Abstract
Long non-coding RNAs (lncRNAs) constitute a significant fraction of the transcriptome, playing important roles in development and disease. However, our understanding of structure-function relationships for this emerging class of RNAs has been limited to secondary structures. Here, we report the 3-D atomistic structural study of epigenetic lncRNA, Braveheart (Bvht), and its complex with CNBP (Cellular Nucleic acid Binding Protein). Using small angle X-ray scattering (SAXS), we elucidate the ensemble of Bvht RNA conformations in solution, revealing that Bvht lncRNA has a well-defined, albeit flexible 3-D structure that is remodeled upon CNBP binding. Our study suggests that CNBP binding requires multiple domains of Bvht and the RHT/AGIL RNA motif. We show that RHT/AGIL, previously shown to interact with CNBP, contains a highly flexible loop surrounded by more ordered helices. As one of the largest RNA-only 3-D studies, the work lays the foundation for future structural studies of lncRNA-protein complexes.
Subject terms: Biophysical chemistry, RNA, Biophysics, Computational biophysics, Molecular biophysics
Many RNA systems possess highly ordered 3-D structures that are essential to their function. Here the authors demonstrate that the long non-coding RNA Braveheart possesses a flexible but defined 3-D structure which is remodeled upon binding the protein CNBP.
Introduction
Long non-coding RNAs (lncRNAs) are emerging as key players in a variety of biological processes, including gene expression, genetic imprinting, histone modification, and chromatin dynamics1. To perform these crucial functions, they interact with proteins, DNA and other RNAs. An understanding of lncRNA-ligand interaction using biochemical and biophysical methods is essential to elucidate the mechanism by which the lncRNAs execute their functions2. However, there are a number of challenges associated with biophysical studies of lncRNAs. Despite the impressive number of annotated transcripts2, only a small number of lncRNAs (e.g., Braveheart3, COOLAIR4, HOTAIR5, ROX16, ROX26, SRA7, XIST repeat A8,9, TancRNA10, and 3′ end of human, zebrafish, and lizard MALAT110) have undergone secondary structure analysis via chemical probing (e.g., selective 2′ hydroxyl acylation analyzed by primer extension, i.e., SHAPE or dimethyl sulfate, i.e., DMS) or low-resolution enzymatic probing2. Akin to early studies of the ribosome, these secondary structures provide the framework for understanding structure–function relationships, producing valuable information like local modularity; however, chemical probing studies yield little information about the overall 3-dimensional (3-D) structure11. As their name suggests, long noncoding RNAs are often large, making their preparation and purification for in vitro studies very challenging. In addition, longer lengths make fold determination more challenging. The lncRNAs are also typically much less abundant than messenger RNAs. For example, only a few copies per cell of immune-gene priming lncRNAs are expressed12. Moreover, lncRNAs usually have a very short half-life (less than 9–12 h, with the exception of MALAT113) making in vivo studies challenging as well. Finally, the large RNA molecules often have flexible regions that further pose challenges for RNA structure determination14.
Although 3-D structures are often essential to establish structure–function relationships, no such studies have been performed for intact, epigenetic long noncoding RNAs, to our knowledge. In fact, a widely held perspective in the RNA community is that lncRNAs tend to be too flexible and unstable for nuclear magnetic resonance (NMR) and crystallization studies. Interestingly, many biologically important RNAs have dynamically changing conformations, making structure determination challenging2,15. However, it has been shown, using chemical probing, that several lncRNAs and portions of lncRNAs adopt well-organized, modular secondary structures3–5,7,9. Furthermore, because SHAPE probing reports on the physical mobility of the backbone for each nucleotide, the many regions of low-SHAPE reactivity in these lncRNA systems demonstrate that these RNAs possess regions with well-defined secondary and possible tertiary interactions16. In addition, when we performed SHAPE and DMS probing of Bvht, reactivities were fairly similar regardless of the probing method3. Likewise, we obtained similar reactivities of probing for the 3′ end of MALAT1, whether we used SHAPE or DMS10. When we probed the steroid receptor RNA activator (SRA), derived secondary structures were fairly similar to each other whether we used SHAPE, DMS, in-line or RNase V17. For COOLAIR, reactivities from both SHAPE and CMCT (1-cyclohexyl-3-(2-morpholi-noethyl) carbodiimide metho-p-toluene sulfonate) are similar to each other4.
These probing data indicate that at least these particular lncRNAs do possess well-organized secondary structures. If these lncRNAs were intrinsically disordered, probing data would have yielded a superposition of all potential structures with prominent protection patterns. The Pyle group also confirmed this phenomenon. For example, DMS and terbium reactivities showed 92–93% agreement with SHAPE data for HOTAIR5. DMS and SHAPE showed similar reactivities for RepA as well9. Building on chemical probing data, the next step is to investigate the 3-D structure of these lncRNAs. One of the well-known lncRNAs, Braveheart (Bvht), binds to cellular nucleic acid binding protein (CNBP) and the SUZ12 component of the PRC2 complex altering chromatin modification17, affecting the expression of many genes that are important for cardiovascular lineage commitment, such as MesP1, GATA4, HAND1, HAND2, NKX2.5, and TBX52. While its secondary structure has been studied3, the 3-D structure of Bvht is unknown.
Regarding 3-D methods in structure determination, high-resolution structure determination methods have several challenges with long RNA molecules and complexes with their interacting partners14. For example, while many excellent NMR studies of biomolecules have been performed18–22, this method typically has been limited to proteins smaller than 50 kDa23 and RNAs smaller than 100–300 nucleotides24. In addition, crystallographic studies of RNA molecules are typically more challenging than DNA and proteins. In particular, as we show in this study, the full-length Bvht lncRNA has a well-defined 3-D structure, but has, at the same time, flexible regions, making it very challenging to trap the molecule in a single conformation, as is required for X-ray crystallography. Of course, there are many X-ray crystallography-based RNA structures. However, except for the ribosome, group I intron and group II intron25–27, these RNAs are relatively small and highly ordered.
On the other hand, small angle X-ray scattering (SAXS) is an excellent alternative method that allows structural studies of fully or partially unfolded proteins and RNAs without being limited by molecular mass14. Therefore, solution scattering is often employed for systems that do not readily crystallize28. SAXS can access large dynamic motions as well. There are several RNA29 and RNA–protein complex structures that have been determined by SAXS14.
To address the above-mentioned need for in-depth structural studies of Bvht, we performed extensive biophysical experiments and computational studies to investigate the three-dimensional structure, primarily based on SAXS. We note that many excellent modeling pipelines have been used for 3-D RNA structure determination30,31. Below, we present a modeling pipeline which is particularly useful for SAXS studies of very large RNA systems. Using this modeling pipeline, we are able to show the 3-D structure of a full lncRNA, Bvht. We note that a partial structure of a lncRNA was reported, e.g., 65 nucleotide long MALAT1 ENE (expression and nuclear retention element) and A-rich tract32. In addition, we show the 3-D structure of Bvht-CNBP complexes. CNBP, a zinc-finger transcription factor, binds Bvht and is important for heart cell lineage differentiation3. This structural study of a lncRNA–protein complex will be informative for further studies, such as the dynamic association between RNA and RNA-binding proteins, a process important in many aspects of the life-cycle of lncRNAs, including their processing, modification, stability, and localization33.
