Abstract
Chediak-Higashi Syndrome (CHS) is a well-characterized, autosomal recessively inherited lysosomal disease caused by mutations in lysosomal trafficking regulator (LYST). The feline model for CHS was originally maintained for ~20 years. However, the colonies were disbanded and the CHS cat model was lost to the research community before the causative mutation was identified. To resurrect the cat model, semen was collected and cryopreserved from a lone, fertile, CHS carrier male. Using cryopreserved semen, laparoscopic oviductal artificial insemination was performed on three queens, two queens produced 11 viable kittens. To identify the causative mutation, a fibroblast cell line, derived from an affected cat from the original colony, was whole genome sequenced. Visual inspection of the sequence data identified a candidate causal variant as a ~20 kb tandem duplication within LYST, spanning exons 30 through to 38 (NM_001290242.1:c.8347-2422_9548 + 1749dup). PCR genotyping of the produced offspring demonstrated three individuals inherited the mutant allele from the CHS carrier male. This study demonstrated the successful use of cryopreservation and assisted reproduction to maintain and resurrect biomedical models and has defined the variant causing Chediak-Higashi syndrome in the domestic cat.
Subject terms: Animal breeding, Medical genomics, Gene duplication
Introduction
Chediak-Higashi syndrome (CHS) (OMIM Accession: 214500) is a rare autosomal recessive disorder characterized in humans by severe immune deficiency, oculocutaneous albinism, bleeding tendencies, recurrent pyogenic infections, progressive neurologic defects and a lymphoproliferative syndrome. The most common cause of death from CHS is from recurrent infections or the development of an accelerated phase with hemophagocytic lymphohistiocytosis. Approximately 90% of deaths occur in the first decade of life, and those who survive into adulthood develop progressive neurological symptoms1. The disease was first described in the 1940s to early 1950s2–6 and has been characterized in a host of diverse species, including cow7–10, mink10–14, killer whale15,16, fox17,18 and domestic cat19–23 (OMIA: 000185-9913, 9733, 494514, 452646, 9685). Continuing studies of CHS models have demonstrated their value in deciphering delta storage pool deficiencies24, heritable platelet disorders25,26 and cellular cytotoxicity27. Bone marrow transplantation has been a viable option for management of human CHS for over 30 years28–31.
The genetic cause for CHS was first defined in rodents32–36, with the locus historically known as beige due to the associated hypopigmentation phenotype33. The human homolog of the mouse beige locus revealed the first causative mutations for CHS in humans37,38 and was defined as lysosomal trafficking regulator (LYST)37. In humans, LYST encodes a 3,801 amino acid protein (11.4 kb transcript), which regulates intracellular protein trafficking to and from the lysosome (GCID:GC01M235824). In many species with CHS, mutations have been consistently identified within LYST. These species include, human39–42, cow43, mouse44,45, rat46,47 and Aleutian mink48. Overall, in the public archive of human genetic variation and phenotypes (clinVar)49, over 40 different pathogenic variants have been identified in LYST in humans, including nonsense and missense mutations, as well as insertions and deletions.
Feline CHS was first noted in a lineage of Persian cats, and as in mice, the disease showed recessive autosomal inheritance and the sentinel presentation was hypopigmentation19 (Fig. 1). The clinical characterization of the feline model for CHS examined neutrophil and platelet functions and auditory and ocular pigmentation abnormalities. Genetic complementation analysis after interspecific somatic cell (fibroblast) hybridization between human and domestic cat cell lines demonstrated a lack of paracrystal formation, indicating homology of the diseases due to similar genetic defects50. However, the exact causative mutation for CHS in the cat model was never determined.
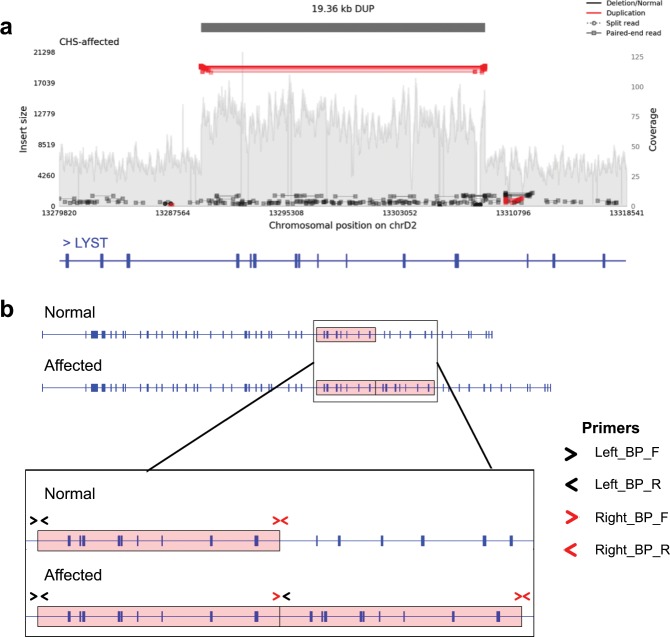
Figure 1.
Chediak-Higashi syndrome in cats at 5.5 months of age. A symptom of Chediak-Higashi syndrome, the affected cat (left) has a much lighter coat color (hypopigmentation) than its non-affected littermate (right). Although both cats are genetically aaB-C-D-E-I-ll, the genotype for a solid black smoke longhaired cat. The affected cat is also photophobic and has pale yellow-green irises, instead of the normal copper color irises of its littermate.
Long-term maintenance of cat heredity disease models, such as CHS, typically requires cats to be managed perpetually as living populations within university research colonies. Since many cat models originate from sporadic, de novo genetic mutations that were identified opportunistically as veterinary clinical cases, the demise of a specific model line may equate with its permanent extinction. However, semen and embryo cryopreservation, combined with artificial insemination or embryo transfer, can be used to preserve cat disease models, as commonly occurs with mouse research models51,52. For the CHS cat model, after nearly 20 years of research, the colonies at Colorado State and Washington State University could no longer be maintained and were lost to the research community. Fortunately, during the dissolution of the former CHS colony, an intact male (Smokey) was donated to the University of California, Davis. Smokey, a 16-year-old carrier for CHS, represented the only viable representative of the cat biomedical model for CHS. Therefore, the feline model for CHS provided an opportunity to apply newly advanced assisted reproductive techniques to resurrect a previously extinct feline disease model.
