Summary
Bacteria masterfully co-opt and subvert host signal transduction. As a paradigmatic example, Salmonella uses two type-3 secretion systems to inject effector proteins that facilitate Salmonella entry, establishment of an intracellular niche, and modulation of immune responses. We previously demonstrated that the Salmonella anti-inflammatory response activator (SarA; Stm2585, GogC, PagJ, SteE) activates the host transcription factor STAT3 to drive expression of immunomodulatory STAT3-targets. Here we demonstrate—by sequence, function, and biochemical measurement—that SarA mimics the cytoplasmic domain of glycoprotein 130 (gp130; IL6ST). SarA is phosphorylated at a YxxQ motif, facilitating binding to STAT3 with greater affinity than gp130. Departing from canonical gp130 signaling, SarA function is JAK-independent but requires GSK-3, a key regulator of metabolism and development. Our results reveal that SarA undergoes host phosphorylation to recruit a STAT3-activating complex, circumventing cytokine receptor activation. Effector mimicry of gp130 suggests GSK-3 can regulate normal cytokine signaling, potentially enabling metabolic/immune crosstalk.
Keywords: SPI-2, gp130, cytokine receptor, SH2, YxxQ, GSK-3, JAK-STAT, SOCS-3, parallel reaction monitoring, PRM
Graphical Abstract
eTOC
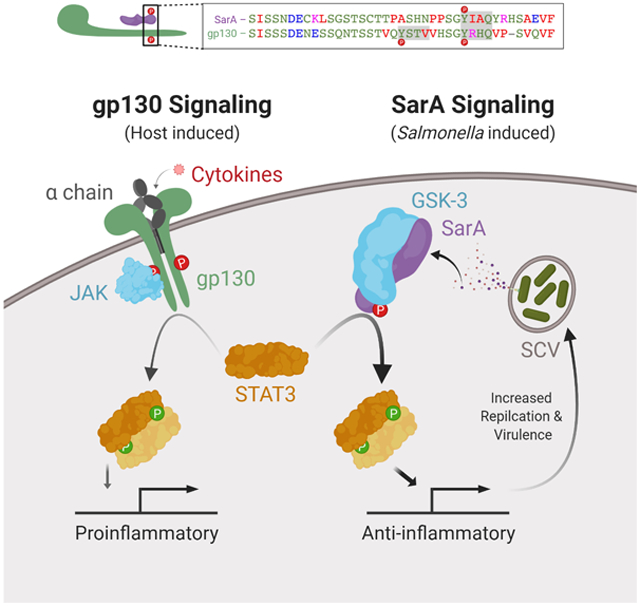
Bacterial effectors manipulate host physiology through diverse mechanisms. Here, Gibbs et al. demonstrate that the secreted Salmonella SarA/SteE effector shares sequence and function with the gp130 cytokine receptor’s intracellular domain. Through a tyrosine-phosphorylated YxxQ motif and recruitment of STAT3 and GSK-3, SarA forms a STAT3-activating complex to reprogram host transcription.
Introduction
Bacteria have evolved an impressive repertoire of mechanisms for regulating host cell biology and signaling through secreted effector proteins (Cui and Shao, 2011; Jimenez et al., 2016). These mechanisms include enzymatically modifying host targets to disrupt cell function, activating host targets to reprogram cellular behavior, and mimicking host signaling.
For Salmonella enterica, secreted effectors are major virulence determinants, which enable Salmonella to cause ~150 million diarrheal illnesses (gastroenteritis), ~0.6 million cases of bacteremia, and ~25 million cases of enteric fever every year, resulting in the highest morbidity of any foodborne pathogen (Kirk et al., 2015). S. enterica’s molecular pathogenesis centers on two type-three secretion systems (T3SS) encoded in Salmonella pathogenicity islands (SPI-1 and SPI-2) (Collazo and Galan, 1996; Galan and Curtiss, 1989; Groisman and Ochman, 1996; Ochman et al., 1996; Shea et al., 1996). The T3SSs deliver many effector proteins that manipulate host cell physiology, enable invasion, and establish a permissive replication niche (Jennings et al., 2017; LaRock et al., 2015).
Previously we showed that the Salmonella effector SarA—also known as Stm2585, GogC (Figueroa-Bossi et al., 2001; Uzzau et al., 2001), PagJ (Navarre et al., 2005), SteE (Niemann et al., 2011)—promotes an anti-inflammatory immune response through tyrosyl phosphorylation and activation of STAT3. We observed SarA-dependent STAT3 activation in B cells, HeLa cells, THP-1 monocytes and mouse spleen during infection (Jaslow et al., 2018). This SarA-dependent reprogramming of host transcription increases intracellular replication and triggers production of the anti-inflammatory cytokine IL-10 in B cells (Jaslow et al., 2018). Recently, SarA has been shown to skew macrophage polarization towards a permissive M2-like phenotype (Stapels et al., 2018). Here, we demonstrate that SarA-mediated STAT3 phosphorylation occurs through SarA imitating the activated intracellular domain of the cytokine receptor common chain gp130 (IL-6 signal transducer; IL6ST; CD130). Specifically, a homologous region in SarA (46% identity) contains a serine-rich stretch and a host-phosphorylated YxxQ motif that facilitates STAT3 binding. Surprisingly, these phosphorylation events are independent of JAK kinases, but require GSK-3, canonically a serine-threonine kinase, that is also present in the SarA-STAT3 complex. Our findings reveal a unique effector mechanism and have important implications for cytokine signaling.
Results
A homologous region in SarA and gp130 are functionally interchangeable
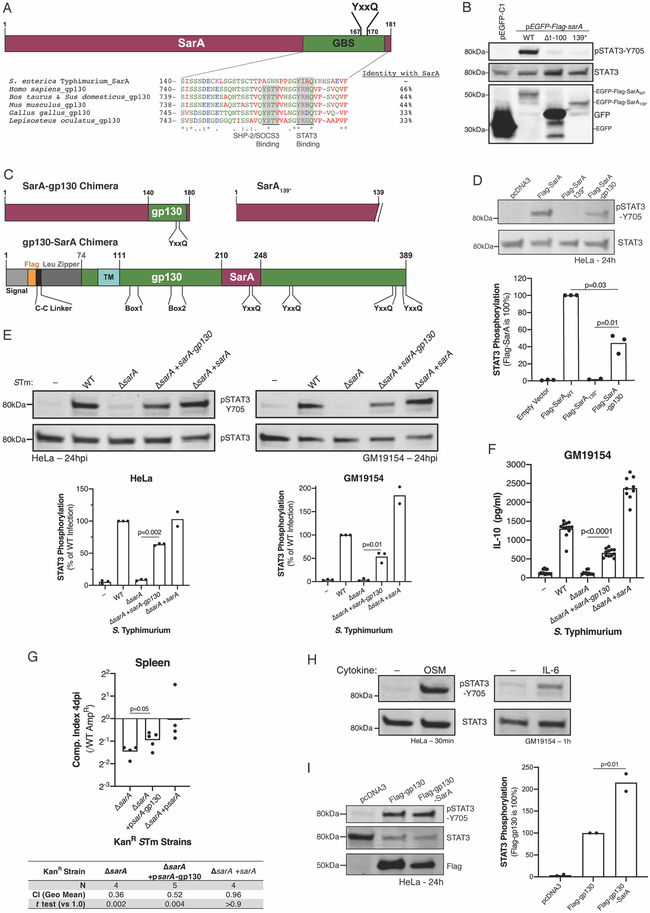
In canonical JAK-STAT signaling, STAT phosphorylation is the culmination of a sequence of tyrosyl phosphorylation events (Stark and Darnell, 2012). As central players, the cytosolic domains of the common chains shared by type-I cytokine receptors (gamma chain, beta chain, and gp130) are tyrosyl phosphorylated to bind JAKs, recruit STATs, and transduce the extracellular signal (Wang et al., 2009). As we previously demonstrated that SarA physically associates with STAT3 and promotes its phosphorylation (Jaslow et al., 2018), we hypothesized that SarA might be performing a function similar to the shared common chains. Therefore, we examined whether SarA shared sequence similarity to the cytosolic portions of each common chain. Pairwise alignment with MATCHER (Rice et al., 2000; Waterman and Eggert, 1987) revealed a segment of 39 amino acids from SarA with high similarity to human gp130 (Figure 1A; similarity score = 50, identity = 46%, similarity = 56%; E-value by pairwise BLAST = 3x10−5). This region is highly conserved among gp130 orthologues (Figure 1A) and contains a YxxQ motif (Y767 in human gp130; Y167 in SarA) that enables direct binding to the SH2 domain of STAT3 (Stahl et al., 1995; Wiederkehr-Adam et al., 2003). Notably, the SarA sequence does not contain a YxxV motif (Y759 in gp130), where the negative regulators SHP-2 (Dittrich et al., 2012) and SOCS3 (Nicholson et al., 2000) bind gp130 to prevent STAT3 activation. We refer to this 39-amino-acid residue stretch of similarity as the Gp130-Binding-of-STAT3 (GBS) sequence. Deletion of SarA’s GBS (SarA139*) prevented STAT3 phosphorylation without altering SarA expression. While the GBS sequence is necessary, N-terminal truncation of SarA (Δ1-100) demonstrates it is insufficient to activate STAT3 (Figure 1B).
