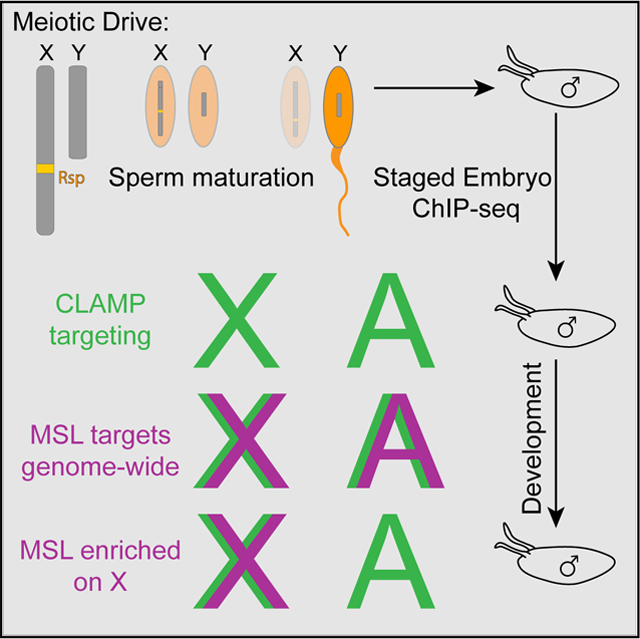
Graphical Abstract
In Brief
Rieder et al. establish a meiotic drive system to study Drosophila X chromosome dosage compensation before the maternal-zygotic transition. This study uncovers another step in the process during which the dosage compensation complex identifies binding sites genome-wide before becoming enriched on the X chromosome.
INTRODUCTION
Chromatin domains are enriched for specific histone modifications that activate or repress transcription, are a common property of metazoan genomes, and are critical for nuclear organization and regulation (Carelli et al., 2017). The formation of chromatin domains is often initiated early during embryogenesis (Evans et al., 2016; Vassetzky et al., 2000). However, little is understood about the dynamics of this essential process.
Strikingly large chromatin domains include the dosage-compensated chromosomes in heterogametic species, in which all of the genes on a chromosome are coordinately regulated. For example, the human female inactive X chromosome is characterized by the heterochromatic marks H3K27 methylation and ubiquitinated H2AK119 (Hall and Lawrence, 2010). Like humans, Drosophila melanogaster also uses an X/Y system of sex determination, but males perform dosage compensation; male Drosophila upregulate their single X chromosome ~ 2-fold to equalize expression with that of females (Hamada et al., 2005; Larschan et al., 2011). The dosage-compensated male X chromosome is marked by acetylated H4K16 (Smith et al., 2001; Turner et al., 1992), which facilitates transcriptional elongation and increased expression (Zippo et al., 2009). Moreover, X chromosome upregulation is conserved across species to balance X chromosome gene expression with that from autosomes and requires the same H4K16ac chromatin mark (Deng et al., 2013).
In Drosophila, the male-specific lethal complex (MSLc) is expressed only in males and accomplishes dosage compensation by specifically targeting the single X chromosome. MSLc includes the histone acetyltransferase males absent on the first (MOF) that deposits the activating H4K16ac mark (Akhtar and Becker, 2000; Smith et al., 2000). MSLc is most highly enriched at ~ 150–300 chromatin entry sites (CESs; also called high-affinity sites) that contain guanosine/adenosine (GA)-rich MSL recognition elements (MREs) (Alekseyenko et al., 2008; Straub et al., 2008; Villa et al., 2016). However, MREs are only 2-fold enriched on the X chromosome compared to autosomes (Kuzu et al., 2016). Furthermore, the targeting of MSLc also requires the six zinc finger transcription factor, chromatin-linked adaptor for MSL proteins (CLAMP) (Soruco et al., 2013), although CLAMP targets MREs genome-wide and is not unique to the X chromosome (Kaye et al., 2017; Rieder et al., 2017; Urban et al., 2017). Therefore, it remains unclear how MSLc targets the X chromosome to establish an active chromatin domain.
Previous models for the formation of the dosage-compensated chromatin domain relied on steady-state patterns in mutant lines, often by examining the larval salivary gland polytene chromosomes. For example, while fully functional MSLc decorates the euchromatic region of the X chromosome, partial complexes are recruited to a subset of X-linked CESs (Deng et al., 2005; Gorman et al., 1995; Gu et al., 1998; Lyman et al., 1997; Palmer et al., 1994; Straub et al., 2008). CESs are also the most highly enriched MSLc binding sites in wild-type flies (Alekseyenko et al., 2008). More recently, an in vitro DNA binding assay and the induction of MSLc in females both uncovered a subset of X-enriched sites called the pioneering sites on the X (PionX) (Villa et al., 2016; Cheetham and Brand, 2018; Schauer et al., 2017; Albig et al., 2019). Finally, when CESs are synthetically inserted onto autosomes, they are targeted by MSLc, which then spreads in cis into neighboring chromatin and in trans to the male X chromosome (Kelley et al., 1999; Larschan et al., 2007).
Based on these observations, several groups proposed a spreading model in which MSLc first targets CESs or X-enriched sites and then spreads in two or three dimensions to active genes on the male X chromosome (Alekseyenko et al., 2013; Lucchesi and Kuroda, 2015; McElroy et al., 2014; Ramírez et al., 2015; Soruco et al., 2013; Straub et al., 2008). However, directly testing this model in vivo is difficult; although dosage compensation is initiated early during development (Franke et al., 1996; Gergen, 1987; Polito et al., 1990; Rastelli et al., 1995), it is challenging to sex embryos pre-zygotic genome activation (ZGA).