Here, semen from the viable CHS carrier of the original CHS colony, was successfully cryopreserved and used for artificial insemination (AI) to produce potential CHS carrier offspring. In addition, whole genome sequencing of fibroblast cell-lines derived from the original CHS cat colony helped identify a candidate causative mutation, a 20 kb tandem segmental duplication within LYST that spanned multiple exons. Finally, viable AI offspring were screened for the causative variant, demonstrating successful resurrection of a previously extinct feline model of a human disease and stability of the LYST mutation.
Materials and Methods
Cat management and sampling
All animal methods were carried out in accordance with PHS Policy on Humane Care and Use of Laboratory Animals, Animal Welfare Act Regulations, and Guide for the Care and Use of Laboratory Animals. Cat housing and husbandry, blood sample collection, assisted reproduction techniques and all other experimental protocols were approved under IACUC research protocols at the following institutions; the University of California- Davis (Protocol no. 16691), University of Missouri (MU) (Protocol no. 8787), and the Cincinnati Zoo & Botanical Garden (Protocol no. 05–064 and 11–102). Primary culture fibroblast cell lines of known carrier and affected cats from the CHS colony were thawed from LN2 storage. Using buccal swabs, DNA was collected from the male sperm donor, his viable offspring, and a random bred negative control. DNA was isolated by standard phenol - chloroform extractions methods53 or using DNeasy blood and tissue kit (Qiagen, Hilden, Germany)
Semen collection and cryopreservation
For semen collection, the CHS-carrier male domestic cat was anesthetized with a combination injection (i.m.) of ketamine (5 mg/kg) and medetomidine (0.04 mg/kg), with anesthetic reversal at the conclusion of the procedure using atipamezole (0.08 mg/kg; i.m.). A rectal probe (1.0 cm diameter, 3 longitudinal electrodes) attached to an electrostimulator (PTE Electronics, Boring, Oregon) was used to deliver three controlled sets of electrical stimuli (2–5 V range; 20 to 30 stimuli/set) as previously described54. Recovered semen from each set was assessed for volume, pH, and presence or absence of spermatozoa. Raw spermic samples were assessed for motility (percent progressively motile; rate of progressive motility (RPM) on scale of 1 to 5) and an aliquot (5 µl) was fixed for later morphological assessment. The remaining semen was diluted 1:1 in culture medium (Feline Optimized Culture Medium with Hepes [FOCMH]55) and maintained in microcentrifuge tubes in the dark at room temperature until completion of semen collection. Spermic samples from each set were combined and then centrifuged (300 g, 10 min). The resulting sperm pellet was resuspended in Test-Egg Yolk (TEY; Irvine Scientific, Santa Ana, CA) containing 0% glycerol to a concentration of 50 × 106 motile sperm/ml, suspended in a foam float in a 300 ml glass container (containing room temperature water), and placed into a refrigerator for 3.5 hours to cool to 5 °C. Cooled spermatozoa were diluted in three steps at 10 minute intervals with an equal volume of TEY (w/ 8% glycerol) to 25 × 106 motile sperm/ml, and aliquots (30 µl each) frozen by pelleting into indentations in dry ice56. Frozen sperm pellets were plunged into liquid nitrogen and transferred into labeled cryovials for ground shipment to the Cincinnati Zoo’s Center for Conservation and Research of Endangered Wildlife (CREW) for storage within liquid nitrogen tanks.
Ovarian synchronization and laparoscopic oviductal artificial insemination (LO-AI)
For ovarian synchronization and LO-AI, three female domestic cats (2–3 years of age) received oral progestin (altrenogest, Regumate®, Intervet/Merck Animal Health, Millsboro DE USA) daily mixed with moist cat food at a standard dosage (0.088 mg/kg body weight) for 39 consecutive days57. After four days of progestin withdrawal, females were treated with a combination regimen of equine chorionic gonadotropin (eCG; 100 IU; i.m.) followed 85 hours later with porcine luteinizing hormone (pLH; 1000 IU; i.m.), as previously described58. At 30–33 hours post-pLH, females were anesthetized and evaluated laparoscopically for the presence and number of mature follicles (>2 mm diameter) and corpora lutea. If mature pre-ovulatory follicles and/or freshly-formed corpora lutea were observed, frozen semen from the CHS-carrier male was thawed by depositing individual sperm pellets (8 pellets/LO-AI) into glass test tubes (1 pellet/tube) containing 100 µl of FOCMH for 30 sec in a 37 °C water bath. The thawed samples were combined and evaluated for sperm motility and concentration, as described above. One aliquot of a frozen-thawed sample was spread onto a microscope slide and later stained with FITC-PNA to assess acrosome status59. The combined sample then was centrifuged (600 g, 8 min), the supernatant removed and the concentrated sperm pellet resuspended in residual FOCMH to a small volume (18–20 µl) for insemination.
For LO-AI, an accessory trocar-cannula was placed into the right abdominal wall and grasping forceps used to secure and evert the ovarian bursa of the left ovary to visualize the oviductal ostium58. A polypropylene IV catheter (18 gauge; 32 mm in length) was inserted through the left abdominal wall perpendicular to the oviductal ostium and a blunted needle (22 gauge, 68 mm in length), containing 9–10 µl of concentrated sperm at its tip, was inserted through the catheter and directed through the oviductal ostium into the lumen of the oviductal ampulla. The sperm sample (9–10 µl) was deposited with slight air pressure delivered by an attached syringe as the insemination needle was slowly withdrawn from the oviduct. The LO-AI procedure was repeated with the right oviduct using a similar approach.
