Abstract
Background
Alzheimer’s disease (AD) is a complex neurological disorder with contributions from genetic and environmental factors. High‐resolution metabolomics (HRM) has the potential to identify novel endogenous and environmental factors involved in AD. Previous metabolomics studies have identified circulating metabolites linked to AD, but lack of replication and inconsistent diagnostic algorithms have hindered the generalizability of these findings. Here we applied HRM to identify plasma metabolic and environmental factors associated with AD in two study samples, with cerebrospinal fluid (CSF) biomarkers of AD incorporated to achieve high diagnostic accuracy.
Methods
Liquid chromatography‐mass spectrometry (LC–MS)‐based HRM was used to identify plasma and CSF metabolites associated with AD diagnosis and CSF AD biomarkers in two studies of prevalent AD (Study 1: 43 AD cases, 45 mild cognitive impairment [MCI] cases, 41 controls; Study 2: 50 AD cases, 18 controls). AD‐associated metabolites were identified using a metabolome‐wide association study (MWAS) framework.
Results
An MWAS meta‐analysis identified three non‐medication AD‐associated metabolites in plasma, including elevated levels of glutamine and an unknown halogenated compound and lower levels of piperine, a dietary alkaloid. The non‐medication metabolites were correlated with CSF AD biomarkers, and glutamine and the unknown halogenated compound were also detected in CSF. Furthermore, in Study 1, the unknown compound and piperine were altered in MCI patients in the same direction as AD dementia.
Conclusions
In plasma, AD was reproducibly associated with elevated levels of glutamine and a halogen‐containing compound and reduced levels of piperine. These findings provide further evidence that exposures and behavior may modify AD risks.
Introduction
Alzheimer’s disease (AD) is a progressive neurological disorder whose onset and progression are influenced by genetic, biological, environmental, and social factors.1 There has been considerable progress in characterizing proteomic changes associated with the core pathology of AD (including Aβ42‐rich neuritic plaques and tau‐rich neurofibrillary tangles).2, 3 However, proteins alone – in the brain or body fluids such as plasma or cerebrospinal fluid (CSF) – are inadequate to characterize metabolic alterations and environmental exposures associated with AD. Targeted metabolomics studies in AD have identified novel risk factors and candidate diagnostic biomarkers,4, 5 but these approaches are limited in the number of metabolites measured. When coupled with only a modest accuracy in clinical diagnosis6 and the presence of pre‐clinical AD in older subjects, these studies are often plagued by false positive or negative findings.
Untargeted metabolomics involves the simultaneous detection of all small molecules in a biofluid – that is, the metabolome – without a priori knowledge of the metabolites involved.7 One untargeted approach, high‐resolution metabolomics (HRM), enables the measurement of thousands of endogenous and exogenous metabolites over eight orders of magnitude.8 We hypothesize that HRM will enable the identification of novel biological and/or man‐made metabolites associated with AD. At the same time, “‐omics” studies in AD research suffer from non‐standardized sample handling, over‐training in a single small cohort, and limited accuracy of the clinical AD diagnosis, with at least 17% of clinically‐probable AD found to have no AD neuropathology on autopsy.6, 9 Along with inter‐individual variability and technical limitations, these factors contribute to the low replication rates of AD metabolomic profiles.5, 10
To identify metabolites whose alterations are consistently associated with AD, we designed an HRM study using plasma samples from subjects with normal cognition (NC), mild cognitive impairment (MCI) and AD dementia. We incorporated CSF AD biomarker information associated with the presence or absence of brain AD pathology11, 12 to achieve high diagnostic accuracy. We then recruited an independent sample of participants to validate our plasma findings, and we used a metabolome‐wide association study (MWAS) approach to identify metabolites consistently altered across the two studies.
Methods
Participants
Subjects for both studies were recruited from the Emory Cognitive Neurology Clinic and the Emory Alzheimer’s Disease Research Center. This study was approved by the Emory University Institutional Review Board. All participants or their legal representatives provided written informed consent. Each subject underwent a detailed evaluation including neurological examination and neuropsychological analysis. Subjects with cognitive impairment or dementia also underwent routine blood tests to rule out common reversible causes of cognitive dysfunction, and brain imaging to rule out structural causes of dementia. Subjects were classified as having NC if there was no subjective cognitive complaint and neuropsychological analysis showed normal cognitive functioning according to age, gender, education, and race and as having MCI13 or AD dementia14 according to NIA‐AA criteria.
Sample collection
Plasma samples were collected and processed as described previously.15 Briefly, 20 mL of whole blood was collected via phlebotomy between 8 am and noon without overnight fasting and centrifuged at 4°C at 1000 x g. Platelet‐rich plasma was immediately removed without disturbing the cellular layers, aliquoted, labeled, frozen, and stored at −80°C until analysis within 2 h of collection. All subjects in the two studies also underwent CSF collection via lumbar puncture with a 24‐gauge atraumatic spinal needle into polypropylene tubes (BD Falcon), immediately aliquoted (0.5 mL), labeled, frozen, and stored at −80°C until analysis within 30 min of collection. CSF AD biomarker analysis was performed as previously described in a Luminex 200 platform, including levels of beta‐amyloid 1‐42 (Aβ42), total tau (t‐Tau), and tau phosphorylated at threonine 181 (p‐Tau181).15 For inclusion into the study, all NC subjects must have t‐Tau/Aβ42 < 0.39 (not consistent with Alzheimer’s disease), and all AD dementia subjects must have t‐Tau/Aβ42 ≥ 0.39 (consistent with Alzheimer’s disease) based on a previously published CSF autopsy study.11 Because only 30–70% of MCI subjects had underlying AD pathology as the cause of their cognitive impairment,16 MCI subjects were classified as likely due to AD (MCI‐AD, t‐Tau/Aβ42 ≥ 0.39) or likely due to a suspected non‐AD pathology (MCI‐SNAP, t‐Tau/Aβ42 < 0.39).
