Abstract
Ferroptosis is an iron-dependent programmed cell death event, whose regulation and physiological significance remain to be elucidated. Analyzing transcriptional responses of mouse embryonic fibroblasts exposed to the ferroptosis inducer erastin, here we found that a set of genes related to oxidative stress protection is induced upon ferroptosis. We considered that up-regulation of these genes attenuates ferroptosis induction and found that the transcription factor BTB domain and CNC homolog 1 (BACH1), a regulator in heme and iron metabolism, promotes ferroptosis by repressing the transcription of a subset of the erastin-induced protective genes. We noted that these genes are involved in the synthesis of GSH or metabolism of intracellular labile iron and include glutamate-cysteine ligase modifier subunit (Gclm), solute carrier family 7 member 11 (Slc7a11), ferritin heavy chain 1 (Fth1), ferritin light chain 1 (Ftl1), and solute carrier family 40 member 1 (Slc40a1). Ferroptosis has also been previously shown to induce cardiomyopathy, and here we observed that Bach1−/− mice are more resistant to myocardial infarction than WT mice and that the severity of ischemic injury is decreased by the iron-chelator deferasirox, which suppressed ferroptosis. Our findings suggest that BACH1 represses genes that combat labile iron-induced oxidative stress, and ferroptosis is stimulated at the transcriptional level by BACH1 upon disruption of the balance between the transcriptional induction of protective genes and accumulation of iron-mediated damage. We propose that BACH1 controls the threshold of ferroptosis induction and may represent a therapeutic target for alleviating ferroptosis-related diseases, including myocardial infarction.
Keywords: cardiovascular disease, cell death, glutathione peroxidase, iron, ischemia, transcription repressor, (BTB domain and CNC homolog 1) BACH1, ferroptosis, iron sequestration, myocardial infarction
Introduction
Ferroptosis is a new form of programmed cell death caused by the iron-dependent accumulation of lipid hydroperoxide (1, 2). As a pathological cell death, ferroptosis causes various oxidative stress-related diseases, including ischemia-reperfusion injury (3–7) and neurodegenerative diseases (8, 9). Ferroptosis also contributes to tumor suppression as a response induced by p53 and is important for organisms in preventing cancer (2, 10–13). Considering the involvement of lipid hydroperoxide, ferroptosis may be executed at the edge of the oxidative stress response. Therefore, ferroptosis may be a regulated process involving the oxidative stress response. However, the regulatory mechanism underlying ferroptosis has yet to be elucidated in full. In particular, it is not known at all how the regulatory genes of ferroptosis are transcriptionally controlled in induction of ferroptosis.
BTB and CNC homology 1 (BACH1)2 is a heme-binding transcription factor required for the proper regulation of the oxidative stress response and metabolic pathways related to heme and iron (14). BACH1 represses Hmox1 encoding heme oxygenase-1 (HO-1), Fth1 and Ftl1 encoding ferritin proteins, Gclm and Gclc encoding glutamate-cysteine ligase modifier and catalytic subunits, and other genes involved in the oxidative stress response (15–17). We hypothesized that BACH1 might regulate ferroptosis by inhibiting the expression of these genes. In addition, although BACH1 is involved in the exacerbation of various diseases involving oxidative stress, such as ischemic heart disease (18), hyperoxic lung injury (19), trinitrobenzene sulfonic acid-induced colitis (20), nonalcoholic steatohepatitis (21), and spinal cord injury (22), it is unknown whether or not they are attributable to ferroptosis. We considered that BACH1 may exacerbate the severity of these diseases through ferroptosis.
To understand the regulatory mechanism underlying ferroptosis, we analyzed the transcriptome response in ferroptotic cells with RNA-Sequence (RNA-Seq). We also examined whether or not BACH1 was involved in the regulation of ferroptosis by comparing ferroptosis and the expression of ferroptosis-induced genes between wildtype (WT) and Bach1−/− mouse embryonic fibroblasts (MEFs). Furthermore, we assessed the influence of BACH1 and ferroptosis on the severity of acute myocardial infarction (AMI) in model mice. We found that BACH1 promoted ferroptosis by directly repressing genes involved in the synthesis of glutathione (GSH) and sequestration of free labile iron. BACH1 also increased the severity of AMI, which was mitigated by the iron chelator deferasirox (DFX). Our findings highlight the coordinated transcriptional response and its regulation by BACH1 upon ferroptosis.
Results
Regulatory genes of ferroptosis show compensatory up-regulation in ferroptotic cells
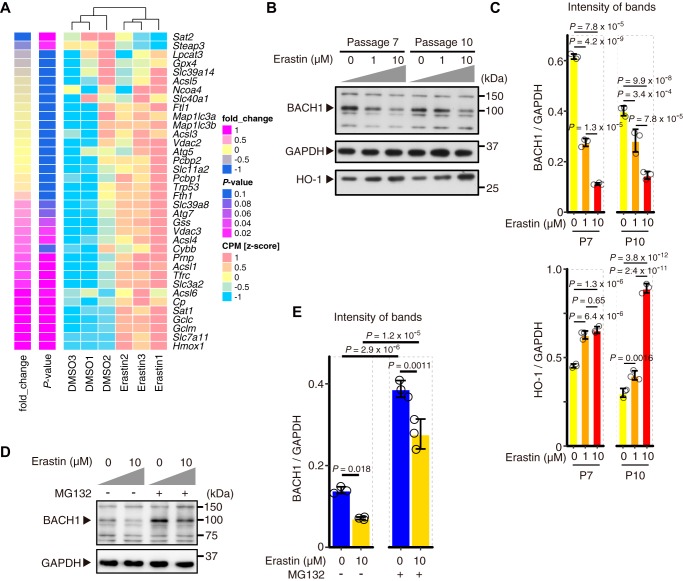
Changes in metabolic and biological processes occur in ferroptotic cells (23, 24). To determine the changes in transcriptome upon ferroptosis, we treated MEFs with erastin, a class I ferroptosis inducer (1, 2), and carried out RNA-seq analyses. Genes related to oxidative stress and iron metabolism showed significant induction in their expression (Fig. 1A). Many of these genes, for example, Slc7a11, Gclm, Gclc, are considered to possess inhibitory effects on ferroptosis (25). Therefore, ferroptosis accompanies the induction of genes that can restrict the execution of ferroptosis.
Figure 1.
Regulatory genes of ferroptosis are up-regulated with decreased BACH1 protein in the induction of ferroptosis. A, RNA-seq was performed in WT MEFs (9th passage: P9) with only DMSO (DMSO group) or DMSO + 3 μm erastin (Erastin group) for 24 h. A heat map of gene expression profiles shows the genes registered to map04216 (Ferroptosis pathway) of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map. The genes were arranged from the bottom in the order of fold-change of erastin group to DMSO group. n = 3 per group. B and C, Western blotting for BACH1, HO-1, and GAPDH of WT MEFs (7th passage: P7, 11th passage: P11) exposed to erastin for 12 h. Representative image (B) and the intensity of bands (C). D and E, Western blotting for BACH1 and GAPDH in WT MEFs (8th passage) exposed to erastin and 25 μm MG132 for 6 h. Representative image (D) and the intensity of bands (E). In A, p value by the differential expression analysis performed on edge R. In C and E, error bars represent S.D. p value by Tukey's test after two-way ANOVA.
Among the induced genes, Hmox1 encoding HO-1 is reported to be associated with ferroptosis (26, 27) and is a negatively regulated target of BACH1 (15). Slc7a11 encodes a component of system xc− (cystine/glutamine transporter) (28, 29) and a well-known regulator of ferroptosis (10). Gclm and Gclc encode glutamate-cysteine ligase modifier and catalytic subunits (30, 31), both considered to suppress ferroptosis by GSH synthesis (25, 32). These genes for the pathway of GSH synthesis are also considered to be targets of BACH1 (17). Indeed, the amount of BACH1 protein was decreased in MEFs exposed to erastin, which was accompanied by the induction of Hmox1 (Fig. 1, B and C, and Fig. S1). Although BACH1 protein accumulated after proteasome inhibition by MG132, the degradation of BACH1 in response to erastin was not completely blocked (Fig. 1D and E). These observations suggest that BACH1 protein is degraded by proteasome-dependent and -independent mechanisms during ferroptosis. With the reduction in BACH1 protein, the production of its mRNA was induced (Fig. S1), suggesting the presence of feedback regulation of BACH1.