For pregnancy diagnosis, the females were assessed for fetal presence, number and viability via transabdominal ultrasound at 23 days post-AI, and pregnant females were reassessed at 41 and 59 days post AI to document fetal growth and viability. Pregnant females were allowed to carry offspring to term and monitored during natural parturition. If dystocia occurred during parturition, the pregnant queen was immediately anesthetized and the offspring were delivered by C-section.
Whole genome sequencing
Cell line DNA from an affected cat (~3 ug) was submitted for whole‐genome sequencing to the McDonnell Genome Institute at Washington University. A TruSeq PCR‐free library (Illumina, San Diego, CA) was constructed with 450 bp insert size. The library was indexed with nine libraries from other cat samples from the 99 Lives project (http://felinegenetics.missouri.edu/99lives)60 pooled together in equal molar ratios based on their concentrations determined by qPCR. The samples were applied to a HiSeq X flow cell to generate ~1 Tb of data as 2 × 150 bp paired‐end reads. Reads were mapped to Felis_catus_9.0 using Burrows-Wheeler Aligner (BWA-MEM) tool61. LYST gene coordinates in Felis_catus_9.0 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000181335.3/) were obtained from NCBI annotation release 104. The sequence reads aligned to the LYST gene were viewed using the integrative genomics viewer (IGV)62 and samplot (https://github.com/ryanlayer/samplot).
PCR validation and Sanger sequencing
Primers were designed using the reference sequence surrounding the 5′, internal, and 3′ breakpoints of the duplicated region (Supplementary Table S1) to validate the segmental duplication in the cell lines and cats produced by AI. The reference sequences surrounding the left and right breakpoints of the duplicated region and the SINEC_Fc reference sequence, which was obtained from Repbase63, were used for primer design for breakpoint validation. Primers Right_BP_F and Left_BP_R spanned the central breakpoint present in the mutant allele. Four primers, when used simultaneously in PCR amplification, were expected to yield two amplicons in homozygous wildtype individuals and three amplicons in individuals carrying at least one copy of the mutant allele. For genotyping by PCR, a reaction volume of 25 µL was prepared with 1 x PCR buffer (1.5 mM MgCl2) (Thomas Scientific, Swedesboro, NJ), 0.6 mM dNTPS (0.15 mM each nucleotide), 0.4 µM left_BP_F primer and right_BP_R1/2 primer, 0.8 µM left_BP_R primer and right_BP_F primer, 1.25 U Choice Taq polymerase (Thomas Scientific) and 25–50 ng template DNA. PCR products were amplified using an Applied Biosystems Veriti thermal cycler (Foster City, CA) with the following conditions: 94 °C for 3:00 min denaturation, followed by 35 cycles at 94 °C for 1:00 min, 58 °C for 1:00 min, and 72 °C for 1:00 min, which ended with 72 °C for 10:00 min and 4 °C hold. For electrophoresis, a 1.25% (w/v) agarose gel in 1x TAE buffer was prepared with samples separated at 70 V for 90 minutes. Each PCR amplicon in a known affected cell line and a random bred negative control was gel extracted and Sanger sequenced at the MU DNA Core on an ABI 3730XL (ABI, Foster City, CA) and assembled into reference-based contigs using Sequencher (GeneCodes, Ann Arbor, MI). Predicted amplicon sequences were extracted from Felis_catus_9.0 and used as the reference for contig assembly.
Results
Semen cryopreservation and laparoscopic oviductal artificial insemination
Electroejaculation of the CHS-carrier male cat recovered a moderate volume (165 µl) of seminal fluid containing highly concentrated spermatozoa (138 × 106 sperm/ml or 22.8 × 106 total sperm). In the raw ejaculate, 70% of spermatozoa were progressively motile with a rate of progressive motility (RPM) of 3.0 (on scale of 1 to 5), and 35% of sperm exhibited normal structural morphology. The major abnormalities included bent tails and bent mid-pieces (~30% of all sperm). Following semen processing and cooling, 32 sperm pellets, each containing ~0.75 × 106 motile sperm, were cryopreserved. Thawing of a single pellet at 10 months after cryopreservation revealed progressive sperm motility of 40%, indicating that each pellet should contain ~0.43 × 106 motile sperm post-thaw. Frozen semen was stored for seven years prior to use with LO-AI procedures.
At the time of laparoscopy, two of the three synchronized female cats exhibited multiple fresh corpora lutea on both ovaries, without the presence of any anovulatory follicles (Table 1). In contrast, the first female evaluated laparoscopically, at 30 hours post-pLH, possessed only mature peri-ovulatory follicles and no corpora lutea. All three females were inseminated bilaterally in both oviducts with frozen-thawed semen from a single ejaculate, with each female receiving a total of 2.2–3.3 million motile sperm (Table 1). Eight semen pellets were thawed for each AI procedure, with half of the concentrated sperm sample deposited into each oviduct. Immediately after thawing, sperm progressive motility for all samples was 40%, with an RPM of 2.5, and acrosome staining of one thawed sample revealed that 32% of sperm had intact acrosomal membranes. For all females, insemination of both oviducts was completed within 20 minutes of semen thawing. Following LO-AI of the anovulatory female (11WBG35), four mature follicles on the right ovary were manually ruptured with a catheter stylet to ensure ovulation and increase the opportunity for fertilization. Ultrasonography at 23 days post-AI determined that both ovulatory females conceived as shown by the presence of gestational sacs with viable fetuses, whereas the anovulatory female showed no evidence of implantations (Table 1). The two pregnant females progressed to term without complication, with fetal development and viability monitored via ultrasonography. One queen (12ODH5), a multiparous female, went into labor at 64 days post-AI, giving birth to five healthy kittens and one stillborn kitten before experiencing dystocia due to presumed uterine inertia. The remaining kittens were delivered via C-section, with 13 kittens born (nine viable, four stillborn). The second queen (12OTN2), a primiparous female, went into labor at 66 days post-AI, but experienced dystocia during parturition of the first kitten, and two viable kittens were delivered by C-section. Viable kittens averaged (+S.D.) 96.7 + 9.0 grams body weight at birth. Fostering of three kittens from the first queen to the second to reduce lactational stress, combined with initial supplemental feeding of kittens, resulted in all 11 viable kittens surviving through weaning. All kittens were transferred from the Cincinnati Zoo & Botanical Garden to MU at four months of age.