Plasma and CSF HRM analysis
Plasma and CSF samples were prepared for HRM using methods detailed elsewhere.17, 18, 19 Briefly, aliquots were removed from storage at −80°C and thawed on ice, upon which 65 μL of biofluid was added to 130 μL of acetonitrile containing a mixture of stable isotopic standards, vortexed, and allowed to equilibrate for 30 min. Proteins were precipitated by centrifuge (16.1g at 4°C for 10 min), and extracts were stored in a refrigerated autosampler. Triplicate 10 μL aliquots were analyzed by reverse‐phase C18 liquid chromatography (Dionex Ultimate 3000) and Fourier transform mass spectrometry (Study 1: Thermo Q‐Exactive; Study 2: Thermo Q‐Exactive HF) in positive electrospray ionization mode, resolution (FWHM) of 70,000 (Study 1) or 120,000 (Study 2) and mass‐to‐charge (m/z) range of 85–1250.20, 21 Samples were grouped by matrix, randomized, and analyzed in batches of 20, with a quality control (QC) pooled reference sample included at the beginning and end of each batch. Raw data files were extracted using apLCMS22 with modifications by xMSanalyzer,23 with each unique mass‐to‐charge (m/z) feature defined by m/z, retention time, and ion abundance. Metabolomics data will be deposited in Metabolomics Workbench (http://www.metabolomicsworkbench.org).
Statistical analysis
Statistical analyses were performed in RStudio v0.99.486.24 Variation in m/z feature intensities related to analytical daily batch was removed using ComBat.25 Prior to analyses, datasets were filtered to remove features with retention times <30 sec.
Associations of m/z features with AD dementia (vs. NC) were assessed using a MWAS approach26, 27 for features present in >80% of study samples. Since the exclusion and/or imputation of non‐detected intensities can result in biased estimates,28 accelerated failure time (AFT) survival models29, 30 were used to model m/z feature intensities as outcomes, which enables the inclusion of all data observations by treating missing values as left‐censored. AFT models were fit using the R package “survival.”31, 32 For each m/z feature, the limit of detection (LOD) was considered to be the lowest detected intensity value. Separately in each study, AFT models, assuming lognormal intensity distributions,33 were constructed for each feature wherein diagnostic status (AD vs. NC) was the primary predictor of intensity, adjusted for sex and age; for the ith individual,
where log(Ti) is the log‐transformed feature intensity, X 1, X 2, and X 3 represent the predictor variables diagnosis (NC vs. AD dementia), sex, and age (continuous 1‐y change), with coefficients β 1, β 2 , and β 3, μ is the intercept term, ε is the error term, and σ is the scale coefficient. The beta coefficient for diagnosis (β 1) can be interpreted as the change in log mean intensity for AD dementia versus NC.
To identify m/z features consistently associated with AD dementia, a fixed effects meta‐analysis was conducted using the “meta” R package.34 A feature was selected for further characterization if it was associated with AD dementia at false discovery rate (FDR) <0.20 in the meta‐analysis and was associated with AD at P < 0.10 in both studies with the same direction of association. An FDR threshold of <0.20 was selected based on previously published metabolomics studies.35, 36, 37 To explore whether these features were iatrogenic metabolites linked to AD medications, regression models were constructed with diagnosis predicting each m/z feature, adjusting for sex, age, and detection (yes/no) for three AD medications ([M + H]+ for memantine (m/z 180.1748) and rivastigmine (m/z 251.1753), [M + H]+ (13C isotope) for donepezil (m/z 381.2254)). A feature would be excluded from further analyses if beta coefficients were attenuated following adjustment for AD medications in one or both studies.
Boxplots were created with “NADA,”38 an R package for censored data. Correlations of AD‐associated features, CSF markers (e.g., tau, Aβ42) and ApoE genotypes were conducted using Spearman rank‐order correlations. Results were combined in a fixed effects meta‐analysis using the “meta” R package.34 Correlations of m/z features between plasma and CSF were conducted using Spearman rank‐order correlations in participants with CSF in Study 1.
Metabolite identification
Detected m/z features were matched between studies based upon m/z (within 5 ppm) and retention times (within 30 sec) using the “xMSanalyzer” R package.23 If one m/z feature matched to multiple m/z features in the other study, the m/z feature with the closest retention time was selected for its match. To cluster m/z signals derived from the same metabolites, we used a custom‐designed application that generates a pseudospectrum for a metabolite feature based on intensity correlations and retention time similarities, then predicts adduct, isotope, and/or fragment identities based on the observed mass differences within each cluster.
Database matching by accurate mass was conducted using the Human Metabolome Database (HMDB)39 with a tolerance of 5 ppm. Searches were conducted using the predicted adduct/isotope/fragment identity from our algorithm; if the algorithm was unable to predict an identity, the database was searched using common adducts ([M + H]+, [M + Na]+, [M + K]+, [M + H‐H2O]+, [M + H‐2H2O]+, [M + ACN+H]+, [M + ACN+Na]+, [M + 2Na‐H]+). Ion dissociation (MS2) studies were completed and spectra characterized using CFM‐ID40 and Sirius 3.0.41 Piperine was confirmed by comparing m/z values, retention times, isotopic distribution, and detected adducts to a piperine reference standard (Sigma Aldrich).
