Abstract
Aberrations in genes coding for subunits of the BAF chromatin remodeling complexes are highly abundant in human cancers. Currently, it is not understood how these loss-of-function mutations contribute to cancer development and how they can be targeted therapeutically. The cancer-type-specific occurrence patterns of certain subunit mutations suggest subunit-specific effects on BAF complex function, possibly by the formation of aberrant residual complexes. Here, we systematically characterize the effects of individual subunit loss on complex composition, chromatin accessibility and gene expression in a panel of knock-out cell lines deficient for 22 BAF subunits. We observe strong, specific and sometimes discordant alterations dependent on the targeted subunit and show that these explain intra-complex co-dependencies, including the synthetic lethal interactions SMARCA4-ARID2, SMARCA4-ACTB and SMARCC1-SMARCC2. These data provide insights into the role of different BAF subcomplexes in genome-wide chromatin organization and suggest approaches to therapeutically target BAF mutant cancers.
Keywords: BAF complex, mammalian SWI/SNF complex, chromatin remodeling, interaction proteomics, ATAC-seq, transcriptomics, synthetic lethality, chromatin accessibility, chromatin binding
Introduction
The human SWI/SNF complexes (BRG1/BRM associated factor (BAF) complexes) are ATP-dependent chromatin remodelers that regulate DNA accessibility dynamically and thereby play important roles in essential cellular processes such as transcription, DNA repair and replication. These large, polymorphic complexes are formed of up to 15 subunits encoded by more than 29 genes1,2. Three major complexes can be discriminated, BAF, PBAF (Polybromo-associated BAF) and non-canonical BAF (ncBAF, GBAF), that exist in multiple compositions as several subunit positions can be occupied alternatively by proteins encoded from paralogous genes. The catalytic ATPase, encoded by SMARCA2 or SMARCA4, is essential for the function of BAF complexes in sliding nucleosomes along the DNA or evicting them from chromatin. Nearly the full remodeling activity of the entire complex is reconstituted in vitro when the ATPase subunits are bound to SMARCB1, SMARCC1, and SMARCC23. A further important complex position, alternatively occupied by ARID1A, ARID1B, or PBAF-specific ARID2, is thought to recruit the complex to chromatin4. Other subunits harbor additional chromatin binding domains, but much less is known about their contribution to complex function. Currently, there is no high resolution structure of the BAF complexes with subunit assignment available.
Mutations in BAF subunits have been observed with high frequency in human cancers5,6. Mostly, these are loss-of-function mutations that result in the loss of the mutated subunits on the protein level. All subunits can be affected, but mutation prevalence is cancer-dependent, suggesting context- and subunit-specific effects on complex function. Currently, it remains unclear how exactly BAF mutations contribute to cancer initiation and/or progression and how they affect BAF complex composition and function. Since loss-of-function mutations are not directly druggable, a focus has been on the identification of synthetic lethalities with BAF mutations. It has been shown that loss of SMARCA4 makes cells dependent on the SMARCA2 gene7–9, and ARID1B becomes essential following ARID1A mutation10–12. These data suggest that paralogous subunits occupying the same key complex positions exert at least in part redundant functions and can compensate for each other.
To systematically evaluate the impact of BAF subunit loss on complex architecture, chromatin accessibility and transcription, we established and comprehensively characterized isogenic mutant cell lines for 22 targetable BAF subunits in HAP1 cells. In these cell lines we further tested all intra-complex synthetic lethalities and identified ARID2 and ACTB as potential therapeutic targets for SMARCA4-mutant cancers as well as SMARCC2 in SMARCC1-mutated cancers and vice versa.
Results
An isogenic cell line panel for loss of individual BAF subunits
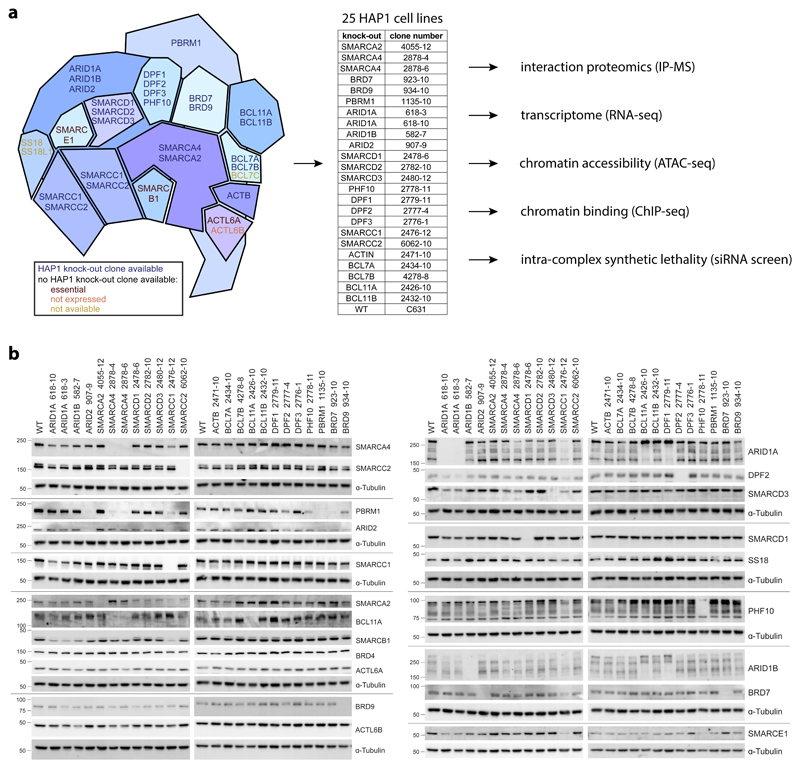
We established and comprehensively characterized a panel of isogenic HAP1 cell lines with individual knock-outs for 22 targetable BAF subunits (Fig. 1a). The knock-outs were validated on the genetic level by mapping the mutation site (Supplementary Table 1), and on the protein level by Western blot (Fig. 1b, Supplementary Note 3.1). In addition to loss of the targeted subunit, we also observed co-regulation of other BAF members in some of these clonal cell lines, for example between PBAF specific subunits. ARID2 knock-out (KO) reduced levels of PBRM1 and BRD7 and a moderate reduction of PHF10. Also BRD7KO, PHF10KO and SMARCA4KO cells had reduced PBRM1 levels.
Fig. 1. An isogenic cell line panel for loss of individual BAF subunits.
(a) Scheme illustrating which BAF subunits are included in the isogenic HAP1 knock-out cell panel and which experiments were applied to these cells. (b) Cropped Western blots for HAP1 wild-type (WT) and different knock-out cells stained for BAF subunits to confirm their knock-outs and check for subunit dependencies.
BAF complex composition changes upon knock-out of single BAF-coding genes
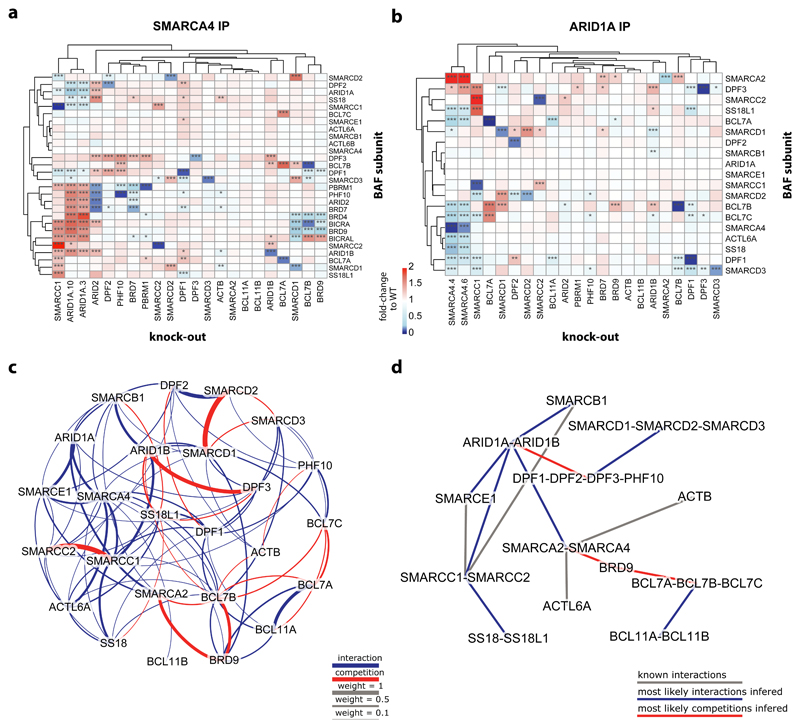
In order to investigate the effect of loss of individual BAF-coding genes on complex composition, we developed a BAF immunoprecipitation (IP) approach with quantitative mass spectrometry as readout. We performed IP for ARID1A, a BAF-specific subunit, and for SMARCA4, a subunit incorporated into all BAF complex subtypes. These bait proteins were enriched in IPs from the different cell lines, except in the respective knock-out cells (Supplementary Fig. 1a). ARID1A levels were additionally reduced in SMARCC1KO cells that express low levels of ARID1A. Furthermore, we did not observe enrichment for the mutually exclusive subunits ARID1B and ARID2 in the ARID1A IP nor for SMARCA2 in the SMARCA4 IP, suggesting paralog specificity of the antibodies used (Supplementary Fig. 1b, c). All other BAF and PBAF members were enriched in the SMARCA4 IP, except ACTL6B, BCL11A and BCL11B that were not or only very lowly expressed in HAP1 cells. As expected, the PBAF-specific subunits PBRM1, PHF10, BRD7 and ARID2 were detected in the SMARCA4 IPs but not in the ARID1A IPs. Also BRD9 was only detected in the SMARCA4 IPs, in line with its incorporation in GBAF/ncBAF complexes that do not contain any ARID subunit13,14.