Table 1.
Ovarian responses and pregnancy results in domestic cats following fixed-time LO-AI with frozen-thawed semen from a Chediak Higashi Syndrome carrier male.
| Female ID | 11WBG35 | 12ODH5 | 12OTN2 |
|---|---|---|---|
| No. ovarian follicles | 14a | 0 | 0 |
| No. corpora lutea | 0 | 52 | 15 |
| No. motile sperm (x106) | 3.3 | 2.2 | 2.6 |
| Pregnancy (Y/N)b | N | Y | Y |
| Gestation length (days) | — | 64 | 66 |
| No. full-term kittens | — | 13 | 2 |
| No. viable kittensc | — | 9 | 2 |
aFour follicles were manually ruptured post-insemination.
bUltrasonography conducted at 23 days post-AI.
cFive kittens were born via vaginal delivery and six kittens were delivered via C-section.
Tandem segmental duplication in LYST is associated with CHS in cats
Whole genome sequencing of a fibroblast cell line derived from a homozygous affected cat from the original CHS colony, produced a mean coverage of 43× . LYST in cats is located at chrD2:13199206–13347218 and predicted to be comprised of 51 exons. Visual analysis of reads mapped to the LYST locus showed increased coverage across several exons that were spanned by discordant read pairs (Fig. 2a). Relative to the cat reference genome, these results were consistent with a tandem segmental duplication encompassing exons 30 through to 38 (NM_001290242.1:c.8347-2422_9548 + 1749dup) (NC_018733.3:g.13289500_13308861dup). In addition, a number of reads near the breakpoints had a read mate that mapped to a SINEC_Fc transposable elements elsewhere in the genome, suggesting the presence of a SINEC_Fc element between the duplicated copies.
Figure 2.
CHS in felines is associated with a 20 kb tandem segmental duplication in LYST. (a) Samplot output showing increased coverage across LYST exons 30 through 38 with reverse orientation read pairs colored in red spanning the duplicated region. (b) Schematic of NM_001290242.1:c.8347-2422_9548 + 1749dup in normal and affected individuals. Introns are depicted as a thin blue line and exons are depicted as thick blue lines. Duplicated region is highlighted in red and is ~20 kb in length. Zoomed in region shows primer pairs overlapping duplication breakpoints. Note, primers Right_BP_F and Left_BP_R produce an amplicon in affected/carrier samples that was absent in normal samples.
To validate the mutation, primers were designed to produce three distinct amplicons, the left and right breakpoints of the duplicated region and a central breakpoint only found in samples carrying the mutant allele. The primers spanning the left breakpoint produce a 462 bp amplicon, while the primers for the right breakpoint produce either a 244 bp or 343 bp amplicon depending on the primer combination used. The primers for the central breakpoint produce a 639 bp product, which contains a SINEC_Fc element (Fig. 2b, Supplementary Table S1) (GenBank id MN240554). PCR validation produced amplicons concordant with the affected status of a positive control fibroblast cell line and normal status of a random bred negative control (Supplementary Fig. S1). Assembled Sanger sequencing for each amplification product was consistent with predicted amplicon sequence. These results demonstrated the presence of the variant allele in a separate affected cat cell line and validate the existence of breakpoints consistent with NM_001290242.1:c.8347-2422_9548 + 1749dup.
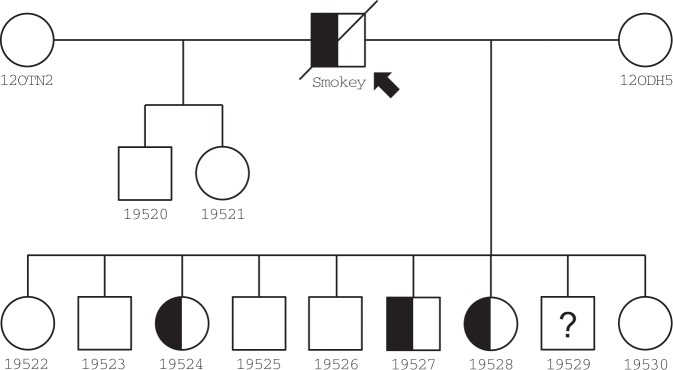
DNA from additional fibroblast cell lines derived from known carriers and affected individuals from the original colony and the offspring produced from AI were genotyped for the presence of NM_001290242.1:c.8347-2422_9548 + 1749dup. The variant allele was present in all fibroblast cell lines. For the 11 viable offspring produced from AI, two females and one male, all from the same litter, contained NM_001290242.1:c.8347-2422_9548 + 1749dup as likely carriers for CHS (Fig. 3, Supplementary Fig. S2). Therefore, these results are consistent with successful resurrection of a previously extinct disease model in cats.
Figure 3.
Pedigree of resurrected feline model of Chediak-Higashi syndrome via assisted reproduction. Half-filled symbols in pedigree represent predicted carrier status based on presence of the mutant allele from genotyping. The founder male Smokey (arrow) is heterozygous, as he did not present with CHS-associated symptoms throughout his life. The founder females, 12OTHN2 and 12ODH5, likely do not carry NM_001290242.1:c.8347-2422_9548 + 1749dup as these females were from a commercial breeding colony of cats. Displayed genotypes were determined using PCR screening, except for 19529, which did not produce any amplicons and whose genotype remains unknown. Lab ID (Fcat) numbers are presented under each individual in the pedigree.