Results
Participant characteristics are presented in Table 1. Study 1 consisted of 41 healthy NC, 45 MCI (20 with MCI‐AD [44% of MCI group]), and 43 AD dementia participants, and Study 2 had 18 NC and 50 AD participants. Plasma metabolomics was run for all participants, while CSF metabolomics was run on a subset of participants in Study 1 (Control, n = 25; MCI, n = 27; AD dementia, n = 26). AD and NC subjects did not differ by sex or age (P > 0.20). As expected, AD dementia participants had lower Aβ42 levels and higher t‐Tau and p‐Tau181 levels (P < 0.0001) and were more likely to have at least one APOE ε4 allele (P = 0.003 in Study 1).
Table 1.
Demographic and clinical data for study samples.
| Study 1 | Control | AD | MCI | Ctrl vs. AD |
|---|---|---|---|---|
| n = 41 | n = 43 | n = 45 | P c | |
| Demographics | ||||
| Male | 11 (27%)a | 16 (37%) | 22 (49%) | 0.43 |
| Age (y) | 67.5 ± 7.3b | 65.9 ± 8.8 | 69.4 ± 6.6 | 0.36 |
| CSF protein biomarkers | ||||
| Aβ42 (pg/mL) | 340 ± 137 | 203 ± 76 | 218 ± 90 | <0.0001 |
| t‐Tau (pg/mL) | 44 ± 24 | 117 ± 70 | 76 ± 67 | <0.0001 |
| p‐Tau181 (pg/mL) | 32 ± 15 | 75 ± 32 | 51 ± 25 | <0.0001 |
| t‐Tau/Aβ42 | 0.14 ± 0.09 | 0.64 ± 0.39 | 0.39 ± 0.33 | <0.0001 |
| APOE genotypes | ||||
| Subjects with data (n) | n = 18 | n = 13 | n = 21 | 0.003 |
| No ε4 alleles | 11 (61%) | 1 (8%) | 10 (48%) | |
| One ε4 allele | 7 (39%) | 8 (62%) | 7 (33%) | |
| Two ε4 alleles | 0 (0%) | 4 (31%) | 4 (19%) | |
| Study 2 | n = 18 | n = 50 | P | |
|---|---|---|---|---|
| Demographics | ||||
| Male | 8 (44%) | 22 (44%) | 1.00 | |
| Age (y) | 61.2 ± 12.6 | 65.1 ± 9.3 | 0.24 | |
| CSF protein biomarkers | ||||
| Aβ42 (pg/mL) | 218 ± 112 | 143 ± 73 | 0.01 | |
| t‐Tau (pg/mL) | 43 ± 30 | 114 ± 54 | <0.0001 | |
| p‐Tau181 (pg/mL) | 27 ± 16 | 56 ± 27 | <0.0001 | |
| t‐Tau/Aβ42 | 0.23 ± 0.13 | 0.93 ± 0.53 | <0.0001 | |
| APOE genotypes | ||||
| Subjects with data (n) | n = 16 | n = 40 | 0.19 | |
| No ε4 alleles | 10 (63%) | 15 (38%) | ||
| One ε4 allele | 5 (31%) | 17 (43%) | ||
| Two ε4 alleles | 1 (6%) | 8 (20%) | ||
N (%) (all such values).
Mean ± SD (all such values).
P‐values for control versus AD comparisons from t‐tests (continuous variables) or chi‐square tests (categorical variables).
MWAS of plasma metabolites
HRM profiling of plasma yielded 2320 and 3855 unique m/z features in Study 1 and Study 2, respectively. To identify metabolites reproducibly associated with AD dementia, we focused on the subset of 968 m/z features that were common to both studies. Within each study, we conducted an MWAS of AD, and we summarized the associations by conducting a meta‐analysis. After adjustment for sex and age, we identified four m/z features consistently associated with AD dementia (meta‐analysis FDR < 0.20 and P < 0.10 in each study; see extended results with relaxed significance thresholds in Table S1). The feature with the strongest association with AD, m/z 251.1753, matched the mass of the [M + H]+ adduct of rivastigmine, an AD medication. To verify that the three remaining m/z features were associated with AD independently of medications, we constructed models for these features adjusting for diagnosis, sex, age, and detection (yes/no) of three common AD medications identified in the unfiltered datasets (rivastigmine, memantine, and donepezil). Associations between AD and the three m/z features were not attenuated after adjustment for medication status, suggesting that their associations were not explained by medication use.
We characterized the identities of the three m/z features (Table 2; Table S2). The m/z feature 129.0661 was identified by MS2 as glutamine. While MS2 fragmentation for m/z 246.9550 was successful, the MS2 spectrum did not have a spectral library match, and m/z 349.1515 could not be identified with MS2 due to low abundance. The most likely accurate mass database match in HMDB for m/z 349.1515 was piperine, which is found in black pepper, Piper nigrum.42 The identification of m/z 349.1515 as piperine was confirmed by accurate mass and retention time matching to a reference standard. The MS1 and MS2 spectra for m/z 246.9550 contained peak patterns consistent with the presence of a halogen isotope (chlorine [Cl] or bromine [Br]). The feature had no matches in HMDB, but a METLIN43 search returned nine compound matches, all classified as toxicants. Among these features, glutamine was positively correlated with CSF t‐Tau and p‐Tau181 levels and the number of ApoE ε4 risk alleles, and piperine and m/z 246.9550 correlated negatively and positively with p‐Tau181 levels, respectively (Table 3).