Results
Effect of Mg2+ on the solution structure of Bvht
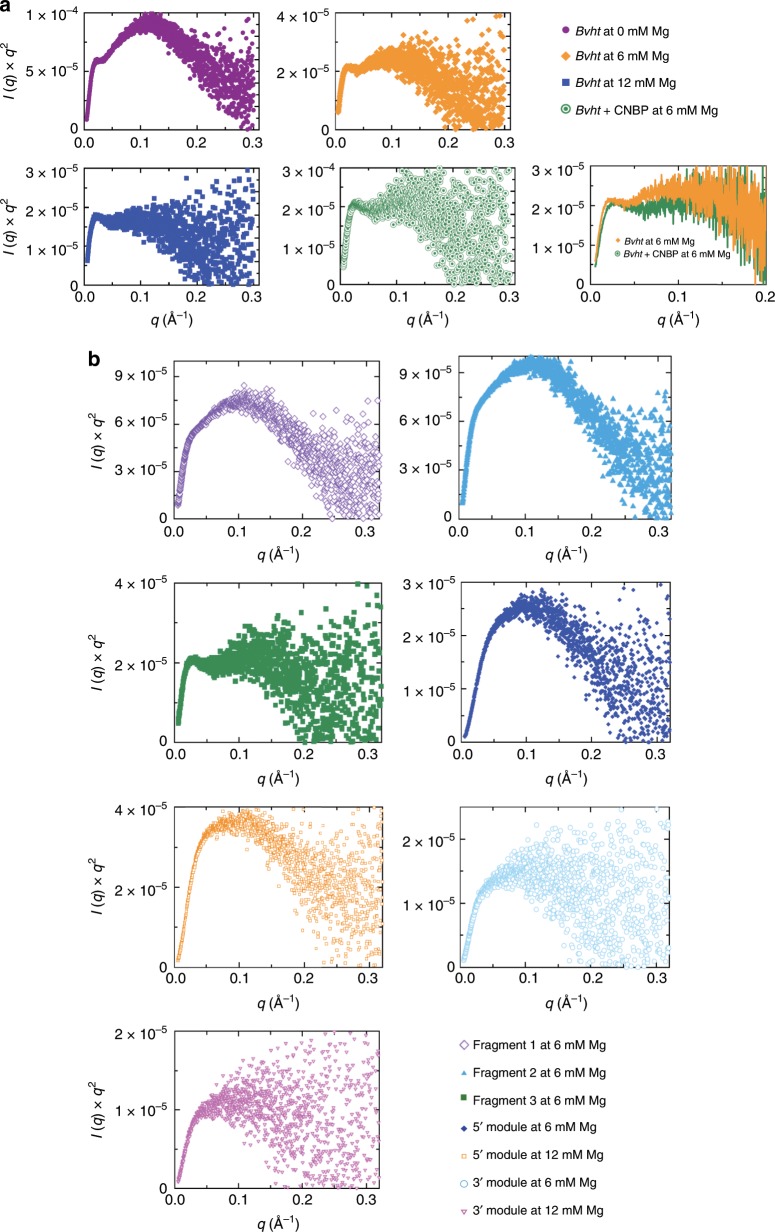
Salt-dependent association can be critical for biological function34. In RNA polymers, divalent magnesium (Mg2+) is essential for folding, higher-order interactions and function35. As such, sensitivity to Mg2+ is an indicator of the presence of tertiary interactions. In fact, due to its small ionic radius, Mg2+ has the highest charge density from all ions in cells36 and is more influential than potassium (K+) for RNA conformation37. Therefore, we determined the effect of Mg2+ concentration on the 3-D solution conformation of Bvht. We collected SAXS data using a size exclusion chromatography device connected in-line with the SAXS instrument (to separate any aggregated/degraded RNA material) for Bvht in 0, 6, and 12 mM MgCl2. We selected data from a monodispersed SEC-SAXS peak and merged them as discussed below (Methods section). The merged SEC-SAXS data along with EMSA data are presented in Fig. 1. The buffer-subtracted merged data were then first analyzed by the Guinier method (plot of (I(q)) vs. (q2)), which allows detection of homogeneity and determination of the radius of gyration (Rg) based on the data from the low angle region38. The Guinier plots presented in Supplementary Fig. 1 display linearity for small q values, suggesting that Bvht samples are aggregation free. Next, we performed Kratky analysis (plot of I(q)q2 vs. q) of SAXS data that allows examination of the folding state of biomolecules39. For example, globular biomolecules will display a bell-shaped distribution. The Kratky plots for Bvht samples under investigation (Fig. 2) suggest that the samples are folded.
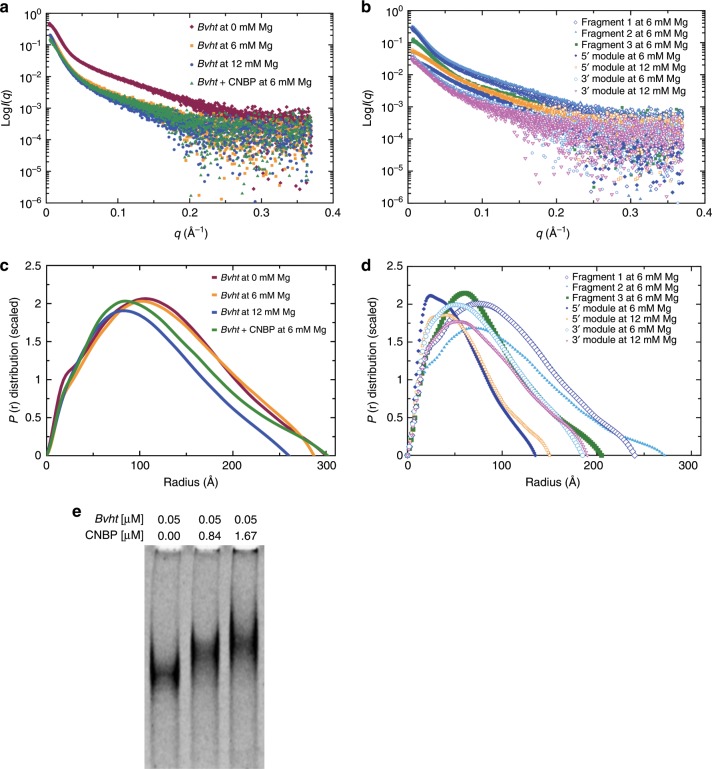
Fig. 1. SAXS and EMSA data for lncRNA Bvht.
a Scattering intensity vs. scattering angle (q = 4πsinθ/λ), indicating the dependence on Mg2+ concentration and effect of Bvht-CNBP complex formation. b Scattering intensity vs. scattering angle for isolated subregions of Bvht lncRNA. c Pair-distance distribution function for full-length Bvht at various Mg2+ concentrations and for Bvht-CNBP complex. d Pair-distance distribution function for subregions of Bvht. e EMSA for full-length Bvht with increasing CNBP concentrations.
Fig. 2. Kratky plots of Bvht.
a Data of the full-length Bvht at various Mg2+ concentrations and of the Bvht-CNBP complex show that overall, all RNA molecules are folded. Upon binding with CNBP, full-length Bvht undergoes a conformation change (bottom right). b Data of Bvht subregions.
It was previously reported that often the large lncRNAs adopt more compact structures with increasing Mg2+ concentration5,9. In order to determine whether Mg2+ has any effect on Bvht conformation in solution, we performed an indirect Fourier transformation to convert the reciprocal-space information of ln(I(q)) vs. q into the real space electron pair-distance distribution function (P(r)) to obtain reliable values of Rg and Dmax (radius at which P(r) approaches to zero) for Bvht samples using the program GNOM. The benefit of using this method over Guinier analysis for Rg determination is that the P(r) method utilizes the entire dataset, whereas the Guinier method that is restricted to the data in the low-q region. Consistent with this observation, the SEC-SAXS data for Bvht suggested that the full-length Bvht becomes more compact as Mg2+ concentration increases from 0 to 12 mM (Table 1). For example, the maximum particle dimension (Dmax) for Bvht solubilized in the absence of Mg2+ was 300 Å, which decreases to 287 and 260 Å in the presence of 6 and 12 mM MgCl2, respectively. Visual inspection also indicates that Bvht conformations tend to be more compact as we increase Mg2+ concentration (Supplementary Fig. 2). This may result from a combination of Mg2+ effects, including increased electrostatic screening, outer sphere coupling Mg2+-RNA interactions and/or specific chelation of Mg2+ by the RNA40–42.
Table 1.
Dependence of structural characteristics of full-length Bvht (636 nucleotides, 206 kDa) on Mg2+ concentration.
| Mg2+ concentration | 0 mM | 6 mM | 12 mM |
|---|---|---|---|
| Rg (Å) | 98.1 ± 0.2 | 99.53 ± 0.2 | 84.8 ± 0.7 |
| Dmax (Å) | 300 | 287 | 260 |
| χ2 | 1.3 | 1.28 | 1.18 |
| NSD | 1.27 ± 0.02 | 1.21 ± 0.04 | 1.24 ± 0.04 |
| Resolution (Å) | 13.9–18.6 | 13.6–35.0 | 14.2–37.2 |
Rg was determined using the P(r) function that utilizes both lower and higher angle datasets for each sample. Dmax is the radius where the P(r) distribution approaches to zero. The χ2 value indicate the goodness of fit between the experimentally collected and model-derived SAXS data, whereas the NSD (normalized spatial discrepancy) describes an agreement between individual models for each dataset. The resolution of models was calculated by phenix.mtriage63. See Methods section for additional detail
Mg2+ dependent conformational changes are more obvious for larger lncRNAs (e.g., 206 kDa for our full-length Bvht3 or up to 700 kDa for other RNAs5,9). For smaller lncRNAs (e.g., 30–69 kDa for Bvht modules), Mg2+ dependent conformational changes are more difficult to observe (Supplementary Fig. 3). For example, when we measured SHAPE reactivities of the 3′ end of human, zebrafish and lizard MALAT1 (46 kDa) at 0 and 6 mM Mg2+, they showed small differences in signal10.
Structures of sub-domains support full-length structure
In addition to Bvht3, other lncRNAs such as COOLAIR4, HOTAIR5, SRA7, and XIST repeat A (RepA)9 fold modularly. Here, small sections of each lncRNA (“modules”) possess secondary structures that fold independently within the lncRNA. Many other lncRNAs likely fold in a modular fashion. This trend seems to enable each lncRNA to contain distinct functional structures. For example, HOTAIR possesses distinct binding domains for PRC2 and LSD1 complexes, making it modular bifunctional RNA43. Modularity in structure would also aid in cotranscriptional recruitment of epigenetic factors to chromatin44, thought to play an important role lncRNA–chromatin interactions. We have identified several modular folds in Bvht by probing the secondary structure of various subregions and comparing the profiles with the lncRNA as a whole. As such, we hypothesized that SAXS-based structures for each module can be determined independently from the whole lncRNA.