Figure 1. SarA functionally mimics part of the gp130 intracellular domain.
(A) The gp130 binding-of-STAT3 (GBS) sequence from S. Typhimurium SarA (amino acids 140-178) aligns with a conserved sequence in gp130 orthologues from human, cow, pig, mouse, chicken, and spotted gar fish. Amino acids are colored by type, and percent identity compared to SarA is listed for each ortholog. (B) SarA lacking the GBS is expressed but fails to drive STAT3-Y705 phosphorylation. HeLas were transfected with 5 μg pEGFP-Flag-sarA WT, Δ1-100 (deletion of SarA’s first 100 amino acids), or 139* (C-terminal truncation removing the GBS) 24 hrs before lysis and immunoblotting. (C) Domain diagrams of SarA-gp130 chimeras. (D) SarA-gp130 chimera drives STAT3 phosphorylation. STAT3 phosphorylation was measured 24 hrs after transfection in three experiments. (E, F) SarA-gp130 chimera under the endogenous promoter complements ΔsarA S Typhimurium induction of (E) STAT3 phosphorylation and (F) IL-10 in HeLa and LCL GM19154 by 24 hpi. No differences were noted in invasion or pyroptosis (Figure S1). IL-10 was measured by ELISA from three experiments. (G) SarA-gp130 chimera partially rescues ΔsarA virulence. Competitive index between AmpR wild type (WT) and KanR strains was measured four days after an intra-peritoneal injection of 1,000 CFUs into C57BL/6J mice. From two experiments. One-sample t tests against zero (no change from 1:1 inoculum) on log10-transformed data. (H) Gp130 ligands drive STAT3 phosphorylation in HeLa and LCL GM19154. Cells were treated with OSM (10 ng/mL) or IL-6 (50 ng/mL) for indicated time. (I) Dimeric gp130-SarA chimera induces more STAT3 phosphorylation than dimeric gp130. HeLa cells were transfected with the indicated constructs and STAT3 phosphorylation was assessed 24 hrs later. Unless noted, all p values are from Welch’s t test.
We next determined whether the gp130 GBS (S740-F777) could functionally substitute for the SarA GBS (Figure 1C). A chimeric expression construct was sufficient to trigger STAT3 phosphorylation in human cells (Figure 1D). We confirmed the activity of the SarA-gp130 chimera by complementing ΔsarA S. Typhimurium with the chimera under the endogenous sarA promoter on a low-copy number plasmid. This complemented strain rescued STAT3 activation (Figure 1E), restored IL-10 induction in an infected lymphoblastoid cell line (LCL) (Figure 1F), and partially rescued ΔsarA S. Typhimurium during competitive infection (Figure 1G).
Conversely, substituting the gp130 GBS with the homologous sequence from SarA also resulted in functional protein. Activation of gp130 normally requires cytokine binding for the active cytokine receptor complex to assemble. As examples, the cytokines OSM (oncostatin M) and IL-6 stimulate STAT3 phosphorylation by, respectively, binding an OSMR and gp130 heterodimer or IL-6R and gp130 heterotetramer (Figure 1H). To circumvent this cytokine requirement and allow assessment of activity in cells with intact gp130, we used a constitutively active gp130, which forces gp130 dimerization through a cysteine linker and the c-Jun leucine zipper (Stuhlmann-Laeisz et al., 2006) (Fig 1C). Replacing the constitutively active gp130 GBS with the corresponding region from SarA (gp130-SarA) resulted in greater activity than the constitutively active gp130 construct alone (Figure 1I). This contrasts with the SarA GBS substitution with the gp130 GBS, which resulted in less activity than wild-type SarA (see Figure 1D). Together, these data show that regions of sequence similarity in SarA and gp130 can functionally substitute for one another and suggest that the SarA GBS sequence is more active.
SarA is phosphorylated at a YxxQ STAT3 binding motif
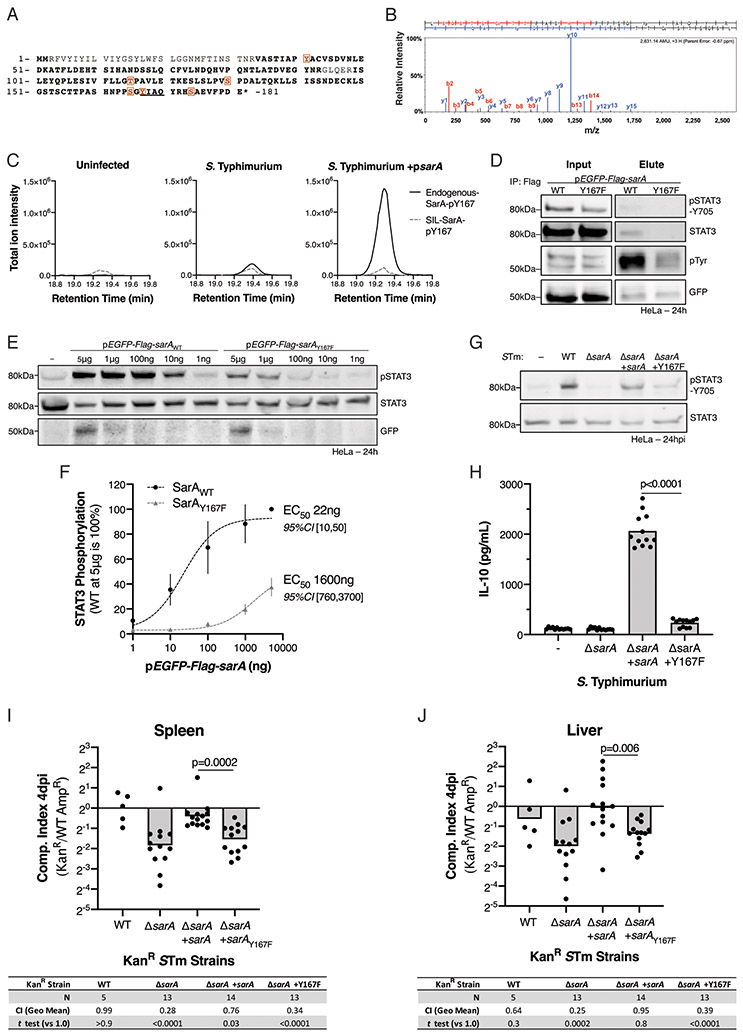
When tyrosyl phosphorylated, the YxxQ motif in gp130 directly binds STAT3 (Stahl et al., 1995; Wiederkehr-Adam et al., 2003). Therefore, we investigated SarA phosphorylation by immunoprecipitation-mass spectrometry (IP-MS). MS analysis revealed that SarA is phosphorylated in HeLa cells at multiple sites when overexpressed (Figure 2A); other modifications, including lysine acetylation and ubiquitination, were not detected. Notably, the highest number of phospho-peptide spectra matched to phospho-Y167 in the SarA YxxQ peptide (Figure 2B). To quantify SarA pY167 phosphopeptide in S. Typhimurium-infected HeLa cells, we used a targeted MS approach with a stable isotope-labeled SarA internal standard. As expected, there was no phospho-SarA detected in the uninfected cells; however, pY167 peptide was detected in cells infected with WT S. Typhimurium and was markedly higher in cells infected with S. Typhimurium expressing SarA from a low-copy number plasmid (Figure 2C). Therefore, SarA Y167 is phosphorylated during Salmonella infection.
Figure 2. Host phosphorylation of SarA-Y167 is required for STAT3 binding and activation.