Discussion
Chediak-Higashi Syndrome is a well-characterized lysosomal disease caused by mutations in lysosomal trafficking regulator (LYST). The classic diagnostic feature of the syndrome is enlarged lysosome-related organelles (LROs) in many cell types, including lysosomes, melanosomes and cytolytic granules. Other diseases associated with the regulation of LRO size and/or vesicle trafficking, such as asthma, urticaria and Leishmania amazonensis infections, could benefit from understanding the molecular function of LYST and identification of its interacting partners may provide therapeutic targets64. Unfortunately, the cat model for CHS was lost to the research community before the causative mutation was discovered.
CHS in the domestic cat was first observed in 1977 in a family of Persian cats, clearly demonstrating an autosomal recessive disease with similar presentation to the human condition19,21,22. Complementation studies suggested the human and cat conditions were likely allelic50. After the CHS colonies at Colorado State and Washington State University were dissolved, years later, the semen donor, Smokey, the last known living representative of this feline model for CHS, was found residing in a University of California, Davis cat colony. However, because of his advanced age and lack of libido, he was unable to naturally breed females. Electroejaculation showed that his testes continued to produce moderate quantities of viable spermatozoa with sufficient progressive motility. Although his sperm morphology was later classified as teratospermic with just 35% normal spermatozoa65, freezing and storage of this ejaculate ensured his genetic potential was preserved, provided the frozen-thawed spermatozoa was capable of fertilizing oocytes in vitro or in vivo. The recent development of LO-AI in cats, combined with the incorporation of oral progestin treatment for precise ovarian synchronization, provided investigators with an efficient strategy for generating viable offspring using this limited genetic resource58,66. As shown by the birth of multiple kittens following LO-AI, post-thaw sperm functionality was adequate to achieve the primary goal of model regeneration, requiring just two to three million motile sperm per procedure. Despite the limited amount of frozen semen, the suboptimal sperm morphology and the relatively low post-thaw sperm motility and acrosomal status, fixed-time LO-AI still proved adequate for model propagation with two of three females conceiving and the delivery of eleven viable kittens. The occurrence of dystocia was likely due to the exceptionally large litter size (13 kittens) and associated prolonged labor in one queen, a multiparous female who had previously given birth to six viable kittens via vaginal delivery during a previous LO-AI pregnancy. The second pregnant female was primiparous and possibly experienced obstructive dystocia due to a relatively narrow birth canal and slightly larger kittens (102–108 grams body weight). Dystocia reportedly occurs in three to six percent of pregnancies in the general cat population67. Generally, pregnancies produced by LO-AI in the Cincinnati Zoo & Botanical Garden’s cat research colony do not appear more prone to dystocia than pregnancies resulting from natural breeding66.
For biomedical research and drug development, the current standard is to use rodent models68. However, 95% of drugs fail at human trials, leading to costs of nearly $2 billion and 10 years of time per FDA drug approval69,70. Large animals, including cats, dogs and pigs, support drug trials, provide additional evidence for deciding to abandon drug development or proceed to human trials. This work provides a “proof-of-principle” demonstrating that cryopreservation and reproductive technologies are now well established in domestic cats, which may lead to the development of genome-edited cats as models for specific diseases. Importantly, future applications of genome-editing will likely provide the best animal model for a given disease and drug development trial, ultimately improving the efficiency and cost effectiveness of the drug development process. This model of feline CHS was selected because no other living animals were carrying the disease at the time the LO-AI procedures were performed. Hence, these ART procedures did not support animal production that could have otherwise been achieved through normal mating. Investigators interested in CHS should now realize the cat model may be available for research. In addition, study results reinforce the need for more routine cryobanking of valuable cat models, since only one CHS carrier cat had cryopreserved gametes available for model rederivation.
CHS in the feline model is likely caused by the identified 20 kb tandem segmental duplication that spans LYST exons 30 through 38 (NM_001290242.1:c.8347-2422_9548 + 1749dup). Discovery of the candidate causative mutation had been hampered by the availability of appropriate feline genomic resources and the complexity of the mutation itself, which required the use of WGS of an affected cat sample to efficiently resolve. A potential mechanism for the mutation is non-allelic homologous recombination, which is known to lead to either deletions or duplications and is implicated in a variety of human genomic disorders71. Examples include Charcot-Marie-Tooth disease type 1 A and Hereditary Neuropathy with Liability to Pressure Palsies72,73. Accumulation of high copy repeated DNA sequences, such as SINEC_Fc elements, around a single locus increase a region’s susceptibility to non-allelic homologous recombination.
One of the limitations of the current genetic analysis is the zygosity of the mutant allele was not determined. Since many molecular techniques require unique priming sites, determining copy number of low copy large tandem repeated sequences is challenging. Also due to the complexity of the mutation, the analyses performed were not able to determine if the haploid copy number of the duplicated region was greater than two copies. Regardless, the NM_001290242.1:c.8347-2422_9548 + 1749dup was amplified in the AI offspring, showing it was successfully inherited. Consequently, more sophisticated techniques for determining the total copy number of the duplicated region, such as RT-PCR, were not required for this analysis.
In this study, two of the primary benefits of assisted reproduction for management of cat hereditary disease models were demonstrated, specifically, 1) the propagation of individuals incapable of natural breeding due to physiological or behavioral incompatibilities, and, 2) the production of offspring following cryopreservation and long-term storage of frozen semen. Furthermore, a candidate causative mutation for the cat model of CHS, NM_001290242.1:c.8347-2422_9548 + 1749dup, was discovered in LYST and used to screen AI offspring to show the feline model for CHS could be successfully resurrected. The allele was successfully inherited from cryopreserved gametes using AI and produced three carrier kittens, allowing for the once extinct feline model for CHS to be fully resurrected. This study supports the use of cryopreservation for long-term maintenance of future feline biomedical models of genetic disease.
Supplementary information
Acknowledgements
Funding for this project was provided in part by the Office of Research Infrastructure Programs/OD R24OD01092, the Winn Feline Foundation (W16-030), University of Missouri Gilbreath McLorn Endowment and donors to the 99 Lives Cat Genome Sequencing Project. We appreciate the laboratory assistance of Thomas R. Juba, MS.