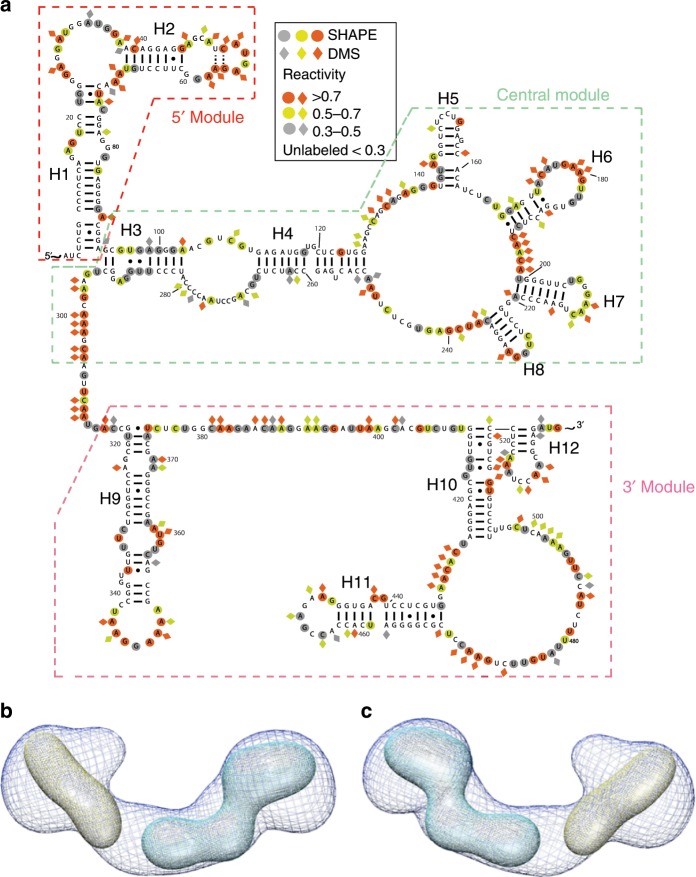
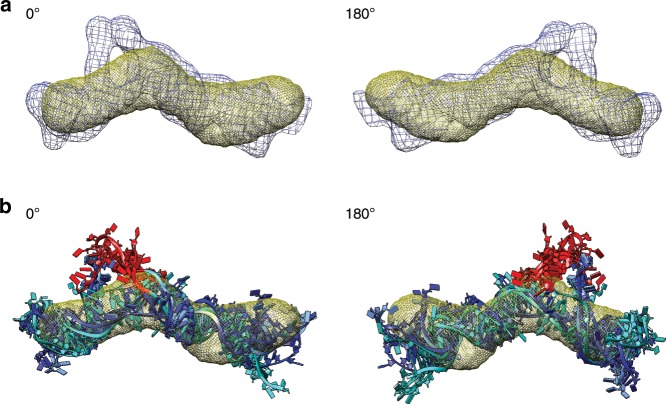
As a positive control to test this hypothesis, we measured SEC-SAXS profiles of modular sub-domains of Bvht (98–224 nts in length) that do not overlap with each other and have modular secondary structures (Fig. 3 and Table 2). The central module (nucleotides # 87–305) did not display mono-dispersity during SEC-SAXS. Therefore, we did not process the data. However, we were able to fit the low-resolution structures for 5′ and 3′ modules of Bvht to the full-length Bvht structure, suggesting that the individual secondary structures of these two modules are consistent with their secondary structures in full-length Bvht.
Fig. 3. Structural studies of Bvht modular sub-domains at 6 mM Mg2+.
a Secondary structures of full-length Bvht based on chemical probing experiments, depicting its modular sub-domains. b Averaged 3-D SAXS solution conformation of full-length Bvht and its modular sub-domains. Blue mesh, averaged solution structure of full-length Bvht; yellow, 5′ module of Bvht; cyan, 3′ module of Bvht. While multiple orientations of the modules fit into the full-length map, we chose orientations most consistent with the connectivity of the secondary structure. c 180° rotated view of (b).
Table 2.
Structural characteristics of independent Bvht modules.
| Samplea | 5′ module | 3′ module | Full-length | |||
|---|---|---|---|---|---|---|
| MW (kDa) | 30.4 | 68.6 | 206 | |||
| MgCl2 concentration | 6 mM | 12 mM | 6 mM | 12 mM | 6 mM | 12 mM |
| Rg (Å) | 42 ± 0.2 | 46 ± 0.2 | 61 ± 0.3 | 63 ± 0.4 | 99.53 ± 0.2 | 85 ± 0.7 |
| Dmax (Å) | 135 | 150 | 185 | 190 | 287 | 260 |
| χ2 | 1.2 | 1.2 | 1.2 | 1.2 | 1.3 | 1.18 |
| NSD | 0.85 ± 0.03 | 0.88 ± 0.02 | 1.06 ± 0.02 | 1.04 ± 0.03 | 1.21 ± 0.04 | 1.24 ± 0.04 |
| Resolution (Å) | 31.3 | 31.8 | 34.9–36.5 | 34.1–36.0 | 13.6–35.0 | 14.2–37.2 |
aCentral module (nucleotides # 87–305 with expected MW of 67.3 kDa) sample did not show mono-dispersity. Therefore, we did not process the data
5′ module encompasses nucleotides # 1–98, 3′ module encompasses nucleotides # 315–538, full-length encompasses nucleotides GG and # 1–634. See Table 1 for more information on SAXS data analysis
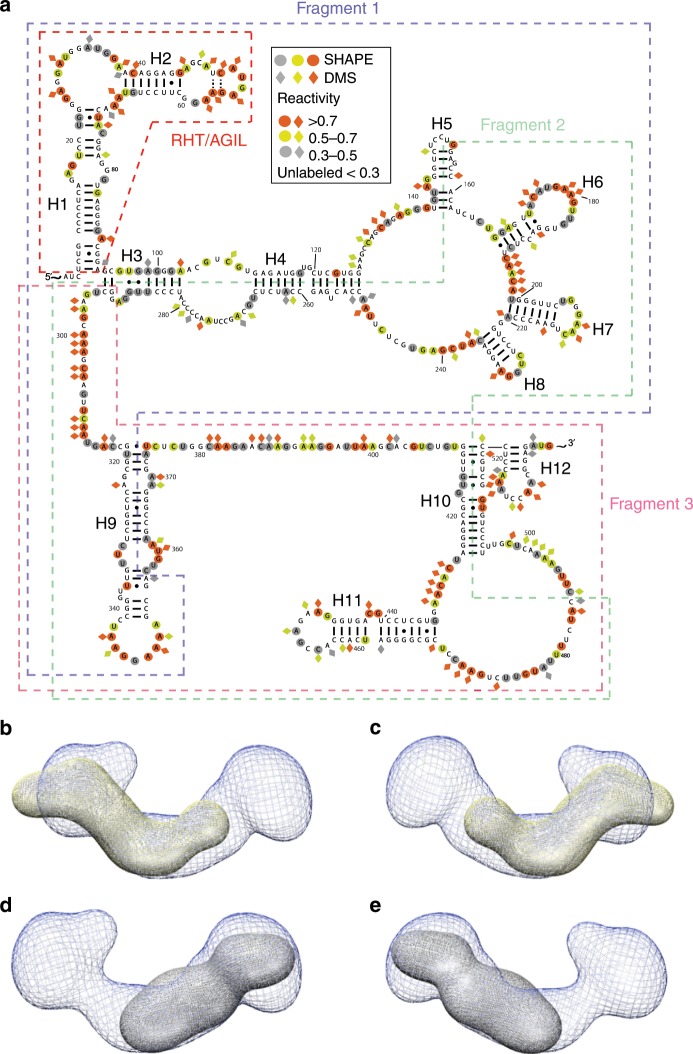
As a negative control, we measured SEC-SAXS profiles of overlapping fragments (344–358 nts in length) of Bvht (Fig. 4 and Table 3). Some of these fragments split helical elements and do not have modular secondary structures (i.e., they do not contain both sides of an RNA double helix). For example, fragment 1 splits helix H9, while fragment 2 splits helices H3, H4, H5 and H10. Therefore, we do not necessarily expect the structures of these fragments to be directly related to the structure, or portions of the structure of the intact, full-length Bvht lncRNA. We find the solution structure of fragment 3 fits nicely into the full-length Bvht solution structure, consistent with the fact that the fragment 3 does contain a modular secondary structure. In the case of fragment 1, the fit is poor, presumably since it only contains approximately half of H9, which may alter the fold relative to the intact Bvht RNA molecule. Fragment 2 contains four split helices. Thus, we expect it to have a dramatically different fold relative to the intact Bvht RNA. Among ~230 total nucleotides, about 38% of it (e.g., ~8 5′ terminal nucleotides and ~80 3′ terminal nucleotides) cannot form base-pairs found in the full-length Bvht.