(A) LC-MS/MS reveals SarA is host phosphorylated in HeLa cells. Orange boxes indicate phosphorylation sites (100% localization probability) and bold represents MS coverage. Confidence of phospho-site localization was determined with the Ascore algorithm. (B) MS/MS spectrum of pY167-containing peptide. Only matching y+ and b+ ions are shown. (C) A targeted MS approach, which uses a stable isotope-labeled internal standard (SIL-SarA), detects endogenous pY167 phosphopeptide in HeLas infected with either S. Typhimurium WT or ΔsarA complemented with sarA on a low-copy plasmid. Total ion chromatograms of matching product ions are shown for the internal standard (dashed grey) and endogenous (solid black) pY167 phosphopeptides. Individual ion traces, including phosphosite-localizing ions, are shown in Figure S2. (D) SarA-Y167 phosphorylation is necessary for STAT3 binding. HeLa cell were lysed 24 hrs after transfection with pEGFP-Flag-sarA, then precipitated with anti-Flag M2 beads. (E,F) SarAY167F mutant induces less STAT3 phosphorylation. STAT3 phosphorylation quantified relative to 5 μg SarAWT transfection in four replicates across four experiments. EC50 calculated from dose-response curve assuming a Hill coefficient of 1. (G,H) SarAY167F fails to complement S. Typhimurium (STm) ΔsarA 24 hpi. LCL GM19154 supernatant IL-10 was measured 24 hpi in twelve replicates across three experiments. (I,J) SarAY167F does not restore virulence in ΔsarA STm at 4 days after an intraperitoneal injection of 1,000 CFUs into C57BL/6J mice. Five experiments. One outlier in ΔsarA +sarAy167F (CI = 39 in spleen and 98 in liver) was identified using Grubbs’ (α = 0.0001) and excluded. P values in tables from one-sample t test against 0 (no change from 1: 1 inoculum) using log10-transformed ratios. Displayed p values are from Welch’s t test.
Mutation of Y167 to the non-phosphorylatable phenylalanine (Y167F) resulted in a large reduction of the phosphotyrosine signal from immunoprecipitated Flag-SarA (Figure 2D). In agreement with the gp130 YxxQ motif’s function, this mutation also impaired SarA binding to STAT3. In contrast to wild-type SarA, immunoprecipitation of SarA Y167F from HeLa cells demonstrated no detectable binding of STAT3 (Figure 2D).
Based on these findings, we hypothesized that tyrosyl phosphorylation at Y167 is crucial for SarA activity. Indeed, expression of SarA Y167F in HeLa cells demonstrated reduced, though not absent activity (Figure 2E). Higher levels of transfection could compensate for low activity, with approximately 70-fold more SarA Y167F plasmid being required for comparable activity to wild-type SarA (Figure 2F). When infecting cells, ΔsarA S. Typhimurium complemented with SarA Y167F reduced STAT3 phosphorylation (Figure 2G) and IL-10 induction (Figure 2H). Crucially, Y167 was necessary for complementation in vivo. In competitive intraperitoneal infection, SarA Y167F was unable to restore ΔsarA S. Typhimurium virulence (Figure 2I, J). These data indicate that a phosphorylated SarA YxxQ motif is crucial for STAT3 recruitment and activation. Further, the inability of Y167F to complement ΔsarA during infection indicates SarA principally affects virulence by activating STAT3.
Phosphorylated SarA peptide containing YxxQ binds STAT3 with greater affinity than gp130 phosphopeptide
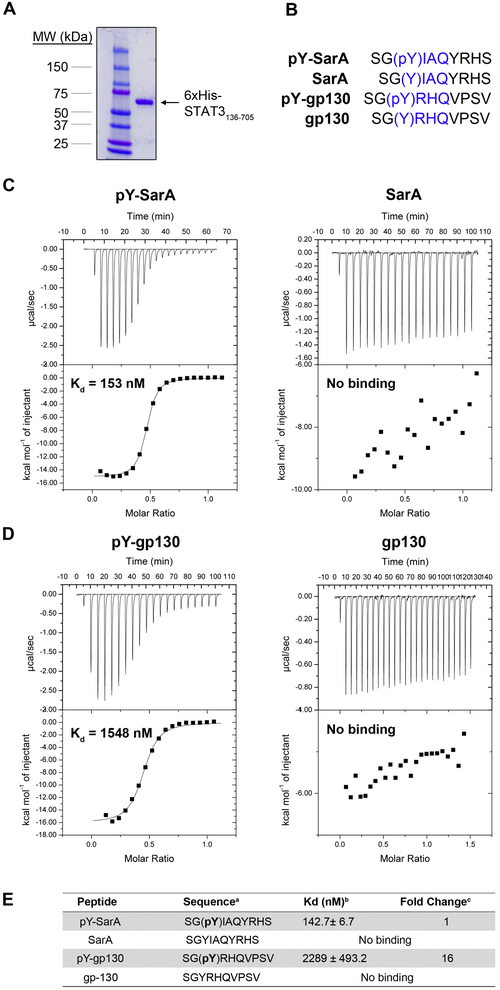
To quantify the importance of SarA Y167 phosphorylation on direct STAT3 interaction, we measured SarA-STAT3 binding affinity using isothermal titration calorimetry (ITC). STAT3 was purified as described (Takakuma et al., 2013) (Figure 3A). We compared STAT3 binding of peptides consisting of ten amino acid residues from within the GBS that spanned the YxxQ motif either with or without tyrosyl phosphorylation (Figure 3B). Strikingly, SarA phospho-peptide bound STAT3 with an affinity of 142.7 ± 6.7 nM, while unphosphorylated peptide had no detectable binding (Figure 3C). Thus, phosphorylation of Y167 is essential for high-affinity binding of STAT3 to the SarA YxxQ motif. Similarly, a gp130 phosphopeptide containing YxxQ bound to STAT3, but its affinity (2289 ± 493.2 nM) was ~16-fold less (Figure 3D, E). This is consistent with a previously reported ID50 of 5.3 μM (Wiederkehr-Adam et al., 2003). Cumulatively, our results show that SarA functionally mimics gp130 by using pYxxQ to bind STAT3.
Figure 3. Quantification of binding affinities of STAT3 to YxxQ containing peptides demonstrates STAT3 binds SarA with greater affinity than gp130.
(A) Purified 6xHis-STAT3136-705. (B) Sequences of the SarA and gp130 peptides used for ITC measurements. YxxQ motifs are highlighted in blue. (C) Y167-phosphorylated SarA peptide binds STAT3 with high affinity. (D) gp130 peptide binds STAT3 in a manner dependent on phosphorylation of Y767. (E) Mean Kd measurements of SarA, pSarA, gp130, and pgp130 binding to STAT3 ± SEM. Fold change represents the ratio of the Kd of the pY-gp130 peptide bound to STAT3 to the Kd of pY-SarA peptide bound to STAT3. Each ITC experiment was repeated 3-5 times with a representative titration curve shown. Thermograms and integrated heats are shown in the top and bottom of each panel, respectively. Solid lines represent the best fit to the corresponding data points using a non-linear least squares fit with the one-set-of-sites model.
SarA requires GSK3 but not JAK for STAT3 activation
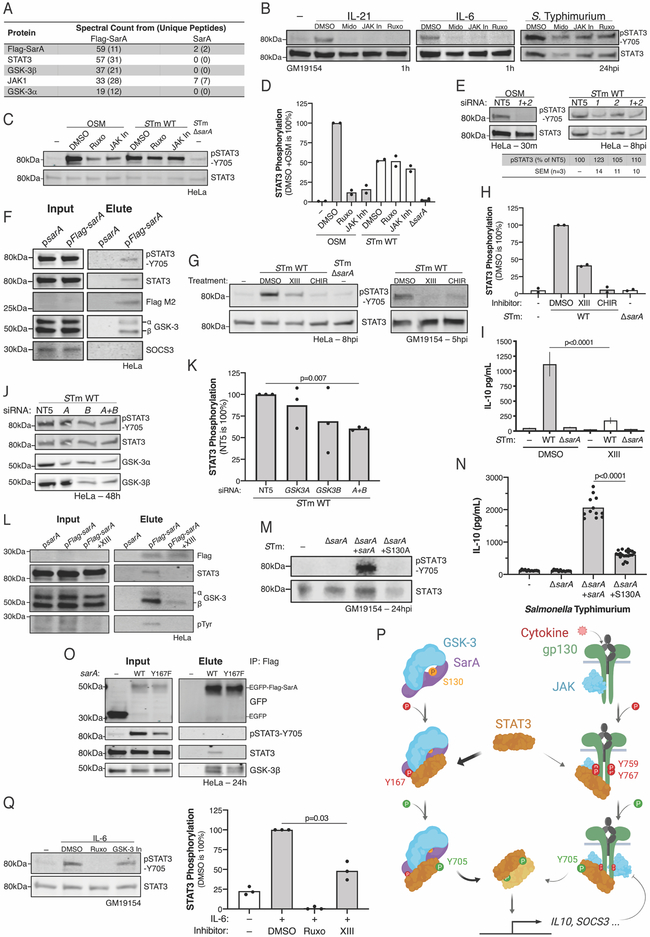
In addition to binding STAT3, the gp130 intracellular domain has at least two additional functions: (1) recruitment of JAK kinases to drive STAT phosphorylation (Lutticken et al., 1994; Stahl et al., 1994) and (2) recruitment of SHP-2 (Dittrich et al., 2012; Schaper et al., 1998; Symes et al., 1997) and SOCS3 (Nicholson et al., 2000) to dampen JAK-STAT signaling (Silver and Hunter, 2010). To determine if SarA has analogous functions, we conducted a discovery IP-MS screen to identify any host kinases or negative regulators that interact with SarA. This screen confirmed SarA binding to STAT3 and suggested additional components of the complex (Figure 4A; Table S1). Notably, SHP-2 (PTPN11) and SOCS3 were not detected in the IP-MS, consistent with SarA’s GBS lacking the YxxV (Y759) motif found in gp130.