Author contributions
L.A.L. and W.F.S. conceived of the idea planned the experiments. Experimental supplies, samples and/or data was provided by L.A.L., D.J.P., M.D.K. and W.F.S. Laboratory experiments were performed by B.G. and R.A.G. Reproductive studies planned and conducted by W.F.S., J.R.H., J.N. and H.L.B. Data analysis was performed by R.M.B., L.A.L. First draft of manuscript written by RMB, L.A.L. and W.F.S. All authors reviewed and approved the manuscript.
Data availability
WGS data can be accessed from the Sequence Read Archive (SRA) under the BioProject accession PRJNA557464.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56896-9.
References
- 1.Ajitkumar, A. & Ramphul, K. In StatPearls. [Internet] (StatPearls Publishing, 2018).
- 2.Chediak MM. [New leukocyte anomaly of constitutional and familial character] Revue d’hematologie. 1952;7:362–367. [PubMed] [Google Scholar]
- 3.Higashi O. Congenital abnormity of peroxidase granules; a case of congenital gigantism of peroxidase granules, preliminary report. The Tohoku journal of experimental medicine. 1953;58:246. doi: 10.1620/tjem.58.246. [DOI] [PubMed] [Google Scholar]
- 4.Higashi O. Congenital gigantism of peroxidase granules; the first case ever reported of qualitative abnormity of peroxidase. The Tohoku journal of experimental medicine. 1954;59:315–332. doi: 10.1620/tjem.59.315. [DOI] [PubMed] [Google Scholar]
- 5.Steinbrink W. Uber eine neue granulations-anomolie der leukocyten. Deutshe Arch. Klin. Med. 1948;193:577–581. [Google Scholar]
- 6.Béguez-César A. Neutropenia cronica maligna familiar con granulaciones atipicas de los leucocitos. Boletin de la Sociedad Cubana. Pediatrics. 1943;15:900–922. [Google Scholar]
- 7.Bell TG, et al. Decreased nucleotide and serotonin storage associated with defective function in Chediak-Higashi syndrome cattle and human platelets. Blood. 1976;48:175–184. doi: 10.1182/blood.V48.2.175.175. [DOI] [PubMed] [Google Scholar]
- 8.Blume RS, Padgett GA, Wolff SM, Bennett JM. Giant neutrophil granules in the Chediak-Higashi syndrome of man, mink, cattle and mice. Canadian journal of comparative medicine. Revue canadienne de medecine comparee. 1969;33:271–274. [PMC free article] [PubMed] [Google Scholar]
- 9.Yamakuchi H, et al. Chediak-Higashi syndrome mutation and genetic testing in Japanese black cattle (Wagyu) Animal genetics. 2000;31:13–19. doi: 10.1046/j.1365-2052.2000.00586.x. [DOI] [PubMed] [Google Scholar]
- 10.Padgett GA, Leader RW, Gorham JR, O’Mary CC. The Familial Occurrence of the Chediak-Higashi Syndrome in Mink and Cattle. Genetics. 1964;49:505–512. doi: 10.1093/genetics/49.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen WG. The Chediak-Higashi syndrome and Aleutian mink. Archives of dermatology. 1967;96:330–331. doi: 10.1001/archderm.1967.01610030108018. [DOI] [PubMed] [Google Scholar]
- 12.Hirano A, Zimmerman HM, Levine S, Padgett GA. Cytoplasmic inclusions in Chediak-Higashi and Wobbler mink. An electron microscopic study of the nervous system. Journal of neuropathology and experimental neurology. 1971;30:470–487. doi: 10.1097/00005072-197107000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Meyers KM, Holmsen H, Seachord CL, Hopkins G, Gorham J. Characterization of platelets from normal mink and mink with the Chediak-Higashi syndrome. American journal of hematology. 1979;7:137–146. doi: 10.1002/ajh.2830070206. [DOI] [PubMed] [Google Scholar]
- 14.Collier LL, Prieur DJ, King EJ. Ocular melanin pigmentation anomalies in cats, cattle, mink, and mice with Chediak-Higashi syndrome: histologic observations. Current eye research. 1984;3:1241–1251. doi: 10.3109/02713688409000828. [DOI] [PubMed] [Google Scholar]
- 15.Ridgway SH. Reported causes of death of captive killer whales (Orcinus orca) Journal of wildlife diseases. 1979;15:99–104. doi: 10.7589/0090-3558-15.1.99. [DOI] [PubMed] [Google Scholar]
- 16.Taylor, R. F. & Farrell, R. K. Light and electron microscopy of peripheral blood neutrophils in a killer whale affected with Chediak-Higashi syndrome. Federation proceedings, 32 (1973).