Fig. 4. Structural studies of Bvht overlapping fragments at 6 mM Mg2+.
a Secondary structures of full-length Bvht and its overlapping fragments. b–e Averaged 3-D SAXS solution conformation of full length Bvht and its overlapping fragments. b Blue mesh, averaged solution structure of full-length Bvht; yellow fragment 1 of Bvht. c 180° rotated view of (b). d Gray, fragment 3 of Bvht. e 180° rotated view of (d).
Table 3.
Structural characteristics of Bvht fragments at 6 mM Mg2+.
| Sample | Fragment 1 | Fragment 2 | Fragment 3 | Full-length |
|---|---|---|---|---|
| MW (kDa) | 116 | 113 | 111 | 206 |
| Rg (Å) | 78.4 ± 0.2 | 81.7 ± 0.7 | 63.9 ± 0.2 | 99.53 ± 0.2 |
| Dmax (Å) | 240 | 271 | 205 | 287 |
| χ2 | 1.7 | 1.4 | 1.4 | 1.3 |
| NSD | 1.27 ± 0.03 | 1.12 ± 0.02 | 1.02 ± 0.02 | 1.21 ± 0.04 |
| Resolution (Å) | 34.1–38.5 | 34.5 | 36.7 | 13.6–35.0 |
Fragment 1 encompasses nucleotides # 1–358, fragment 2 encompasses nucleotides # 156–504, Fragment 3
encompasses nucleotides # 291–588 and # 589−634 is 3′ structure cassette for primer extension after SHAPE and DMS probing. See Table 1 for more information on SAXS data analysis
The pair-distance distribution function (P(r)) plots presented in Fig. 1d have skewed bell shape curves with extended tails, indicating that these Bvht fragments generally adopt extended structures. For example, Bvht fragments (MW ~111–116 kDa) have an Rg of ~64–82 Å and Dmax of ~205–271 Å. In contrast, the full-length Bvht (206 kDa MW) has an Rg of ~85–99 Å and Dmax of ~260–300 Å. These results suggest that each fragment tends to extend flexibly rather than collapsing in a globular fashion.
We also found that fragment 3 of Bvht is significantly more compact than other fragments. Fragments 1 and 2 were quite similar to each other in terms of their solution parameters (Rg, Dmax)). However, fragment 3 has a smaller volume than these two other fragments (Table 3). This shows that Bvht fragments do not have a monotonic relationship between the volume and nucleotide length unlike a series of riboswitches34. Chen et al. previously merged SAXS-based Rg values and reported the monotonic relationship in different riboswitches (e.g., Rg values increase fairly linearly with increasing nucleotide length)34.
Ensemble of 3-D models is consistent with SAXS data
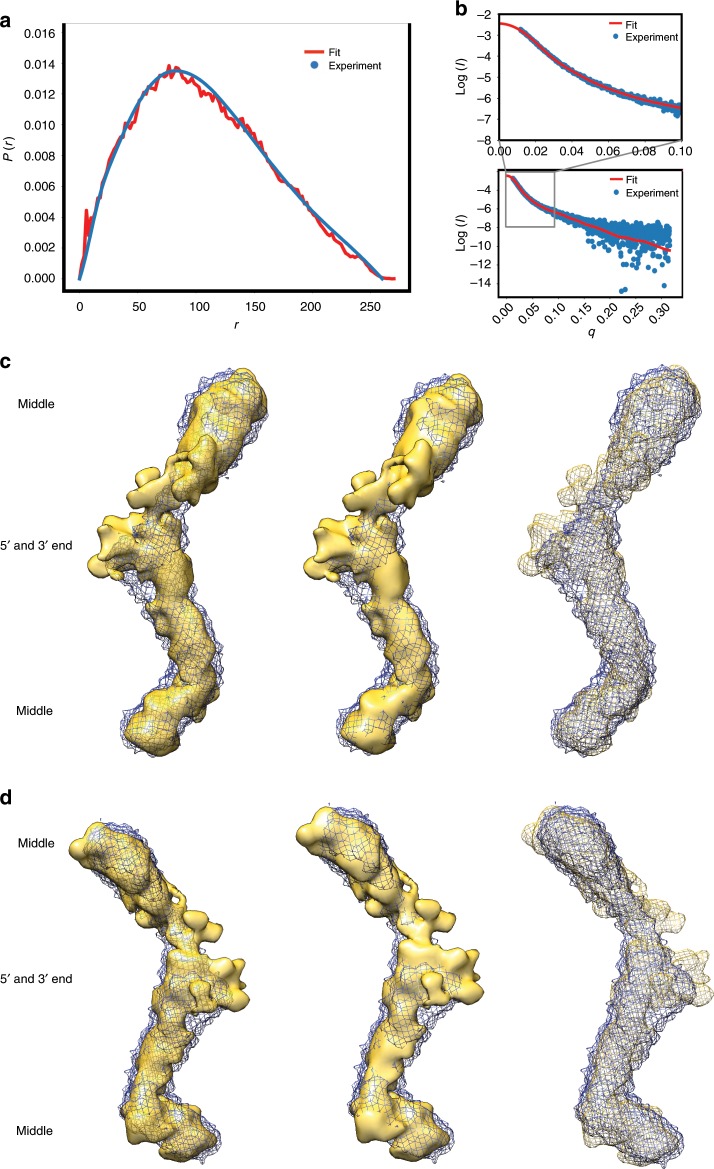
Modeling SAXS data with atomistic structures presents a different set of challenges relative to crystallography as we can only obtain low-resolution structural information from solution scattering data (for our cases, 13.4–38.5 Å resolution). Using the RNA modeling program, ERNWIN45, we have produced an ensemble of atomistic RNA structures highly consistent with our SAXS data (Fig. 5, Supplementary Fig. 4 and Supplementary Fig. 5). We also used Bvht secondary structure information as restraints (Figs. 3 and 4). We modeled full-length Bvht atomistic models (Supplementary Movie 1, Supplementary Fig. 6 and Supplementary Fig. 7) and selected an ensemble of models that fit with the pair-distance distribution function, and with raw scattering data (with χ values of 1.7–2.6, and an average value of 2.1), consistent with our former SAXS based computational approach29. When superimposing 30 top-ranked models, identifying the most densely populated regions, and comparing this to the SAXS-derived solution structures, we find that the computational structures closely match with the SAXS-derived low-resolution solution structures (Fig. 5). The close agreement not only gives us confidence in our 3-D models, but also in our 2-D secondary structure, upon which the 3-D models are based. In addition, using ERNWIN, we modeled atomistic structures of the 5′ module of Bvht (Fig. 6), which also agree well with the pair-distance distribution function. The simulated annealing model building approach from SAXS data suggests that Bvht has flexible regions, leading to minor variation in each of the low-resolution atomistic structures we calculated. To account for this intrinsic flexibility, we optimized the pair-distance distribution of the ensemble as opposed to individual structures during structure prediction, sampling multiple individual trajectories.
Fig. 5. Ensemble of atomistic models of Bvht matches measured SAXS data of Bvht at 12 mM Mg2+.
a Pair-distance distribution function displays agreement between experimental data (blue) and data back-calculated from computational models (red). b Scattering intensity vs. angle indicates agreement between experimental and model derived data. c A comparison of averaged solution structures of full-length Bvht derived from SAXS data (meshed blue) and an ensemble of computationally derived atomistic models (yellow) shows agreement (Supplementary Fig. 6 and Supplementary Fig. 7). SAXS measured structure was rendered with two different surface opacities and with wire mesh to clearly display surfaces of both SAXS measured solution structure and solution structure of ensemble atomistic model (left, low opacity; middle, high opacity; right, yellow wire mesh). d 180° view of (c). See Supplementary Movie 1 for 360° rotation.
Fig. 6. Superposition of atomistic models of Bvht 5′ module with experimental structure (yellow mesh).
a Ensemble of simulated structures of the five top χ ranked atomistic models of Bvht 5′ module (blue mesh) superimposed with SAXS structure (yellow mesh). b Five top χ ranked atomistic models of Bvht 5′ module at 6 mM Mg2+ (cyan to dark blue gradient). As in the full-length Bvht, atoms outside of SAXS-derived solution structure may indicate flexible regions. Most of this “flexible” region is the 11-nucleotide region (colored red) in the RHT/AGIL motif known to be essential to bind CNBP3. See Supplementary Movie 3 for more information.