Analyses of the relative abundance of the detected subunits revealed knock-out-specific alteration of complex compositions (Fig. 2a/b, Supplementary Fig. 1d). None of the tested knock-outs led to a complete disruption of the BAF complexes in contrast to an observed dissociation of the SWI/SNF complex upon loss of Snf5, Snf6, and Snf12 in yeast15,16. This is likely due to the evolution of modularity of the mammalian SWI/SNF complex. For example, Snf12 has three orthologs in humans, namely SMARCD1, SMARCD2, and SMARCD3, and our data suggest that the loss of either is compensated by increased incorporation of a paralog-encoded protein. Snf6 does not have an ortholog in humans and SMARCB1, the ortholog of Snf5, could not be tested as the gene is essential in HAP1 cells17.
Fig. 2. BAF complex composition changes upon knock-out of single BAF-coding genes.
(a) Heatmap showing enrichment of BAF subunits in SMARCA4 immunoprecipitation in the different HAP1 knock-out cells relative to their enrichment in HAP1 WT cells. Hierarchical clustering of the knock-outs and the BAF subunits was done based on Euclidean distance. n = 2 biological independent experiments. Significant changes were calculated as described in the Supplementary Note and resulting FDR values are indicated by * (* < 1%, ** < 0.1%, *** < 0.01%). (b) Same as in a) for the results of the ARID1A IP-MS. (c) Weighted frequencies of all pairwise interactions (blue) or competitions (red) between the subunits of the BAF complexes as inferred from simulations based on the ARID1A IP-MS data. The weight of an interaction or competition is given by the normalized sum of the fitnesses of all simulated graphs in which it was observed. A cut-off of weight > 0.1095 was used, corresponding to the minimal weight at which all subunits form a single connected graph. (d) Putative organization of the BAF complexes obtained by combining interactions and competition classes with direct experimental evidence and most likely interactions inferred from the simulations based on the ARID1A IP-MS data (using a cut-off of weight > 0.42 to result in a single connected graph).
Loss of one protein was often compensated by increased incorporation of its paralogous proteins into the complex, for example for ARID1A-ARID1B-ARID2, SMARCA2-SMARCA4, SMARCC1-SMARCC2, SMARCD1-SMARCD2-SMARCD3, DPF1-DPF2-DPF3-PHF10, and BCL7A-BCL7B-BCL7C. This argues for the mutually exclusive incorporation of these subunits to fulfill the requirement to keep the respective complex position occupied. While in most instances such compensatory effects occurred in a mutual fashion irrespective of which subunit was targeted, there are also cases where these mechanisms appear unidirectional.
Clustering of the subunits by their relative abundance in SMARCA4 IPs revealed clear co-regulation of the PBAF-specific subunits ARID2, PBRM1, PHF10 and BRD7. They were all increased in complexes isolated from ARID1AKO cells, whereas they were lost in ARID2KO and to a lesser degree in BRD7KO clones. Loss of PHF10 only resulted in a mild reduction of PBRM1 incorporation whereas PBRM1 loss did not affect the level of any other PBAF-specific subunit in the complexes. These data suggest a clear hierarchy in the assembly of the PBAF complexes: ARID2>BRD7>PHF10>PBRM1. This observation is in line with a previous report showing that ARID2 is important for the stability of PBRM1 and the PBAF complex and not vice versa18. Furthermore, we found that BRD7KO cells expressed less PBRM1 on the protein level and also showed reduced enrichment of specific PBAF-members in SMARCA4 IPs, suggesting feedback mechanisms by destabilization of non-incorporated proteins.
The multiple positions that can alternatively be occupied by different subunits suggest that these complexes can theoretically exist in thousands of different configurations and it is unknown which of these exist in human cells. In order to simulate which subunits directly lead to the increased (‘interaction’) and decreased (‘competition’) incorporation of other subunits into the BAF complexes, we implemented a genetic algorithm that tested all pairwise combinations using the IP-MS data as input. We first investigated the BAF complexes using the ARID1A IP-MS data (Fig. 2c,d, Supplementary Fig. 2a,b). The computational analysis confirmed very strong competition between SMARCD1-SMARCD2-SMARCD3 and SMARCC1-SMARCC2. It further suggested competition between ARID1B-DPF3 and SMARCA2-BRD9-BCL7B. We then repeated the analysis with just the SMARCA4 IP-MS data and with all data combined providing input for all possible BAF and PBAF complexes that contain either SMARCA4 and/or ARID1A (Supplementary Fig. 2c-h). In addition to the competitions already found in the ARID1A-containing BAF analysis, these data revealed competition between BCL7A-BCL7B-BCL7C, DPF1-DPF2, DPF3-PBRM1, and ARID1A-ARID1B-ARID2. The analyses further provide strong support for the interaction between SMARCC1-ARID1A and between the PBAF-specific subunits ARID2-BRD7-PHF10-PBRM1.
We performed the IP experiments under stringent conditions, but were still able to identify some interaction partners of the BAF complexes (Supplementary Fig. 3a). Among the most strongly enriched interaction partners found in the SMARCA4 IP were Bromodomain-containing Protein 4 (BRD4) and BRD4 Interacting Chromatin Remodeling Complex Associated Protein (Like) (BICRA and BICRAL). They were not identified in the ARID1A IPs. Moreover, the interaction with all three proteins was lost in SMARCD1KO cells, and BRD4 and BICRA enrichment was also reduced in BRD9KO and BCL7BKO cells (Fig. 2a, Supplementary Fig. 3b). Interestingly, a common feature of these three cell lines is decreased incorporation of BRD9 into the complexes, in line with the specificity of these proteins for the recently described GBAF/ncBAF complex13,14,19 and the presence of a functional SMARCD1-BRD9-BICRA (GLTCR1) module in fitness correlation network analyses from RNAi screens20. Accordingly, we observed stronger enrichment of BRD4 and BICRA when we used an antibody against BRD9 for IP, compared to ARID1A, ARID2 or common BAF subunit IPs (Supplementary Fig. 3c).
Knock-out of single BAF-coding genes alters global chromatin accessibility
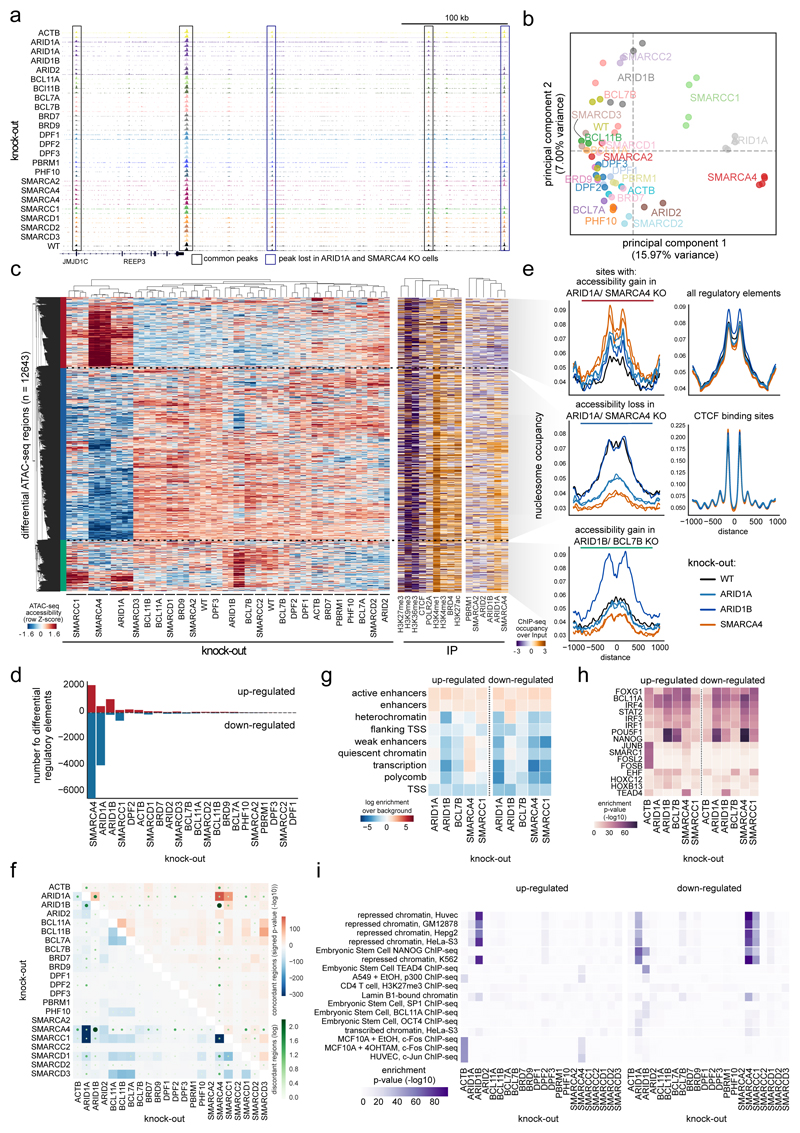
We next systematically investigated the functional consequences of the loss of individual BAF-coding genes on chromatin accessibility by applying the assay for transposase accessible chromatin with high-throughput sequencing (ATAC-seq) in the HAP1 wild-type (WT) and knock-out cells. The overall quality of the samples and the global distribution of open chromatin regions were highly comparable across all samples (Supplementary Fig. 4a-e, Supplementary Table 2). In general, chromatin accessibility was observed mainly at 5’ UTRs, transcription start sites (TSS), enhancers and at regions harboring marks that are associated with active chromatin (Supplementary Fig. 4f-h).