- 17.Sjaastad OV, Blom AK, Stormorken H, Nes N. Adenine nucleotides, serotonin, and aggregation properties of platelets of blue foxes (Alopex lagopus) with the Chediak-Higashi syndrome. American journal of medical genetics. 1990;35:373–378. doi: 10.1002/ajmg.1320350312. [DOI] [PubMed] [Google Scholar]
- 18.Fagerland JA, Hagemoser WA, Ireland WP. Ultrastructure and stereology of leukocytes and platelets of normal foxes and a fox with a Chediak-Higashi-like syndrome. Veterinary pathology. 1987;24:164–169. doi: 10.1177/030098588702400210. [DOI] [PubMed] [Google Scholar]
- 19.Kramer JW, Davis WC, Prieur DJ. The Chediak-Higashi syndrome of cats. Laboratory investigation; a journal of technical methods and pathology. 1977;36:554–562. [PubMed] [Google Scholar]
- 20.Parker MT, Collier LL, Kier AB, Johnson GS. Oral Mucosa Bleeding Times of Normal Cats and Cats with Chediak-Higashi Syndrome or Hageman Trait (Factor XII Deficiency) Veterinary clinical pathology / American Society for Veterinary Clinical Pathology. 1988;17:9–12. doi: 10.1111/j.1939-165X.1988.tb00479.x. [DOI] [PubMed] [Google Scholar]
- 21.Prieur DJ, Collier LL. Inheritance of the Chediak-Higashi syndrome in cats. The Journal of heredity. 1981;72:175–177. doi: 10.1093/oxfordjournals.jhered.a109467. [DOI] [PubMed] [Google Scholar]
- 22.Prieur DJ, Collier LL. Neutropenia in cats with the Chediak-Higashi syndrome. Canadian journal of veterinary research = Revue canadienne de recherche veterinaire. 1987;51:407–408. [PMC free article] [PubMed] [Google Scholar]
- 23.Wardrop KJ, Dhein CR, Prieur DJ, Meyers KM. Evaluation of hepatic and renal function in cats with chediak-higashi syndrome. Veterinary clinical pathology / American Society for. Veterinary Clinical Pathology. 1987;16:40–44. doi: 10.1111/j.1939-165X.1987.tb00459.x. [DOI] [PubMed] [Google Scholar]
- 24.Masliah-Planchon J, Darnige L, Bellucci S. Molecular determinants of platelet delta storage pool deficiencies: an update. British journal of haematology. 2013;160:5–11. doi: 10.1111/bjh.12064. [DOI] [PubMed] [Google Scholar]
- 25.Nurden A, Nurden P. Advances in our understanding of the molecular basis of disorders of platelet function. Journal of thrombosis and haemostasis: JTH. 2011;9(Suppl 1):76–91. doi: 10.1111/j.1538-7836.2011.04274.x. [DOI] [PubMed] [Google Scholar]
- 26.Nurden AT, Freson K, Seligsohn U. Inherited platelet disorders. Haemophilia: the official journal of the World Federation of Hemophilia. 2012;18(Suppl 4):154–160. doi: 10.1111/j.1365-2516.2012.02856.x. [DOI] [PubMed] [Google Scholar]
- 27.Sieni E, et al. Familial hemophagocytic lymphohistiocytosis: a model for understanding the human machinery of cellular cytotoxicity. Cellular and molecular life sciences: CMLS. 2012;69:29–40. doi: 10.1007/s00018-011-0835-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White JG, Hess RA, Gahl WA, Introne W. Rapid ultrastructural detection of success or failure after bone marrow transplantation in the Chediak-Higashi syndrome. Platelets. 2013;24:71–74. doi: 10.3109/09537104.2011.654293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rihani R, et al. Unrelated cord blood transplantation can restore hematologic and immunologic functions in patients with Chediak-Higashi syndrome. Pediatric transplantation. 2012;16:E99–E105. doi: 10.1111/j.1399-3046.2010.01461.x. [DOI] [PubMed] [Google Scholar]
- 30.Eapen M, et al. Hematopoietic cell transplantation for Chediak-Higashi syndrome. Bone marrow transplantation. 2007;39:411–415. doi: 10.1038/sj.bmt.1705600. [DOI] [PubMed] [Google Scholar]
- 31.Kazmierowski JA, Elin RJ, Reynolds HY, Durbin WA, Wolff SM. Chediak-Higashi syndrome: reversal of increased susceptibility to infection by bone marrow transplantation. Blood. 1976;47:555–559. doi: 10.1182/blood.V47.4.555.555. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura M, et al. Beige rat: a new animal model of Chediak-Higashi syndrome. Blood. 1989;74:270–273. doi: 10.1182/blood.V74.1.270.270. [DOI] [PubMed] [Google Scholar]
- 33.Bennett JM, Blume RS, Wolff SM. Characterization and significance of abnormal leukocyte granules in the beige mouse: a possible homologue for Chediak-Higashi Aleutian trait. The Journal of laboratory and clinical medicine. 1969;73:235–243. [PubMed] [Google Scholar]
- 34.Renshaw HW, Davis WC. Growth characteristics of bone marrow cells from beige mutant, the mouse homologue of the Chediak-Higashi syndrome of man, propagated in semisolid agar cultures. In vitro. 1975;11:5–13. doi: 10.1007/BF02615316. [DOI] [PubMed] [Google Scholar]
- 35.Brandt EJ, Swank RT. The Chediak-Higashi (beige) mutation in two mouse strains. Allelism and similarity in lysosomal dysfunction. The American journal of pathology. 1976;82:573–588. [PMC free article] [PubMed] [Google Scholar]
- 36.Masui N, et al. The rat lysosomal trafficking regulator (Lyst) gene is mapped on the telomeric region of chromosome 17. Experimental animals / Japanese Association for Laboratory Animal Science. 2003;52:89–91. doi: 10.1538/expanim.52.89. [DOI] [PubMed] [Google Scholar]
- 37.Barbosa MD, et al. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature. 1996;382:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagle DL, et al. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nature genetics. 1996;14:307–311. doi: 10.1038/ng1196-307. [DOI] [PubMed] [Google Scholar]
- 39.Weisfeld-Adams JD, et al. Atypical Chediak-Higashi syndrome with attenuated phenotype: three adult siblings homozygous for a novel LYST deletion and with neurodegenerative disease. Orphanet journal of rare diseases. 2013;8:46. doi: 10.1186/1750-1172-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaya Z, et al. A novel single point mutation of the LYST gene in two siblings with different phenotypic features of Chediak Higashi syndrome. Pediatric blood & cancer. 2011;56:1136–1139. doi: 10.1002/pbc.22878. [DOI] [PubMed] [Google Scholar]
- 41.Zarzour W, et al. Two novel CHS1 (LYST) mutations: clinical correlations in an infant with Chediak-Higashi syndrome. Molecular genetics and metabolism. 2005;85:125–132. doi: 10.1016/j.ymgme.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Certain S, et al. Protein truncation test of LYST reveals heterogenous mutations in patients with Chediak-Higashi syndrome. Blood. 2000;95:979–983. [PubMed] [Google Scholar]