Figure 4. SarA interaction with GSK-3, but not JAK1, is required for STAT3 activation.
(A) IP-MS identified SarA-STAT3 signaling complex components. Peptide spectral counts from the indicated number of unique peptides. (B-D) JAK inhibitors block gp130-ligands’ induction of STAT3 phosphorylation without affecting SarA-mediated STAT3 phosphorylation in LCL (B) and in HeLa (C, D). LCL GM19154 was pre-treated for 1 hr with 50 μM Midostaurin (Mido), 50 μM JAK Inhibitor I (JAK In), or 50 μM Ruxolitinib (Ruxo) before 1 hr stimulation with IL-6 (50ng/ml) or IL-21 (50ng/ml) or infection with S. Typhimurium (STm). HeLa cells were identically pre-treated before 30 min stimulation with OSM (10ng/mL) or infection with STm. (E) RNAi confirms STAT3 phosphorylation by SarA is JAK independent, unlike OSMR signaling. HeLas were pre-treated with JAK1 and/or JAK2 siRNA for 48 hrs before infection with STm or stimulation with OSM (10ng/mL) for indicated time. qPCR confirmed knockdown exceeded 3-fold each time. (F) Co-immunoprecipitation (coIP) confirms GSK-3α and β, but not SOCS3, interact with SarA. HeLa cells were transfected with codon-optimized pcDNA3.1-sarA or pcDNA3.1-Flag-sarA. (G-I) GSK-3 inhibition demonstrates that GSK-3 enzymatic activity is necessary for SarA-induced STAT3 phosphorylation in HeLa and LCL GM19154. HeLas were pre-treated with 15 μM GSK-3 Inhibitor XIII (XIII) or 15 μM CHIR-98014 (CHIR) for 1 hr before STm infection (MOI 5) and harvested 8 hpi. LCLs were pre-treated for 1 hr with 12 μM GSK-3 Inhibitor XIII or 500 nM CHIR-98014 before STm infection (MOI 30) and lysed 5 hpi. IL-10 in LCL supernatant (I) is eight replicates across three experiments. P value from Sidak’s post-hoc comparison of WT infection after two-way ANOVA (STm genotype, treatment, and interaction term all significant with p<0.0001). Bars are mean ±SEM. (J, K) Knockdown of GSK3A and GSK3B confirms partial dependence of SarA on GSK-3. HeLa were pre-treated identically to D. (L) GSK-3 inhibitor blocks SarA interaction with STAT3 and GSK-3, which prevent STAT3 and SarA phosphorylation. coIP performed like E with 5 μM GSK-3 Inhibitor XIII added 1 h after transfection. (M, N) S130A mutation reduces complementation during STm infection. P value from Welch’s t test. (O) SarA Y167F still associates with GSK-3. Anti-Flag M2 coIP and blotting performed same as figure 2D. (P) Model of the SarA-GSK-3 complex activating STAT3 compared to JAK1-gp130 activation of STAT3. (Q) GSK-3 inhibition reduces IL-6-induced STAT3 phosphorylation in LCLs. GM19154 was pre-treated with 15 μM GSK-3 inhibitor XIII or 50 μM Ruxolitinib for 1 hr before stimulation with IL-6 (100ng/mL) for 1 hr. Blot is representative of three experiments which are quantified relative to uninhibited IL-6 treatment. P value from Welch’s t test on log transformed ratio.
JAK1 peptides were enriched in the Flag-SarA IP-MS analysis (Figure 4A). This is despite the absence of “Box1” and “Box2” motifs in SarA, which are required for gp130 binding JAK1 (Narazaki et al., 1994). However, JAK activity was not necessary for SarA-dependent STAT3 phosphorylation. Treatment of infected LCLs with three different JAK inhibitors had minimal effect on STAT3 phosphorylation following S. Typhimurium infection, despite completely blocking IL-6- or IL-21-stimulated STAT3 phosphorylation (Figure 4B). We confirmed this result in infected HeLa cells, where JAK inhibitors reduced OSM-stimulated STAT3 phosphorylation without markedly inhibiting SarA-dependent STAT3 phosphorylation (Figure 4C, D). Similarly, RNAi depletion of JAK1 & JAK2 did not affect STAT3 activation following S. Typhimurium infection (Figure 4E). These data did not agree with established mechanisms of STAT activation but were consistent with previous reports of Salmonella-mediated STAT3 activation being JAK-independent (Hannemann et al., 2013).
An inspection of the interactome data for alternative kinases revealed relatively high levels of GSK-3α and GSK-3β (Figure 4A). This was particularly intriguing because STAT3 activation by IFN receptor stimulation is regulated by GSK-3 (Beurel and Jope, 2008). Further, SarA contains a phosphorylated SLPV(pS)P GSK-3 binding motif (see Figure 2A). The presence of GSK-3 in the SarA-STAT3 complex was verified by Flag-SarA immunoprecipitation and immunoblotting for GSK-3α and GSK-3β, while no interaction with SOCS3 was observed (Figure 4F). Unlike treatment with JAK inhibitors, treating infected cells with GSK-3 inhibitors reduced SarA-dependent STAT3 phosphorylation (Figure 4G, H) and IL-10 production (Figure 4I). The functional importance of GSK-3 was confirmed using RNAi against GSK3A and/or GSK3B, with combined knockdown having the greatest effect (Figure 4J, K).
We next examined if GSK-3 activity is required for formation of the SarA signaling complex by inhibiting GSK-3 during Flag-SarA immunoprecipitation. This GSK-3 inhibition reduced GSK-3 recruitment to SarA and abrogated both STAT3-SarA interaction and SarA phosphorylation (Figure 4L). These data indicate that GSK-3 enzymatic activity is necessary (1) to recruit GSK-3 to the SarA-STAT3 complex, (2) for SarA tyrosyl phosphorylation, and thereby, (3) for SarA interaction with STAT3.
To test if the putative GSK-3-binding motif in SarA, SLPV(pS)P, regulates GSK-3 recruitment, we mutated SarA S130, which is serine phosphorylated in host cells, to alanine. This mutation did not affect STAT3 phosphorylation following SarA overexpression (Figure S3); however, complementation with SarA S103A on low-copy number bacterial plasmid during ΔsarA S. Typhimurium infection reduced STAT3 phosphorylation (Figure 4M) and IL-10 production (Figure 4N). Thus, similar to the SarA Y167F mutation, S130A has a partial decrease in activity that can be most clearly observed in the context of Salmonella infection. In contrast to S130A, Y167F still binds to GSK-3 (Figure 4O).
Our results suggest a model in which SarA binds to GSK-3, with serine phosphorylation of SarA S130 facilitating this GSK-3 recruitment (Figure 4P). Then GSK-3 promotes the phosphorylation of SarA’s YxxQ motif within a larger stretch of homology to the gp130 cytokine receptor common chain. Binding of STAT3 is facilitated by both SarA’s YxxQ motif and GSK-3 activity.
A role for GSK-3 in gp130 signaling
While GSK-3 has been reported to regulate STAT3 activation in response to interferons (Beurel and Jope, 2008), it is not known if GSK-3 regulates signaling through the gp130 cytokine receptor family. Stimulation of LCLs with IL-6, which signals through gp130, produced robust STAT3 activation. JAK inhibitors caused a near complete block of STAT3 phosphorylation in this context, while GSK-3 inhibitors also showed a significant, partial decrease (Figure 4Q). Thus, GSK-3 may be broadly relevant for cytokine receptor signaling.
Discussion
Our work shows that SarA mimics the activated, phosphorylated cytokine co-receptor, gp130, to reprogram host cells. SarA has evolved more robust activity than gp130, due to greater affinity for STAT3 and a lack of recruitment of the negative regulators SHP-2 and SOCS3. We are not aware of other examples of a bacterial effector mimicking a cytokine receptor, although viral mimics exist (Alcami, 2003).