CNBP binding requires multiple structural domains of Bvht
CRISPR/Cas9 genome editing studies demonstrated that the right-hand-turn (RHT)/5′ asymmetric G-rich internal loop (AGIL) motif in Bvht is essential for cardiovascular lineage commitment. Interestingly, CNBP (a zinc-finger transcription factor) antagonizes this function by binding to the AGIL motif of Bvht3. Accordingly, we were curious to know if binding with CNBP affects Bvht conformation. Therefore, we performed SAXS with the CNBP sample as well. The Guinier plot presented in Supplementary Fig. 8 displays linearity for small q values suggesting that CNBP samples are aggregation free. Although it is relatively small, the increase in Dmax for the protein–RNA system relative to Bvht RNA alone indicates that Bvht and CNBP formed a complex (Table 4). In addition, we observed by EMSA that Bvht migrates more slowly with higher CNBP concentrations (Fig. 1e). While we are hesitant to overinterpret the SAXS results due to dynamic conformational changes and low-resolution of SAXS data, we observed that CNBP binding is more evident with full-length Bvht than its fragments/modules (Fig. 7, Supplementary Fig. 9 and Supplementary Fig. 10). We believe that this small structural difference by CNBP binding results from weak interaction between fragment 1 of Bvht and CNBP. Fragments 2 and 3 of Bvht also showed weak interaction with CNBP (Supplementary Table 1).
Table 4.
Dependence of Bvht structures on CNBP protein (22 kDa) binding (6 mM Mg2+).
| Sample | Fragment 1 of Bvht | Full-length Bvht | ||
|---|---|---|---|---|
| CNBP presence | −CNBP | +CNBP | −CNBP | +CNBP |
| MW (kDa) | 116 | 138 | 206 | 228 |
| Rg (Å) | 78.4 ± 0.2 | 82.4 ± 0.5 | 99.53 ± 0.2 | 95.1 ± 0.4 |
| Dmax (Å) | 240 | 270 | 287 | 301 |
| χ2 | 1.15 | 1.7 | 1.3 | 1.3 |
| NSD | 1.20 ± 0.03 | 1.14 ± 0.04 | 1.21 ± 0.04 | 1.29 ± 0.03 |
| Resolution (Å) | 34.1–38.5 | 32.1 | 13.6–35.0 | 13.4–17.7 |
See Table 1 for more information on SAXS data analysis
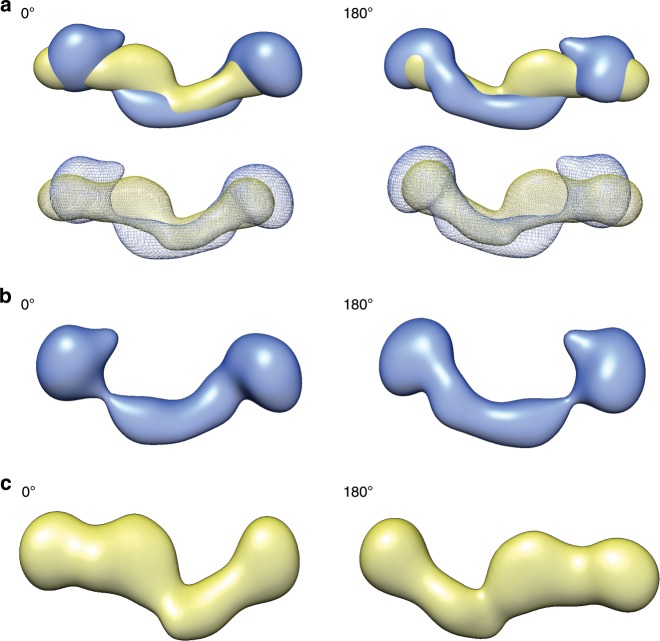
Fig. 7. Solution conformation of Bvht and its complex with CNBP.
a Blue, averaged solution structure of Bvht only; yellow, averaged solution structure of Bvht-CNBP complex. b Bvht only. c Bvht-CNBP complex. Individual solution structures are presented in Supplementary Fig. 9.
Our data also suggests that CNBP facilitates compaction of the full-length Bvht RNA (e.g., Rg in Table 4, Fig. 1). Based on pair-distance distribution analysis, we confirmed that upon interaction with CNBP, Bvht undergoes conformational changes. This compaction due to CNBP-binding is consistent with overall compaction observed in previous riboswitch studies (e.g., ligand-bound riboswitches are generally more compact than the free riboswitches34). Interestingly, the compaction of Bvht by CNBP and the EMSA binding results, when taken together with the module analysis, suggest that CNBP interacts with at least two distinct sites on the Bvht RNA, which, in turn, lead to a more compact state.
In summary, as we compare the low-resolution structures of Bvht only and Bvht-CNBP complex, we observed an obvious shape change for the full-length Bvht-CNBP complex, while the fragment 1 RNA–CNBP complex did not show a significant difference relative to the fragment 1 only solution structure, suggesting that intact, full-length Bvht is required for efficient CNBP binding.
Discussion
In the epigenetics and pharmaceutical communities, there has been great interest in the question: do lncRNAs have well-defined structures? It has not been clear whether the majority of lncRNAs are disordered, extended or compact46. Due to their large structures (200–100,000 nts) and dynamic binding function with partner molecules (including proteins), it is sometimes assumed that lncRNA structures are generally too disordered, or highly flexible to be studied by high-resolution structural determination methods. However, based on secondary structure determination for nine different lncRNAs, it is evident that these lncRNAs do contain modular structures. In fact, regarding RNA systems in general, there are other RNAs of similar sizes (e.g., group I and II intron26,27,47) with well-defined high-resolution crystallographic and cryo-EM structures. Large RNA–protein complexes, such as the ribosome and spliceosome have also yielded high-resolution cryo-EM structures26,48. Therefore, the key outstanding issue is whether or not there are specific lncRNAs that exist with well-defined structures comparable to other large RNAs with well-defined structures. Our SAXS study of Bvht lays the foundational step in this direction, revealing that this RNA does possess a 3-D structure and this 3-D structure may play a role in function. Specifically, the dependence of the physical size, as estimated by Rg and Dmax, of the full-length Bvht RNA on Mg2+ concentration is clear evidence of the presence of tertiary contacts. In addition, we find that Bvht directly binds the zinc-finger protein CNBP and that the conformational ensemble of the Bvht RNA is significantly altered upon protein binding. The fact that our SEC-SAXS data on sub-domains of Bvht (e.g., 5′ and 3′ modules, MW: 30–69 kDa) did not show Mg2+-dependent changes in conformation (P(r) distribution shapes are similar between 6 and 12 mM Mg2+ in Fig. 1) suggests that Mg2+ may mediate inter-domain RNA–RNA tertiary interactions in the case of the full-length Bvht system.
Our SAXS studies of sub-regions of Bvht, in addition to the full-length RNA, are consistent with a modular construction of the full 3-D fold, supporting our previous 2-D chemical probing study of Bvht, also showing the structure to be modular. Our SAXS data are consistent with the conformational ensemble of this lncRNA containing relatively rigid, modular subsections, connected by flexible regions. We expect that our experimental strategy will be easily applicable to other lncRNAs with modular secondary structures.
To date, the only information available for the Bvht–CNBP complex has been that functional in vivo CNBP binding requires the 5′ asymmetric G-rich internal loop (RHT/AGIL motif) of Bvht3. As mentioned, our in vitro data suggests that for high-affinity binding, CNBP requires the full-length Bvht, including the 5′ module (which contains the RHT/AGIL motif), the central module (which contains the multiway junction), and the 3′ module. In addition, EMSA experiments suggest that fragment 1 interacts with CNBP as well (Supplementary Table 1). Interestingly, while the fragment 1 of Bvht alone (positions 1–358) was enough for CNBP binding (Table 4), the 5′ module of Bvht (positions 1–98) alone was not enough for the CNBP binding when we analyzed this interaction using SEC-SAXS. These data suggest that both the RHT/AGIL motif (positions 27–37) and other structural elements (perhaps the multiway junction in position rage 38–358) are required for CNBP binding. Specifically, CNBP binding to Bvht requires both the 5′ module and either the central module or 3′ module of Bvht. This finding, combined with the modular, but flexible nature of Bvht is consistent with a functional role of the conformational heterogeneity that may be required for the efficient binding of proteins.
In addition, CNBP appears to bind to full-length and fragment 1 of Bvht with equimolar ratio, respectively, based on the fact that we loaded Bvht and CNBP with equimolar ratio and in light of their Dmax values (Table 4). To date, knowledge about stoichiometry between lncRNAs and their protein binding partners has been quite sparse3,17. We note that our estimation of the equimolar binding of Bvht and CNBP is suggestive rather than definitive for the following reasons. It is known that RNA and DNA scatter more strongly than proteins. Therefore, we would not be surprised even if we cannot clearly observe CNBP with this low resolution of SAXS (~13.4–38.5 Å). Furthermore, in light of previous size exclusion chromatography studies49, we know that RNAs tend to have much larger effective Rg than proteins do. In addition, CNBP (21.5 kDa MW) is much smaller than Bvht (206 kDa MW).