These observations suggest that, when cells are exposed to erastin, the expression of genes that counteract ferroptosis is induced in part by a reduction in BACH1 protein and that the amount or activity of BACH1 and the kinetics of its feedback regulation may influence ferroptosis by suppressing this counteracting subprogram of ferroptosis.
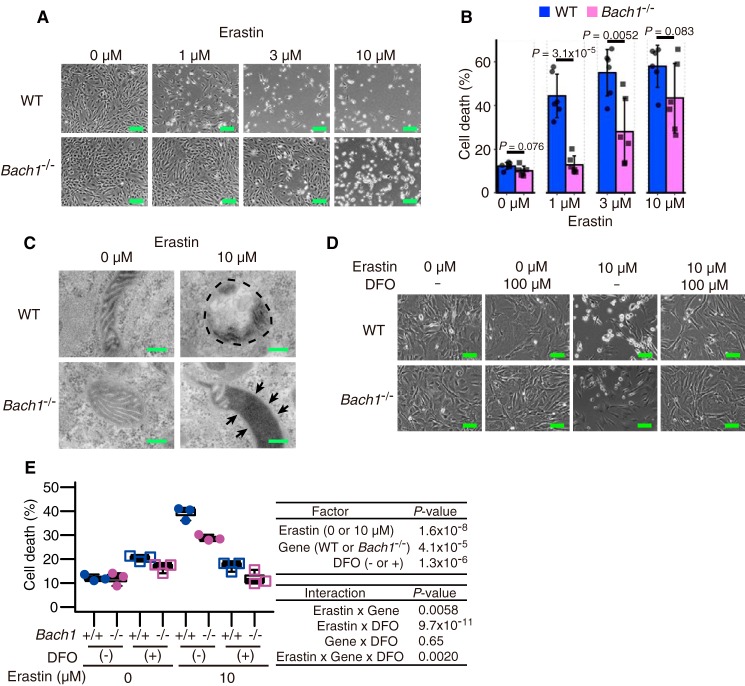
BACH1 promotes ferroptosis
To clarify whether or not BACH1 regulates ferroptosis, we treated WT and Bach1−/− MEFs with erastin, stained them with propidium iodide (PI) and annexin V, and compared the cell death by a flow cytometry analysis (Fig. S2). Bach1−/− MEFs showed less cell death in response to erastin than WT cells (Fig. 2, A and B). When the erastin-treated MEFs were observed with a transmission electron microscope, mitochondrial spheroids, which reflect oxidative stress (33), were frequently observed in WT MEFs (Fig. 2C). Although mitochondrial spheroids were not clear in Bach1−/− MEFs exposed to erastin, condensed matrix structures, which are also observed as parts of mitochondrial spheroids (33), were observed (Fig. 2C). These results may reflect lower oxidative stress and higher resistance to erastin of Bach1−/− MEFs than WT MEFs. The cell death in our experiments was inhibited by the iron chelator deferoxamine (DFO) (Fig. 2, D and E), confirming that this death was ferroptosis. These results showed that BACH1 promoted ferroptosis in MEFs.
Figure 2.
BACH1 promotes ferroptosis. A and B, optical microscope image (A) and quantification of cell death by flow cytometer (B) of WT and Bach1−/− MEFs (11th passage: P11) exposed to erastin for 24 h. Scale bars in A represent 100 μm. C, transmission electron microscope image of WT and Bach1−/− MEFs (P9) exposed to erastin for 10 h. The portion surrounded by dashed line: a mitochondrial spheroid. Arrow: mitochondrial inner membrane-matrix condensation. Scale bars represent 200 nm. D and E, optical microscope image (D) and quantification of cell death by flow cytometer (E) of WT and Bach1−/− MEFs (P12) exposed to erastin and DFO for 24 h. Scale bars in D represents 200 μm. A, B, D, and E, representative of three independent experiments. Error bars of B represent S.D. The box and whisker plots of E show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. p value of B by t test. p value of E by three-way ANOVA.
It should be noted that Bach1−/− MEFs lost their resistance to ferroptosis under high doses of erastin (Fig. 2, A and B). Although decreased BACH1 by erastin is considered as a protective response to ferroptosis (Fig. 1, A–C), there is also its limitation to the inhibition of cell death. However, at least at the early stage, the reduction in BACH1 protein may be a part of the ferroptosis program, and BACH1 may set the threshold for ferroptosis. Execution of ferroptosis may be determined by the basal amount of BACH1 and how rapidly it is degraded in response to ferroptosis inducers.
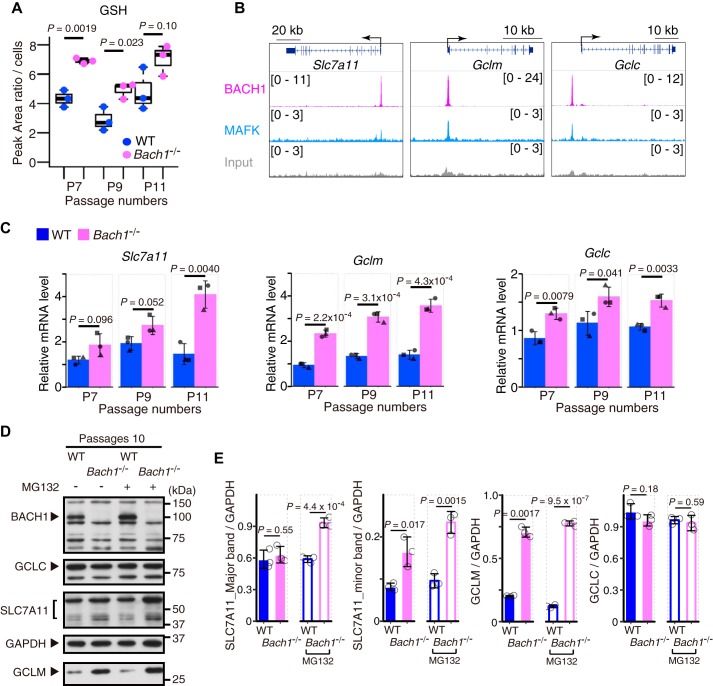
BACH1 represses the expression of genes involved in the GSH synthesis pathway
GSH is the key regulator of ferroptosis (1, 2) and the genes involved in the pathway of GSH synthesis were up-regulated as BACH1 protein decreases (Fig. 1, A–C). Therefore, BACH1 may decrease GSH by repressing the expression of these genes. To investigate this possibility, we measured the intracellular GSH concentrations in WT and Bach1−/− MEFs. Because the sensitivity to ferroptosis decreased as we passaged MEFs (Fig. S3, A and B), we performed the measurement of GSH and the subsequent analysis of gene expression at three different stages of primary culture (7th, 9th, and 11th passage; P7, P9, and P11). The amounts of GSH at P7 and P9 were significantly higher in Bach1−/− MEFs than in WT cells and a similar trend was observed at P11 (Fig. 3A), suggesting that BACH1 promoted ferroptosis by reducing GSH within cells.
Figure 3.
BACH1 decreases GSH by repressing the transcription of Slc7a11, Gclm, and Gclc. A, intracellular concentration of GSH in WT and Bach1−/− MEFs (7th, 9th, and 11th passage: P7, P9, and P11) by UHPLC/MS/MS. B, ChIP-seq analysis of the binding of BACH1, MAFK for gene regions of Slc7a11, Gclm, and Gclc in M1 cells. C, qPCR analysis for Slc7a11, Gclm, and Gclc mRNA relative to Actb mRNA in WT and Bach1−/− MEFs (P7, P9, P11). n = 3 of independent lots of MEFs per genotype. D and E, Western blotting for BACH1, SLC7A11, GCLM, GCLC, and GAPDH in WT and Bach1−/− MEFs (P10) exposed to 25 μm MG132. Representative image (D) and the intensity of bands (E). The box and whisker plots of A show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. Error bars of C and E represent S.D. p value of A, C, and E by t test.