While many genomic sites were accessible in all cell lines, we observed also knock-out-specific differences at specific loci (Fig. 3a). This prompted us to systematically assess global similarity in chromatin accessibility across the various knock-out cells (Fig. 3b, Supplementary Fig. 4i). These analyses revealed that there are differences in chromatin accessibility for all knock-out clones compared to WT cells with the strongest alterations in the SMARCC1KO, ARID1AKO and SMARCA4KO clones. This picture was confirmed by the locus-specific analysis of regions significantly de-regulated for each knock-out: SMARCA4KO, ARID1AKO, and SMARCC1KO cells grouped together and showed, compared to WT cells, reduced accessibility at many regions across the genome (blue cluster), and accessibility gain at fewer other regions (red cluster) (Fig. 3c,d). This is in line with observations in colorectal cancer cells, where knock-down of ARID1A led to reduced accessibility at many genomic loci11. In contrast, ARID1BKO cells gained chromatin accessibility at numerous genomic loci compared to WT cells (green cluster). In line with these findings, the total open chromatin fraction was reduced in the SMARCA4KO, ARID1AKO, and SMARCC1KO cells and increased in ARID1BKO cells compared to WT cells (Supplementary Fig. 4j). By leveraging the paired-end ATAC-seq data, we also investigated how nucleosome positions in these regions changed (Fig. 3e). Sites with reduced accessibility upon knock-out gained nucleosome signal at the center of ATAC-seq peaks compared to WT cells and vice versa, providing functional implications of nucleosome positioning changes on chromatin accessibility.
Fig. 3. Knock-out of single BAF-coding genes alters global chromatin accessibility.
(a) Example browser tracks of ATAC-seq data. (b) Principal component analysis of the ATAC-seq data for individual replicates using all regions in which a peak was called in any sample (154533 regions). KO gene is indicated. (c) Left: Clustering of samples and regions significantly different between any KO compared to WT ATAC-seq data. Hierarchical clustering was performed with Pearson correlation as distance measure between accessibility values transformed with a Z-score per row. Right: ChIP-seq enrichment for indicated proteins at differential ATAC-seq regions in WT cells. (d) Number of up -and down-regulated regions in each KO compared to WT cells. (e) Nucleosome occupancy as determined with the NucleoATAC method for selected cell lines are shown around indicated genomic sites. (f) The percentage of regions that are changing in two KOs concordantly compared to WT are shown in blue for down-regulated regions and in red for up-regulated regions. The number of disconcordantly changing regions are indicated with green dots. (g-i) Enrichment in (g) chromatin states, (h) transcription factor motif and (i) ChIP-seq data from the ENCODE and Blueprint consortia, as determined by the location overlap analysis (LOLA) method, of up- and down-regulated regions in indicated KO cells. For panels f), h) and i), significance of overlap was assessed with a one-sided Fisher’s Exact Test and no adjustments were made for multiple comparisons. All panels were derived from at least n = 2 biologically independent ATAC-seq experiments.
In order to assess the chromatin context of the sites that changed accessibility, we performed ChIP-seq in HAP1 WT cells and associated them to the differential ATAC-seq regions (Fig. 3c, Supplementary Table 3). The sites of the blue and green clusters showed enrichment for members of the BAF chromatin complexes as well as for enhancer marks in HAP1 WT cells, indicating that preferentially BAF-bound active regulatory regions are changing accessibility when a specific BAF subunit is lost and confirming a function of the BAF complex not only at transcription start sites, but also at distal regions11,12,21,22. On the other hand, the sites of the red cluster were rather enriched for inactive histone modifications such as H3K27me3 and H3K9me3 and not bound by the BAF complexes under WT condition. These sites, however, are bound by ARID1A-containing BAF complexes in SMARCA4KO cells, which show the strongest gain of chromatin accessibility in these regions (Supplementary Fig. 5). Together, the data provide strong evidence that intact BAF complexes containing SMARCA4, ARID1A, and SMARCC1 are important for the recruitment of BAF complexes to cell-specific enhancer sites and their activation. Our data further hint that BAF and PBAF complexes occupy different genomic loci (Supplementary Fig. 6a-g). While the BAF complex is preferentially bound to active enhancers (marked with H3K4me1 and H3K27ac), the PBAF-enriched sites are either active (marked with H3K4me3/H3K36me3/POLR2A) or inactive (marked with H3K27me3/H3K9me3) regions.
Analyses of shared and discordantly changing accessibility across the different cell lines illustrated again the highly similar changes in SMARCA4KO, SMARCC1KO, and ARID1AKO cells that are distinct from the accessibility changes in the other cell lines (Fig. 3f, Supplementary Fig. 7a). Furthermore, knock-out of mutually exclusive subunits resulted in similar as well as discordant changes at certain genomic loci. For example, accessibility at some genomic loci is changed in the opposite direction in ARID1AKO vs. ARID1BKO cells, in line with published data23. Accordingly, the regions of altered accessibly in the two different knock-outs showed opposite patterns for their localization across the genome, transcription factor (particularly pluripotency factors) and ChIP-seq signal enrichment (Fig. 3g-i, Supplementary Fig. 7b). Overall, the data suggest that the structural compensation of mutually exclusive subunits may not necessarily rescue the function of the lost protein as they might have partly distinct regulatory roles.
Expression changes correlate with altered chromatin accessibility
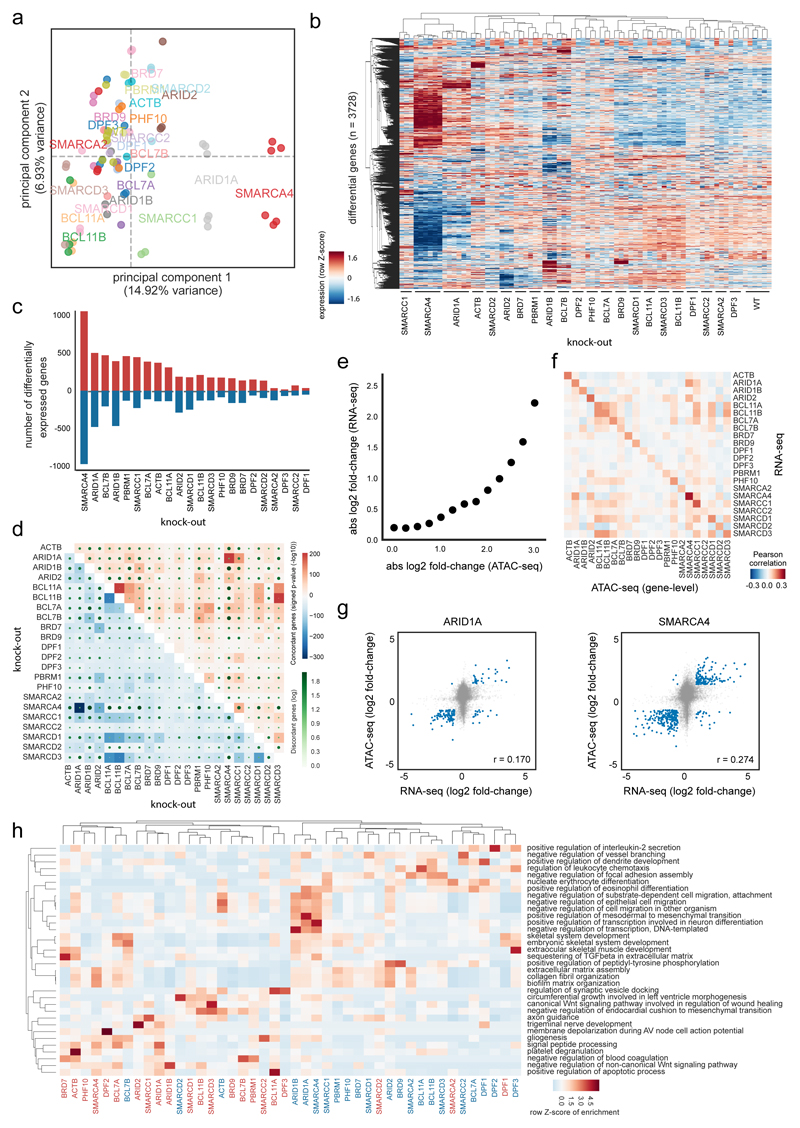
To test whether the observed alterations in chromatin accessibility go along with changes in gene expression, we measured the transcriptome of all BAF mutant HAP1 clones using RNA sequencing (Supplementary Table 4). We first analyzed the BAF member expression pattern across the different samples, suggesting a transcriptional co-regulation of some subunits (Supplementary Fig. 8).
Investigating the global gene expression changes, the knock-out cells showing similar changes in chromatin accessibility also clustered together in regard to their transcriptomes (Fig. 4a,b). This was especially obvious for the knock-out cells with prominent accessibility changes, namely SMARCA4KO, ARID1AKO, and SMARCC1KO cells that also showed the most differentially expressed genes (Fig. 4c, Supplementary Fig. 9a). In general, nearly similar numbers of up- and down-regulated genes were observed in each cell line. Despite overall coordination in expression changes between the knock-out cell lines, we also found a small number of genes with opposite directions of change, especially compared to the SMARCA4KO cells (Fig. 4d).
Fig. 4. Expression changes correlate with altered chromatin accessibility.