To overcome this obstacle, we used a meiotic drive system to generate male- and female-enriched pools of embryos and performed chromatin immunoprecipitation sequencing (ChIP-seq) for CLAMP, MSLc, and the H4K16ac chromatin mark at precise embryonic stages surrounding the initiation of dosage compensation. This system of sexing embryos before ZGA represents a powerful tool to measure the recruitment dynamics of the dosage compensation complex. We identified the following multi-stage process for targeting MSLc: (1) CLAMP targets loci genome-wide; (2) in an intermediate step, MSLc identifies loci that are bound by CLAMP genome-wide, but not at CESs; and(3) MSLc becomes more enriched at CESs and less enriched at other CLAMP binding sites. Overall, we provide insight into the dynamic mechanism by which a large transcription complex identifies its binding sites during early development.
RESULTS AND DISCUSSION
A Meiotic Drive System Generates Sexed Embryos before ZGA
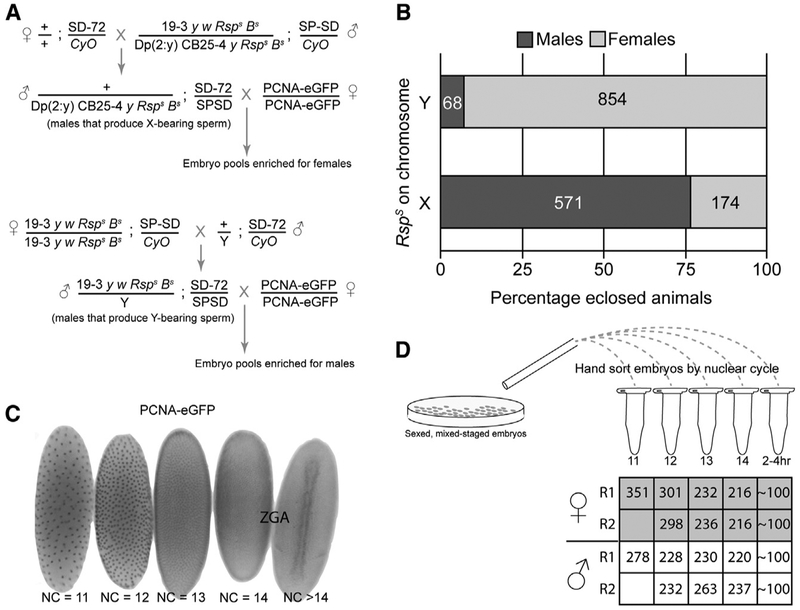
A significant impediment to observing the initial steps of dosage compensation is the inability to sex the Drosophila embryo before ZGA, when sex-specific reporter transgenes are activated (Schauer et al., 2017). To overcome this obstacle, we used the segregation distorter (SD) meiotic drive system (Larracuente and Presgraves, 2012). In this system, expression of the segregation distorter gene product (Sd), a mislocalized form of RanGAP, interacts with sensitive alleles of the Responder (Rsp) locus, a large pericentromeric stretch of satellite DNA. In its naturally occurring form, both the Sd and Rsp loci reside on chromosome 2, and Sd selfishly enriches its own transmission by preventing the maturation of spermatids carrying Rsp-bearing chromosomes.
Although the mechanism of SD remains unknown, the Sd and Rsp loci can be placed on ectopic chromosomes with the same effect. We therefore used existing Drosophila melanogaster stocks in which Sd remains on chromosome 2, but sensitive alleles of the Rsp locus (RspS) now reside on the X or Y chromosomes (Cheng et al., 2016; Polito et al., 1990; Walker et al., 1989). Males produced from this crossing strategy (Figure 1A) produce predominantly X or Y chromosome-bearing sperm, depending on their genotypes, and sire a preponderance of progeny of the corresponding sex (Figure 1B). If dosage compensation was significantly affected by the autosomal SD system, then a strong male-specific viability defect would be expected, but the autosomal SD system does not cause a strong sex bias (Denell et al., 1969). Recently, meiotic drive has also been used to sex select mouse embryos, but this system has not yet been used to study dosage compensation (Umehara et al., 2019).
Figure 1. Validation of a Meiotic Drive System to Generate Sexed Embryos.
(A) Crossing scheme using SD meiotic drive system to generate sex-enriched pools of embryos expressing PCNA-EGFP. Rsps is a sensitive allele of the Rsp locus. SPSD and SD-72 are strong Sd alleles, and we found the drive to be strongest when the SD alleles were in trans.(B) Percentage of adult animals eclosed of each sex due to crossing schemes in (A). The numbers within the bars represent the total number of animals counted.(C) Staging embryos by nuclear cycle (NC) using the PCNA-EGFP transgene. Zygotic genome activation (ZGA) occurs during NC14.(D) Workflow to sort embryos for small-scale ChIP-seq. We hand sorted between 200 and 351 embryos for each biological replicate. We performed two biological replicates for male and female embryos between NC12 and NC14 and one replicate for NC11. To represent a “late” stage of dosage compensation, we chose ~100 2- to 4-h embryos.