The evolution of effectors to be more effective than their mammalian counterparts is a recurring theme in bacterial pathogenesis. Yersinia secretes a phosphatase (YopH), structurally similar to eukaryotic phosphatases (Stuckey et al., 1994), that has the greatest catalytic activity of any characterized phosphatase (Sun et al., 2003; Zhang et al., 1992). Similarly, the SPI-2 effector SseJ outcompetes host effectors for binding to RhoA (LaRock et al., 2012). This highlights the selective pressure for pathogen effectors to drive supraphysiological responses that enable replication and dissemination whereas normal signaling has pressure toward a more measured and regulated response that enables homeostasis and a return to a healthy state.
In light of previous gp130 studies, SarA’s ability to activate STAT3 more robustly than normal gp130 signaling implies SarA drives a non-canonical, anti-inflammatory response. Normally, IL-6 stimulation of the gp130/STAT3 pathway results in rapid but relatively short-lived activation of STAT3 and pro-inflammatory gene expression (El Kasmi et al., 2006; Jones, 2005). However, mutation of the SOCS3/SHP-2 binding site of gp130 or deletion of SOCS3 disrupts a key negative feedback loop (Silver and Hunter, 2010). This results in prolonged and sustained gp130-mediated activation of STAT3 that switches the normally pro-inflammatory IL-6 signal to an anti-inflammatory response akin to IL-10 signaling (El Kasmi et al., 2006; Yasukawa et al., 2003). Our results indicate SarA has evolved more robust recruitment of STAT3 than gp130 while avoiding negative regulation by lacking the SOCS3/SHP-2 binding site. This activation likely mimics the constitutively active gp130 to provide a sustained STAT3 response that is anti-inflammatory in nature. This idea is supported by SarA driving expression of anti-inflammatory target genes including IL-10, SOCS3, BCL-3, and others (Jaslow et al., 2018) and SarA-induced polarization of macrophages to an IL4RA-expressing M2-like phenotype (Stapels et al., 2018).
Additionally, by shifting the dependence of signal transduction from JAK to GSK-3, SarA circumvents the dependence on cytokine activation and instead relies on a kinase that is highly active at baseline in unstimulated cells (Wang et al., 2014). While our data demonstrates a strong dependence on GSK-3 for SarA activation, our experiments with GSK-3 inhibitors on gp130 signaling demonstrate a moderate phenotype. We hypothesize that GSK-3 plays an accessory role in gp130 signaling, whereas SarA has evolved to co-opt GSK-3 into a more central role. While GSK-3 is canonically a S/T kinase, tyrosyl auto-phosphorylation (Y279 for GSK-3α and Y216 for GSK-3β) does enhance activity (Cole et al., 2004; Lochhead et al., 2006). Indeed, complementary work has shown GSK-3 to be the cognate tyrosine kinase of both SarA and STAT3 (Panagi et al., this issue). Their work also underscores the broad relevance of SarA, as they characterize SarA-STAT3 activation in primary murine macrophages. The ability for GSK-3 to regulate gp130, a type-I cytokine receptor, in addition to its known regulation of the type-II cytokine receptor IFNγ (Beurel and Jope, 2008) and TLR response (Martin et al., 2005), suggests a wider role for GSK-3 in immune regulation and may present an opportunity for metabolic and immune cross-talk during infection and autoimmunity. This warrants further investigation.
Finally, our findings may provide one mechanism by which Salmonella infection affects risk of cancer and autoimmunity. Specific epithelial and immune cell subtypes that have been reprogrammed by SarA may continue to impact the immune system, either through prolonged STAT3 activation or downstream consequences. Future work will focus on these possible long-term consequences of Salmonella infection on immune cell function in infection, autoimmunity, and cancer.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information, as well as plasmids and bacterial strains generated for this study, are available by request from the Lead Contact, Dennis Ko (dennis.ko@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Cells
Lymphoblastoid cell lines (LCLs; EBV-immortalized B cells) were from the Coriell Institute. HeLa cells were from the Duke Cell Culture Facility (originally from ATCC). LCLs were maintained at 37°C in a 5% CO2 atmosphere and were grown in RPMI 1640 media (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin-G, and 100 mg/ml streptomycin. HeLa cells were grown in DMEM supplemented with 10% FBS, 100 U/ml penicillin-G, and 100 mg/ml streptomycin. Infection assays were carried out in the same media but without pen/strep and phenol red. For generation of a stable isotope-labeled SarA standard, cells were conditioned for >6-doublings in DMEM for SILAC (Invitrogen) supplemented with 25 mg/L of 13C615N2-Lys and 13C615N2-Arg, 10 mg/L L-Pro and 10% dialyzed FBS (Sigma). Cells were verified as mycoplasma free by the Universal Mycoplasma Detection Kit (ATCC).
Bacteria
Salmonella enterica serovar Typhimurium (STm) strain 14028s and other strains (Table S2) were grown at 37°C and 250 rpm in Luria-Bertani (LB) broth (Thermo Fisher Scientific). To quantify invasion during invasion assays, Salmonellae were tagged with an inducible GFP plasmid [pMMB67GFP from (Pujol and Bliska, 2003)], which carries GFP under an IPTG-inducible promoter and is maintained with 100 μg/mL ampicillin. For SPI-1 induction, overnight cultures were diluted 1:33 and grown for another 2hrs and 40min before infection. For bacterial complementation, SarA and its endogenous promoter (+786 bp 5’) were cloned into a pWSK129 (Wang and Kushner, 1991) and maintained with 50 μg/mL kanamycin. For complementation with the SarA-gp130 chimera, the native bacterial sequence with the E. coli expression optimized gp130 segment (aa 740-777) was cloned into pUC57-Kan with the same endogenous promoter as pWSK129-sarA.
METHOD DETAILS
Sequence comparisons
Pairwise alignment of SarA vs. cytosolic domains of type I cytokine receptor common chains— gp130 (IL6ST) 642-918, beta common chain (CSF2RB) 461-897, gamma common chain (IL2RG) 284-369—was carried out using MATCHER (Rice et al., 2000; Waterman and Eggert, 1987). Gap open penalty and gap extend were set to 12 and 4. CLUSTAL Omega (Sievers et al., 2011) alignment was carried out with default parameters with EBI online tools (Madeira et al., 2019). Pairwise NCBI BLASTp of the 39 amino acid stretch of similarity was carried out with default parameters (Johnson et al., 2008).
Plasmids
Plasmids are listed in Table S3. GenScript supplied the pcDNA3.1-Flag-sarA and pcDNA3.1-Flag-sarA-gpl30 chimera codon-optimized for mammalian expression as well as the pUC57-sarA-gpl30 optimized for E. coli expression. Site-directed mutagenesis was carried out using Quikchange Lightning Site-Directed Mutagenesis Kit (Agilent, # 210518) (primers in Table S4).
Cellular treatments
JAK was inhibited by 1 hr pre-treatment with 50 μM JAK1 Inhibitor I (SCBT sc-204021) or 50 μM Ruxolitinib (APExBIO A3012) before cytokine stimulation or infection. GSK-3 was inhibited by pre-treatment at indicated concentrations with CHIR-98014 (Selleckchem S2745) or GSK-3 Inhibitor XIII (SCBT sc-203987) for 1 hr before infection or stimulation. Recombinant human cytokines, OSM (PeproTech 300-10), IL-6 (PeproTech 200-06), and IL-21 (PeproTech 200-21), were resuspended in water and incubated with cells for indicated time and concentration.
Transfections
HeLa cells were seeded at 1.75 x 105 cells per well in 6-well TC-treated plates 24 hours before transfection. Transfection was accomplished with Lipofectamine 3000 (ThermoFisher, Catalog #L3000008). Cells were harvested 24 hours post transfection.