(a) Principal component analysis of RNA-seq data in HAP1 WT and KO cells (KO gene is indicated) using all expressed genes. (b) Clustering of samples based on differentially expressed genes between any KO and WT cells (KO gene is indicated). Hierarchical clustering was performed with Pearson correlation as distance measure between expression values transformed with a Z-score per row. (c) Number of up- and down-regulated genes per KO. (d) The percentage of genes that change in two KOs concordantly compared to WT are shown in blue for down-regulated and in red for up-regulated genes. The number of disconcordantly changing genes are indicated with green dots. Significance of overlap was assessed with a one-sided Fisher’s Exact Test without adjustments for multiple comparisons. (e) Relationship of expression and ATAC-seq signal aggregated per genes across all KOs. Genes were binned into groups based on ATAC-seq change. (f) Pearson correlation of log fold-changes in KO versus WT cells (KO gene indicated) between RNA-seq and ATAC-seq. (g) Scatter plot and Pearson correlation coefficient for HAP1 ARID1AKO and SMARCA4KO cells between RNA-seq and ATAC-seq log fold-changes relative to HAP1 WT cells. (h) Over-representation enrichment analysis of Gene Ontology terms for up- or down-regulated genes in different HAP1 KO cells. A row Z-score on the –log10(P value) of a one-sided Fisher’s Exact Test (Enrichr software) is displayed without adjustments for multiple comparisons. RNA-seq data of all panels are derived from n = 3 biologically independent experiments per clone.
To test an association between ATAC-seq and RNA-seq data, we next compared the changes in the ATAC-seq enrichment to the expression alterations of the associated genes and found an overall good correlation (Fig. 4e-g). This suggests a causative relationship between chromatin remodeling and the resulting cellular expression status. We then analyzed the transcriptome changes regarding enrichment for transcription factor binding sites and gene ontology terms. For example, genes down-regulated in SMARCA4KO, ARID1AKO or ARID1BKO cells were related to cell migration and transcription regulation (Fig. 4h, Supplementary Fig. 9b). In general, the down- and up-regulated genes in ARID1AKO and SMARCA4KO cells were enriched for similar gene ontology (GO) terms (Supplementary Fig. 10).
Systematic targeting of multiple BAF subunits identifies novel intra-complex synthetic lethalities
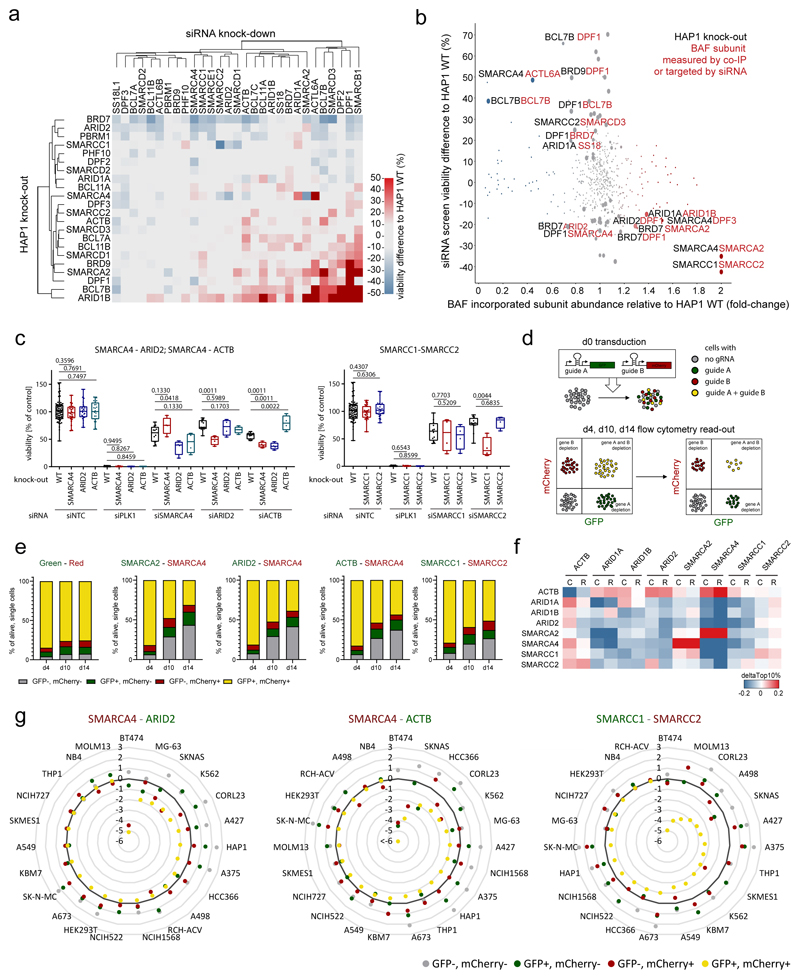
We next applied the fully characterized cell line panel to identify intra-complex synthetic lethality with BAF mutations. We therefore assembled pools of siRNAs targeting all BAF subunits individually and all possible combinations of alternative subunits. We then transfected all HAP1 clones with these siRNA pools and measured viability following 5 days in cell culture (Supplementary Table 5). While transfection of the non-targeting control (NTC) did not reduce the viability of the cells, knock-down of Polo Like Kinase 1 (PLK1), whose inhibition has been shown to induce apoptosis in leukemic cells24–26, led to cell death across all cell lines included in the screen (Supplementary Fig. 11a). This argues for similar efficiency of RNAi mediated knock-down across all included cell lines. Unbiased clustering of cell viability data revealed that the sensitivities and resistances of the BAF mutant cell lines can in part be explained by the alterations we observed in BAF complex composition and in part by the chromatin and transcriptome changes (Fig. 5a, b, Supplementary Fig. 11a-d). For example, ARID2KO, BRD7KO, and PBRM1KO cells were most sensitive, while BCL7BKO and ARID1BKO clones, for which we had observed similar changes in gene expression, were resistant to further siRNA knock-down of many BAF subunits.
Fig. 5. Systematic targeting of multiple BAF subunits identifies novel intra-complex synthetic lethalities.
(a) Heatmap showing viability difference between HAP1 knock-out cell lines (y-axis) and HAP1 WT cells upon single siRNA treatment (x-axis). (b) Effect on viability in different knock-out cells (black) upon siRNA treatment (red) (siRNA screen data, y-axis) is compared to the change of subunit incorporation (red) in different knock-out cells (black) relative to their incorporation in WT cells (IP-MS data, x-axis). (c) Boxplot showing viability 5 d after siRNA knock-down for three selected synthetic lethalities. Mann-Whitney test was used to calculate significant viability difference in knock-out cells relative to wild-type cells upon the same siRNA treatment (n ≥ 4 independent measurements from two biologically independent experiments). First and third quartiles are denoted by lower and upper hinges. Whiskers extend to smallest and largest values. (d) Scheme explaining CRISPR/Cas9 multicolor competition assay. (e) Multicolor competition assay in HAP1 cells. Bar graphs show percentage of the four different cell populations 4, 10 and 14 days after transduction of the gRNAs targeting the genes indicated above (Green, Red: vectors only with the fluorescent marker). (f) Heatmap showing if cells expressing low levels of the gene (left) are more sensitive to CRISPR (C) or RNAi (R) targeting a BAF subunit (top) as cells that express high levels of this gene. The analyses were done with public data (depmap: CRISPR (Avana) Public 18Q4; Combined RNAi). (g) Multicolor competition assay results for three synthetic lethalities across various cell lines. log2(d14/d4) is displayed for each population. Targeted subunits are indicated above.
The three cell lines that showed strong loss of chromatin accessibility did not show co-clustering in the siRNA screen, but they were all sensitive to knock-down of their respective paralog: SMARCA4KO cells required SMARCA2 for their survival, ARID1AKO cells were sensitive to ARID1B knock-down, both in line with previous reports7–12, but also SMARCC1KO cells were sensitive to loss of SMARCC2. The subunits SMARCC1 and SMARCC2, despite high sequence homology, are thought to both be incorporated into BAF complexes simultaneously27, but we observe the same signatures for these proteins as for the alternative subunits SMARCA2-SMARCA4 and ARID1A-ARID1B. Therefore, our data suggest to re-evaluate whether these subunits are incorporated into BAF complexes as monomer, homodimer, and/or heterodimer. In addition, we observed that SMARCA4KO cells were sensitive to knock-down of ARID2, ACTB, and SMARCB1. SMARCA2KO cells had reduced viability upon knock-down of PBRM1, and DPF2KO cells were sensitive to knock-down of SMARCA4 and ACTL6A.
We then assessed changes in viability of BAF mutant cell lines relative to HAP1 WT cells in relation to changes in complex compositions observed by co-immunoprecipitation (Fig. 5b). We observed that subunits that are incorporated with increased abundance in BAF complexes following a particular knock-out often constitute vulnerabilities of these cell lines. For example, in SMARCA4KO cell lines there is increased complex incorporation of SMARCA2, and siRNA-mediated targeting of SMARCA2 specifically caused a growth disadvantage in SMARCA4KO cells. Also the following subunits show increased BAF complex incorporation and evidence of requirements for cell viability: ARID1B in ARID1AKO cells, SMARCC2 in SMARCC1KO cells, DPF1 in ARID2KO and BRD7KO cells, SMARCA2 in BRD7KO cells, and DPF3 in SMARCA4KO cells. On the other hand, cell lines were often relatively more resistant to knock-down of subunits that showed reduced complex incorporation in a particular knock-out. This is most prominently exemplified by the loss of ACTL6A in SMARCA4 mutant complexes and the resistance of SMARCA4KO cells to ACTL6A knock-down, which goes along with the described direct binding of ACTL6A to SMARCA428. Additional examples are BCL7B and BRD7 in DPF1KO cells, DPF1 in BRD9KO and BCL7BKO cells, and SMARCD3 in SMARCC2KO cells.