Table 2.
Non‐medication plasma metabolite features reproducibly associated with AD from MWAS.
| Feature | Study 1 | Study 2 | Meta‐analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| m/z a | RTa | Metabolite | Est (SE) | P | Est (SE) | P | Est (SE) | P | FDR |
| 129.0661 | 89 | Glutamine | 0.22 (0.11) | 0.04 | 0.31 (0.13) | 0.02 | 0.25 (0.08) | 0.002 | 0.07 |
| 246.9550 | 127 | Unknown | 0.41 (0.17) | 0.02 | 0.38 (0.21) | 0.07 | 0.40 (0.14) | 0.003 | 0.08 |
| 349.1515 | 80 | Piperine | −0.59 (0.31) | 0.06 | −0.89 (0.49) | 0.07 | −0.68 (0.27) | 0.01 | 0.18 |
m/z and retention time (RT, in seconds) reflect the mean values in Studies 1 and 2.
Table 3.
Spearman correlationsa of AD‐associated non‐medication plasma features with CSF protein biomarkers of AD and APOE‐ε4 genotype.
| Aβ42 (pg/mL) | t‐Tau (pg/mL) | p‐Tau181 (pg/mL) | APOE (number of ε4 alleles) | |
|---|---|---|---|---|
| Glutamine | −0.15* | 0.21† | 0.18† | 0.33† |
| m/z 246.9550 | −0.16† | 0.13 | 0.19† | 0.09 |
| Piperine | −0.01 | −0.16* | −0.17† | 0.04 |
Correlations reflect results from a fixed effects meta‐analysis of the partial Spearman correlations, adjusted for sex and age, between the listed variables in Studies 1 and 2; *P < 0.10; † P < 0.05.
Metabolite alterations in MCI
We explored whether the three metabolites were altered in MCI subjects in Study 1. Two metabolites, m/z 246.9550 and piperine, were associated with MCI (vs. NC) in AFT models adjusted for sex and age, both in the same direction as their associations with AD dementia. Stratification by MCI‐SNAP and MCI‐AD did not reveal differences in the feature intensities between the two subgroups (Fig. 1).
Figure 1.
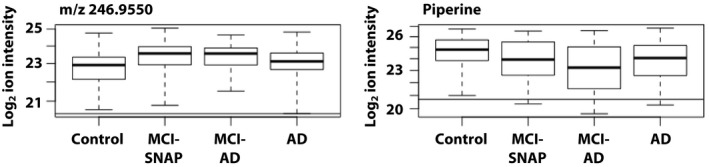
Boxplots of plasma features altered in MCI and AD in Study 1. Figure displays boxplots of log2‐transformed feature intensities by diagnosis for metabolites altered in both MCI and AD in Study 1. Horizontal lines show the lowest detectable ion intensity for the corresponding feature.
Correlation of metabolites between plasma and CSF
To determine the plausibility of plasma metabolites influencing the brain, we checked whether the features were detectable in CSF. We identified matches for glutamine and m/z 246.9550; plasma and CSF levels were modestly correlated for glutamine (rho = 0.26, P = 0.03) but not for m/z 246.9550 (rho = −0.04, P = 0.72). CSF levels of these two features were not associated with AD (P > 0.05; data not shown).
Discussion
Untargeted metabolomics has the promise to identify novel compounds involved in AD pathogenesis, but inconsistencies in clinical diagnoses, sample handling protocols, and analytical methods affect the generalizability of metabolomics studies. Here, we identified plasma and CSF metabolites associated with biomarker‐supported AD in two studies that we recruited and analyzed independently. In plasma, we replicated associations with glutamine, a previously‐identified metabolite, and we found novel metabolites increased (unknown halogen‐containing compound) and decreased (piperine) in AD dementia. Glutamine and the halogen‐containing compound were also detectable in CSF, supporting their entry into or synthesis in the central nervous system. In sum, our findings suggest that AD dementia is associated with reproducible alterations of plasma metabolites of endogenous and exogenous origin.
Previous studies have examined untargeted metabolomic profiles of AD dementia with varying approaches and results (summarized in Table 4). Several untargeted metabolomics and lipidomics studies have identified AD‐associated phospholipid alterations.44, 45, 46, 47 In most of these studies, the primary outcome was classification accuracy, which relies on machine learning algorithms that run the risk of overfitting. Only one study48 included Discovery and Validation studies similar to our design, which identified three metabolites across the two samples. To the best of our knowledge, our study is the first to employ HRM in two independent studies to identify metabolites reproducibly associated with biomarker‐supported AD dementia. However, replication in another geographical region is needed for generalization of our findings beyond the southeastern United States.
Table 4.