We combined the SD sexing system with a maternally inherited proliferating cell nuclear antigen (PCNA)-EGFP reporter transgene (Blythe and Wieschaus, 2016), which allowed us to precisely stage embryos by nuclear cycle (NC), representing a developmental time course of ~1 h surrounding ZGA (Figure 1C). These data demonstrate that, when combined, the SD meiotic drive and PCNA-EGFP reporter systems represent a powerful tool to both stage and sex embryos before ZGA. We hand sorted fixed embryos produced from these crosses (Figure 1A) and performed low-input ChIP-seq (Figure 1D) (Blythe and Wieschaus, 2015) for factors associated with dosage compensation.
MSLc Identifies the Male X Chromosome in a Multistage Process
We previously demonstrated that the CLAMP protein, which targets CESs and other GA-rich sequences genome-wide, facilitates MSLc recruitment and dosage compensation (Kaye et al., 2018; Kuzu et al., 2016; Soruco et al., 2013). In addition, CLAMP is maternally deposited into the oocyte, while MSLc does not assemble until later in development (Graveley et al., 2011). We therefore hypothesized that CLAMP identifies its binding sites before MSLc. To test this hypothesis, we performed ChIP-seq on male-enriched embryo pools for CLAMP, MSL3 (a component of MSLc), and the H4K16ac mark, and on female-enriched embryo pools for CLAMP (Rieder et al., 2017) and H4K16ac. MSLc does not assemble in female embryos due to post-transcriptional repression of the structural MSL2 component by the master sex regulator Sex-lethal (Kelley et al., 1997). It is possible that contamination from spurious females in the male population alters the binding pattern of CLAMP, but not of MSLc because MSLc does not form in females.
To perform small-scale ChIP-seq, we used 200–351 embryos for each NC (Figure 1D). In addition, we performed ChIP-seq on ~ 100 sexed, mixed-stage embryos aged to 2–4 h after egg lay, representing a post-ZGA stage (Gergen, 1987). Because hand sorting is laborious, we were limited in the number of biological replicates that we could perform; we performed two biological replicates for each sex and time point, except for NC11 and the 2- to 4-h time point, for which we performed one biological replicate for each sex. In total, we hand sorted >3,500 embryos. To ensure that we used only the highest-quality data, we selected the best biological replicate based on which replicate had the highest overall scores for PCR bottlenecking coefficients 1 and 2 (measures of approximate library complexity) (Bailey et al., 2013; Landt et al., 2012) (Table S1). The overlap between peaks in selected samples is shown in Figure S1.
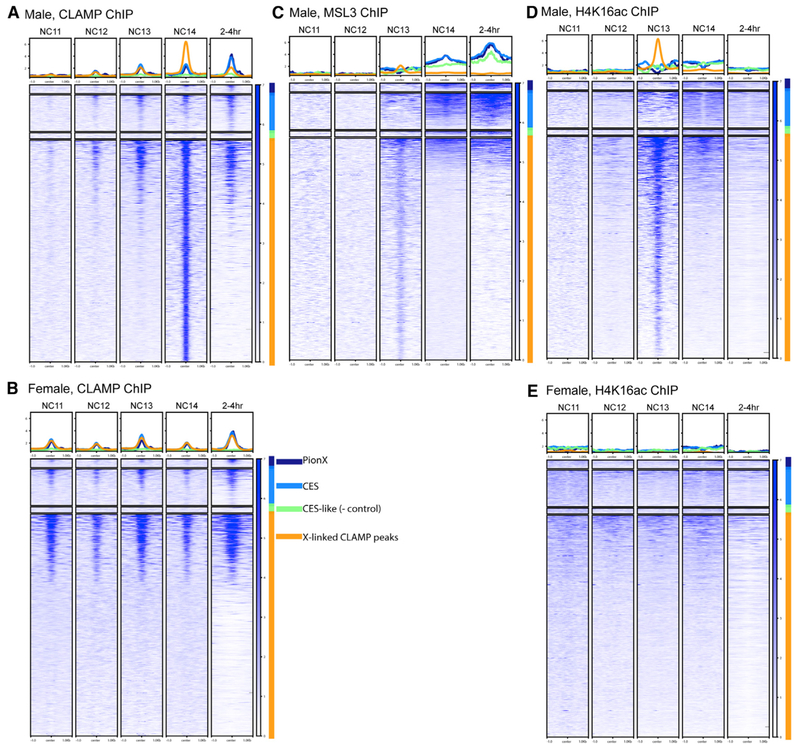
To visualize dynamic recruitment throughout early development, we plotted heatmaps of the highest-quality replicates to measure occupancy at several classes of loci. We examined MSLc and CLAMP binding to several classes of sites that have been previously identified as involved in dosage compensation:(1) PionX sites interact with MSL complex in vitro and were identified in vivo using a cell culture-induction system in which MSLc is induced in female cells (Villa et al., 2016); (2) non-PionX CESs associate with MSLc in vivo (Soruco et al., 2013) but not in vitro (Villa et al., 2016); (3) CES-like sites do not recruit MSLc in cell lines but have similar properties such as being located in gene bodies and having the H3K36me3 chromatin mark (Soruco et al., 2013); and (4) CLAMP binding sites at the onset of ZGA are identified in male NC14 (the present study).