Intra-complex synthetic lethalities alter BAF complex composition
To choose the most robust intra-complex synthetic lethal pairs, we next analyzed the data for reciprocal synthetic lethalities observed in both cell line/knock-down combinations. There, the following pairs stood out: SMARCA4-ACTB, SMARCA4-ARID2, and SMARCC1-SMARCC2 (Fig. 5c). Growth curves by quantifying nuclei after Hoechst staining following siRNA treatment confirmed slower growth under the synthetic lethal conditions (Supplementary Fig. 11e). We next validated these synthetic interactions using CRISPR/Cas9-mediated gene targeting as alternative approach to siRNA mediated knock-down. Therefore, we applied a multicolor competition assay with flow cytometry as a readout (Fig. 5d, Supplementary Fig. 11f, g). While transduction of the vectors without any gRNA barely showed a change of the different populations with time, the population of cells that contained both gRNAs of the new synthetic lethalities decreased over time nearly as strong as when the well-described synthetic lethal genes SMARCA4 and SMARCA2 were targeted (Fig. 5e). These data independently validate and confirm all three synthetic lethalities under conditions of full genetic knock-out.
We next explored biochemical relationships between the synthetic lethal subunits that may explain the phenotype. We used siRNA to knock-down the synthetic lethal subunit in the corresponding HAP1 knock-out cell line for the pairs SMARCA4-ARID2, SMARCA4-ACTB, and SMARCC1-SMARCC2 and analyzed cell cycle distribution and complex composition (Supplementary Fig. 12). When we targeted SMARCC1 in SMARCC2KO cells and vice versa, we observed strong reductions of the global protein levels of key BAF subunits SMARCA4, SMARCB1, ARID1A, and SMARCD1 (Supplementary Fig. 12a). This is in line with similar observations in HEK293T cells13, published during the revision of our manuscript. Therefore, near-complete destabilization of BAF complexes is likely causative for reduced viability and proliferation when both SMARCC1 and SMARCC2 are lost (Supplementary Fig. 12b). For the other two synthetic lethalities, we investigated the BAF complex compositions in the combined versus single perturbed condition by SMARCC1 IP-MS (Supplementary Fig. 12c). The data revealed that both constitutive and acute loss of SMARCA4 resulted in increased ACTB incorporation into BAF complexes. Similar to other examples where increased complex incorporation results in increased dependency on a particular subunit, also ACTB then specifically becomes essential to the viability of SMARCA4KO cells. Analyzing the SMARCC1 IP-MS data further enabled us to assess the effects of SMARCA4 loss on incorporation of PBAF specific subunits, information that cannot be obtained from the SMARCA4 and ARID1A-IPs. Besides reduced incorporation of ACTL6, BCL7 and DPF1 into BAF complexes, SMARCA4 KO resulted in strong loss of PBRM1 expression and complex incorporation (Fig. 1b, Supplementary Fig. 12c), whereas the other PBAF subunits ARID2, BRD7 and PHF10 were present at normal or slightly increased levels. When ARID2 was depleted in SMARCA4KO cells, these other PBAF-specific subunits were lost in addition to PBRM1. Overall, the changes we observed in complex composition after targeting of two subunits correlated well with the expected changes based on the network that we previously generated from the IP-MS data across the different knock-out cells (Supplementary Fig. 12d). We further validated that HAP1 SMARCA4 KO cells contain PBAF complexes with less PBRM1 by ARID2 IP (Supplementary Fig. 12e). Moreover, combined perturbation of SMARCA4 and ARID2 led to accessibility changes of regions related to cell death and gene expression alterations of cell cycle genes (Supplementary Fig. 13). In summary, these data suggest that in the case of SMARCA4-ARID2 synthetic lethality, the effects of the single lost subunits add up to result in more severe perturbation of BAF complexes and consequent detrimental chromatin and gene expression changes.
Validation of intra-complex synthetic lethalities in other cell lines
Finally, we tested whether the synthetic lethalities are conserved across different cell types by first analyzing publically available datasets on gene essentiality in cancer cell lines (depmap: CRISPR (Avana) Public 18Q4) (Supplementary Fig. 14)29–34. As only few cancer cell lines included in the dataset harbor mutations in the synthetic lethal subunits SMARCC1, SMARCC2, ARID2, ACTB and the functional consequences of these mutations are often uncharacterized, we compared the sensitivity of the top 10% versus the bottom 10% BAF subunit expressing cells lines to depletion of the other synthetic lethal BAF subunits by CRISPR or RNAi (Fig. 5f). The analyses revealed high conservation of the SMARCA4-SMARCA2 synthetic lethality, frequent occurrence of ARID1A-ARID1B, SMARCC1-SMARCC2, and SMARCA4-ACTB synthetic lethality, while SMARCA4-ARID2 synthetic lethality seemed not widely conserved in other cell lines. Weaknesses of these analyses are the arbitrary thresholds for high/low expressing cells and the low variability of expression levels across cell lines for some BAF genes, such as SMARCC1, SMARCC2 or ARID2. Therefore, we directly tested the three novel synthetic lethality candidates in 23 different cell lines originating from multiple tissues using the multicolor competition assay (Fig. 5g, Supplementary Fig. 15a, Supplementary Table 1). Some of these cell lines (NB4, NCIH727, A498, HEK293T, RCH-ACV, NCIH522) appeared mostly resistant to all perturbations, including the well-established SMARCA2-SMARCA4 synthetic lethality (Supplementary Fig. 15b-d), possibly in part due to low editing efficiency in certain cell lines (Supplementary Note 2.1). The fractions of SMARCA4-ARID2 double targeted cells and of SMARCA4-ACTB double targeted cells each were depleted more than two-fold over a 14-day period in approximately one third of the cell lines tested. In contrast to the homozygous SMARCA4-mutant cell lines, depletion of ARID2 or ACTB alone was not able to reduce cell viability of heterozygous SMARCA4 mutant cells (CORL23), suggesting that complete loss of SMARCA4 is necessary for these synthetic lethalities. These data validate the two synthetic lethalities, but also highlight their context-dependency. For the SMARCC1-SMARCC2 pair we observed strong synthetic lethality in fourteen of the tested cell lines, to a similar level and in the same cell lines as the known SMARCA4-SMARCA2 vulnerability. Importantly, in SKMES1-cells that harbor a SNP in SMARCC1 resulting in very low SMARCC1 expression, targeting of SMARCC2 alone depletes cells nearly as efficiently as concomitant targeting of SMARCC1-SMARCC2. In summary, these data confirm that additional cell lines respond to combined SMARCA4-ARID2 and SMARCA4-ACTB targeting and indicate that SMARCC1-SMARCC2 is a highly conserved and strong synthetic lethality that can be exploited in cancers cells with low expression of either subunit.
Discussion
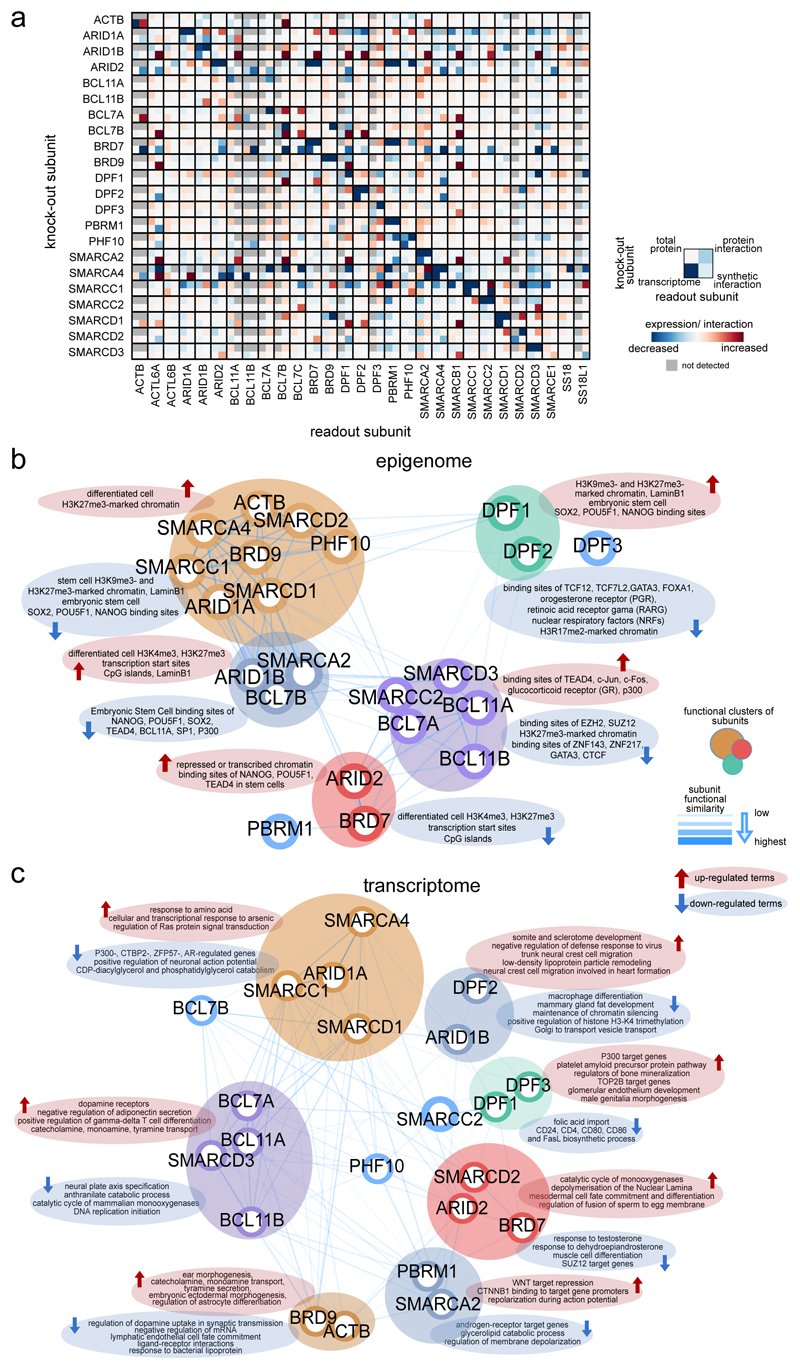
Loss-of-function mutations of different subunits of the BAF chromatin remodeling complexes have been detected in many human cancers, however, the cellular consequences and differences between the loss of individual subunits are hardly understood. Here we present a systematic investigation of single BAF subunit loss in human cells. The study provides a basis for understanding subunit dependencies on several layers (Fig. 6a), the role of single subunits in BAF function (Fig. 6b,c) and the cellular consequences that occur in BAF-mutated cancers. It suggests that preferred configurations of the human SWI/SNF complexes beyond BAF/PBAF/ncBAF exist and their distribution is altered by loss of single subunits. BAF mutations ultimately result in alterations of chromatin accessibility and gene expression, strongly dependent on the subunit that is lost. Most prominent are the global reduction of chromatin accessibility in ARID1AKO, SMARCC1KO, and SMARCA4KO cells and an increase in accessibility following mutation of ARID1B. This revealed similar cellular consequences upon loss of subunits that occupy different positions and functions in the complex, while mutually exclusive subunits were functionally not completely redundant. By observing clonal cell lines, we have described the long-term changes following subunit loss allowing complex rewiring and homeostasis to select for viability and proliferation, processes that may also occur in carcinogenesis. Furthermore, we observe that chromatin accessibility five days after SMARCA4 knock-down is already very similar to the changes observed in SMARCA4KO cells. Future work will extend these studies to include kinetic resolution immediately after subunit loss thus allowing the investigation of processes related to tumor initiation.