Regarding RNA–protein interactions, the CNBP protein is believed to function mainly inside the cell, binding to nucleic acids and controlling transcription and translation50. Our study provides an important stepping stone in understanding Bvht and CNBP function at the molecular level. While our study has helped to elucidate the Bvht–CNBP interaction, a more thorough investigation will be required to delineate the exact interaction points on RNA and protein, as well as the role of dynamics in the interaction. As CNBP aids in transcriptional control, future studies of lncRNA–chromatin interactions and the role of lncRNAs in chromatin looping may shed more light on CNBP function44. For example, it will be more informative once we learn the binding ratios between different lncRNAs and nucleosomes/chromatin. All these efforts will contribute to better decipher biological functions of noncoding RNAs.
While we find Bvht to have a defined 3-D structure and a highly organized secondary structure, it is also flexible. This behavior is not unlike the SAM-I riboswitch, which also has well-defined secondary and tertiary structure (in particular, a high-resolution crystal structure in the ligand-bound state), but is highly flexible in its apo state. In fact, many structured RNAs can adapt multiple conformations and are highly flexible1,2,35. For example, even with 300 kV cryo-electron microscopy (cryo-EM) using an energy filter, Zhang et al. could obtain only a ~9 Å resolution map of 30 kDa RNA (47 nucleotide dimer)51. In their report, the internal structural flexibility of the RNA limited the cryo-EM resolution and this hypothesis is supported by molecular dynamics simulation. With respect to these findings, it is not surprising that the Bvht RNA (206 kDa MW) would have more flexible regions than this 30 kDa RNA, as shown in our atomistic models (Supplementary Movie 2, Supplementary Fig. 6). Indeed, Zhang et al. summarized that relatively large RNAs (e.g., >200 kDa) may have flexible conformations. Even the 116 kDa MW RNA (fragment 1 of Bvht) shows flexible conformations (Supplementary Fig. 11). These dynamic conformations are what may confer diverse biological influences of RNAs, such as transient binding.
The flexibility of the Bvht lncRNA emphasizes the importance and advantage of using SAXS to investigate 3-D structures of lncRNAs. The structural information on flexible regions that are often not resolved by X-ray crystallography is apparent in SAXS-based low-resolution studies. Our study allowed us to compare representatives of individual conformational clusters to evaluate the nonuniqueness of the SAXS-based reconstruction to decipher whether there are different conformations of the lncRNA52. SAXS also provides estimates of shape parameters such as Rg and Dmax of biological macromolecules in solution14 (Tables 1–4). In addition, we emphasize that SAXS-based structure data shows more physiologically relevant structures, avoiding packing effects present in crystallographic studies. In fact, it is also known that a “true solution” state (e.g., NMR) differs from even a “frozen-solution” state (cryo-EM)51. While the intrinsic flexibility of Bvht likely provides the dominant contribution to its high Rg values (for example, Bvht fragment 1 of 358 nt has a 78.4 Å Rg), the fact that SAXS studies are performed in solution also contributes. For comparison with other RNA crystal structures of similar molecular weight, the group II intron lariat (PDB id 4R0D, 622 nts), has a Rg of only 41 Å. In a second example, 500–700 nt subregions of crystal structures of the small subunit of the ribosome (e.g., PDB id 4GKK) have Rg values between 39 and 57 Å, resulting from the tightly packed tertiary folds of ribosomal RNA. Finally, SAXS allows the use of more physiological buffers in real-time (chemical additives for crystallization and surfactant and carbon for cryo-EM grid optimization are not required), allowing the observation of structural changes resulting predominantly from certain conditions (such as Mg2+ concentration differences) more clearly. Similarly, we are confident that our samples were monodisperse and homogeneous (interpretation of SAXS data itself would have been extremely difficult if it were polydisperse).
Regarding secondary structure, RNA secondary structure prediction accuracy can be greatly improved by covariation-based constraints7,9. However, compared to protein-coding genes, long noncoding RNAs tend to have weaker sequence conservation53 and identifying homologs can be challenging7. Recently developed covariance tools54 reveal structure conservation in lncRNAs55. However, in the case of Bvht, no homolog has been identified to date. Our SAXS study helps to confirm the lncRNA secondary structure determined by 3S SHAPE and DMS chemical probing experiments. In particular, our ensemble of atomistic 3-D models of the Bvht RNA is highly consistent with the SAXS data. Since the 3-D models are based on the secondary structure, the SAXS experiments bolster the secondary structure. Our approach of using chemical probing and SAXS for modeling 3-D lncRNA structures complements a wide variety of approaches used for RNA modeling30,31,56–59. Indeed, it has been difficult to predict 3-D structures accurately without secondary structural constraints60,61.
Overall, we have shown that physiologically relevant three-dimensional SAXS-based structures of long noncoding RNAs can be determined in spite of their considerable length and flexible conformations. Our approach for this characterization is unique since it combines several biophysical/computational methodologies in an analogous fashion to Huang et al.62: (i) in vitro transcription of long noncoding RNA, (ii) SEC-SAXS experiments to study solution structures, (iii) computational structure determination using SAXS and secondary structure information as restraints, (iv) transformation of SAXS DAMFILT files into cryo-EM style maps for superposition, (v) construction of simulated solution structures from atomistic cartesian coordinates using in-house PHENIX63 scripts, and (vi) resolution estimation and flexible fitting using programs which were developed for cryo-EM map applications. Our atomistic model is also the longest isolated RNA (e.g., 636 nucleotides) to date, with the next longest RNA being 622–625 nucleotides to our knowledge27,64. To corroborate our findings, we performed EMSA analysis and used measured SHAPE reactivities. Our approach is broadly applicable to other RNA systems and lays the foundation for similar studies in the widely expanding classes of long noncoding RNAs, viral RNAs and mRNA–protein complexes.
Methods
Sample preparation
We prepared RNA samples using snapcool refolding65 immediately before experimental characterization (e.g., EMSA and SAXS). The full sequence of Bvht is in Supplementary Note 1. The CNBP coding sequence (Supplementary Note 2) was cloned into pET-28a (MilliporeSigma) with a C-terminal 6-histidine tag and expressed in the ArcticExpress (Agilent) strain of E. coli. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at 0.4–0.8 OD600 to induce expression for either 3 h at 37 °C or overnight at 13 °C. Cell pellets were sonicated, and after additional centrifugation, the supernatant was applied to a Ni-NTA column (GE Healthcare).
Electrophoretic mobility shift assay (EMSA)
We performed EMSA to study Bvht–CNBP complex migration using a 6% polyacrylamide gel containing 0.5× TBE (45 mM Tris, 45 mM Boric acid, 1 mM EDTA disodium salt, pH 8.3) and 2 mM MgCl2. EMSA was performed on ice at 130 V for 4 h or 100 V for 6 h. Gels were stained with ethidium bromide for 5 min and destained with water several times. Gel images were scanned with a Hitachi FMBioII fluorescence imager (532 nm laser excitation, 605 nm bandpass emission filter) at 100 µm resolution and −2.85 mm focus. The original image of the Fig. 1 gel is in Supplementary Fig. 12.
SAXS experiment
In order to collect data for the monodispersed sample, devoid of any high-molecular-weight aggregates or degraded material, we performed SAXS data collection using a SEC66, controlled by an Agilent HPLC-SAXS set-up at the B21 beamline, Diamond Light Source (Didcot, UK). An Agilent 1200 (Agilent Technologies, Stockport, UK) in-line HPLC system was connected to a specialized flow cell and an absorbance detector. 50 µL of each sample (Bvht at 0 mM Mg2+, Bvht at 6 mM Mg2+, Bvht at 12 mM Mg2+, and Bvht + CNBP at 6 mM Mg2+) was injected into a Shodex KW403-4F SEC column (Showa Denko America Inc.) which had been pre-equilibrated with sample buffer (50 mM HEPES-KOH, 100 mM KCl, pH 7.6, and either 0, 6, or 12 mM MgCl2). Injected sample concentrations are in Supplementary Table 2. The SEC-separated sample was exposed to X-rays, followed by data collection every 3 s.
SAXS data processing
Using ScÅtter67, the sample peak regions that were selected were then buffer subtracted and merged using either ScÅtter or Primus in the ATSAS suite68. The CRYSOL, DAMAVER, DAMCLUST, DAMMIN, GNOM, SUPCOMB, programs in the ATSAS suite were used. Molecular physical properties were calculated using software modules from ATSAS. Guinier and Kratky analyses were performed to ensure that samples are homogenous and well-folded, respectively. The GNOM program was used to determine Dmax and Rg by calculating the pair-distance distribution P(r) plot29,69,70. We estimate Dmax in accordance with Trewhella et al.39 and note that the general decrease in Dmax for full-length Bvht with increasing Mg2+ concentration is also observed with complementary methods (e.g., analytical ultracentrifugation) for other lncRNAs5,9 (see Supplementary Table 3 for other details). The low-resolution structures were calculated using the DAMMIN program71. For each sample, >15 low-resolution models were calculated, followed by alignment and averaging of each set of models using the program DAMAVER to obtain a representative shape. We also performed a clustering calculation to identify likely clusters of full-length RNA in 6 mM MgCl2 buffer using the DAMCLUST program. We converted the reconstructed bead models into electron density maps (as in cryo electron microscopy) with the program Situs72. We used a Gaussian kernel width of 6 Å and a voxel spacing of 1 Å.