We observed MSLc targeting in multiple stages. As we hypothesized, CLAMP identifies binding sites in both sexes before MSLc (Figures 2A and 2B). Before MSLc identifies CESs in NC14, CLAMP is modestly de-enriched at CESs compared to other CLAMP binding sites. Concurrently, MSLc identifies other CLAMP binding sites genome-wide and is not restricted to the X chromosome (Figures 2C and S2), an observation that is supported by male H4K16ac ChIP (Figure 2D). H4K16ac patterns in males mirror those of MSLc throughout development as would be expected because the MOF component of MSLc deposits the H4K16ac mark (Smith et al., 2005). Because H4K16ac can be deposited by a different transcription complex in females (non-specific lethal complex) (Raja et al., 2010), its occupancy pattern differs between males and females (Figures 2D and 2E) and is consistent with previous reports in cultured female Kc cells (Gelbart et al., 2009).
Figure 2. Staged, Sexed ChIP-Seq Heatmaps at X-Linked CLAMP Sites.
Data are mapped over X-linked CLAMP sites and broken into categories, including PionX sites (Villa et al., 2016) (n = 55; dark blue), CESs (Soruco et al., 2013; Alekseyenko et al., 2008) (n = 234; light blue), and other X-linked CLAMP peaks from the male NC14 sample (n = 1,417; orange). CES-like sites represent negative control sites (Soruco et al., 2013; Alekseyenko et al., 2008) (n = 38; green).
(A) CLAMP ChIP-seq from male embryos.
(B) CLAMP ChIP-seq from female embryos.
(C) MSL3 ChIP-seq from male embryos.
(D) H4K16ac ChIP-seq from male embryos.
(E) H4K16ac ChIP-seq from female embryos.
See also Figures S1, S2, and S3.
After MSLc identifies CESs, CLAMP also becomes enriched at CESs, specifically in males, which is consistent with the documented synergy between the two factors (Albig et al., 2019; Larschan et al., 2012). Individual profiles surrounding selected CESs (Figure S3) are consistent with heatmaps. Overall, we observe a dynamic pattern of MSLc targeting in which it identifies CLAMP binding sites throughout the genome, followed by specific targeting to CESs. We do not observe a time point at which PionX sites (Villa et al., 2016) are specifically enriched for MSLc, compared to other CESs.
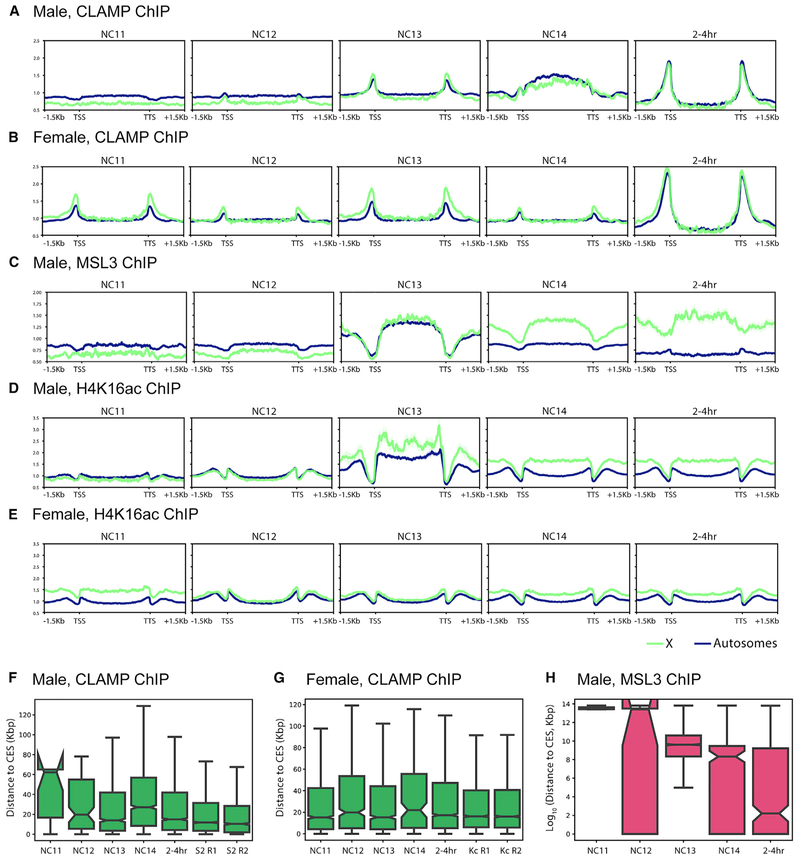
To define how CLAMP, MSLc, and H4K16ac occupancies change over time at both genes on the X chromosome and autosomes, we generated average gene profiles. CLAMP is similarly enriched on the X chromosome (Figures 3A and 3B) and autosomes (Figures S2A and S2B) in both males and females, although CLAMP becomes enriched at CESs compared to other binding sites after 2 h (Figures 2A and 2B). CLAMP average gene profiles are similar in males and females until NC14, when the profiles diverge (Figures 3A and 3B), which is consistent with the previously documented synergy between CLAMP and MSLc only in males (Soruco et al., 2013). In contrast, MSLc becomes more enriched at CESs and less enriched at other CLAMP binding sites over time (Figures 2C and 3C). MSLc and H4K16ac (Figure 3D) are enriched over gene bodies in males, consistent with previous reports in cell culture (Alekseyenko et al., 2006). Furthermore, the X enrichment of MSLc and H4K16ac increases over time in males but not in females (Figures 2C, 2D, 3C, and 3D).
Figure 3. CLAMP and MSL3 Gene Profiles and Distance to CESs over Development.