Fig. 6. Integrative view of BAF complex subunit dependencies and functional similarity.
(a) Joint visualization of effects of single BAF subunit perturbation on the remaining complex subunits. Data from four assays have been used: 1) Transcriptional changes compared to WT cells (log2 fold-change); 2) total protein changes relative to WT cells as measured by Western blot; 3) Subunit incorporation into BAF complexes relative to WT cells as measured by IP-MS; 4) Synthetic interactions via siRNA knock-down in knock-out cells compared to the WT background (% difference to WT). Not detected or performed measurements are shown in grey. (b,c) Functional similarity of BAF complex subunits assessed through similarity of enriched terms in each KO cells compared to WT cells. In b), differential chromatin regions (measured by ATAC-seq, n = 2 biologically independent experiments), whereas in c), differentially expressed genes (measured by RNA-seq, n = 3 biologically independent experiments) were used. The strength of similarity is reflected in the 2D proximity between subunits and in the width and transparency of the edges connecting them. Distances between subunits were derived by the Pearson correlation of enrichment significance (one-sided Fisher’s Exact test P value without adjustments for multiple comparisons). In the case of epigenome similarity, enriched terms from the LOLA tool were used, whereas for transcriptome various ontology, pathway and perturbation terms from the Enrichr tool were used. Top enriched terms specific to each cluster of subunits were extracted by comparing the mean enrichment values across subunits in the same cluster versus all other subunits.
In addition to the known intra-complex synthetic lethalities ARID1A-ARID1B10–12 and SMARCA4-SMARCA27–9, we identified SMARCC1-SMARCC2, SMARCA4-ACTB and SMARCA4-ARID2 as prominent synthetic lethal interactions. Integrative analyses of effects on complex composition, chromatin accessibility and gene expression in the synthetic lethal conditions enabled us to provide mechanistic hypotheses for explaining these intra-complex co-dependencies. Future studies will further characterize these synthetic lethalities, including their dependence on the levels and catalytic activities of core ATPases. Finally and most importantly, steps need to be taken to test whether these results can be translated in vivo. In addition to genetic models, the development of pharmacological tools to perturb specific BAF subunits will dramatically accelerate these approaches.
Overall, our study characterizes the impact of individual subunits on BAF complex compositions, their functions and intra-complex dependencies, providing a basis and potential novel targets towards the goal of developing targeted treatments for BAF-mutated cancers.
Methods
Cell culture
HAP1 wild-type and knock-out cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Thermo Fisher Scientific, 21980-032) supplemented with 10% heat inactivated Fetal Bovine Serum (FBS, Thermo Fisher Scientific, 10500) and 1% Pen Strep (100 units/ml Penicillin, 100 μg/ml Streptomycin, Thermo Fisher Scientific, 15140-122). For the multicolor competition assay, following Cas9-expressing cells were used: A549 cells were cultured in F-12K Nut Mix (Thermo Fisher Scientific, 21127-022) supplemented with 10% FBS; HEK293T, MG-63 and SKNAS cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, 41965-039) with 10% FBS; KBM7 and HAP1 in IMDM with 10% FBS; MOLM13, NB4, RCH-ACV, A673, SK-N-MC, BT474, K562, THP1, A498, NCIH727 (NCI-H727 [H727] ATCC® CRL-5815TM, ATCC), A427, A375, COR-L23 (Sigma-Aldrich, 92031919-1VL), NCIH1568, NCIH522, and HCC366 were cultured in RPMI Medium 1640 (Thermo Fisher Scientific, 21875-034) with 10% FBS, and SK-MES-1 (ATCC® HTB-58™, ATCC) in Lonza™ BioWhittaker™ Minimal Essential Medium Eagle (EMEM, Lonza, 12-662F) with 1% GlutaMAX™-I (Thermo Fisher Scientific, 35050061) and 10% FBS. A list of all used cell lines is provided in Supplementary Table 1.
siRNA screen
ON-TARGETplus SMARTpool siRNA (pool of four siRNAs targeting the same gene; Dharmacon, see Supplementary Table 1) were transferred to the screening plates using an Echo 550 liquid handler (Labcyte) and dried. 30-90 min prior to seeding of the cells, the appropriate amount of transfection reagent in Opti-MEM I Reduced Serum Medium (Thermo Fisher Scientific, 31985070) was added to the siRNAs. In particular, 500 HAP1 cells were seeded per well of a 96-well plate and 0.0125 μM siRNA was transfected with 0.075% Dharmafect 1 (Dharmacon, T-2001-03). Cell viability was measured 5 days after siRNA transfection using CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7570). After equilibration of the plates and the reagent to room temperature, the CellTiter-Glo reagent was added to the wells using a Multidrop™ Combi Reagent Dispenser (Thermo Fisher Scientific). The readout was performed after 20 min incubation using a 2104 EnVision™ Multilabel Plate Reader (PerkinElmer).
Western Blot
Cell pellets were lysed rotating at 4°C for 1 h in RIPA buffer (50 mM Tris pH 8, 1% NP-40, 0.5% Sodiumdeoxycholate, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 1% Glycerol, 2.5 mM MgCl2, 2 mM Na3VO4) containing 1× cOmplete™, EDTA-free Protease Inhibitor Cocktail (Sigma, 4693132001). Protein content of the supernatant after centrifuging for 10 min, 4°C, 13,000 rpm was measured using Bradford assay (AppliChem, A6932). Equal amounts of protein were loaded on acrylamide gels using 4× SDS loading buffer (0.2 M Tris pH 6.8, 40% glycerol, 4% SDS, Bromphenol Blue, 0.04% β-Mercaptoethanol) as well as a protein ladder (Precision Plus Protein™ All Blue Prestained Protein Standards, Bio-Rad, 1610373). After gel electrophoresis the proteins were transferred to Immobilon-FL PVDF membranes (Merck, IPFL00005; pre-wetted in 100% methanol). Transfer conditions were 1.5 h 125 mA for 12% acrylamide gels and 1.5 h 200 mA for 7% acrylamide gels. After blocking in TBST + 1% casein, the membranes were incubated over-night with primary antibody in 5% milk in TBST (Supplementary Table 1). The next day, membranes were washed in TBST, incubated with fluorescently labeled secondary antibodies diluted in TBST + 1% casein for 1 h at RT, washed again in TBST and imaged on a Bio-Rad ChemiDoc™ MP. Image analysis was performed using Bio-Rad Image Lab software. In cases where chemiluminescence detection was used, proteins were transferred to Amersham Protran 0.45 μm nitrocellulose membranes (GE Healthcare, 10600002), membranes were blocked in 5% milk in TBST and secondary antibodies were diluted in TBST. Membranes were then developed using Clarity Western ECL Substrate (Bio-Rad, #170-5060) and imaged on a Bio-Rad ChemiDoc™ MP.