By revisiting our previous data of ChIP with sequencing (ChIP-Seq) of BACH1 in mouse myeloblast M1 cells (34, 35), we found peaks of BACH1 and its partner MAFK in the regulatory regions of genes encoding molecules for GSH synthesis, including Gclm, Gclc, and Slc7a11 (Fig. 3B). Furthermore, by comparing the expression of these genes in WT and Bach1−/− MEFs by quantitative PCR (qPCR), the expression of all of these genes was confirmed to be higher in Bach1−/− MEFs than in WT cells (Fig. 3C). These results suggested that BACH1 bound to the regulatory regions of these genes to repress their expression.
A comparison of the protein amounts of SLC7A11, GCLM, and GCLC in MEFs by Western blotting revealed that more GCLM protein was present in Bach1−/− MEFs than in WT cells (Fig. 3, D and E, and Fig. S4, A and B). Although the amounts of SLC7A11 protein were similar in WT and Bach1−/− MEFs (Fig. 3, D and E, and Fig. S4, A and B), more SLC7A11 protein was present in Bach1−/− MEFs than in WT cells when they were treated with proteasome inhibitor MG132 (Fig. 3, D and E). These observations suggest that the amount of SLC7A11 protein is further tuned by proteasomal-mediated degradation. There were no marked differences in the amount of GCLC protein with or without MG132 (Fig. 3, D and E, and Fig. S4, A and B). BACH1 may affect the expression of GCLC protein under certain circumstances. Given these results, we surmised that BACH1 decreased the amount of GSH by repressing the expression of Gclm and in part Slc7a11.
Knockdown of the genes in GSH synthesis pathway and Hmox1 increases the sensitivity of ferroptosis
We next examined whether or not the resistance of Bach1−/− MEFs against ferroptosis was actually dependent on the increased expression of the genes involved in the GSH synthesis pathway. Although it is not always statistically significant, knockdown of any of Slc7a11, Gclm, and Gclc resulted in slight but reproducible increases in ferroptosis in both WT and Bach1−/− MEFs (Figs. S5, A–C; S6, A–C; and S7, A–C). These results show that the genes involved in the GSH synthesis pathway have inhibitory effects against ferroptosis and suggest that BACH1 promotes ferroptosis by repressing their expression. We next examined the effect of knockdown of Hmox1. WT MEFs became more sensitive to ferroptosis by knockdown of Hmox1 than cells with control knockdown (Figs. S5D, S6D, and S7D). We thus concluded that HO-1 works as an inhibitor of ferroptosis under our experimental conditions. However, the effect of HO-1 to accelerate ferroptosis has also been reported (7, 26). The function of HO-1 in ferroptosis might differ depending on the situations of cells. Importantly, knockdown of Slc7a11, Gclm, Gclc, or Hmox1 did not decrease the observed differences in ferroptosis between WT and Bach1−/− MEFs (Fig. S7). These results suggest that the role of BACH1 in promoting ferroptosis depends on the repression of multiple genes involved in ferroptosis.
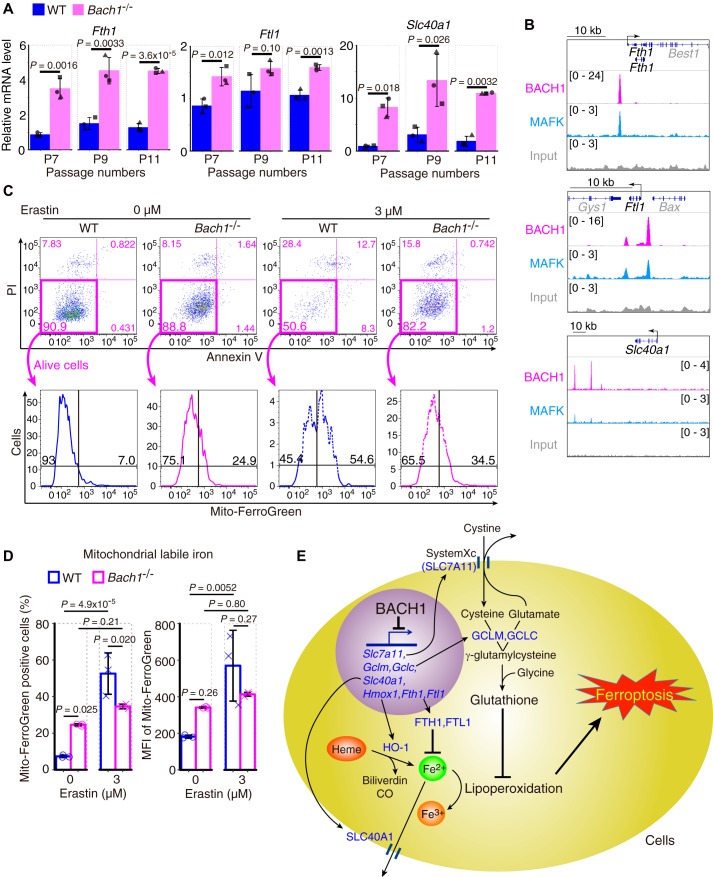
BACH1 accelerates ferroptosis by suppressing labile iron metabolism
To explore other target genes of BACH1 in the regulation of ferroptosis, we examined genes involved in the regulation of iron metabolism (Fth1, Ftl1, Slc40a1, Tfrc, Mfn2, and Fxn), heavy metal stress (Mt1), and lipoperoxidation (Gpx4). Some of these genes were up-regulated in response to erastin (see Fig. 1A). Among these genes, ferritin genes (Fth1 and Ftl1) and the ferroportin gene (Slc40a1) were dramatically up-regulated in Bach1−/− MEFs (Fig. 4A), and binding peaks of BACH1 and MAFK were observed near their regulatory regions (Fig. 4B). In contrast, the expression of Tfrc, Mfn2, Fxn, Mt1, and Gpx4 was only mildly increased in Bach1−/− MEFs (Fig. S8A). There were no strong binding peaks of BACH1 or MAFK in the regulatory regions of these genes (Fig. S8B). Considering that both ferritin and ferroportin reduce the availability of free labile iron and are known to inhibit ferroptosis (36–38), these results suggest that BACH1 promotes ferroptosis by repressing the transcription of ferritin and ferroportin genes.
Figure 4.
BACH1 increases mitochondrial labile iron during ferroptosis by repressing the transcription of genes of ferritin and ferroportin. A, qPCR analysis for Fth1, Ftl1, and Slc40a1 mRNA relative to Actb mRNA in WT and Bach1−/− MEFs (7th, 9th, and 11th passage: P7, P9, and P11). n = 3 of independent lots of MEFs per genotype. B, ChIP-seq analysis of the binding of BACH1, MAFK for gene regions of Fth1, Ftl1, and Slc40a1 in M1 cells. C and D, WT and Bach1−/− MEFs (P8) were exposed to erastin for 24 h and afterward, stained by Mito-FerroGreen, a fluorophore. Representative flow cytometry images (C) showing the strategy that was implemented for the sorting of Mito-FerroGreen positive cells. The positive cells were judged as mitochondrial-labile iron positive cells. Quantification of mitochondrial-labile iron (D). MFI, mean fluorescence intensity. E, conceptual diagram. Error bars of A and D represent S.D. p value of A by t test. p value of D by Tukey's test after two-way ANOVA.