Summary of untargeted metabolomics studies of AD in human biofluids.
| Study | Diagnostic groups | Replication cohort | Confirmation of AD pathology | Specimen(s) | Platform | Analytical approach | Metabolite results |
|---|---|---|---|---|---|---|---|
| Orešič et al. 201147 | MCI (n = 143), AD (n = 47), Control (n = 46) | None | Imaging, CSF biomarkers | Serum | GC‐MS | Penalized GLM, logistic regression | Molecular signature of AD of three metabolites (PC (16:0/16:0); unidentified carboxylic acid; 2,4‐dihydroxybutanoic acid) |
| Ibáñez et al. 201210 | AD (n = 25), MCI‐AD (n = 13), MCI‐SNAP (n = 24), Control (n = 23) | None | Imaging, CSF biomarkers | CSF | CE‐MS | PCA, LDA | 14 key metabolites that predicted progression to AD (including choline, dimethylarginine, arginine, valine, proline, serine, histidine, creatine, carnitine, and suberylglycine) |
| Trushina et al. 201372 | MCI (n = 15), AD (n = 15), Control (n = 15) | None | None | Plasma, CSF | LC–MS | ANOVA, PCA, OPLS‐DA | 342 plasma and 351 CSF features altered across AD, MCI, and control groups (22% putatively identified) |
| Motsinger‐Reif et al. 201354 | AD (n = 40), Control (n = 38) | None | CSF biomarkers | CSF | GC–MS, LC–MS | Stepwise logistic regression | Two discriminant features for AD vs. control |
| Cui et al. 201448 | AD (n = 46), Control (n = 37) | AD (n = 63), Control (n = 67) | None | Serum, urine | LC–MS | OPLS‐DA, ANCOVA, logistic regression | Three metabolites (serum palmitic amide and lysoPC(18:2), urine 5‐L‐glutamylglycine) with consistent prediction across studies |
| Graham et al. 201573 | MCI (n = 16), MCI‐AD (n = 19), Control (n = 37) | None | None | Plasma | LC–MS | OPLS‐DA, t‐tests | 263 metabolites altered in MCI vs. control, 162 metabolites altered in MCI‐AD vs. control (putatively identified) |
| Morris et al. 201874 | AD (n = 64), Control (n = 62) | None | None | Serum | LC–MS | PLS‐DA, Mann–Whitney U‐tests | Poor classification of AD vs. control; ability to distinguish type 2 diabetes patients in controls but not in AD |
| Pena‐Bautista et al. 201975 | MCI‐AD (n = 29), Control (n = 29) | None | Imaging, CSF biomarkers | Plasma | LC–MS | Elastic net | 53 discriminant features; confirmed identity for choline |
| Habartová et al. 201976 | AD (n = 20), Control (n = 13) | None | Imaging | Plasma | LC–MS | LDA | Seven features altered in AD vs. control (putatively identified) |
AD, Alzheimer’s disease; MCI, mild cognitive impairment; SNAP, suspected non‐Alzheimer’s pathophysiology; CSF, cerebrospinal fluid; CE‐MS, capillary electrophoresis‐mass spectrometry; LC–MS, liquid chromatography‐mass spectrometry; GC‐MS, gas chromatography‐mass spectrometry; PCA, principal component analysis; LDA, linear discriminant analysis; PET, positron emission tomography; ANOVA, analysis of variance; GLM, generalized linear model; OPLS‐DA, orthogonal partial least squares discriminant analysis; ANCOVA, analysis of covariance; PLS‐DA, partial least squares discriminant analysis.
Consistent with previous targeted metabolomics studies,4, 49, 50, 51, 52, 53, 54, 55 we identified elevated plasma glutamine in AD dementia. In our studies, glutamine was positively associated with APOE‐ε4 status and CSF t‐Tau and p‐Tau181 levels. Glutamine is a precursor to several excitatory (glutamate and aspartate) and inhibitory (neurotransmitter γ‐amino butyric acid, or GABA) neurotransmitters.56 Previous studies found that mice with knocked‐in apoE ε4 have elevated brain glutamine57 and greater susceptibility to excitotoxicity58 than APOE ε3 knock‐in mice. However, human studies examining glutamine levels in AD dementia compared to NC have generated mixed findings.48, 55, 59 Importantly, glutamine can readily pass through the blood‐brain barrier, and we found a modest correlation between plasma and CSF glutamine levels. While it is possible that memantine (a non‐competitive NMDA receptor antagonist) might influence glutamine levels, effect estimates for plasma glutamine and memantine predicting AD dementia were unchanged in models containing both metabolites simultaneously, suggesting these metabolites were independently related to AD. Thus, our current findings add to the body of evidence implicating glutamine dysregulation and excitotoxicity in AD. Future studies should investigate whether plasma glutamine can serve as a biomarker to identify AD patients susceptible to excitotoxicity.
Piperine, which was found at reduced levels in AD, is a bioactive dietary compound found at high levels in black pepper (Piper nigrum).60 The compound was negatively associated with CSF p‐Tau181 and was also reduced in MCI. Piperine has a range of physiological effects, including antioxidant,61 antinflammatory,62 and anti‐secretase63 activities directly relevant to AD. Piperine has been shown to be neuroprotective in AD mouse models,64, 65, 66, 67, 68 but to our knowledge, this is the first report of an association between piperine and AD in a human study. Although we are unable to draw conclusions about the direction of the association, these findings warrant further investigation given the alkaloid’s low risks and costs, as well as its ability to enhance absorption of other neuroprotective compounds.69, 70
Identification of m/z features remains a bottleneck in HRM. Identification is particularly difficult for low‐abundance metabolites since MS2 spectral matching is challenging71 and spectral information may not be present in public databases for less‐common compounds. For example, m/z 246.9550 matched nine toxicants in METLIN, but none had actual or in silico MS2 spectra, and only a small proportion had standards that were available for purchase. However, the metabolite’s association with AD dementia in two independent study samples and its detectable CSF levels support the need for future work to identify the chemical and examine its role in AD susceptibility or pathogenesis.