Nuclear extraction and immunoprecipitation
Cells were harvested by scraping and washed several times with cold PBS. Cell pellets were then resuspended in 3× volume of Buffer N (300 mM sucrose, 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1 mM DTT, 0.75 mM spermidine, 0.15 mM spermine, 0.1% NP-40, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, TPCK, protease inhibitor), vortexed and incubated on ice for 5 min. The supernatant contained the cytoplasmic fraction. Nuclear pellets were resuspended in Buffer N and pelleted by centrifugation two more times. Pellets were resuspended in 1× Buffer C420 (20 mM HEPES pH 7.9, 420 mM NaCl, 25% glycerol, 1 mM EDTA, 1 mM EGTA, 0.1 mM DTT, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, TPCK, protease inhibitor) and shaked in a Thermomixer at 4°C, 1,400 rpm for 30 min. Salt concentration of the supernatant obtained after centrifugation was reduced to 150 mM by diluting the sample with HEPES I (20 mM HEPES pH 7.9, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF). NP-40 (final concentration 0.1%), 20 U/ml benzonase (Novagene, D00150432) and 50 ng/ml RNase A (Sigma, R4875) was added and the samples incubated for 15 min at 4°C. Protein amounts were quantified using Bradford assay (AppliChem, A6932). Protein lysates were incubated overnight at 4°C with SMARCA4 (Abcam, ab110641, GR150844-22), ARID1A (Abcam, ab182560, GR269670-9), SMARCC1 (Cell signaling, 11956S, Lot2), SMARCC2 (Cell signaling, 12760, Lot1), ARID2 (Thermo Fisher Scientific, PA5-35857, RJ2288871K), SMARCE1 (Abcam, ab137081, GR106541-6), SMARCB1 (Abcam, ab126734, YI080106CS), or BRD9 (Abcam, ab137245, GR257571-5) antibody. Samples were incubated for 3 h with washed Protein G dynabeads (Thermo Fisher Scientific, 10004D). Captured protein complexes were washed three-times with Buffer I (50 mM HEPES pH 7.9, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, TPCK, protease inhibitors) and twice with Buffer II (50 mM HEPES pH 7.9, 150 mM NaCl, 5 mM EDTA). Proteins were eluted from the beads using SDS buffer (50 mM HEPES pH 7.9, 150 mM NaCl, 5 mM EDTA, 2% SDS). For preparation of the mass spectrometry samples, condition of the mass spectrometry runs and the data analyses, please see Supplementary Note.
Multicolor competition assay
gRNA oligonucleotides were phosphorylated, annealed and ligated into BbsI-linearized pKLV2-U6gRNA5(BbsI)-PGKpuro2AZsG-W (Addgene, #67975) or pKLV2-U6gRNA5(BbsI)-PGKpuro2AmCherry-W (Addgene, #67977) vectors. One Shot Stbl3 Chemically Competent Escherichia coli (Invitrogen, C7373-03) were used for transformation and Qiagen kits for the purification of the plasmids. Virus was produced with HEK293T cells transfected with psPAX2 (Addgene, 12260), pMD2.G (Addgene, 12259) and the gRNA plasmid using polyethylenimine (PEI, Polysciences, 23966-1). Adherent target cells were seeded the day before transduction with the virus and the help of 8 μg/ml polybrene (Santa Cruz, SC-134220). Suspension cells were seeded in medium with polybrene, virus was added and the cells were spin transduced for 45 min at 2,000 rpm. Final gRNAs are listed in Supplementary Table 1. Samples were measured with a BD LSRFortessa™ flow cytometer (BD Biosciences) using B/E Alexa Fluor 488 (GFP) and YG/D PE Texas Red (mCherry) 4 d, 10 d and 14 d after transduction.
Cell cycle FACS analysis
EdU cell cycle analyses were performed with the Click-iT™ Plus EdU Alexa FluorTM 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific, C10633) according to manufacturer’s instructions. In brief, cells were treated for 1 h with 10 μM EdU and then harvested by trypsinization. 1 million cells were fixed in Click-iT fixative for 15 min at room temperature, followed by a 15 min incubation at room temperature in Click-iT saponin-based permeabilization and wash reagent. The fixed and permeabilized cells were incubated for 1 h at 37°C with Roche In Situ Cell Death Detection Kit (Sigma-Aldrich, 12156792910). Subsequently, the Click-It reaction was performed according to manufacturer’s protocol in a total volume of 100 μl (2 μl Copper protectant, 0.5 μl Fluorescent dye picolyl azide, 0.2 μl secondary antibody, 10 μl reaction buffer additive, 87.3 μl PBS) for 1 h at room temperature in the dark. After two washes with Click-It saponin-bases permeabilization and wash reagent, cells were resuspended in PBS with 5 μg/ml DAPI. Samples were measured with a BD LSRFortessa™ (BD Biosciences) using V/C Horizon V450 (DAPI), B/E Alexa Fluor 488 (EdU), and YG/D PE-Texas Red (TMR-red).
ATAC-seq
ATAC-seq was performed according to Buenrostro et al. with small adaptations 35. In brief, 50,000 cells were resuspended in 25 μl transposase reaction mix (0.05% digitonin, 1× TD buffer, 0.08% TDE1 (Nextera DNA Library Preparation Kit, Illumina, FC-121-1031)) and incubated for 30 min, 37°C, 300 rpm. Then DNA was purified using MinElute kit (Qiagen, 28004) and eluted in 11 μl elution buffer. 1 μl was used to determine cycle number for PCR using a qPCR approach. The remaining 10 μl were complemented with 1× NEBnext High-Fidelity PCR master mix (New England BioLabs, M0541), 1.25 μM index primer 1 and 1.25 μM index primer containing a barcode (Supplementary Table 1). PCR was performed: 5 min 72°C, 30 sec 98°C, X cycles of 10 sec 98°C + 30 sec 63°C + 1 min 72°C, 1 min 72°C and then cleaned-up using Agencourt AMPure XP beads (Beckman Coulter, A63880). The libraries were checked on a Bioanalyzer instrument using High Sensitivity DNA Chips (Agilent), quantified using Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, Q32854) and pooled. Libraries were sequenced using the Illumina HiSeq3000/4000 platform with the 75 bp paired-end configuration.
RNA-seq
RNA was isolated using the RNeasy Mini kit (Qiagen, 74106). The amount of total RNA was quantified using Qubit 2.0 Fluorometric Quantitation system (Life Technologies) and the RNA integrity number (RIN) was determined using Experion Automated Electrophoresis System (Bio-Rad). RNA-seq libraries were prepared with TruSeq Stranded mRNA LT sample preparation kit (Illumina) using Sciclone and Zephyr liquid handling robotics (PerkinElmer). Library amount was quantified using Qubit 2.0 Fluorometric Quantitation system (Life Technologies) and the size distribution was assessed using Experion Automated Electrophoresis System (Bio-Rad). Libraries were pooled, diluted and sequenced using the Illumina HiSeq3000/4000 platform with the 50 bp single-read configuration.
ChIP-seq
Cells were fixed in 1% formaldehyde (Thermo Fisher Scientific, 28908) for 10 minutes at room temperature. The fixation was quenched using 0.125 M glycine pH 2.5 for 5 min at 4°C. After several washes with PBS, the cells were collected by scraping. Cells were then resuspended in buffer L1 (50 mM HEPES KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 10% glycerol, 0.5% NP-40, 0.25% Triton-X 100) and incubated 10 min on ice. After centrifugation, the pellet was resuspended and incubated for 5 min on ice in buffer L2 (200 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0). The pellet was resuspended in buffer L3 (1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 10 mM Tris pH 8.0, 100 mM NaCl, 0.1% Na-deoxycholate, 0.17 mM N-Lauroyl sarcosine, 1× protease inhibitor) and washed twice in shearing buffer (10 mM Tris-HCl, pH 7.8, 1 mM EDTA pH 8.0, 0.1% SDS). Then the chromatin was sheared in harsh shearing buffer (10 mM Tris-HCl pH 7.8, 2 mM EDTA pH 8, 0.25% SDS, 1× protease inhibitor) using a Covaris S220 (duty cycle: 5%, intensity: 4, peak incident power: 140 W, cycles per burst: 200, time: 30 min). Supernatant after centrifugation was diluted 1:1.5 in equilibration buffer (10 mM Tris-HCl pH 8.0, 223 mM NaCl, 1.66% Triton X-100, 0.166% DOC, 1 mM EDTA pH 8, 1× protease inhibitor). Antibody incubation was performed over night at 4°C, followed by a three hours incubation with dynabeads protein G (Thermo Fisher Scientific, 10004D). The beads were then washed twice in RIPA-LS (10 mM Tris-HCl pH 8.0, 140 mM NaCl, 1 mM EDTA pH 8.0, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton x-100), RIPA-HS (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 500 mM NaCl, 1% Triton x-100, 0.1% SDS, 0.1% DOC) and then RIPA-LiCl/Doc (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% DOC). After a transfer of the beads in TE to a new tube, the bound chromatin were eluted in elution buffer (1% SDS, 0.1 M NaHCO3) twice for 20 minutes at room temperature. Eluted material was incubated with RNase for 30 minutes at 37°C, with proteinase K for 2.5 h at 55°C and decrosslinked overnight at 65°C. DNA was then extracted using phenol-chloroform, precipitated and then dissolved in TE buffer. Libraries were prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, E7645S) and sequenced using the Illumina HiSeq3000/4000 platform with the 50 bp single-read configuration.
Histone and CTCF ChIP-seq data were generated using the ChIPmentation method as described previously36.
Pre-processing of the ATAC-seq and ChIP-seq data
Reads were trimmed using Skewer37 and aligned to the GRCh37/hg19 assembly of the human genome using Bowtie238 with the ‘-very-sensitive’ parameter. Duplicate reads were removed using sambamba39, and only properly paired reads with mapping quality > 30 and alignment to the nuclear genome were kept. All downstream analyses were performed on the filtered reads. For visualization exclusively, we generated genome browser tracks with the genomeCoverageBed command in BEDTools40 and normalized such that each value represents the read count per base pair per million mapped and filtered reads. This was done for each sample individually and for replicates merged. ATAC-seq peak calling was performed with MACS241 using the ‘-nomodel’ and ‘-extsize 147’ parameters, and peaks overlapping blacklisted features as defined by the ENCODE project42 were discarded. For ChIP-seq and ChIPmentation, library quality was assessed with the phantomPeakQualtools scripts43, and we used HOMER findPeaks44 in “factor” mode to call peaks with matched IgG controls as background. We used ChromHMM45 to segment the genome into 12 states using 6 ChIP-seq marks (H3K27ac, H3K27me3, H3K36me3, H3K4me3, H3K4me1, H3K9me3) and ATAC-seq, creating HAP1-specific chromatin states.