To examine this hypothesis, we measured mitochondrial labile iron in WT and Bach1−/− MEFs with a fluorophore Mito-FerroGreen (39). Bach1−/− MEFs contained more mitochondrial labile iron than WT without erastin (Fig. 4, C and D). It is probably due to the effects of HO-1 overexpression. Although prominent increases in labile iron and cell death were observed in WT MEFs in response to erastin, these alterations were much less in Bach1−/− MEFs (Fig. 4, C and D, and Fig. S9, A and B). These results suggest BACH1 increases mitochondrial labile iron by repressing the transcription of ferritin and ferroportin during ferroptosis. These findings, along with the regulation of GSH synthesis pathway by BACH1, suggest that BACH1 accelerates ferroptosis by decreasing the intracellular activity of GSH and increasing the oxidative activity of labile iron (Fig. 4E).
BACH1 aggravates acute myocardial infarction by promoting ferroptosis
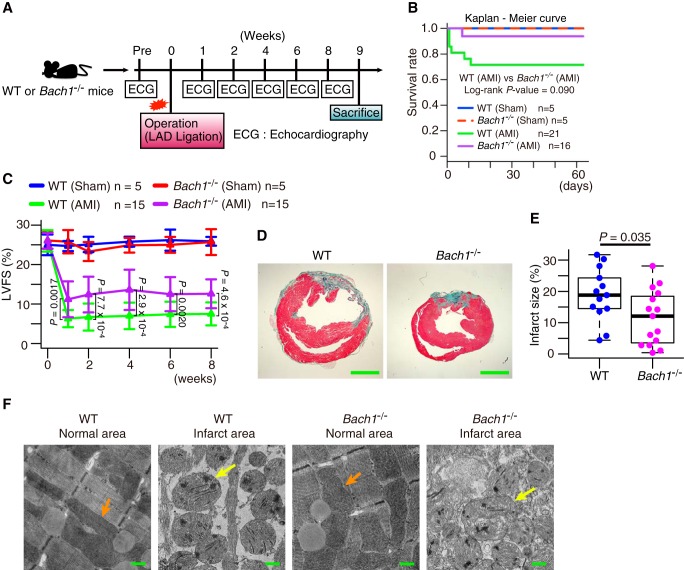
Finally, we tried to examine whether or not the promotion of ferroptosis by BACH1 is involved in pathological changes in vivo. It has long been known that iron and lipid peroxidation are involved in oxidative stress-related cell death, including ischemia-reperfusion injury (40–42). Furthermore, there are several reports showing that ferroptosis is involved in ischemia-reperfusion injury in the heart (3, 4, 7). Therefore, we used an AMI model based on left anterior descending coronary artery (LAD) ligation (43, 44) (Fig. 5A). In this model, Bach1−/− mice showed less severe injuries than WT mice as judged by the post-operative survival rate and an evaluation of the cardiac function with echocardiography (Fig. 5, B and C, Fig. S10, A–C, and Movie S1, A–D). The infarct area on pathological specimens was also smaller in Bach1−/− mice than in WT mice (Fig. 5, D and E). These results suggest that BACH1 exacerbates the pathology of AMI.
Figure 5.
BACH1 aggravates AMI. A, experimental process. B, Kaplan-Meier curve of each group. C, left ventricular fractional shortening (LVFS) on echocardiogram. D and E, mice was dissected after 9 weeks from operation. D, representative photographs of heart sections stained with elastica Masson staining. E, infarct size to left ventricular section. Scale bars in D represent 2 mm. F, mice was dissected next day from operation. Transmission electron microscope image of normal and infarct area of hearts of mice next day from operation. Orange arrow: normal mitochondria. Yellow arrow: mitochondrial spheroid-like abnormality. Scale bars represent 500 nm. Error bars of C represent S.D. The box and whisker plots of E show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. p value of B by log-rank test between WT (AMI) and Bach1−/− (AMI). p value of C by Tukey-Kramer method after two-way ANOVA. p value of E by t test.
To investigate whether or not ferroptosis is involved in the pathology, we observed the myocardial infarct regions using a transmission electron microscope. Mitochondrial spheroid-like abnormality was observed in both WT and Bach1−/− mice (Fig. 5F). This alteration was consistent with the findings of MEFs exposed to erastin (Fig. 2C). We then investigated whether or not the pathological changes could be improved by administering DFX (Fig. 6A), which is a clinically used iron chelator. First, we confirmed that it inhibited ferroptosis in MEFs (Fig. S10, D and E). Although there was no improvement in the survival rates in WT or Bach1−/− mice (Fig. 6B), an improvement in the cardiac function on echocardiography was observed in the DFX group, which was more prominent in the WT mice than Bach1−/− mice (Fig. 6, C and D, and Fig. S10, F–K). The DFX group of WT mice showed a reduction in the infarct area; however, no such effect was noted in Bach1−/− mice (Fig. 6, E and F). These results suggest that BACH1 exacerbates the pathology of AMI by promoting ferroptosis.
Figure 6.
An iron chelator DFX alleviates AMI. A, experimental process. B, Kaplan-Meier curve of each group. C and D, left ventricular fractional shortening (LVFS) of WT mice (C) and Bach1−/− mice (D) on echocardiogram. E and F, mice were dissected after 9 weeks from operation. E, representative photographs of heart sections stained with elastica Masson staining. F, infarct size to left ventricular section. Scale bars in E represent 2 mm. Error bars of C and D represent S.D. The box and whisker plots of F show the 25th and 75th percentile quartiles and median values (center black line) and maximum and minimum values of the data. p value of C, D, and F by t test.
Discussion
Although genes involved in ferroptosis are being discovered (25), how their expression is regulated during ferroptosis remains unclear. In this study, we found that many of the inhibitory genes of ferroptosis were coordinately up-regulated upon induction of ferroptosis with erastin (Fig. 1A). Such a coordinated response may be a mechanism for restricting ferroptosis. We further showed that BACH1 directly counteracted this coordinated response of genes, including Hmox1, Slc7a11, Gclm, Gclc, Fth1, Ftl1, and Slc40a1 (Figs. 3, B and C, and 4, A and B), which are involved in the metabolism of GSH or labile iron. The protein amount of BACH1 was reduced upon the induction of ferroptosis (Fig. 1B). Our results suggest that BACH1 is degraded in part by proteasome during ferroptosis (Fig. 1, D and E). BACH1 is known to be degraded by proteasome under oxidative stress including heme (45–49), therefore the similar mechanism may work in ferroptosis. BACH1 is known to repress the expression of Slc40a1 in macrophages (50). Therefore, reduction of the BACH1 protein level may trigger the coordinated induction of these genes as a subprogram of the initial phase of ferroptosis program. Cells can then integrate distinct signals leading to BACH1 degradation, and thus judge whether or not they should undergo ferroptosis. Thus, BACH1 sets the threshold for whether or not ferroptosis occurs in response to lipid peroxide synthesized.
NRF2 is known to activate some of the genes that are repressed by BACH1, including Hmox1, Slc7a11, Gclm, and Gclc (51–56). Even though NRF2 increases the intracellular GSH amount, it only weakly protects cells from ferroptosis (57). Other reports have shown that NRF2 can inhibit ferroptosis (27, 58, 59). It is known Hmox1 is induced by hypoxia inducible factor-1α (Hif-1α) (60, 61) and some reports suggest the possibility that Hif-1α also inhibits ferroptosis (32, 62). Therefore, ferroptosis execution may depend on the initial amounts and kinetics of the induction or reduction of these transcription factors. This mechanism may extend our understanding of the regulation of ferroptosis, wherein ferroptosis is a cell death programmed at the level of the gene regulatory network.
We showed that GSH was higher in Bach1−/− MEFs than WT cells (Fig. 3A). Our results strongly suggest that BACH1 decreases intracellular GSH by repressing the expression of Gclm, Gclc, and Slc7a11 (Fig. 3, B and C). Indeed, the protein amount of GCLM was higher in Bach1−/− MEFs than in WT cells (Fig. 3, D and E, and Fig. S4, A and B). However, the protein amounts of GCLC and SLC7A11 were similar between WT and Bach1−/− MEFs (Fig. 3, D and E, and Fig. S4, A and B). Cells may have additional mechanisms to tune strictly the protein amounts of GCLC and SLC7A11, managing the intracellular GSH amount and maintaining homeostasis. We found that SLC7A11 was further regulated by proteasomal degradation (Fig. 3, D and E). This observation suggests that the decision to undergo ferroptosis may be made based upon whether or not cells can induce efficiently inhibitory proteins like SLC7A11. Cells with higher amounts of SLC7A11 may likely be protected from ferroptosis. Gclc and Slc7a11 may be critical factors for cells, with the transcriptional regulation by BACH1 and additional layers of regulation, although these points will need to be explored in further studies.