Our study had several strengths, including the inclusion of multiple biofluids, two independent samples, and application of the HRM platform. Among the three non‐medication metabolites, two were associated with MCI in Study 1 regardless of cause, suggesting a role in brain vulnerability or degeneration not specific to AD. We also acknowledge limitations beyond those inherent to observational studies, sample size concerns, and generalizability beyond the study’s geographic region. First, it is possible that AD treatment alters non‐neurological metabolic pathways. We could not confirm the identities of two features through MS2. We did not have lifestyle history to corroborate past exposures, and we did not have sufficient information (e.g., half‐life) on their associated metabolites to build a temporal relationship between exposure and disease onset. Additionally, the analysis of non‐fasting plasma samples may have introduced noise into the metabolite measurements. Nevertheless, our HRM workflow identified endogenous and environmental metabolites reproducibly altered in AD dementia, providing confirmatory and novel findings for hypothesis generation for future in vitro mechanistic studies and in vivo observational studies.
Conflict of Interest
The authors declare no actual or potential conflicts of interest.
Supporting information
Table S1. Metabolites associated with AD dementia in Studies 1 and 2 with P < 0.20 and consistent direction of association.
Table S2. Summary of MS1 and MS2 results for m/z 129.0667, 246.9550, and 349.1515.
Acknowledgments
This study was supported by K23 AG042856, R21 AG043885, P50 AG025688, T32 ES012870, and P30 ES019776 from the National Institutes of Health.
Funding Information
This study was supported by K23 AG042856, R21 AG043885, P50 AG025688, T32 ES012870, and P30 ES019776 from the National Institutes of Health.
Funding Statement
This work was funded by National Institute of Environmental Health Sciences grants P30 ES019776 and T32 ES012870; National Institute on Aging grants K23 AG042856, P50 AG025688, and R21 AG043885.
Contributor Information
Megan M. Niedzwiecki, Email: megan.niedzwiecki@mssm.edu.
William T. Hu, Email: wthu@emory.edu.
References
- 1. Reitz C. Toward precision medicine in Alzheimer's disease. Ann Transl Med 2016;4:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu WT, Chen‐Plotkin A, Arnold SE, et al. Novel CSF biomarkers for Alzheimer's disease and mild cognitive impairment. Acta Neuropathol 2010;119:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu WT, Holtzman DM, Fagan AM, et al. Plasma multianalyte profiling in mild cognitive impairment and Alzheimer disease. Neurology 2012;79:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richardson JR, Roy A, Shalat SL, et al. Elevated serum pesticide levels and risk for Alzheimer disease. JAMA Neurol 2014;71:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mapstone M, Cheema AK, Fiandaca MS, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 2014;20:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beach TG, Monsell SE, Phillips LE, Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J Neuropathol Exp Neurol 2012;71:266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alonso A, Marsal S, Julia A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uppal K, Walker DI, Liu K, et al. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol 2016;29:1956–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zierer J, Menni C, Kastenmuller G, Spector TD. Integration of 'omics' data in aging research: from biomarkers to systems biology. Aging Cell 2015;14:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ibanez C, Simo C, Barupal DK, et al. A new metabolomic workflow for early detection of Alzheimer's disease. J Chromatogr A 2013;1302:65–71. [DOI] [PubMed] [Google Scholar]
- 11. Shaw LM, Vanderstichele H, Knapik‐Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol 2009;65:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seeburger JL, Holder DJ, Combrinck M, et al. Cerebrospinal fluid biomarkers distinguish postmortem‐confirmed Alzheimer's disease from other dementias and healthy controls in the OPTIMA cohort. J Alzheimers Dis 2015;44:525–539. [DOI] [PubMed] [Google Scholar]
- 13. Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011;7:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS‐ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology 1984;34:939–944. [DOI] [PubMed] [Google Scholar]
- 15. Hu WT, Watts KD, Shaw LM, et al. CSF beta‐amyloid 1–42 ‐ what are we measuring in Alzheimer's disease? Ann Clin Transl Neurol 2015;2:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li JQ, Tan L, Wang HF, et al. Risk factors for predicting progression from mild cognitive impairment to Alzheimer's disease: a systematic review and meta‐analysis of cohort studies. J Neurol Neurosurg Psychiatry 2016;87:476–484. [DOI] [PubMed] [Google Scholar]
- 17. Park YH, Lee K, Soltow QA, et al. High‐performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012;295:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soltow Q, Strobel FH, Mansfield KG, et al. High‐performance metabolic profiling with dual chromatography‐Fourier‐transform mass spectrometry (DC‐FTMS) for study of the exposome. Metabolomics 2013;9(Suppl 1):S132-S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Go YM, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high‐resolution metabolomics applications to exposome research. Toxicol Sci 2015;148:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Accardi CJ, Walker DI, Uppal K, et al. High‐resolution metabolomics for nutrition and health assessment of armed forces personnel. J Occup Environ Med 2016;58:S80–S88. [DOI] [PubMed] [Google Scholar]
- 21. Walker DI, Pennell KD, Uppal K, et al. Pilot metabolome‐wide association study of benzo(a)pyrene in serum from military personnel. J Occup Environ Med. 2016;58:S44–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu T, Park Y, Li S, Jones DP. Hybrid feature detection and information accumulation using high‐resolution LC‐MS metabolomics data. J Proteome Res 2013;12:1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uppal K, Soltow QA, Strobel FH, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large‐scale, non‐targeted metabolomics data. BMC Bioinform 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Team R. RStudio: integrated development for R. Boston, MA: RStudio, Inc., 2015. [Google Scholar]
- 25. Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high‐throughput experiments. Bioinformatics 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holmes E, Loo RL, Stamler J, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008;453:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bictash M, Ebbels TM, Chan Q, et al. Opening up the "Black Box": metabolic phenotyping and metabolome‐wide association studies in epidemiology. J Clin Epidemiol 2010;63:970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hrydziuszko O, Viant MR. Missing values in mass spectrometry based metabolomics: an undervalued step in the data processing pipeline. Metabolomics 2012;8(S1):161–174. [Google Scholar]
- 29. Tekwe CD, Carroll RJ, Dabney AR. Application of survival analysis methodology to the quantitative analysis of LC‐MS proteomics data. Bioinformatics 2012;28:1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor SL, Leiserowitz GS, Kim K. Accounting for undetected compounds in statistical analyses of mass spectrometry ‘Omic Studies. Stat Appl Genet Mol Biol. 2013;12:703–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Therneau T. A Package for Survival Analysis in S. version 2.38 ed. 2015.