- 43.Kunieda T, Nakagiri M, Takami M, Ide H, Ogawa H. Cloning of bovine LYST gene and identification of a missense mutation associated with Chediak-Higashi syndrome of cattle. Mammalian genome: official journal of the International Mammalian Genome. Society. 1999;10:1146–1149. doi: 10.1007/s003359901181. [DOI] [PubMed] [Google Scholar]
- 44.Kato K, et al. Genetic deletion of mouse platelet glycoprotein Ibbeta produces a Bernard-Soulier phenotype with increased alpha-granule size. Blood. 2004;104:2339–2344. doi: 10.1182/blood-2004-03-1127. [DOI] [PubMed] [Google Scholar]
- 45.Spritz RA. Genetic defects in Chediak-Higashi syndrome and the beige mouse. Journal of clinical immunology. 1998;18:97–105. doi: 10.1023/A:1023247215374. [DOI] [PubMed] [Google Scholar]
- 46.Masui N, et al. An allele-specific genotyping method for rat lyst (lysosomal trafficking regulator) gene. Experimental animals / Japanese Association for Laboratory Animal Science. 2004;53:77–80. doi: 10.1538/expanim.53.77. [DOI] [PubMed] [Google Scholar]
- 47.Mori M, et al. A new beige mutant rat ACI/N-Lystbg-Kyo. Experimental animals / Japanese Association for Laboratory Animal Science. 2003;52:31–36. doi: 10.1538/expanim.52.31. [DOI] [PubMed] [Google Scholar]
- 48.Anistoroaei R, Krogh AK, Christensen K. A frameshift mutation in the LYST gene is responsible for the Aleutian color and the associated Chediak-Higashi syndrome in American mink. Animal genetics. 2013;44:178–183. doi: 10.1111/j.1365-2052.2012.02391.x. [DOI] [PubMed] [Google Scholar]
- 49.Landrum MJ, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penner JD, Prieur DJ. Interspecific genetic complementation analysis with fibroblasts from humans and four species of animals with Chediak-Higashi syndrome. American journal of medical genetics. 1987;28:455–470. doi: 10.1002/ajmg.1320280223. [DOI] [PubMed] [Google Scholar]
- 51.Mazur P, Leibo S, Seidel GE., Jr Cryopreservation of the germplasm of animals used in biological and medical research: importance, impact, status, and future directions. Biology of reproduction. 2008;78:2–12. doi: 10.1095/biolreprod.107.064113. [DOI] [PubMed] [Google Scholar]
- 52.Swanson WF. Research in nondomestic species: experiences in reproductive physiology research for conservation of endangered felids. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2003;44:307–316. doi: 10.1093/ilar.44.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green, M. R. & Sambrook, J. Molecular cloning. A Laboratory Manual 4th (2012).
- 54.Howard JG, Brown JL, Bush M, Wildt DE. Teratospermic and normospermic domestic cats: ejaculate traits, pituitary-gonadal hormones, and improvement of spermatozoal motility and morphology after swim-up processing. Journal of andrology. 1990;11:204–215. [PubMed] [Google Scholar]
- 55.Herrick JR, et al. Toward a feline-optimized culture medium: impact of ions, carbohydrates, essential amino acids, vitamins, and serum on development and metabolism of in vitro fertilization-derived feline embryos relative to embryos grown in vivo. Biology of reproduction. 2007;76:858–870. doi: 10.1095/biolreprod.106.058065. [DOI] [PubMed] [Google Scholar]
- 56.Herrick JR, et al. In vitro fertilization and sperm cryopreservation in the black-footed cat (Felis nigripes) and sand cat (Felis margarita) Biology of reproduction. 2010;82:552–562. doi: 10.1095/biolreprod.109.081034. [DOI] [PubMed] [Google Scholar]
- 57.Stewart RA, et al. Oral progestin priming increases ovarian sensitivity to gonadotropin stimulation and improves luteal function in the cat. Biology of reproduction. 2012;87:137. doi: 10.1095/biolreprod.112.104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conforti VA, et al. Laparoscopic oviductal artificial insemination improves pregnancy success in exogenous gonadotropin-treated domestic cats as a model for endangered felids. Biology of reproduction. 2013;89:4. doi: 10.1095/biolreprod.112.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vick MM, Bateman HL, Lambo CA, Swanson WF. Improved cryopreservation of domestic cat sperm in a chemically defined medium. Theriogenology. 2012;78:2120–2128. doi: 10.1016/j.theriogenology.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Mauler D, et al. Precision medicine in cats: Novel Niemann‐Pick type C1 diagnosed by whole‐genome sequencing. Journal of veterinary internal medicine. 2017;31:539–544. doi: 10.1111/jvim.14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 (2013).
- 62.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji, X., Chang, B., Naggert, J. K. & Nishina, P. M. In Retinal Degenerative Diseases 745–750 (Springer, 2016).
- 65.Pukazhenthi BS, Wildt DE, Howard JG. The phenomenon and significance of teratospermia in felids. Journal of reproduction and fertility. Supplement. 2001;57:423–433. [PubMed] [Google Scholar]
- 66.Swanson WF. Practical application of laparoscopic oviductal artificial insemination for the propagation of domestic cats and wild felids. Reproduction, Fertility and Development. 2019;31:27–39. doi: 10.1071/RD18350. [DOI] [PubMed] [Google Scholar]
- 67.Johnston, S. D., Root Kustritz, M. V. & Olson, P. S. Canine and feline theriogenology. (2001).
- 68.Ericsson AC, Crim MJ, Franklin CL. A brief history of animal modeling. Missouri medicine. 2013;110:201. [PMC free article] [PubMed] [Google Scholar]
- 69.Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. Jama. 2006;296:1727–1732. doi: 10.1001/jama.296.14.1731. [DOI] [PubMed] [Google Scholar]
- 70.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. Journal of health economics. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 71.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raedt TD, et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nature genetics. 2006;38:1419–1423. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- 73.Lindsay SJ, Khajavi M, Lupski JR, Hurles ME. A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am J Hum Genet. 2006;79:890–902. doi: 10.1086/508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS data can be accessed from the Sequence Read Archive (SRA) under the BioProject accession PRJNA557464.