(A–E) Average X-linked (green) and autosome-linked (blue) gene profiles over developmental time (shading represents SE). Male, CLAMP ChIP (A); female, CLAMP ChIP (B); male, MSL3 ChIP (C); male, H4K16ac ChIP (D); and female, H4K16ac ChIP (E). TSS, transcription start site; TTS, transcription termination site. (F and G) Distance to CES of (F) male CLAMP peaks and (G) female CLAMP peaks. For comparison, we have included previously published CLAMP ChIP-seq data from cultured male S2 cells and female Kc cells (two biological replicates each from Soruco et al., 2013).(H) Distance to CESs of male MSL3 peaks. For all box and whisker plots, the 95% confidence interval is shown with a notch around the median line; whiskers represent 1.5 interquartile range (IQR), outliers have been omitted.
We also investigated the distances of CLAMP and MSL3 peaks to the nearest CESs over developmental time. In both males and females, CLAMP peaks reside very close to CESs, even at the earliest time points (Figures 3F and 3G). However, male MSL3 peaks grow closer to CESs throughout development (Figure 3H). These data suggest that CLAMP targets CESs in both sexes early during development before the MSLc complex identifies CESs. Initially, MSLc identifies CLAMP binding sites genome-wide that are not located at CESs. Next, synergy between CLAMP and MSLc occurs specifically at CESs, and likely other factors such as 1.688 repeat elements (Joshi and Meller, 2017) enrich MSLc occupancy specifically at CESs.
In this study, we identified an unexpected step in the process of MSLc identifying the X chromosome: the association of MSLc with CLAMP binding sites genome-wide at NC13. A key question is how does MSLc become restricted to the X chromosome after identifying CLAMP binding sites genome-wide? MSLc and CLAMP can function synergistically, which has been defined in vivo (Soruco et al., 2013) and in vitro (Albig et al., 2019). The sites where MSL and CLAMP function synergistically often have binding motifs that deviate from the MSL complex consensus sequence (MSL recognition element; MRE) (Alekseyenko et al., 2008) and specific DNA shape properties that require synergy between both factors for stable binding (Albig et al., 2019). However, similar sequences exist throughout the genome, and therefore sequence alone is not sufficient to define CESs. MREs are often more clustered at CES compared to other sites in the genome (Kuzu et al., 2016), and it is possible that clustering and CLAMP multimerization promote the ability of MSLc to specifically associate with CESs.
We observed that CLAMP is de-enriched at CES compared to other sites in the genome early in development, despite the known synergy between CLAMP and the MSL complex at these sites (Soruco et al., 2013; Albig et al., 2019). Furthermore, MSLc and CLAMP associate at the same autosomal sites before they co-occupy CESs. The delayed synergy between CLAMP and MSLc at CESs may be caused by several mechanisms: (1) other factors at CESs such as GAGA factor (GAF) may compete with CLAMP to reduce its enrichment early in development. GAF can compete with CLAMP to bind to CESs and can promote MSLc recruitment, although not as well as CLAMP (Kaye et al., 2017).(2) CLAMP may be titrated away from CESs early in development, as it may associate with its other binding partners such as insulator proteins (Kaye et al., 2017; Bag et al., 2019) or components of the spliceosome (Urban et al., 2017). (3) CESs cluster together in three-dimensional space (Ramírez et al., 2015) and contain multiple closely spaced CLAMP binding sites (Soruco et al., 2013). Also, CLAMP is part of two insulator complexes (Kaye et al., 2017; Bag et al., 2019). Therefore, CLAMP may multimerize to mediate three-dimensional organization at CESs such that CLAMP multimers are buried within a dynamic three-dimensional structure and are not accessible by CLAMP antibodies.
Overall, we define a multi-step process by which MSLc first identifies thousands of CLAMP-occupied sites throughout the genome before becoming enriched at its strongest X-linked target sites (CESs). Synergistic interactions between CLAMP and MSLc (Albig et al., 2019; Soruco et al., 2013) are likely to enhance the occupancy of both factors at CESs. We hypothesize that additional factors promote the specific interaction between MSLc and CESs, including direct binding of MSL2 to DNA mediated by DNA shape (Albig et al., 2019), specific three-dimensional conformation surrounding CESs (Ramírez et al., 2015), and 1.688 repetitive elements (Joshi and Meller, 2017). In the future, it will be possible to use the meiotic drive system to define the contribution of each of these factors to MSLc targeting in a developmental context.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Leila Rieder (leila.rieder@emory.edu). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We maintained flies on standard cornmeal sucrose media at 24 °C. Meiotic drive fly stocks (both gifts from Cynthia Staber), +; SD72/CyO and 19–3, yw, Rsp[s]-B[s]/Dp(2:y)CB25–4, y+, Rsp[s]B[s]; SPSD/CyO (Bloomington BSC64332) are published in Rieder et al. (2017). To obtain female embryos, we mated +; SD72/CyO females to 19–3, yw, Rsp[s]-B[s]/Dp(2:y) CB25–4, y+, Rsp[s]B[s]; SPSD/CyO males to obtain +/Dp(2:y) CB25–4, y+, Rsp[s]B[s]; SPSD/SD72 males. To obtain male embryos, we mated +; SD72/CyO males to 19–3, yw, Rsp[s]-B[s]/Dp(2:y) CB25–4, y+, Rsp[s]B[s]; SPSD/CyO females to obtain 19–3, yw, Rsp[s]-B[s]/Y; SPSD/SD72 males. We mated males of both genotypes to yw; attP2{PCNA-EGFP} virgin females (Blythe and Wieschaus, 2016).