Immunoblotting
Cells were lysed with RIPA lysis buffer: 50mM Tris-HCl pH 7.4, 150mM NaCl, 0.1% SDS, 0.5% NaDeoxycholate, 1% Triton X-100, cOmplete Mini protease inhibitor cocktail (Millipore Sigma #11836170001), 10mM NaF, and 1mM Na Orthovanadate. SDS-PAGE was performed using Mini-PROTEAN TGX Stain-Free Precast 4-20% gels (Bio-Rad #456-8096). The gels’ stain-free dye was activated by a 5 min UV exposure and protein was transferred to Immun-Blot low-fluorescence PVDF membrane (Bio-Rad #162-0264) using Hoefer TE77X. Primary antibodies used were: anti-FLAG M2 (Sigma F3165, RRID:AB_259529), anti-pY705-STAT3 clone D3A7 (CST #9145, RRID:AB_2491009), anti-STAT3 clone 124H6 (CST #9139, RRID:AB_331757), anti-GFP clone D5.1 (CST #2956, RRID:AB_1196615), anti-GSK-3α clone D80E6 (CST #4337, RRID:AB_10859910), anti-GSKβ clone D5C5Z (CST #12456, RRID:AB_2636978), and anti-pTyr-1000 monoclonal pool (CST #8954, RRID:AB_2687925). Blots were developed using LI-COR infrared secondary antibodies (IRDye 800CW Donkey anti-Rabbit IgG and IRDye 680LT Donkey anti-Mouse IgG) and imaged on a LI-COR Odyssey Classic. Total protein was measured following a 30 sec UV exposure. Band integrated intensity was quantified using LI-COR Odyssey Imaging System Software v3.0. Background was subtracted using the median of the top and bottom of the band. STAT3 phosphorylation was calculated as pSTAT3-Y705 integrated intensity over total STAT3 integrated intensity and then set relative to a control as specified in the figure. Statistics calculated on log transformation of the experimental to control ratio.
Infection assays
Assaying infection of LCLs and HeLa cells was done as previously described (Ko et al., 2009). Overnight cultures of Salmonella in LB media were sub-cultured 1:33 and grown for 160 minutes at 37°C and 250 rpm. LCLs at 100,000 in 100 μl were infected at multiplicity of infection (MOI) 30 in 96-well non-TC plates. HeLa cells at 300,000 in 2 mL were infected at MOI 5 in 6-well TC-treated plates. Gentamicin was added 1 hpi at 50 μg/mL to kill the extracellular bacteria. For 24 hr incubations, gentamicin was diluted to 15 μg/mL at 2 hpi. To induce GFP, IPTG was added 75 min prior to the desired timepoint. Infection and cell death were measured with a Guava EasyCyte Plus flow cytometer (Millipore). Cell death was measured by 7AAD (7-aminoactinomycin D; Enzo Life Sciences) staining. IL-10 protein in the LCL supernatant at 24 hpi was measured by human IL-10 ELISA (R&D systems Catalog #DY217B).
Immunoprecipitation
HeLa cells were seeded at 1 x 106 cells per 10 cm dish 24 hours before transfection with codon-optimized pcDNA3.1-Flag-sarA (GenScript) or pcDNA3.1-sarA as described above. Three plates per condition were lysed in 1mL of 0.1% Triton X-100, 50mM Tris pH 7.4, 150mM NaCl, cOmplete Mini protease inhibitor cocktail (Millipore Sigma #11836170001), 10mM NaF, and 1mM Na Orthovanadate. Lysate was incubated with anti-FLAG M2 magnetic beads (Millipore Sigma #M8823) for 4 hrs while rotating at 4°C then washed 3x with 10x volumes of lysis buffer using a DynaMag Spin Magnet (Invitrogen #12320D). Bound protein was eluted with 45 μl of 0.1M Glycine-HCl buffer pH 3 and neutralized with 5 μl of 0.5M Tris-HCl pH 7.4 1.5M NaCl. Eluted proteins were analyzed by immunoblot.
Mass spectrometry
Immunoprecipitates were separated by SDS-PAGE (NuPage 4-20% Bis-Tris) for 5 min followed by Colloidal Blue staining. The entirety of the stained region was excised and subjected to in-gel digestion with trypsin (Wilm et al., 1996). Lyophilized peptides were reconstituted in 10 μl of 1% (v/v) TFA and 2% (v/v) acetonitrile (MeCN) and analyzed by liquid chromatography, tandem mass spectrometry (LC-MS/MS) using a Waters NanoACQUITY interfaced to a Thermo Q-Exactive HF MS. Briefly, the LC used a trapping configuration followed by reversed-phase separation on a 75 μM x 25 cm analytical column using a 5-30% MeCN gradient over 90 min; and MS used Top12 data-dependent acquisition method (MS1 120,000 resolution, AGC target 1E6 and 50 ms max. IT; MS2 30,000 resolution, AGC target 5E4 and 45 ms max. IT). Mascot generic files were created from the raw data using Thermo Proteome Discoverer and database searching was performed using Mascot 2.4 (Matrix Sciences) using a Swissprot human database (downloaded on 08/16/16) appended with Flag-SarA sequence (20,204 total entries). Search parameters included: 5 ppm MS1 and 0.02 Da MS2 tolerance; fixed carbamidomethyl(C); and variable deamidation(NQ), oxidation(M), acetylation(protein N-termini) and phosphorylation(S/T/Y). Data was annotated at a 1% peptide false discovery in Scaffold (Proteome Software). Confidence of site localization was evaluated using the Ascore algorithm (Beausoleil et al., 2006) in Scaffold PTM (Proteome Software).
For targeted quantification of the SarA pYXXQ peptide, stable isotope-labeled SarA (SIL-SarA) was purified from HeLa cells grown in SILAC DMEM media and transfected with pcDNA3.1-Flag-sarA as described above. One-third of the SIL-SarA from a single 15 cm dish of transfected HeLa cells was spiked into 400 μg of HeLa cell lysate (uninfected, or infected with WT or SarA- overexpressing Salmonella) followed by digestion with 20 μg Sequencing Grade Modified Trypsin (Promega) for 2 h using an S-Trap mini device (Protifi) using the manufacturers protocol. Phosphopeptides were enriched with titanium oxide as previously described (Foster et al., 2017). After lyophilization, peptides were reconstituted in 12 μl of 1% TFA/2% MeCN containing 10 mM citrate, and 4 μl was analyzed by parallel reaction monitoring (PRM). The LC separation was as described above except that a 30 min gradient from 5-30% MeCN was used. The MS analysis used a Fusion Lumos MS (Thermo). A precursor (MS1) scan was collected from 0-30 min at 60,000 resolution, AGC target 2E5 and 50 ms max. IT; Orbitrap tMS2 scans were collected from 16-25 min using a 1.2 m/z isolation width, 120,000 resolution, AGC target 1E5, 246 ms max. IT and HCD fragmentation with a stepped collision energy of 30 ± 5%. The inclusion list targeted LSGSTSC[+57.021464]TTPASHNPPSGY[+79.966331]IAQYR (light; 878.05477 m/z; charge state 3) and LSGSTSC[+57.021464]TTPASHNPPSGY[+79.966331]IAQYR (heavy; 881.39086 m/z; charge 3). Data was analyzed in Skyline as previously described (Foster et al., 2017).
Protein expression and purification
Codon-optimized human STAT3 136-705 (hSTAT3136-705) was cloned into the pET28a vector, in-frame with the N-terminal 6xHis tag (GenScript). This clone was transformed into the E. coli OverExpress C41(DE3) strain (Lucigen). Expression of hSTAT3136-705 was induced by growing cultures to an OD of 0.6 and inducing with 0.2 M IPTG for 3 hours at 30°C (Takakuma et al., 2013). Cultures were centrifuged at 4000 rpm for 10 minutes at 4°C and the resulting bacterial cell pellets were stored in −80°C. Cell pellets were lysed using a microfluidizer (Microfluidics) in lysis buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl and 5 mM β-ME). Clarified cell lysate was obtained by centrifugation of lysed cells at 12,000 rpm for 20 minutes at 4°C. 6xHis-hSTAT3136-705 was purified from the lysate using Ni-NTA (Qiagen) affinity chromatography, the resin of which was pre-equilibrated with lysis buffer. After lysate loading, the resin was washed with buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM β-ME and 10 mM imidazole. The protein was eluted in a stepwise manner using elution buffer (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM β-ME, 10% glycerol) containing concentrations of imidazole ranging from 20 mM to 250 mM. Germane fractions of the 6xHis-hSTAT3136-705 were combined and furthered purified using a Superdex S200 gel filtration column (GE Healthcare) in gel filtration buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 5 mM β-ME, 10% glycerol).
Isothermal Titration Calorimetry
6xHis-hSTAT3136-705 and all tested peptides (Genscript) were diluted in gel filtration buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl, 5 mM β-ME, 10% glycerol). Each ITC run included an initial 2 μL injection, followed by 20-25 injections of 8 μL of 600 μM peptide into a cell containing 6xHis-hSTAT3136-705 at 60 μM (monomeric concentration). ITC measurements were performed at 25°C using a VP-ITC (Malvern Panalyticals) with a stirring speed of 307 rpm. After data from the relevant controls were subtracted, the data were analyzed using the manufacturer supplied software package, Origin 7.0 (OriginLab Corp). The first data point from each experiment is omitted from the analysis.