Reports on the function of HO-1 are conflicting, with studies conversely describing it to promote or inhibit ferroptosis (7, 26, 27, 63). These discrepant findings may be because HO-1 degrades prooxidant heme to produce not only the radical scavengers biliverdin and bilirubin but also free iron that mediates ferroptosis through Fenton reaction (14, 25). Therefore, to allow HO-1 to function effectively as an anti-oxidative stress enzyme, it is essential to suppress the reactivity of labile iron derived from heme. We showed that BACH1 represses the expression of the genes of ferritin and ferroportin (Fig. 4, A and B), both of which reduce the intracellular availability of labile iron. By increasing the expression of not only HO-1 but also ferritin and ferroportin during the induction of ferroptosis (Fig. 1A), the prooxidant activities of heme and heme-derived free iron can be suppressed efficiently, thus protecting cells from ferroptosis. Conversely, BACH1 represses the expression of ferritin and ferroportin in addition to HO-1, thus effectively promoting ferroptosis (Fig. 4E). Indeed, we showed Bach1−/− MEFs were more resistant to the elevation of mitochondrial labile iron than WT MEFs during ferroptosis (Fig. 4, C and D). Based on the present and previous findings, we proposed a model in which BACH1 accelerates ferroptosis by suppressing two major intracellular counteracting mechanisms against ferroptosis: the GSH synthesis pathway and the system for the sequestration of labile iron (Fig. 4E).
In addition, we showed that ferroptosis was involved in the pathology of not only ischemia-reperfusion injury (3, 4, 7) but also AMI. The severity of AMI was improved by the iron chelator, DFX particularly in WT mice (Fig. 6, C–F). The peripheral areas of AMI are naturally reperfused by angiogenesis, where ferroptosis is likely induced. Our results here suggest that the therapeutic effect of DFX is expected in AMI and ischemia-reperfusion injury. Necroptosis is also reportedly involved in cardiac ischemic disease (64, 65). Therefore, the double inhibition of ferroptosis and necroptosis may lead to a more effective treatment of AMI. In addition, this study suggests that Bach1−/− mice are more resistant to AMI than WT mice because of their lower rate of ferroptosis than in WT mice (Figs. 5 and 6). BACH1 may be a potential therapeutic target of AMI in the future.
Ferroptosis is thought to play a major role in cancer suppression (2, 10). Our results suggest that cancer cells may acquire resistance against ferroptosis by decreasing BACH1 protein, thus eluding elimination by ferroptosis. We previously reported that BACH1 promotes the proliferation of MEFs transformed with H-RasV12 and their tumor formation in a mouse transplantation model (66). Recently, BACH1 was found to promote the proliferation and/or metastasis of breast cancer, ovarian cancer, and lung cancer cells (48, 49, 67–70). BACH1 is therefore considered to have dual functions in cancers: promoting cell proliferation and cell death through ferroptosis. Cancer cells may adapt to their surrounding environment by changing the expression of BACH1; cancer cells may highly express BACH1 during stages of proliferation and metastasis but may reduce their levels of BACH1 under stress conditions, such as toxicity due to anti-cancer drugs. Such flexibility in the amount of BACH1 protein expressed may enhance the malignancy of cancer cells. Therapy that targets this flexibility, such as the down-regulation of BACH1 in response to erastin, may expand the field of potential cancer treatments.
Experimental procedures
Mice
The generation of Bach1−/− mice on the C57BL/6J background was described previously (15). Mice 13 weeks of age were analyzed for models of AMI. Animals were euthanized by cervical dislocation under anesthetic inhalation overdose with isoflurane before anatomy. These mice were bred at the animal facility of Tohoku University. Mice were housed under specific pathogen-free conditions. All experiments performed in this study were approved by the Institutional Animal Care and Use Committee of the Tohoku University Environmental and Safety Committee.
Mice models of AMI
Induction of AMI was performed as described previously (43, 44). The mice were subjected to ligation of the proximal LAD to induce AMI. They were randomly assigned to sham or AMI (Fig. 5A), DMSO, or DFX groups (Fig. 6A). To follow up the time course of LV function after AMI, we performed transthoracic two-dimensional echocardiography. For histological analysis and analysis with transmission electron microscope, the heart was divided along the short axis at the center of the infarct.
Histopathological analysis
Excised hearts were fixed with 4% paraformaldehyde for histological and immunohistochemical examination. After 24–48 h of fixation and dehydration through increasing concentrations of ethanol, the tissue specimens were embedded in paraffin and sliced at 3 μm in thickness. The sections were used for Elastica-Masson staining. The extent of infarct area was calculated as a rate of fibrotic area using the following formula: fibrotic area/(LV free wall + interventricular septum) × 100 (%) with use of Photoshop software (Adobe).
Transmission electron microscopy
Cells and hearts were treated in 2.5% glutaraldehyde in 0.1 m cacodylate buffer (pH 7.4) for at least 24 h, and washed with 0.1 m cacodylate buffer 4 times and then treated with 1% OsO4 in 0.1 m cacodylate buffer for 90 min. After dehydration through an ethanol series (50–100% ethanol), cells were embedded in Epon resin. Thin sections were cut with a microtome (Leica EM UC-7), stained with 2% uranyl acetate and 0.4% lead citrate, and examined and photographed under a transmission electron microscope (Hitachi High-Technologies H-7600).
Isolation and culture of MEFs
MEFs were derived from 13.5-day-old embryos of WT or Bach1−/− mice. Following removal of the head and organs, embryos were rinsed with PBS (Nissui, Tokyo, Japan), minced and digested with trypsin (0.05% (v/v) solution containing 0.53 mm EDTA) (Gibco), and 1.8 mg/ml of DNase I (Roche, Basel, Switzerland) in PBS and incubated for 60 min at 37 °C. Trypsin was inactivated by addition of Dulbecco's modified Eagle's medium with high glucose (Gibco) containing 10% (v/v) fetal bovine serum (Sigma-Aldrich), 1× minimal essential medium nonessential amino acids (Gibco), and 0.1 mm 2-mercaptoethanol (Sigma-Aldrich). MEFs from a single embryo were plated into a 100-mm diameter culture dish and incubated at 37 °C in 3% oxygen (1st passage: P1). MEFs from embryos of homosexual littermates were mixed at 2nd passage (P2) and stocked.
MEFs were maintained at 37 °C in culture medium (Dulbecco's modified Eagle's medium with (Gibco) containing 10% fetal bovine serum (Sigma-Aldrich), 1× minimal essential medium nonessential amino acids (Gibco), penicillin/streptomycin (100 units/ml and 100 μg/ml each) (Gibco) and 0.1 mm 2-mercaptoethanol (Sigma-Aldrich)) in 3% oxygen for experiments. The number of passages were recorded for each lot of MEFs. From 5th to 11th passage MEFs were used for all experiments.
Reagents
Erastin, DMSO, and DFO were purchased from Sigma-Aldrich. MG132 was purchased from Calbiochem (San Diego, CA). DFX was transferred as raw material from Novartis Pfarma (Basel, Switzerland). S-Adenosylmethionine-13C5,15N, and GSH-13C2,15N were purchased from Taiyo Nissan Corp. (Tokyo, Japan) and used as internal standard for MS. Methanol, acetonitrile, and ammonium hydroxide for MS were purchased from Kanto Chemical (Tokyo, Japan). Ammonium bicarbonate (1 mol/liter) for MS was purchased from Cell Science and Technology Institute, Inc. (Miyagi, Japan). Formic acid for MS was purchased from Wako Pure Chemical Industries (Osaka, Japan).