Resolution of SAXS based Solution Structure
Exact resolution estimation is difficult with typical SAXS measurements (e.g., SAXS “resolution” is ambiguous, not directly related to 2π/q). We can estimate the resolution of our SAXS based solution structures at ~13.6–37.2 Å (DAMAVER derived solution structures tend to have higher resolution than DAMCLUST ones) using phenix.mtriage63. Although the phenix.mtriage is being actively used for cryo-electron microscopy maps, we found that it is applicable to our SAXS based solution structure as well. For example, when we filtered the volume with a gaussian by UCSF Chimera73, it reasonably reflected decreased resolution. Although we estimate the resolution, SAXS based solution structural data should not be overinterpreted.
Atomistic structure modeling of Bvht
To model atomistic RNA structures, we used a two-step procedure that mimics hierarchical folding11: starting from the published secondary structure of Bvht, we inserted a few additional base-pairs with RNAfold74. Published SHAPE and DMS data3 were used as soft constraints. The reason that we added more base-pairs on top of published base-pairs is that it is unlikely that long single stranded regions would not form at least some noncanonical on- and off-interactions. We then assembled known RNA fragments using a Monte Carlo algorithm to build the tertiary structure. The idea of fragment assembly is well established75 and has been used to predict models matching SAXS data by Dzananovic et al.29. In ERNWIN, we use fragments for secondary structure elements (hairpins, interior loops, multiloop segments), extracted from the representative set of RNA containing PDB structures76 with the help of our Python RNA structure library Forgi77. After every sampling step, we used the correlation between the pair-wise distance distribution function of the proposed tertiary structure and the experimental SAXS distance distribution function, derived with GNOM. To save computation time, the pair-wise distance distribution of our models was calculated using only one point per nucleotide. After sampling was complete, we used the established tool CRYSOL to further filter the predicted structures for the top-ranked χ value. In contrast to our estimated pair-wise distance distribution function used during sampling, this program takes all atoms and the hydration layer into account. To save computation time, we only evaluated every 1000th structure using CRYSOL. The best presented atomistic models for full-length Bvht at 12 mM Mg2+ have a χ value better (lower) than 1.75. We also carried out the same sampling procedure for an alternative secondary structure and for a null hypothesis system, random secondary structures (predicted from di-nucleotide shuffled sequences at higher temperatures, to roughly match the number of base-pairs in the Bvht secondary structure), as a control. Control structures have poorer χ values (Supplementary Fig. 5).
Alignment and visualization details
We used SUPCOMB to align SAXS-based solution structures and models to improve chirality correctness. For Fig. 5c, d, we superimposed all 30 atomistic models with equal weight using UCSF Chimera (e.g., File −> Save PDB −> Save multiple models in a single file) saving an NMR style pdb file that uses a model number. When we aligned superimposed models to averaged solution structures, we used “Fit in map” of UCSF Chimera73. All solution structures, atomistic models, and movies were visualized by UCSF Chimera.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are grateful to all members of the B21 beamline at Diamond Light Source (Didcot, UK) for help with SEC-SAXS experiments. We would also like to thank all members of the PHENIX team for their help with cryo-EM software development and usage. This work was supported by NIH NIGMS GM110310 and LANL LDRD. TRP acknowledges Canada Research Chair program. TM was paid through the NSERC Discovery grant to TRP. Public release of this article is authorized by LA-UR-19-30307. We acknowledge support from LANL Institutional Computing.
Author contributions
K.S., T.P., and I.H. conceived and designed the project. D.K. and S.H. prepared the samples. T.P. and T.M. conducted SAXS experiments and analyzed the data. B.T. and D.K. modeled the atomistic structures. D.K. wrote the initial draft and all authors contributed to the paper preparation and interpretation of data.
Data availability
All of our SAXS data used for modeling have been deposited to the small angle scattering biological data bank (https://www.sasbdb.org/project/939/x6kirb9f97/)78. All other data including atomistic structures and cloning constructs will be made available from the corresponding author upon reasonable request.
Code availability
All codes (e.g., ERNWIN and in-house python codes for SAXS data processing) are available at https://github.com/pkerpedjiev/ernwin and https://github.com/kimdn/in_house_codes_for_SAXS.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Trushar R. Patel, Email: trushar.patel@uleth.ca
Karissa Y. Sanbonmatsu, Email: kys@lanl.gov
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-13942-4.
References
- 1.Wang Chenguang, Wang Lianzong, Ding Yu, Lu Xiaoyan, Zhang Guosi, Yang Jiaxin, Zheng Hewei, Wang Hong, Jiang Yongshuai, Xu Liangde. LncRNA Structural Characteristics in Epigenetic Regulation. International Journal of Molecular Sciences. 2017;18(12):2659. doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens L, Rühle F, Stoll M. LncRNA secondary structure in the cardiovascular system. Noncoding RNA Res. 2017;2:137–142. doi: 10.1016/j.ncrna.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue Z, et al. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell. 2016;64:37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawkes EJ, et al. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 2016;16:3087–3096. doi: 10.1016/j.celrep.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somarowthu S, et al. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilik IA, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang R, Moss WN, Rutenberg-Schoenberg M, Simon MD. Probing Xist RNA structure in cells using targeted structure-seq. PLoS. Genet. 2015;11:1–29. doi: 10.1371/journal.pgen.1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Somarowthu S, Pyle AM. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat. Chem. Biol. 2017;13:282–289. doi: 10.1038/nchembio.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B, et al. Identification and characterization of a class of MALAT1-like genomic loci. Cell Rep. 2017;19:1723–1738. doi: 10.1016/j.celrep.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel BC, Flamm C, Hofacker IL. RNA structure prediction: from 2D to 3D. Emerg. Top. Life Sci. 2017;1:275–285. doi: 10.1042/ETLS20160027. [DOI] [PubMed] [Google Scholar]
- 12.Fanucchi S, Mhlanga MM. Lnc-ing trained immunity to chromatin architecture. Front. Cell. Dev. Biol. 2019;7:1–7. doi: 10.3389/fcell.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018;11:63. doi: 10.1186/s13045-018-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel TR, et al. Structural studies of RNA-protein complexes: a hybrid approach involving hydrodynamics, scattering, and computational methods. Methods. 2017;118–119:146–162. doi: 10.1016/j.ymeth.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Ponce-Salvatierra A, et al. Computational modeling of RNA 3D structure based on experimental data. Biosci. Rep. 2019;39:BSR20180430. doi: 10.1042/BSR20180430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dekoster GT, Delaney KJ, Hall KB. A compare-and-contrast NMR dynamics study of two related RRMs: U1A and SNF. Biophys. J. 2014;107:208–219. doi: 10.1016/j.bpj.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duchardt-Ferner E, et al. What a difference an OH makes: conformational dynamics as the Basis for the ligand specificity of the neomycin-sensing riboswitch. Angew. Chem. Int. Ed. 2016;55:1527–1530. doi: 10.1002/anie.201507365. [DOI] [PubMed] [Google Scholar]
- 20.LeBlanc RM, Longhini AP, Tugarinov V, Dayie TK. NMR probing of invisible excited states using selectively labeled RNAs. J. Biomol. NMR. 2018;71:165–172. doi: 10.1007/s10858-018-0184-3. [DOI] [PubMed] [Google Scholar]
- 21.Keyhani S, Goldau T, Blümler A, Heckel A, Schwalbe H. Chemo-enzymatic synthesis of position-specifically modified RNA for biophysical studies including light control and NMR spectroscopy. Angew. Chem. Int. Ed. 2018;57:12017–12021. doi: 10.1002/anie.201807125. [DOI] [PubMed] [Google Scholar]
- 22.Schlagnitweit J, Steiner E, Karlsson H, Petzold K. Efficient detection of structure and dynamics in unlabeled RNAs: the SELOPE approach. Chemistry. 2018;24:6067–6070. doi: 10.1002/chem.201800992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frueh DP, Goodrich AC, Mishra SH, Nichols SR. NMR methods for structural studies of large monomeric and multimeric proteins. Curr. Opin. Struct. Biol. 2013;23:734–739. doi: 10.1016/j.sbi.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnwal RP, Yang F, Varani G. Applications of NMR to structure determination of RNAs large and small. Arch. Biochem. Biophys. 2017;628:42–56. doi: 10.1016/j.abb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown A, Shao S. Ribosomes and cryo-EM: a duet. Curr. Opin. Struct. Biol. 2018;52:1–7. doi: 10.1016/j.sbi.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 26.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 27.Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014;514:193. doi: 10.1038/nature13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franke D, Jeffries CM, Svergun DI. Machine learning methods for X-ray scattering data analysis from biomacromolecular solutions. Biophys. J. 2018;114:2485–2492. doi: 10.1016/j.bpj.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzananovic E, et al. Impact of the structural integrity of the three-way junction of adenovirus VAIRNA on PKR inhibition. PLoS ONE. 2017;12:1–21. doi: 10.1371/journal.pone.0186849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain S, Laederach A, Ramos SBV, Schlick T. A pipeline for computational design of novel RNA-like topologies. Nucleic Acids Res. 2018;46:7040–7051. doi: 10.1093/nar/gky524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Z, Westhof E. RNA structure: advances and assessment of 3D structure prediction. Annu. Rev. Biophys. 2017;46:483–503. doi: 10.1146/annurev-biophys-070816-034125. [DOI] [PubMed] [Google Scholar]
- 32.Brown JA, et al. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Pollack L. SAXS studies of RNA: structures, dynamics, and interactions with partners. Wiley Interdiscip. Rev. RNA. 2016;7:512–526. doi: 10.1002/wrna.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abeysirigunawardena SC, Woodson SA. Differential effects of ribosomal proteins and Mg2+ ions on a conformational switch during 30S ribosome 5′-domain assembly. RNA. 2015;21:1859–1865. doi: 10.1261/rna.051292.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierhaus KH. Mg2+, K+, and the Ribosome. J. Bacteriol. 2014;196:3817–3819. doi: 10.1128/JB.02297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen SH, Wong K-P. The role of magnesium and potassium ions in the molecular mechanism of ribosome assembly: Hydrodynamic, conformational, and thermal stability studies of 16 S RNA from Escherichia coli ribosomes. Arch. Biochem. Biophys. 1986;249:137–147. doi: 10.1016/0003-9861(86)90568-0. [DOI] [PubMed] [Google Scholar]
- 38.Guinier, A. & Fournet, G. Small-Angle Scattering of X-Rays. (John Wiley & Sons, Inc., New York Chapman & Hall Ltd., London, 1955).