RNAi experiments
HeLa cells at 300,000 in 6-well TC-treated plates were treated for 2 days with 0.33 μM total siRNA from either non-targeting siGENOME (Dharmacon) siRNA #5 (NT5; #D-001210-05) or SMARTpool directed against human GSK3A (#M-003010-03) and/or GSK3B (#M-003009-01), or JAK1 (#M-003145-02) and/or JAK2 (#M-003146-02). Infections were conducted as described above.
Mouse Infections
Salmonellae were cultured as above then washed with PBS. Litter and sex matched C57BL/6J mice from the Duke DLAR breeding core were infected by intraperitoneal injection of 1,000 CFUs in a 1:1 mixture of Amp resistant WT S. Typhimurium (+pWSK29) and the indicated Kan resistant strain (+pWSK129 constructs) when 10 to 18 weeks old. Spleens and livers were collected four days post infection, homogenized by bead beating with ZrO beads (GlenMills #7305-000031) in a Mini-BeadBeater-24 (Biospec #112011). Appropriate volumes were plated on LB agar with Amp (100 μg/mL) or Kan (50 μg/mL) to calculate Kan/Amp CFU ratios. One-way and Welch’s t tests were performed on log-transformed ratios. Infections were approved by Duke IACUC (protocol #A145-18-06).
QUANTIFICATION AND STATISTICAL ANALYSIS
Descriptive statistics and visualization
Descriptive statistics were performed with GraphPad Prism 8 (GraphPad Software, US). The size of each study or number of replicates, along with the statistical tests performed can be found in figure legends. Numerical data are presented as the mean ± SEM (standard error of mean).
DATA AND CODE AVAILABILITY
The published article includes all datasets generated during this study.
Supplementary Material
Figure S1. Complementation with endogenous chimera does not affect Salmonella invasion or host cell pyroptosis, Related to Figure 1E. Invasion and pyroptosis of GM19154 was measured 3 hpi after 70 min of IPTG treatment to induce GFP in infected cells and 5 min of 7AAD staining to mark dead cells. Percent GFP+ and 7AAD+ quantified by flow cytometry.
Figure S2. Targeted quantification of pTyr167 peptide by parallel reaction monitoring, Related to Figure 2C. (A) The endogenous pTyr167 peptide was identified on the basis of the labeled y- and b-ions, which are as follows: y10+, 1231.5507 m/z; y8+, 1037.4452 m/z; y7+, 950.4132 m/z; y19++, 1093.9699 m/z; y15++, 869.3909 m/z; y13++, 785.3459 m/z; y10++, 616.2790 m/z; b4+, 345.1769 m/z. (B-C) Ion chromatograms of endogenous (light) pTyr167 peptide enriched from HeLa cells infected with (B) WT Salmonella or (C) SarA-overexpressing Salmonella. Ions are labeled as in (A). Phosphosite-localizing ions (y7 and y8) are labeled.
Figure S3. Overexpression of SarA S130A induces STAT3 phosphorylation with similar efficiency as wild-type SarA, Related to Figure 4M. HeLa cells transfected with increasing pEGFP-Flag-sarA (1 ng to 5 μg) 24 hrs before lysis and immunoblotting. Wild-type SarA had an EC50 of 22 ng (95% CI [10,50]) while the S130A mutant had a similar EC50 of 30 ng (95% CI [13,71]). Dots are means from four experiments; bars are SEM.
Table S2. Bacterial strains, Related to STAR Methods.
Table S3. Plasmids, Related to STAR Methods.
Table S4. Primers, Related to STAR Methods.
Table S1. SarA vs. Flag-SarA IP-MS results, Related to Figure 4A.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-FLAG M2 | Sigma | F3165, RRID:AB_259529 |
| anti-pY705-STAT3 clone D3A7 | CST | #9145, RRID:AB_2491009 |
| anti-STAT3 clone 124H6 | CST | #9139, RRID:AB_331757 |
| anti-GFP clone D5.1 | CST | #2956, RRID:AB_1196615 |
| anti-GSK-3α clone D80E6 | CST | #4337, RRID:AB_10859910 |
| anti-GSK-3β clone D5C5Z | CST | #12456, RRID:AB_2636978 |
| and anti-pTyr-1000 monoclonal pool | CST | #8954, RRID:AB_2687925 |
| Bacterial and Virus Strains | ||
| See Table S2 for strains derived from S. enterica serovar Typhimurium 14028s | ATCC | 14028; CDC 6516-60 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| JAK1 Inhibitor I | SCBT | sc-204021 |
| Ruxolitinib | APExBIO | A3012 |
| CHIR-98014 | Selleckchem | S2745 |
| GSK-3 Inhibitor XIII | SCBT | Sc-203987 |
| OSM | PeproTech | 300-10 |
| IL-6 | PeproTech | 200-06 |
| IL-21 | PeproTech | 200-21 |
| Critical Commercial Assays | ||
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | 210518 |
| Lipofectamine 3000 | ThermoFisher | #L3000008 |
| human IL-10 ELISA | R&D systems | #DY217B |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Lymphoblastoid cell lines | Coriell | GM19154 |
| HeLa cells | Duke Cell Culture Facility, originally from ATCC | CCL-2 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Jackson lab | 000664 |
| Oligonucleotides | ||
| See Table S4 for list of primers. | IDT | See Table S4. |
| Recombinant DNA | ||
| See Table S3 for list of plasmids. | GenScript or this manuscript | See Table S3. |
| Software and Algorithms | ||
| Matcher | Rice et al., 2000; Waterman and Eggert, 1987 | https://www.ebi.ac.uk/Tools/psa/emboss_matcher/ |
| CLUSTAL Omega | Sievers et al., 2011 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Pairwise NCBI BLASTp | Johnson et al., 2008 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| LI-COR Odyssey Imaging System Software | LI-COR | v3.0 |
| Scaffold Proteome Software | Proteome Software | http://www.proteomesoftware.com/ |
| Scaffold PTM | Proteome Software | http://www.proteomesoftware.com/ |
| Skyline | MacCoss Lab Software | https://skyline.ms/project/home/software/skyline/begin.view |
| Origin 7.0 | OriginLab Corp. | https://www.originlab.com/ |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/ |
| Other | ||
Highlights.
The bacterial effector SarA shares sequence and function with gp130 cytokine coreceptor
Phosphorylation of SarA’s YxxQ motif facilitates direct binding to STAT3
SarA driven STAT3 activation requires interaction with active GSK-3
SarA has greater affinity for STAT3 than gp130 and lacks the SOCS3 interaction
Acknowledgments
KDG, SLJ, JSB, RG, and DCK were supported by R01AI118903. KDG, EJW, JSB, and DCK were supported by R21AI146520. KDG was supported by F31AI136313. JSB was supported by F31AI143147. We thank Dr. Samuel Miller and members of the Ko lab for useful discussion. We thank the Duke School of Medicine for providing access to the Proteomics and Metabolomics Shared Resource and acknowledge the National Institutes of Health for funding the acquisition of the Fusion Lumos Tribid (Thermo) instrument used for targeted MS analysis (Grant 1S10OD0224999-01). We thank Dominick J. Jenkins for assistance in carrying out immunoblots.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcami A (2003). Viral mimicry of cytokines, chemokines and their receptors. Nature reviews Immunology 3, 36–50. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, and Gygi SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Beurel E, and Jope RS (2008). Differential regulation of STAT family members by glycogen synthase kinase-3. J Biol Chem 283, 21934–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A, Frame S, and Cohen P (2004). Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. The Biochemical journal 377, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo CM, and Galan JE (1996). Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun 64, 3524–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, and Shao F (2011). Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem Sci 36, 532–540. [DOI] [PubMed] [Google Scholar]
- Dittrich A, Quaiser T, Khouri C, Gortz D, Monnigmann M, and Schaper F (2012). Model-driven experimental analysis of the function of SHP-2 in IL-6-induced Jak/STAT signaling. Mol Biosyst 8, 2119–2134. [DOI] [PubMed] [Google Scholar]
- El Kasmi KC, Holst J, Coffre M, Mielke L, de Pauw A, Lhocine N, Smith AM, Rutschman R, Kaushal D, Shen Y, et al. (2006). General nature of the STAT3-activated anti-inflammatory response. J Immunol 177, 7880–7888. [DOI] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Uzzau S, Maloriol D, and Bossi L (2001). Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Molecular microbiology 39, 260–271. [DOI] [PubMed] [Google Scholar]
- Foster MW, Gwinn WM, Kelly FL, Brass DM, Valente AM, Moseley MA, Thompson JW, Morgan DL, and Palmer SM (2017). Proteomic Analysis of Primary Human Airway Epithelial Cells Exposed to the Respiratory Toxicant Diacetyl. J Proteome Res 16, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, and Curtiss R 3rd (1989). Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86, 6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, and Ochman H (1996). Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87, 791–794. [DOI] [PubMed] [Google Scholar]
- Hannemann S, Gao B, and Galén JE (2013). Salmonella modulation of host cell gene expression promotes its intracellular growth. PLoS Pathog 9, e1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaslow SL, Gibbs KD, Fricke WF, Wang L, Pittman KJ, Mammel MK, Thaden JT, Fowler VG Jr., Hammer GE, Elfenbein JR, et al. (2018). Salmonella Activation of STAT3 Signaling by SarA Effector Promotes Intracellular Replication and Production of IL-10. Cell Rep 23, 3525–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings E, Thurston TLM, and Holden DW (2017). Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences. Cell Host Microbe 22, 217–231. [DOI] [PubMed] [Google Scholar]