Sample preparation for UHPLC/MS/MS
MEFs (3–8 × 106 cells for each lot) were suspended in 100 μl of methanol containing the internal standards (0.2 μg/ml of S-adenosylmethionine-13C5,15N for positive ion mode (Pos) and 1 μg/ml of GSH-13C2,15N for negative ion mode (Neg)), and were homogenized by mixing for 30 s followed by sonication for 10 min. After centrifugation at 16,400 × g for 20 min at 4 °C followed by deproteinization, 3 μl of each extract was analyzed by ultra HPLC triple quadrupole MS (UHPLC/MS/MS).
UHPLC/MS/MS analysis
The UHPLC/MS/MS analysis was performed on an AcquityTM Ultra Performance LC I-class system (Waters Corp., Milford, UK) interfaced to a Waters Xevo TQ-S MS/MS system equipped with electrospray ionization. The MS/MS was performed using the multiple reaction monitoring mode with a scan time of 1 ms for each compound. The transitions of the precursor ion to the product ion, cone voltage, and collision energy are listed in Table S1. The other settings are as follows: 3.5 kV (Pos) or 2.5 kV (Neg) capillary voltage, 30 V cone voltage, 50 V source offset, source temperature at 150 °C, 150 liter/h of cone gas (N2) flow rate, desolvation temperature at 450 °C, 1000 liter/h of desolvation gas flow, 0.15 min/ml of collision gas flow, 7.00 bar nebulization gas (N2) flow. LC separation was performed as described before (71), using a normal-phase column (ZIC-pHILIC; 100 × 2.1 mm inner diameter, 5-μm particle size; Sequant, Darmstadt, Germany) with a gradient elution using solvent A (10 mmol/liter of NH4HCO3, adjusted to pH 9.2 using ammonia solution) and B (acetonitrile) at 300 μl/min: 99 to 70% B from 0.5 to 4.0 min, 70 to 1% B from 4.0 to 6.5 min, 1% B for 2.5 min, and 99% B for 9 min until the end of the run. The oven temperature was 20 °C. The data were collected using the MassLynx version 4.1 software (Waters Corp.) and the ratio of the peak area of analyte to the internal standard was analyzed by Traverse MS (Reifycs Inc., Tokyo, Japan).
RNA interference
All siRNAs (siControl: Stealth RNAiTM siRNA Negative Control, Med GC; siGclm #1: MSS204722; siGclm #3: MSS204724; siSlc7a11 #1: MSS218649; siSlc7a11 #2: MSS218650; siGclc #2: MSS204720; siGclc #3: MSS204721; siHmox1 #1: MSS247281; siHmox1 #3: MSS274857) were obtained from Invitrogen. Sequences of the siRNAs are described in Table S2. 2 × 106 cells of MEFs were transfected with 1.2 nm siRNAs using Amaxa Nucleofector II (Lonza, Basel, Switzerland) and MF 1 Nucleofector kit (Lonza) according to the manufacturer's protocols. After transfection, MEFs were passaged to dishes or culture plate with culture medium.
Western blotting
Cells were trypsinized, pelleted, and washed twice in PBS. Cells were lysed beyond 5 min in SDS sample buffer (62.5 mm Tris-HCl (pH 6.8), 1% (v/v) 2-mercaptoethanol, 1% (w/v) SDS, 10% (w/v) glycerol, 0.02% (w/v) bromphenol blue). Lysates were resolved on 7.5–10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Millipore). The antibody for detection of β-actin (sc-1616) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The antibody for detection of HO-1 (ADI-SPA-896) was purchased from Enzo Life Science (New York). The antibodies for GAPDH (ab8245), Gclc (ab53179), and Gclm (ab124827) were purchased from Abcam (Cambridge, UK). The antibody for Slc7a11 (119-11215) was purchased by RayBiotech (Norcross, GA). The antibody for BACH1 was described previously (15). For the quantification of signals, all samples to be compared were run on the same gel. Bands were quantified using ImageJ (72, 73). All bands to be compared were quantified on the same image and were within the linear range of detection of the software.
Quantitative PCR with reverse transcription
Total RNA was purified with RNeasy plus micro kit or RNeasy plus mini kit (Qiagen, Hilden, Germany). Complementary DNA was synthesized by a SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed using LightCycler Fast Start DNA Master SYBR Green I, and LightCycler nano (Roche) or LightCycler 96 (Roche). mRNA transcript abundance was normalized to that of Actb. Sequences of the qPCR primers are described in Table S3.
Administration of erastin and cell death assessment by flow cytometry
Before administration of erastin, the medium was exchanged to the experimental medium (culture medium without 2-mercaptoethanol and penicillin/streptomycin). Erastin was dissolved in DMSO and administered to experimental medium with DMSO. The concentration of DMSO was adjusted among each samples. Cell death was assessed 24 h after administration of erastin. PI and Annexin V staining were used for assessment of cell death. APC-Annexin V was purchased from BD Bioscience (Franklin Lakes, NJ). MEFs were stained by APC-Annexin V according to the manufacturer's protocols. PI was added to aliquot (1 μg/ml) before flow cytometry. The MEFs were sorted with a FACS Aria II (BD) and analyzed by FlowJo software (Tree Star, Ashland, OR). MEFs of positive, of whether at least Annexin V or PI, was assessed as dead cells. The gating strategy for assessing dead cells (Figs. 2, B and E; 4, C and D; and Figs. S3B, S7, A–D, S9B, and S10E) was shown in Fig. S2.
ChIP-Seq
We used ChIP-seq data of BACH1 and MAFK in the M1 cell line from GEO (Gene Expression Ominibus) data set GSE79139 that deposited for our previous report (34, 35).
Detection of mitochondrial labile iron
To detect mitochondrial labile iron, a fluorophore Mito-FerroGreen (Dojindo, Kumamoto, Japan) (39) was used according to the manufacturer's protocol. MEFs on 12-well plates (Corning, NY) were washed three times with Hank's balanced salt solution (HBSS) (Gibco) to remove the residual medium. Then MEFs were treated with 5 μm Mito-FerroGreen with HBSS for 30 min at 37 °C. After incubation, Mito-FerroGreen was removed by washing twice with HBSS. MEFs were stained by APC-Annexin V and PI, and sorted with a FACS Aria II (BD Bioscience) and analyzed by FlowJo software (Tree Star). MEFs that were negative for both Annexin V and PI were sorted as alive cells (Fig. 4, C and D). The gating strategy was shown in Fig. S2.
RNA-Seq
Total RNA was purified using an RNeasy plus mini kit (Qiagen). To remove ribosomal RNA (rRNA), 4 μg of the total RNA was treated with a GeneRead rRNA Depletion kit (Qiagen) and then with an RNeasy MiniElute kit (Qiagen) for cleanup. For fragmentation, 100 ng of the rRNA-depleted RNA was incubated at 95 °C for 10 min and purified by a Magnetic Beads Cleanup Module (Thermo Fisher Scientific, Carlsbad, CA). The libraries were constructed with an RNA-seq library kit version 2 (Thermo Fisher Scientific) on ABI library builder (Thermo Fisher Scientific), and was barcoded with Ion Xpress RNA-seq BC primer (Thermo Fisher Scientific). The library fragments with a size range of 100–200 bp were selected with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). Templates were prepared on the Ion Chef system using an Ion PI Hi-Q Chef kit (Thermo Fisher Scientific) and sequencing was performed on an Ion Proton system using the Ion PI Hi-Q sequencing kit (Thermo Fisher Scientific) and the PI v3 chip (Thermo Fisher Scientific). The sequence data were obtained as fastq files. The sequence data were aligned to reference hg19 using the RNASeq Analysis plugin from Ion torrent suite software (Thermo Fisher Scientific). Mapped reads were counted for each gene using HTSeq version 0.9.1 htseq-count. The differential expression analysis was performed on edge R version 3.16.5 after removal of low count lead genes using three biological replicates for each condition (less than 5 leads per gene in the sample and counts per million mapped reads (cpm) of 1 or less).