- 32. Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. New York: Springer, 2000. [Google Scholar]
- 33. Dinse GE, Jusko TA, Whitt IZ, et al. Associations between selected Xenobiotics and antinuclear antibodies in the National Health and Nutrition Examination Survey, 1999–2004. Environ Health Perspect 2016;124:426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwarzer G. meta: an R package for meta‐analysis. R News 2007;7:40–45. [Google Scholar]
- 35. Walker DI, Uppal K, Zhang L, et al. High‐resolution metabolomics of occupational exposure to trichloroethylene. Int J Epidemiol 2016;45:1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee SJ, Woo SI, Ahn SH, et al. Functional interpretation of metabolomics data as a new method for predicting long‐term side effects: treatment of atopic dermatitis in infants. Sci Rep 2014;4:7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao Q, Moore SC, Keadle SK, et al. Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. Int J Epidemiol 2016;45:1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee L. NADA: Nondetects and Data Analysis for Environmental Data. R package version 1.6‐1 ed. 2017.
- 39. Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0–the human metabolome database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Allen F, Pon A, Wilson M, et al. CFM‐ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res 2014;42(W1):W94–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Böcker S, Dührkop K. Fragmentation trees reloaded. J Cheminformatics 2016;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Genest C, Smith DM, Pepper CDG. Pepper analysis, a critical study of two procedures for the determination of piperine in black and white pepper. J Agr Food Chem 1963;11:508–512. [Google Scholar]
- 43. Smith CA, O'Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit 2005;27:747–751. [DOI] [PubMed] [Google Scholar]
- 44. Han X, Rozen S, Boyle SH, et al. Metabolomics in early Alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE 2011;6:e21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato Y, Suzuki I, Nakamura T, et al. Identification of a new plasma biomarker of Alzheimer's disease using metabolomics technology. J Lipid Res 2012;53:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ibáñez C, Simó C, Barupal DK, et al. A new metabolomic workflow for early detection of Alzheimer's disease. J Chromatogr A 2013;1302:65–71. [DOI] [PubMed] [Google Scholar]
- 47. Oresic M, Hyotylainen T, Herukka SK, et al. Metabolome in progression to Alzheimer's disease. Transl Psychiatry 2011;1:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cui Y, Liu X, Wang M, et al. Lysophosphatidylcholine and amide as metabolites for detecting alzheimer disease using ultrahigh‐performance liquid chromatography‐quadrupole time‐of‐flight mass spectrometry‐based metabonomics. J Neuropathol Exp Neurol 2014;73:954–963. [DOI] [PubMed] [Google Scholar]
- 49. Kaddurah‐Daouk R, Zhu H, Sharma S, et al. Alterations in metabolic pathways and networks in Alzheimer's disease. Transl Psychiatry. 2013;3:e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Czech C, Berndt P, Busch K, et al. Metabolite profiling of Alzheimer's disease cerebrospinal fluid. PLoS ONE 2012;7:e31501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaddurah‐Daouk R, Rozen S, Matson W, et al. Metabolomic changes in autopsy‐confirmed Alzheimer's disease. Alzheimers Dement 2011;7:309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tukiainen T, Tynkkynen T, Mäkinen V‐P, et al. A multi‐metabolite analysis of serum by 1H NMR spectroscopy: Early systemic signs of Alzheimer’s disease. Biochem Biophys Res Comm 2008;375:356–361. [DOI] [PubMed] [Google Scholar]
- 53. Graham SF, Holscher C, Green BD. Metabolic signatures of human Alzheimer’s disease (AD): 1H NMR analysis of the polar metabolome of post‐mortem brain tissue. Metabolomics 2014;10(4:744-753). [Google Scholar]
- 54. Motsinger‐Reif AA, Zhu H, Kling MA, et al. Comparing metabolomic and pathologic biomarkers alone and in combination for discriminating Alzheimer’s disease from normal cognitive aging. Acta Neuropathol Commun 2013;1:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. González‐Domínguez R, García‐Barrera T, Gómez‐Ariza JL. Metabolite profiling for the identification of altered metabolic pathways in Alzheimer's disease. J Pharm Biomed Anal 2015;107:75–81. [DOI] [PubMed] [Google Scholar]
- 56. Albrecht J, Sidoryk‐Węgrzynowicz M, Zielińska M, Aschner M. Roles of glutamine in neurotransmission. Neuron Glia Biol 2010;6:263–276. [DOI] [PubMed] [Google Scholar]