Pre-processing of the RNA-seq data
Base calls provided by the Illumina Realtime Analysis software were converted into BAM format using Illumina2bam and demultiplexed using BamIndexDecoder (https://github.com/wtsi-npg/illumina2bam). Reads were trimmed with Trimmomatic46 and aligned to the GRCh37/hg19 assembly of the human genome using Bowtie147 with the following parameters: “-q -p 6 -a -m 100 -minins 0 -maxins 5000 -fr -sam -chunkmbs 200”. Duplicate reads were removed with Picard’s MarkDuplicates utility with standard parameters before transcript quantification with BitSeq48 using the Markov chain Monte Carlo method and standard parameters. To obtain gene-level quantifications, we assigned the expression values of its highest expressed transcript to each gene. Differential gene-level expression between each knockout and wild-type was performed using DESeq249 from the raw count data with a significance threshold of 0.05.
Bioinformatic analyses
A consensus map of chromatin accessibility sites was created by merging a 1-kb window around the summit of ATAC-seq peaks from all samples using the BEDTools merge command40. The chromatin accessibility of each region in each sample was quantified using Pysam, counting the number of reads from the filtered BAM file that overlapped each region. To normalize read counts across samples, we performed quantile normalization using the normalize.quantiles function from the preprocessCore package in R. We annotated each region with the identity of and distance to the nearest transcription start site and the overlap with Ensembl gene annotations (promoters were defined as the 2,500-bp region upstream of the transcription start site). Annotation with chromatin states was based on the 12-state ChromHMM genome segmentation on ChIP-seq marks.
Unsupervised analysis was performed with the scikit-learn library50 for principal component analysis (sklearn.decomposition.PCA) applied to the chromatin accessibility values in the consensus map for all ATAC-seq samples. Supervised analysis was performed using DESeq249, comparing chromatin accessibility between each knockout and wild-type using the raw count data. We considered a region differential if having a FDR-adjusted P value smaller than 0.01 and absolute log2 fold-change higher than 1. Region set enrichment analysis was performed on the significant regions of each group using LOLA51 with its core databases: transcription factor binding sites from ENCODE 42, tissue clustered DNase hypersensitive sites52, the CODEX database53, UCSC Genome Browser annotation tracks54, the Cistrome database55 and data from the BLUEPRINT project56. Motif enrichment analysis was performed with HOMER findMotifs44 the AME tool from the MEME suite57 using 250 bp sequences centered on the chromatin-accessible regions.
To assess the relationship between chromatin accessibility differences and nucleosome occupancy, data were pooled between replicates and we used NucleoATAC58 with default parameters to output nucleosome signal, call nucleosome dyads and nucleosome-free regions. To get a complementary view on the relation of ATAC-seq signal to nucleosome position, we further split the ATAC-seq fragments in bins depending on their size: nucleosome-free fragments up to 100 bp; nucleosome-fragments between 180 bp and 247 bp. We then quantified bulk ATAC-seq signal, the NucleoATAC smooth nucleosome occupancy signal, NucleoATAC predicted dyad positions and signal from nucleosome-free fragments and nucleosome-associated fragments in the sets of differential chromatin regions discovered with DESeq2, in the center of CTCF binding sites and center of all regions in the consensus chromatin accessibility region set.
In figures where log2 fold-change of chromatin accessibility is reduced to one value per gene, we took the maximum or minimum value of change for the regulatory elements associated with a gene if all values agreed in the direction of change (positive or negative) by a two-thirds majority or otherwise the mean of all values.
To detect differential BAF- vs PBAF-bound sites, we used the ChIP-seq peaks from samples of BAF complex members (ARID1A, ARID1B, ARID2, PBRM1, SMARCA2 and SMARCA4) and created a consensus regions set as for ATAC-seq data. Signal was quantified and normalized in the same fashion. We calculated mean signal intensity and log2 fold-change between ARID2 and ARID1A ChIPs and standardized the fold-change in 250 bins along the mean. We then fit a Bivariate Gaussian Kernel (scipy.stats.gaussian_kde) on the two variables and selected differential sites as the ones in the 5th percentile of density and with absolute normalized log2 fold-change higher than 1.5.
Nomenclature
For clarity, gene names were also used for proteins. A list of alternative names can be found in the Supplementary Note.
Statistics and reproducibility
The statistical tests used are described in the respective figure legends and method sections. Additional information regarding sample size and statistics is available in Supplementary Table 6.
Supplementary Material
Acknowledgements
We thank the Biomedical Sequencing Facility, the Proteomics and Metabolomics Facility and the Platform Austria for Chemical Biology (PLACEBO) at CeMM for their support in generating and analyzing the next generation sequencing, proteomics or screening data, respectively. We gratefully acknowledge Horizon Discovery for providing us the HAP1 cell lines, and Boehringer Ingelheim, the Superti-Furga laboratory (CeMM) and Winter laboratory (CeMM) for various cancer cell lines. We appreciate the experimental support by Jana Block and Derya Donertas. Research in the Kubicek laboratory is supported by the Austrian Federal Ministry for Digital and Economic Affairs and the National Foundation for Research, Technology, and Development, the Austrian Science Fund (FWF) F4701 and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-CoG-772437). C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement no. 679146).
Footnotes
Reporting Summary
Further information on research design can be found in the Life Sciences Reporting Summary linked to this article.
Data, software and code availability
Next generation sequencing data were deposit to the NCBI GEO repository: GSE108390. Mass spectrometry data were deposited to PRIDE: PXD013102. Furthermore, the processed data and software used for analyses are available at http://baf-complex.computational-epigenetics.org
Author Contributions
S.S., G.B., M.P. and S.K. planned the study and designed the experiments; S.S., K.R., M.H., T.P., K.P., C.S., A.R. and B.B. performed the experiments; A.F.R., P.M., L.V., S.S., and S.K. analyzed the data; S.S. and S.K. wrote the manuscript; S.K., M.P., G.B., A.M., C.B., and J.M. supervised the work; S.K. provided funding
Competing Interests Statement
G.B. and M.P. are employees of Boehringer Ingelheim RCV GmbH & Co KG.
References
- 1.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–82. [PMC free article] [PubMed] [Google Scholar]
- 2.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 4.Chandler RL, et al. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–80. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges C, Kirkland JG, Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci U S A. 2014;111:3128–33. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson BG, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34:1136–44. doi: 10.1128/MCB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oike T, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res. 2013;73:5508–18. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 10.Helming KC, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–4. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelso TWR, et al. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife. 2017;6 doi: 10.7554/eLife.30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathur R, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. doi: 10.1038/ng.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mashtalir N, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. 2018;175:1272–1288 e20. doi: 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alpsoy A, Dykhuizen EC. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem. 2018;293:3892–3903. doi: 10.1074/jbc.RA117.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta A, et al. Composition and Function of Mutant Swi/Snf Complexes. Cell Rep. 2017;18:2124–2134. doi: 10.1016/j.celrep.2017.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen P, et al. Loss of Snf5 Induces Formation of an Aberrant SWI/SNF Complex. Cell Reports. 2017;18:2135–2147. doi: 10.1016/j.celrep.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–6. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 18.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michel BC, et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol. 2018;20:1410–1420. doi: 10.1038/s41556-018-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan J, et al. Interrogation of Mammalian Protein Complex Structure, Function, and Membership Using Genome-Scale Fitness Screens. Cell Syst. 2018;6:555–568 e7. doi: 10.1016/j.cels.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XF, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nature Genetics. 2017;49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodges HC, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 2018;25:61–72. doi: 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raab JR, Resnick S, Magnuson T. Genome-Wide Transcriptional Regulation Mediated by Biochemically Distinct SWI/SNF Complexes. PLoS Genet. 2015;11:e1005748. doi: 10.1371/journal.pgen.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valianou M, et al. Pharmacological inhibition of Polo-like kinase 1 (PLK1) by BI-2536 decreases the viability and survival of hamartin and tuberin deficient cells via induction of apoptosis and attenuation of autophagy. Cell Cycle. 2015;14:399–407. doi: 10.4161/15384101.2014.986394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang NN, et al. Molecular targeting of the oncoprotein PLK1 in pediatric acute myeloid leukemia: RO3280, a novel PLK1 inhibitor, induces apoptosis in leukemia cells. Int J Mol Sci. 2015;16:1266–92. doi: 10.3390/ijms16011266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao CL, et al. Downregulation of PLK1 by RNAi attenuates the tumorigenicity of esophageal squamous cell carcinoma cells via promoting apoptosis and inhibiting angiogenesis. Neoplasma. 2015;62:748–55. doi: 10.4149/neo_2015_089. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 28.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 29.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFarland JM, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Cell Line Encyclopedia, C. & Genomics of Drug Sensitivity in Cancer, C. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528:84–7. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers RM, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald ER, 3rd, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell. 2017;170:577–592 e10. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Tsherniak A, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576 e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21 29 1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–4. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 12 1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–41. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–31. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaus P, Honkela A, Rattray M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28:1721–8. doi: 10.1093/bioinformatics/bts260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 51.Sheffield NC, Bock C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 2016;32:587–9. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheffield NC, et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–88. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez-Castillo M, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2015;43:D1117–23. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–81. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–6. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 57.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schep AN, et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25:1757–70. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.