Statistics
For all experiments, differences of data sets were considered statistically significant when p values were lower than 0.05. Statistical comparisons were performed using the t test in comparison between the two groups, and one-, two-, or three-way ANOVA followed by Tukey's test or Tukey-Kramer method in comparison among multiple groups. For the t test, Student's t test was used when the S.D. of the groups was not significantly different by f test. Welch's t test was used when the S.D. of the groups was significantly different by f test.
Author contributions
H. N., M. M., and K. I. conceptualization; H. N., H. K., K. S., M. S., and Y. I. resources; H. N. formal analysis; H. N. validation; H. N., M. M., and D. S. investigation; H. N., M. M., T. S., D. S., H. S., and K. I. methodology; H. N. writing-original draft; M. M. and K. I. funding acquisition; K. I. supervision; K. I. project administration; K. I. writing-review and editing.
Supplementary Material
Acknowledgments
We thank members of the Department of Biochemistry, Tohoku University Graduate School of Medicine, for discussions and support and the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support. We thank Novartis for the raw material transfer of DFX for this study.
This work was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science 19K07680 and 16K07108 (to M. M.) and 15H02506, 24390066, 21249014, and 18H04021 (to K. I.) and Agency for Medical Research and Development Grant JP16gm050001 (to K. I.). H. Nishizawa received DFX as raw material from Novartis Pharma for this study. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S10, Tables S1–S3, and Movie S1
The RNA-seq data has been deposited at the GEO database under accession code GSE131444.
- BACH1
- BTB and CNC homology 1
- HO-1
- heme oxygenase-1
- MEF
- mouse embryonic fibroblast
- AMI
- acute myocardial infarction
- DFX
- deferasirox
- PI
- propidium iodide
- DFO
- deferoxamine
- qPCR
- quantitative PCR
- LAD
- left anterior descending coronary artery
- UHPLC
- ultra HPLC
- P7
- passage 7
- HBSS
- Hank's balanced salt solution
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- RNA-Seq
- RNA-sequence.
References
- 1. Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. 3rd, Stockwell B. R. (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang W. S., SriRamaratnam R., Welsch M. E., Shimada K., Skouta R., Viswanathan V. S., Cheah J. H., Clemons P. A., Shamji A. F., Clish C. B., Brown L. M., Girotti A. W., Cornish V. W., Schreiber S. L., and Stockwell B. R. (2014) Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao M., Monian P., Quadri N., Ramasamy R., and Jiang X. (2015) Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell 59, 298–308 10.1016/j.molcel.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba Y., Higa J. K., Shimada B. K., Horiuchi K. M., Suhara T., Kobayashi M., Woo J. D., Aoyagi H., Marh K. S., Kitaoka H., and Matsui T. (2018) Protective effects of the mechanistic target of rapamycin against excess iron and ferroptosis in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 314, H659–H668 10.1152/ajpheart.00452.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Linkermann A., Bäasen J. H., Darding M., Jin M. K., Sanz A. B., Heller J. O., De Zen F., Weinlich R., Ortiz A., Walczak H., Weinberg J. M., Green D. R., Kunzendorf U., and Krautwald S. (2013) Two independent pathways of regulated necrosis mediate ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 110, 12024–12029 10.1073/pnas.1305538110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linkermann A., Skouta R., Himmerkus N., Mulay S. R., Dewitz C., De Zen F., Prokai A., Zuchtriegel G., Krombach F., Welz P. S., Weinlich R., Vanden Berghe T., Vandenabeele P., Pasparakis M., Bleich M., et al. (2014) Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 111, 16836–16841 10.1073/pnas.1415518111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang X., Wang H., Han D., Xie E., Yang X., Wei J., Gu S., Gao F., Zhu N., Yin X., Cheng Q., Zhang P., Dai W., Chen J., Yang F., Yang H. T., Linkermann A., Gu W., Min J., and Wang F. (2019) Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 116, 2672–2680 10.1073/pnas.1821022116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chiang G. C., Mao X., Kang G., Chang E., Pandya S., Vallabhajosula S., Isaacson R., Ravdin L. D., Alzheimer's Disease Neuroimaging Initiative, and Shungu D. C. (2017) Relationships among cortical glutathione levels, brain amyloidosis, and memory in healthy older adults investigated in vivo with 1H-MRS and Pittsburgh compound-B PET. Am. J. Neuroradiol. 38, 1130–1137 10.3174/ajnr.A5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Di Domenico F., Tramutola A., and Butterfield D. A. (2017) Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of Alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 111, 253–261 10.1016/j.freeradbiomed.2016.10.490 [DOI] [PubMed] [Google Scholar]
- 10. Jiang L., Kon N., Li T., Wang S. J., Su T., Hibshoosh H., Baer R., and Gu W. (2015) Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62 10.1038/nature14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S. E., Zhang L., Ma K., Riegman M., Chen F., Ingold I., Conrad M., Turker M. Z., Gao M., Jiang X., Monette S., Pauliah M., Gonen M., Zanzonico P., Quinn T., Wiesner U., Bradbury M. S., and Overholtzer M. (2016) Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 11, 977–985 10.1038/nnano.2016.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Viswanathan V. S., Ryan M. J., Dhruv H. D., Gill S., Eichhoff O. M., Seashore-Ludlow B., Kaffenberger S. D., Eaton J. K., Shimada K., Aguirre A. J., Viswanathan S. R., Chattopadhyay S., Tamayo P., Yang W. S., Rees M. G., et al. (2017) Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 10.1038/nature23007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hangauer M. J., Viswanathan V. S., Ryan M. J., Bole D., Eaton J. K., Matov A., Galeas J., Dhruv H. D., Berens M. E., Schreiber S. L., McCormick F., and McManus M. T. (2017) Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 10.1038/nature24297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Igarashi K., and Watanabe-Matsui M. (2014) Wearing red for signaling: the heme-bach axis in heme metabolism, oxidative stress response and iron immunology. Tohoku J. Exp. Med. 232, 229–253 10.1620/tjem.232.229 [DOI] [PubMed] [Google Scholar]
- 15. Sun J., Hoshino H., Takaku K., Nakajima O., Muto A., Suzuki H., Tashiro S., Takahashi S., Shibahara S., Alam J., Taketo M. M., Yamamoto M., and Igarashi K. (2002) Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21, 5216–5224 10.1093/emboj/cdf516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hintze K. J., Katoh Y., Igarashi K., and Theil E. C. (2007) Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, β-globin, and NADP(H) quinone (oxido) reductase 1. J. Biol. Chem. 282, 34365–34371 10.1074/jbc.M700254200 [DOI] [PubMed] [Google Scholar]
- 17. Warnatz H. J., Schmidt D., Manke T., Piccini I., Sultan M., Borodina T., Balzereit D., Wruck W., Soldatov A., Vingron M., Lehrach H., and Yaspo M. L. (2011) The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem. 286, 23521–23532 10.1074/jbc.M111.220178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yano Y., Ozono R., Oishi Y., Kambe M., Yoshizumi M., Ishida T., Omura S., Oshima T., and Igarashi K. (2006) Genetic ablation of the transcription repressor Bach1 leads to myocardial protection against ischemia/reperfusion in mice. Genes Cells 11, 791–803 10.1111/j.1365-2443.2006.00979.x [DOI] [PubMed] [Google Scholar]