- 57. Dumanis SB, DiBattista AM, Miessau M, et al. APOE genotype affects the presynaptic compartment of glutamatergic nerve terminals. J Neurochem 2013;124:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buttini M, Masliah E, Yu GQ, et al. Cellular source of apolipoprotein E4 determines neuronal susceptibility to excitotoxic injury in transgenic mice. Am J Pathol 2010;177:563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Timmer NM, Herbert MK, Claassen JA, et al. Total glutamine synthetase levels in cerebrospinal fluid of Alzheimer's disease patients are unchanged. Neurobiol Aging 2015;36:1271–1273. [DOI] [PubMed] [Google Scholar]
- 60. Chavarria D, Silva T, Magalhães e Silva D, et al. Lessons from black pepper: piperine and derivatives thereof. Expert Opin Ther Pat 2016;26:245–264. [DOI] [PubMed] [Google Scholar]
- 61. Mittal R, Gupta RL. In vitro antioxidant activity of piperine. Methods Find Exp Clin Pharmacol 2000;22:271–274. [DOI] [PubMed] [Google Scholar]
- 62. Mujumdar AM, Dhuley JN, Deshmukh VK, et al. Anti‐inflammatory activity of piperine. Jpn J Med Sci Biol 1990;43:95–100. [DOI] [PubMed] [Google Scholar]
- 63. Murata K, Matsumura S, Yoshioka Y, et al. Screening of beta‐secretase and acetylcholinesterase inhibitors from plant resources. J Nat Med 2015;69:123–129. [DOI] [PubMed] [Google Scholar]
- 64. Chonpathompikunlert P, Wattanathorn J, Muchimapura S. Piperine, the main alkaloid of Thai black pepper, protects against neurodegeneration and cognitive impairment in animal model of cognitive deficit like condition of Alzheimer's disease. Food Chem Toxicol 2010;48:798–802. [DOI] [PubMed] [Google Scholar]
- 65. Mahdy K, Shaker O, Wafay H, et al. Effect of some medicinal plant extracts on the oxidative stress status in Alzheimer’s disease induced in rats. Eur Rev Med Pharmacol Sci 2012;16(Suppl 3):31–42. [PubMed] [Google Scholar]
- 66. Hritcu L, Noumedem JA, Cioanca O, et al. Methanolic extract of piper nigrum fruits improves memory impairment by decreasing brain oxidative stress in amyloid beta(1–42) rat model of Alzheimer’s disease. Cell Mol Neurobiol 2014;34:437–449. [DOI] [PubMed] [Google Scholar]
- 67. Banji D, Banji OJF, Dasaroju S, Kumar Ch K . Curcumin and piperine abrogate lipid and protein oxidation induced by d‐galactose in rat brain. Brain Res 2013;1515:1–11. [DOI] [PubMed] [Google Scholar]
- 68. Elnaggar YS, Etman SM, Abdelmonsif DA, Abdallah OY. Intranasal Piperine‐loaded chitosan nanoparticles as brain‐targeted therapy in Alzheimer's disease: optimization, biological efficacy, and potential toxicity. J Pharm Sci 2015;104:3544–3556. [DOI] [PubMed] [Google Scholar]
- 69. Arcaro CA, Gutierres VO, Assis RP, et al. Piperine, a natural bioenhancer, nullifies the antidiabetic and antioxidant activities of curcumin in streptozotocin‐diabetic rats. PLoS ONE 2014;9:e113993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rinwa P, Kumar A. Piperine potentiates the protective effects of curcumin against chronic unpredictable stress‐induced cognitive impairment and oxidative damage in mice. Brain Res. 2012;1488:38–50. [DOI] [PubMed] [Google Scholar]
- 71. Tautenhahn R, Cho K, Uritboonthai W, et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol 2012;30:826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Trushina E, Dutta T, Persson X‐MT, et al. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and alzheimer’s disease using metabolomics. PLoS ONE 2013;8:e63644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Graham SF, Chevallier OP, Elliott CT, et al. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L‐arginine metabolism in mild cognitive impairment subjects converting to Alzheimer’s disease. PLoS ONE 2015;10:e0119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Morris JK, Piccolo BD, Shankar K, et al. The serum metabolomics signature of type 2 diabetes is obscured in Alzheimer's disease. Am J Physiol Endocrinol Metab 2018;314:E584–E596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pena‐Bautista C, Roca M, Hervas D, et al. Plasma metabolomics in early Alzheimer's disease patients diagnosed with amyloid biomarker. J Proteomics 2019;200:144–152. [DOI] [PubMed] [Google Scholar]
- 76. Habartova L, Hrubesova K, Syslova K, et al. Blood‐based molecular signature of Alzheimer's disease via spectroscopy and metabolomics. Clin Biochem 2019;72:58–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Metabolites associated with AD dementia in Studies 1 and 2 with P < 0.20 and consistent direction of association.
Table S2. Summary of MS1 and MS2 results for m/z 129.0667, 246.9550, and 349.1515.