METHOD DETAILS
Meiotic drive validation
K is the proportion of SD-bearing progeny compared to total progeny (Ganetzky, 1977; Gell and Reenan, 2013). In this case, k is expressed as the number of non-Rsp-bearing progeny, or the number of the expected sex, compared to total progeny (Figure 1B). We conducted k-tests at 24 °C based on Gell and Reenan (2013) at by crossing either +/Dp(2:y) CB25–4, y+, Rsp[s]B[s]; SPSD/SD72 males or 19–3, yw, Rsp[s]-B[s]/Y; SPSD/SD72 males to yw; attP2{PCNA-EGFP} virgin females.
Embryo fixation and sorting
We collected embryos on apple juice plates with yeast paste at 24 °C. We performed 0–4hr timed lays and fixed embryos according to Blythe and Wieschaus (2015). We then hand-sorted embryos using a Zeiss Discovery.V8 microscope under GFP excitation using an X-CITE 120Q stereo light source. We pooled 200–351 embryos (NC 11–14, Figure 1D). For 2–4hr embryos, we used approximately 100 mixed stage embryos. We froze embryos at −80 °C until further processing.
Chromatin immunoprecipitation (ChIP)-sequencing
We performed ChIP as in Blythe and Wieschaus (2015) using 2 mL of rabbit anti-CLAMP antibody, 2 mL of polyclonal rabbit IgG (Millipore-Sigma, 12–370), 3 mL of rabbit polyclonal anti-H4K16ac (Millipore-Sigma, 07–329) or 4 mL of a 1:10 dilution of goat anti-MSL3 serum (gift from Mitzi Kuroda) per sample. We synthesized libraries using the NEBNext ChIP-seq kit (New England Biosystems, E6240L) and sequenced libraries on an Illumina HiSeq 2500 in 2 × 3 100-bp or 2 × 3 150-bp mode. ChIP-seq data is deposited at NCBI GEO: GSE133637.
QUANTIFICATION AND STATISTICAL ANALYSIS
We mapped sequencing reads to release 6 of the Drosophila melanogaster genome (dm6) using Bowtie2 (version 2.3.0) (Langmead and Salzberg, 2012) with default parameters. We identified reads with a MAPQ < 30 and removed PCR duplicate reads using Picard MarkDuplicates (version 2.9.2) (Picard Toolkit. 2019. Broad Institute, GitHub Repository. http://broadinstitute.github.io/picard/; Broad Institute) using SAMtools (version 1.9) (Li et al., 2009). We used MACS2 (version 2.1.1) (Zhang et al., 2008) to identify peaks with the following parameters:–nomodel -B -SPMR -keep-dup all -g dm. We used input for peak calling in all samples with the exception of male 2–4 hour time point. We used a narrow peak calling with a q cutoff of 0.01 for CLAMP ChIPs and broad peak calling with a q cutoff of 0.05 for MSL3 and H4K16ac ChIPs; we also used–extsize of 147 for H4K16ac ChIPs. We used MACS2 to generate fold enrichment tracks for each ChIP. We generated average profiles using deepTools (version 3.1.0) (Ramírez et al., 2014). We calculated distances to the nearest CES using bedtools (version 2.27.1) (Quinlan and Hall 2010). We determined peak overlaps using Intervene (version 0.5.8) (Khan and Mathelier, 2017).
DATA AND CODE AVAILABILITY
Female embryo CLAMP ChIP-seq data are deposited in NCBI GEO: GSE119448 (Rieder et al., 2017). Cell culture CLAMP ChIP-seq data are deposited in NCBI GEO: GSE39271 (Soruco et al., 2013). ChIP-seq data (this study) is deposited at NCBI GEO: GSE133637. We lifted CES locations (Alekseyenko et al., 2008; Soruco et al., 2013) over to dm6 using the UCSC liftOver tool.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CLAMP antibody | Erica N. Larschan | RRID:AB_2195548 |
| Goat anti-MSL3 serum | Mitzi Kuroda | RRID:AB_2147786 |
| rabbit polyclonal anti-H4K16ac | Millipore-Sigma | 07–329; RRID:AB_310525 |
| polyclonal rabbit IgG | Millipore-Sigma | 12–370; RRID:AB_145841 |
| Critical Commercial Assays | ||
| NEBNext ChIP-seq kit | New England Biolabs | E6240L |
| Deposited Data | ||
| Female embryo CLAMP ChIP-seq | NCBI GEO | GSE119448 |
| Cell culture CLAMP ChIP-seq | NCBI GEO | GSE39271 |
| Male embryo CLAMP ChIP-seq | NCBI GEO | GSE133637 |
| Experimental Models: Organisms/Strains | ||
| +; SD72/CyO | Cynthia Staber, Stowers Institute | N/A |
| 19–3, yw, Rsp[s]-B[s]/Dp(2:y)CB25–4, y+, Rsp[s]B[s]; SPSD/CyO | Bloomington Stock Center | BSC64332 |
| yw; attP2{PCNA-EGFP} | Blythe and Wieschaus, 2016 | N/A |
| Software and Algorithms | ||
| Bowtie2 | Langmead and Salzberg, 2012 | version 2.3.0 |
| Picard MarkDuplicates | Picard Toolkit.2019. Broad Institute, GitHub Repository. http://broadinstitute.github.io/picard/; Broad Institute | version 2.9.2 |
| SAMtools | Li et al., 2009 | version 1.9 |
| MACS2 | Zhang et al., 2008 | version 2.1.1 |
| deepTools | Ramírez et al., 2014 | version 3.1.0 |
| bedtools | Quinlan and Hall, 2010 | version 2.27.1 |
| Intervene | Khan and Mathelier, 2017 | version 0.5.8 |
Highlights.