- 39.Trewhella J, et al. 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: An update. Acta Crystallogr. Sect. D Struct. Biol. 2017;73:710–728. doi: 10.1107/S2059798317011597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J. Am. Chem. Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allnér O, Nilsson L, Villa A. Magnesium ion–water coordination and exchange in biomolecular simulations. J. Chem. Theory Comput. 2012;8:1493–1502. doi: 10.1021/ct3000734. [DOI] [PubMed] [Google Scholar]
- 42.Trachman RJ, Draper DE. Comparison of interactions of diamine and Mg2+ with RNA tertiary structures: similar versus differential effects on the stabilities of diverse RNA folds. Biochemistry. 2013;52:5911–5919. doi: 10.1021/bi400529q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai M-C, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689 LP–689693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Böhmdorfer G, Wierzbicki AT. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerpedjiev P, Honer Zu Siederdissen C, Hofacker IL. Predicting RNA 3D structure using a coarse-grain helix-centered model. RNA. 2015;21:1110–1121. doi: 10.1261/rna.047522.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novikova IV, Hennelly SP, Tung C-S, Sanbonmatsu KY. Rise of the RNA Machines: exploring the structure of long non-coding RNAs. J. Mol. Biol. 2013;425:3731–3746. doi: 10.1016/j.jmb.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 47.Belfort Marlene, Lambowitz Alan M. Group II Intron RNPs and Reverse Transcriptases: From Retroelements to Research Tools. Cold Spring Harbor Perspectives in Biology. 2019;11(4):a032375. doi: 10.1101/cshperspect.a032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, et al. Structures of the human spliceosomes before and after release of the ligated exon. Cell Res. 2019;29:274–285. doi: 10.1038/s41422-019-0143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim I, McKenna SA, Viani Puglisi E, Puglisi JD. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margarit Ezequiel, Armas Pablo, García SiburuNicolás, Calcaterra NB. CNBP modulates the transcription ofWnt signaling pathway components Ezequiel. Biochim. Biophys. Acta. 2014;1839:1151–1160. doi: 10.1016/j.bbagrm.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 51.Zhang K, et al. Structure of the 30 kDa HIV-1 RNA dimerization resource structure of the 30 kDa HIV-1 RNA dimerization signal by a hybrid cryo-EM, NMR, and molecular dynamics approach. Struct. Des. 2018;26:490–498. doi: 10.1016/j.str.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petoukhov, M. V & Tuukkanen, A. SAS-Based Structural Modelling and Model Validation. in Biological Small Angle Scattering: Techniques, Strategies and Tips (eds. Chaudhuri, B., Muñoz, I. G., Qian, S. & Urban, V. S.) 87–105 (Springer, Singapore 2017). [DOI] [PubMed]
- 53.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Rivas E, Clements J, Eddy SR. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat. Methods. 2017;14:45–48. doi: 10.1038/nmeth.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavares RCA, Pyle AM, Somarowthu S. Phylogenetic analysis with improved parameters reveals conservation in lncRNA structures. J. Mol. Biol. 2019;431:1592–1603. doi: 10.1016/j.jmb.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dallaire P, Major F. Exploring alternative RNA structure sets using MC-Flashfold and db2cm. Methods Mol. Biol. 2016;1490:237–251. doi: 10.1007/978-1-4939-6433-8_15. [DOI] [PubMed] [Google Scholar]
- 57.Wu MJ, Andreasson JOL, Kladwang W, Greenleaf W, Das R. Automated design of diverse stand-alone riboswitches. ACS Synth. Biol. 2019;8:1838–1846. doi: 10.1021/acssynbio.9b00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miao Z, et al. RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017;23:655–672. doi: 10.1261/rna.060368.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu X, Zhao C, Chen S-J. VfoldLA: a web server for loop assembly-based prediction of putative 3D RNA structures. J. Struct. Biol. 2019;207:235–240. doi: 10.1016/j.jsb.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding F, Lavender CA, Weeks KM, Dokholyan NV. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods. 2012;9:603–608. doi: 10.1038/nmeth.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cesari A, et al. Fitting corrections to an RNA force field using experimental data. J. Chem. Theory Comput. 2019;15:3425–3431. doi: 10.1021/acs.jctc.9b00206. [DOI] [PubMed] [Google Scholar]
- 62.Huang W, Ravikumar KM, Parisien M, Yang S. Theoretical modeling of multiprotein complexes by iSPOT: Integration of small-angle X-ray scattering, hydroxyl radical footprinting, and computational docking. J. Struct. Biol. 2016;196:340–349. doi: 10.1016/j.jsb.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Afonine PV, et al. New tools for the analysis and validation of Cryo-EM maps and atomic models. Acta Crystallogr. Sect. D Struct. Biol. 2018;D74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan RT, et al. Structural basis for the second step of group II intron splicing. Nat. Commun. 2018;9:4676. doi: 10.1038/s41467-018-06678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hennelly SP, Sanbonmatsu KY. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011;39:2416–2431. doi: 10.1093/nar/gkq1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meier M, et al. Structure and hydrodynamics of a DNA G-quadruplex with a cytosine bulge. Nucleic Acids Res. 2018;46:5319–5331. doi: 10.1093/nar/gky307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reuten R, et al. Structural decoding of netrin-4 reveals a regulatory function towards mature basement membranes. Nat. Commun. 2016;7:13515. doi: 10.1038/ncomms13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franke D, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deo S, et al. Activation of 2′ 5′-oligoadenylate synthetase by stem loops at the 5′-end of the West Nile virus genome. PLoS ONE. 2014;9:e92545. doi: 10.1371/journal.pone.0092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernal I, et al. Molecular organization of soluble type III secretion system sorting platform complexes. J. Mol. Biol. 2019;431:3787–3803. doi: 10.1016/j.jmb.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 71.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wriggers W. Using situs for the integration of multi-resolution structures. Biophys. Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersen EF, et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Lorenz R, Luntzer D, Hofacker IL, Stadler PF, Wolfinger MT. SHAPE directed RNA folding. Bioinformatics. 2016;32:145–147. doi: 10.1093/bioinformatics/btv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl Acad. Sci. U. S. A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leontis, N. B. & Zirbel, C. L. Nonredundant 3D Structure Datasets for RNA Knowledge Extraction and Benchmarking. (Springer, Berlin, Heidelberg, 2012).
- 77.Thiel BC, Beckmann IK, Kerpedjiev P, Hofacker IL. 3D based on 2D: calculating helix angles and stacking patterns using forgi 2.0, an RNA Python library centered on secondary structure elements. F1000Research. 2019;8:287. doi: 10.12688/f1000research.18458.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2014;43:D357–D363. doi: 10.1093/nar/gku1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All of our SAXS data used for modeling have been deposited to the small angle scattering biological data bank (https://www.sasbdb.org/project/939/x6kirb9f97/)78. All other data including atomistic structures and cloning constructs will be made available from the corresponding author upon reasonable request.
All codes (e.g., ERNWIN and in-house python codes for SAXS data processing) are available at https://github.com/pkerpedjiev/ernwin and https://github.com/kimdn/in_house_codes_for_SAXS.