- Jimenez A, Chen D, and Alto NM (2016). How Bacteria Subvert Animal Cell Structure and Function. Annu Rev Cell Dev Biol 32, 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, and Madden TL (2008). NCBI BLAST: a better web interface. Nucleic acids research 36, W5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA (2005). Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol 175, 3463–3468. [DOI] [PubMed] [Google Scholar]
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, et al. (2015). World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med 12, e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, Shukla KP, Fong C, Wasnick M, Brittnacher MJ, Wurfel MM, Holden TD, O'Keefe GE, Van Yserloo B, Akey JM, et al. (2009). A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease. Am J Hum Genet 85, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock DL, Brzovic PS, Levin I, Blanc MP, and Miller SI (2012). A Salmonella typhimurium-translocated glycerophospholipid:cholesterol acyltransferase promotes virulence by binding to the RhoA protein switch regions. J Biol Chem 287, 29654–29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock DL, Chaudhary A, and Miller SI (2015). Salmonellae interactions with host processes. Nat Rev Microbiol 13, 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, and Cleghon V (2006). A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Molecular cell 24, 627–633. [DOI] [PubMed] [Google Scholar]
- Lutticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga T, et al. (1994). Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science 263, 89–92. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic acids research 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narazaki M, Witthuhn BA, Yoshida K, Silvennoinen O, Yasukawa K, Ihle JN, Kishimoto T, and Taga T (1994). Activation of JAK2 kinase mediated by the interleukin 6 signal transducer gp130. Proc Natl Acad Sci U S A 91, 2285–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, and Libby SJ (2005). Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Molecular microbiology 56, 492–508. [DOI] [PubMed] [Google Scholar]
- Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, et al. (2000). Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proc Natl Acad Sci U S A 97, 6493–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann GS, Brown RN, Gustin JK, Stufkens A, Shaikh-Kidwai AS, Li J, McDermott JE, Brewer HM, Schepmoes A, Smith RD, et al. (2011). Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect Immun 79, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Soncini FC, Solomon F, and Groisman EA (1996). Identification of a pathogenicity island required for Salmonella survival in host cells. Proceedings of the National Academy of Sciences 93, 7800–7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, and Bliska JB (2003). The ability to replicate in macrophages is conserved between Yersinia pestis and Yersinia pseudotuberculosis. Infect Immun 77, 5892–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, and Bleasby A (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 76, 276–277. [DOI] [PubMed] [Google Scholar]
- Schaper F, Gendo C, Eck M, Schmitz J, Grimm C, Anhuf D, Kerr IM, and Heinrich PC (1998). Activation of the protein tyrosine phosphatase SHP2 via the interleukin-6 signal transducing receptor protein gp130 requires tyrosine kinase Jak1 and limits acute-phase protein expression. The Biochemical journal 335 (Pt 3), 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, and Holden DW (1996). Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proceedings of the National Academy of Sciences 93, 2593–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JS, and Hunter CA (2010). gp130 at the nexus of inflammation, autoimmunity, and cancer. J Leukoc Biol 88, 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S, et al. (1994). Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science 263, 92–95. [DOI] [PubMed] [Google Scholar]
- Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE Jr., and Yancopoulos GD (1995). Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science 267, 1349–1353. [DOI] [PubMed] [Google Scholar]
- Stapels DAC, Hill PWS, Westermann AJ, Fisher RA, Thurston TL, Saliba AE, Blommestein I, Vogel J, and Helaine S (2018). Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362, 1156–1160. [DOI] [PubMed] [Google Scholar]
- Stark GR, and Darnell JE (2012). The JAK-STAT pathway at twenty. In Immunity, pp. 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, and Saper MA (1994). Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature 370, 571–575. [DOI] [PubMed] [Google Scholar]
- Stuhlmann-Laeisz C, Lang S, Chalaris A, Krzysztof P, Enge S, Eichler J, Klingmuller U, Samuel M, Ernst M, Rose-John S, et al. (2006). Forced dimerization of gp130 leads to constitutive STAT3 activation, cytokine-independent growth, and blockade of differentiation of embryonic stem cells. Mol Biol Cell 77, 2986–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JP, Wu L, Fedorov AA, Almo SC, and Zhang ZY (2003). Crystal structure of the Yersinia protein-tyrosine phosphatase YopH complexed with a specific small molecule inhibitor. J Biol Chem 278, 33392–33399. [DOI] [PubMed] [Google Scholar]
- Symes A, Stahl N, Reeves SA, Farruggella T, Servidei T, Gearan T, Yancopoulos G, and Fink JS (1997). The protein tyrosine phosphatase SHP-2 negatively regulates ciliary neurotrophic factor induction of gene expression. Curr Biol 7, 697–700. [DOI] [PubMed] [Google Scholar]
- Takakuma K, Ogo N, Uehara Y, Takahashi S, Miyoshi N, and Asai A (2013). Novel multiplexed assay for identifying SH2 domain antagonists of STAT family proteins. PLoS One 8, e71646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, and Bossi L (2001). Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A 98, 15264–15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kumar A, Lamont RJ, and Scott DA (2014). GSK3beta and the control of infectious bacterial diseases. Trends Microbiol 22, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, and Garcia KC (2009). Structural biology of shared cytokine receptors. Annual review of immunology 27, 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MS, and Eggert M (1987). A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J Mol Biol 197, 723–728. [DOI] [PubMed] [Google Scholar]
- Wiederkehr-Adam M, Ernst P, Muller K, Bieck E, Gombert FO, Ottl J, Graff P, Grossmuller F, and Heim MH (2003). Characterization of phosphopeptide motifs specific for the Src homology 2 domains of signal transducer and activator of transcription 1 (STAT1) and STAT3. J Biol Chem 275, 16117–16128. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, and Mann M (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, et al. (2003). IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol 4, 551–556. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Clemens JC, Schubert HL, Stuckey JA, Fischer MW, Hume DM, Saper MA, and Dixon JE (1992). Expression, purification, and physicochemical characterization of a recombinant Yersinia protein tyrosine phosphatase. J Biol Chem 267, 23759–23766. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Complementation with endogenous chimera does not affect Salmonella invasion or host cell pyroptosis, Related to Figure 1E. Invasion and pyroptosis of GM19154 was measured 3 hpi after 70 min of IPTG treatment to induce GFP in infected cells and 5 min of 7AAD staining to mark dead cells. Percent GFP+ and 7AAD+ quantified by flow cytometry.
Figure S2. Targeted quantification of pTyr167 peptide by parallel reaction monitoring, Related to Figure 2C. (A) The endogenous pTyr167 peptide was identified on the basis of the labeled y- and b-ions, which are as follows: y10+, 1231.5507 m/z; y8+, 1037.4452 m/z; y7+, 950.4132 m/z; y19++, 1093.9699 m/z; y15++, 869.3909 m/z; y13++, 785.3459 m/z; y10++, 616.2790 m/z; b4+, 345.1769 m/z. (B-C) Ion chromatograms of endogenous (light) pTyr167 peptide enriched from HeLa cells infected with (B) WT Salmonella or (C) SarA-overexpressing Salmonella. Ions are labeled as in (A). Phosphosite-localizing ions (y7 and y8) are labeled.
Figure S3. Overexpression of SarA S130A induces STAT3 phosphorylation with similar efficiency as wild-type SarA, Related to Figure 4M. HeLa cells transfected with increasing pEGFP-Flag-sarA (1 ng to 5 μg) 24 hrs before lysis and immunoblotting. Wild-type SarA had an EC50 of 22 ng (95% CI [10,50]) while the S130A mutant had a similar EC50 of 30 ng (95% CI [13,71]). Dots are means from four experiments; bars are SEM.
Table S2. Bacterial strains, Related to STAR Methods.
Table S3. Plasmids, Related to STAR Methods.
Table S4. Primers, Related to STAR Methods.
Table S1. SarA vs. Flag-SarA IP-MS results, Related to Figure 4A.
Data Availability Statement
The published article includes all datasets generated during this study.