Establishment of a system to study dosage compensation before zygotic transcription starts
The Drosophila DCC binds genome-wide before enrichment at X chromosome binding sites
The CLAMP transcription factor binds to chromatin before zygotic transcription starts
ACKNOWLEDGMENTS
We thank C. Staber and the Bloomington Drosophila Stock Center for the SD stocks and S. Blythe for the PCNA-EGFP stock. We thank Mitzi Kuroda for the goat anti-MSL3 serum. We thank the Hart lab at Brown University and the Dunn lab, formerly at Brown, now at Yale University, for use of their stereomicroscopes, and S. Siebert for assistance in taking the embryo image in Figure 1C. We thank Dr. Marcela Soruco, Dr. Mattew Booker, Dr. Guray Kuzu, and Dr. Michael Tolstorukov for their important contributions. This work was supported by NIH F32GM109663, K99HD092625, and R00HD092625 to L.E.R.; NIH R35GM126994 to E.N.L.; and a HHMI Gilliam fellowship and a NSF GRFP grant to W.T.J.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.095.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akhtar A, and Becker PB (2000). Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375. [DOI] [PubMed] [Google Scholar]
- Albig C, Tikhonova E, Krause S, Maksimenko O, Regnard C, and Becker PB (2019). Factor cooperation for chromosome discrimination in Drosophila. Nucleic Acids Res. 47, 1706–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Ellison CE, Gorchakov AA, Zhou Q, Kaiser VB, Toda N, Walton Z, Peng S, Park PJ, Bachtrog D, and Kuroda MI (2013). Conservation and de novo acquisition of dosage compensation on newly evolved sex chromosomes in Drosophila. Genes Dev. 27, 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, and Kuroda MI (2006). High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20, 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee O-K, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, and Kuroda MI (2008). A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag I, Dale RK, Palmer C, and Lei EP (2019). The zinc-finger protein CLAMP promotes gypsy chromatin insulator function in Drosophila 132, jcs226092 10.1242/jcs.226092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, Madrigal P, Taslim C, and Zhang J (2013). Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput. Biol 9, e1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2015). Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2016). Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5, e20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, Sharma G, and Ahringer J (2017). Broad Chromatin Domains: An Important Facet of Genome Regulation. BioEssays 39, 1700124. [DOI] [PubMed] [Google Scholar]
- Cheetham SW, and Brand AH (2018). RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat. Struct. Mol. Biol 25, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Kuppanda N, Aldrich JC, Akbari OS, and Ferree PM (2016). Male-Killing Spiroplasma Alters Behavior of the Dosage Compensation Complex during Drosophila melanogaster Embryogenesis. Curr. Biol 26, 1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denell RE, Judd BH, and Richardson RH (1969). Distorted sex ratios due to segregation distorter in Drosophila melanogaster. Genetics 61, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Ma W, Nguyen DK, Hiatt JB, Noble WS, Shendure J, and Disteche CM (2013). Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev. Cell 25, 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Rattner BP, Souter S, and Meller VH (2005). The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev 122, 1094–1105. [DOI] [PubMed] [Google Scholar]
- Evans KJ, Huang N, Stempor P, Chesney MA, Down TA, and Ahringer J (2016). Stable Caenorhabditis elegans chromatin domains separate broadly expressed and developmentally regulated genes. Proc. Natl. Acad. Sci. USA 113, E7020–E7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Dernburg A, Bashaw GJ, and Baker BS (1996). Evidence that MSL-mediated dosage compensation in Drosophila begins at blastoderm. Development 122, 2751–2760. [DOI] [PubMed] [Google Scholar]
- Ganetzky B (1977). On the components of segregation distortion in Drosophila melanogaster. Genetics 86, 321–355. [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, and Kuroda MI (2009). Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat. Struct. Mol. Biol 16, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell SL, and Reenan RA (2013). Mutations to the piRNA pathway component aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster. Genetics 193, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen JP (1987). Dosage Compensation in Drosophila: Evidence That daughterless and Sex-lethal Control X Chromosome Activity at the Blastoderm Stage of Embryogenesis. Genetics 117, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M, Franke A, and Baker BS (1995). Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121, 463–475. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Szauter P, and Lucchesi JC (1998). Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet 22, 56–64. [DOI] [PubMed] [Google Scholar]
- Hall LL, and Lawrence JB (2010). XIST RNA and architecture of the inactive X chromosome: implications for the repeat genome. Cold Spring Harb. Symp. Quant. Biol 75, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, and Kuroda MI (2005). Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19, 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi SS, and Meller VH (2017). Satellite Repeats Identify X Chromatin for Dosage Compensation in Drosophila melanogaster Males. Curr. Biol 27, 1393–1402.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye EG, Booker M, Kurland JV, Conicella AE, Fawzi NL, Bulyk ML, Tolstorukov MY, and Larschan E (2018). Differential Occupancy of Two GA-Binding Proteins Promotes Targeting of the Drosophila Dosage Compensation Complex to the Male X Chromosome. Cell Rep 22, 3227–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye EG, Kurbidaeva A, Wolle D, Aoki T, Schedl P, and Larschan E (2017). Drosophila Dosage Compensation Loci Associate with a Boundary-Forming Insulator Complex. Mol. Cell. Biol 37, e00253–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, and Kuroda MI (1999). Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98, 513–522. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, and Kuroda MI (1997). Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387, 195–199. [DOI] [PubMed] [Google Scholar]
- Khan A, and Mathelier A (2017). Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzu G, Kaye EG, Chery J, Siggers T, Yang L, Dobson JR, Boor S, Bliss J, Liu W, Jogl G, et al. (2016). Expansion of GA Dinucleotide Repeats Increases the Density of CLAMP Binding Sites on the X-Chromosome to Promote Drosophila Dosage Compensation. PLoS Genet. 12, e1006120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, and Presgraves DC (2012). The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics 192, 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, and Kuroda MI (2007). MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 28, 121–133. [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, and Kuroda MI (2011). X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Soruco MML, Lee O-K, Peng S, Bishop E, Chery J, Goebel K, Feng J, Park PJ, and Kuroda MI (2012). Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet. 8, e1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, and Kuroda MI (2015). Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol 7, a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, and Kuroda MI (1997). Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147, 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy KA, Kang H, and Kuroda MI (2014). Are we there yet? Initial targeting of the Male-Specific Lethal and Polycomb group chromatin complexes in Drosophila. Open Biol. 4, 140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MJ, Richman R, Richter L, and Kuroda MI (1994). Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 8, 698–706. [DOI] [PubMed] [Google Scholar]
- Polito C, Pannuti A, and Lucchesi JC (1990). Dosage compensation in Drosophila melanogaster male and female embryos generated by segregation distortion of the sex chromosomes. Dev. Genet 11, 249–253. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja SJ, Charapitsa I, Conrad T, Vaquerizas JM, Gebhardt P, Holz H, Kadlec J, Fraterman S, Luscombe NM, and Akhtar A (2010). The nonspecific lethal complex is a transcriptional regulator in Drosophila. Mol. Cell 38, 827–841. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Düdar F, Diehl S, Grüning BA, and Manke T (2014). deep-Tools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42 (W1), W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Lingg T, Toscano S, Lam KC, Georgiev P, Chung H-R, Lajoie BR, de Wit E, Zhan Y, de Laat W, et al. (2015). High-Affinity Sites Form an Interaction Network to Facilitate Spreading of the MSL Complex across the X Chromosome in Drosophila. Mol. Cell 60, 146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Richman R, and Kuroda MI (1995). The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev 53, 223–233. [DOI] [PubMed] [Google Scholar]
- Rieder LE, Koreski KP, Boltz KA, Kuzu G, Urban JA, Bowman SK, Zeidman A, Jordan WT 3rd, Tolstorukov MY, Marzluff WF, et al. (2017). Histone locus regulation by the Drosophila dosage compensation adaptor protein CLAMP. Genes Dev. 31, 1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer T, Ghavi-Helm Y, Sexton T, Albig C, Regnard C, Cavalli G, Furlong EEM, and Becker PB (2017). Chromosome topology guides the Drosophila Dosage Compensation Complex for target gene activation. EMBO Rep. 18, 1854–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Allis CD, and Lucchesi JC (2001). Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem 276, 31483–31486. [DOI] [PubMed] [Google Scholar]
- Smith ER, Cayrou C, Huang R, Lane WS, Côté J, and Lucchesi JC (2005). A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol 25, 9175–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, and Lucchesi JC (2000). The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol 20, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soruco MML, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, et al. (2013). The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 27, 1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, and Becker PB (2008). The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 4, e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, and Lavender J (1992). Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69, 375–384. [DOI] [PubMed] [Google Scholar]
- Umehara T, Tsujita N, and Shimada M (2019). Activation of Toll-like receptor 7/8 encoded by the X chromosome alters sperm motility and provides a novel simple technology for sexing sperm. PLoS Biol. 17, e3000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JA, Urban JM, Kuzu G, and Larschan EN (2017). The Drosophila CLAMP protein associates with diverse proteins on chromatin. PLoS One 12, e0189772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassetzky Y, Hair A, and Méchali M (2000). Rearrangement of chromatin domains during development in Xenopus. Genes Dev. 14, 1541–1552. [PMC free article] [PubMed] [Google Scholar]
- Villa R, Schauer T, Smialowski P, Straub T, and Becker PB (2016). PionX sites mark the X chromosome for dosage compensation. Nature 537, 244–248. [DOI] [PubMed] [Google Scholar]
- Walker ES, Lyttle TW, and Lucchesi JC (1989). Transposition of the Responder element (Rsp) of the Segregation distorter system (SD) to the X chromosome in Drosophila melanogaster. Genetics 122, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, and Oliviero S (2009). Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138, 1122–1136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Female embryo CLAMP ChIP-seq data are deposited in NCBI GEO: GSE119448 (Rieder et al., 2017). Cell culture CLAMP ChIP-seq data are deposited in NCBI GEO: GSE39271 (Soruco et al., 2013). ChIP-seq data (this study) is deposited at NCBI GEO: GSE133637. We lifted CES locations (Alekseyenko et al., 2008; Soruco et al., 2013) over to dm6 using the UCSC liftOver tool.