- 19. Ito M., Nagano N., Arai Y., Ogawa R., Kobayashi S., Motojima Y., Go H., Tamura M., Igarashi K., Dennery P. A., and Namba F. (2017) Genetic ablation of Bach1 gene enhances recovery from hyperoxic lung injury in newborn mice via transient upregulation of inflammatory genes. Pediatr. Res. 81, 926–931 10.1038/pr.2017.17 [DOI] [PubMed] [Google Scholar]
- 20. Harusato A., Naito Y., Takagi T., Uchiyama K., Mizushima K., Hirai Y., Higashimura Y., Katada K., Handa O., Ishikawa T., Yagi N., Kokura S., Ichikawa H., Muto A., Igarashi K., and Yoshikawa T. (2013) BTB and CNC homolog 1 (Bach1) deficiency ameliorates TNBS colitis in mice: role of M2 macrophages and heme oxygenase-1. Inflamm. Bowel Dis. 19, 740–753 10.1097/MIB.0b013e3182802968 [DOI] [PubMed] [Google Scholar]
- 21. Inoue M., Tazuma S., Kanno K., Hyogo H., Igarashi K., and Chayama K. (2011) Bach1 gene ablation reduces steatohepatitis in mouse MCD diet model. J. Clin. Biochem. Nutri. 48, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanno H., Ozawa H., Dohi Y., Sekiguchi A., Igarashi K., and Itoi E. (2009) Genetic ablation of transcription repressor Bach1 reduces neural tissue damage and improves locomotor function after spinal cord injury in mice. J. Neurotrauma 26, 31–39 10.1089/neu.2008.0667 [DOI] [PubMed] [Google Scholar]
- 23. Shimada K., Skouta R., Kaplan A., Yang W. S., Hayano M., Dixon S. J., Brown L. M., Valenzuela C. A., Wolpaw A. J., and Stockwell B. R. (2016) Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 12, 497–503 10.1038/nchembio.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang W. S., Kim K. J., Gaschler M. M., Patel M., Shchepinov M. S., and Stockwell B. R. (2016) Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 113, E4966–E4975 10.1073/pnas.1603244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stockwell B. R., Friedmann Angeli J. P., Bayir H., Bush A. I., Conrad M., Dixon S. J., Fulda S., Gascon S., Hatzios S. K., Kagan V. E., Noel K., Jiang X., Linkermann A., Murphy M. E., Overholtzer M., et al. (2017) Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwon M. Y., Park E., Lee S. J., and Chung S. W. (2015) Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget 6, 24393–24403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., and Tang D. (2016) Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology (Baltimore, MD) 63, 173–184 10.1002/hep.28251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sato H., Tamba M., Kuriyama-Matsumura K., Okuno S., and Bannai S. (2000) Molecular cloning and expression of human xCT, the light chain of amino acid transport system xc. Antioxid. Redox Signal. 2, 665–671 [DOI] [PubMed] [Google Scholar]
- 29. Sato H., Shiiya A., Kimata M., Maebara K., Tamba M., Sakakura Y., Makino N., Sugiyama F., Yagami K., Moriguchi T., Takahashi S., and Bannai S. (2005) Redox imbalance in cystine/glutamate transporter-deficient mice. J. Biol. Chem. 280, 37423–37429 10.1074/jbc.M506439200 [DOI] [PubMed] [Google Scholar]
- 30. Telorack M., Abplanalp J., and Werner S. (2016) Low levels of glutathione are sufficient for survival of keratinocytes after UV irradiation and for healing of mouse skin wounds. Arch. Dermatol. Res. 308, 443–448 10.1007/s00403-016-1660-9 [DOI] [PubMed] [Google Scholar]
- 31. Fan X., Liu X., Hao S., Wang B., Robinson M. L., and Monnier V. M. (2012) The LEGSKO mouse: a mouse model of age-related nuclear cataract based on genetic suppression of lens glutathione synthesis. PloS One 7, e50832 10.1371/journal.pone.0050832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miess H., Dankworth B., Gouw A. M., Rosenfeldt M., Schmitz W., Jiang M., Saunders B., Howell M., Downward J., Felsher D. W., Peck B., and Schulze A. (2018) The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 37, 5435–5450 10.1038/s41388-018-0315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ding W. X., Li M., Biazik J. M., Morgan D. G., Guo F., Ni H. M., Goheen M., Eskelinen E. L., and Yin X. M. (2012) Electron microscopic analysis of a spherical mitochondrial structure. J. Biol. Chem. 287, 42373–42378 10.1074/jbc.M112.413674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ebina-Shibuya R., Watanabe-Matsui M., Matsumoto M., Itoh-Nakadai A., Funayama R., Nakayama K., Muto A., and Igarashi K. (2016) The double knockout of Bach1 and Bach2 in mice reveals shared compensatory mechanisms in regulating alveolar macrophage function and lung surfactant homeostasis. J. Biochem. 160, 333–344 10.1093/jb/mvw041 [DOI] [PubMed] [Google Scholar]
- 35. Ebina-Shibuya R., Matsumoto M., Kuwahara M., Jang K. J., Sugai M., Ito Y., Funayama R., Nakayama K., Sato Y., Ishii N., Okamura Y., Kinoshita K., Kometani K., Kurosaki T., Muto A., et al. (2017) Inflammatory responses induce an identity crisis of alveolar macrophages, leading to pulmonary alveolar proteinosis. J. Biol. Chem. 292, 18098–18112 10.1074/jbc.M117.808535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y. Q., Chang S. Y., Wu Q., Gou Y. J., Jia L., Cui Y. M., Yu P., Shi Z. H., Wu W. S., Gao G., and Chang Y. Z. (2016) The protective role of mitochondrial ferritin on erastin-induced ferroptosis. Front. Aging Neurosci. 8, 308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hou W., Xie Y., Song X., Sun X., Lotze M. T., Zeh H. J. 3rd, Kang R., and Tang D. (2016) Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geng N., Shi B. J., Li S. L., Zhong Z. Y., Li Y. C., Xua W. L., Zhou H., and Cai J. H. (2018) Knockdown of ferroportin accelerates erastin-induced ferroptosis in neuroblastoma cells. Eur. Rev. Med. Pharmacol. Sci. 22, 3826–3836 [DOI] [PubMed] [Google Scholar]
- 39. Hirayama T., Kadota S., Niwa M., and Nagasawa H. (2018) A mitochondria-targeted fluorescent probe for selective detection of mitochondrial labile Fe(II). Metallomics 10, 794–801 10.1039/C8MT00049B [DOI] [PubMed] [Google Scholar]
- 40. Starke P. E., and Farber J. L. (1985) Endogenous defenses against the cytotoxicity of hydrogen peroxide in cultured rat hepatocytes. J. Biol. Chem. 260, 86–92 [PubMed] [Google Scholar]
- 41. Kehrer J. P. (2000) The Haber-Weiss reaction and mechanisms of toxicity. Toxicology 149, 43–50 10.1016/S0300-483X(00)00231-6 [DOI] [PubMed] [Google Scholar]
- 42. Rauen U., Petrat F., Sustmann R., and de Groot H. (2004) Iron-induced mitochondrial permeability transition in cultured hepatocytes. J. Hepatol. 40, 607–615 10.1016/j.jhep.2003.12.021 [DOI] [PubMed] [Google Scholar]
- 43. Abarbanell A. M., Herrmann J. L., Weil B. R., Wang Y., Tan J., Moberly S. P., Fiege J. W., and Meldrum D. R. (2010) Animal models of myocardial and vascular injury. J. Surg. Res. 162, 239–249 10.1016/j.jss.2009.06.021 [DOI] [PubMed] [Google Scholar]
- 44. Shindo T., Ito K., Ogata T., Hatanaka K., Kurosawa R., Eguchi K., Kagaya Y., Hanawa K., Aizawa K., Shiroto T., Kasukabe S., Miyata S., Taki H., Hasegawa H., Kanai H., and Shimokawa H. (2016) Low-intensity pulsed ultrasound enhances angiogenesis and ameliorates left ventricular dysfunction in a mouse model of acute myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 36, 1220–1229 10.1161/ATVBAHA.115.306477 [DOI] [PubMed] [Google Scholar]
- 45. Ogawa K., Sun J., Taketani S., Nakajima O., Nishitani C., Sassa S., Hayashi N., Yamamoto M., Shibahara S., Fujita H., and Igarashi K. (2001) Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20, 2835–2843 10.1093/emboj/20.11.2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zenke-Kawasaki Y., Dohi Y., Katoh Y., Ikura T., Ikura M., Asahara T., Tokunaga F., Iwai K., and Igarashi K. (2007) Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol. Cell. Biol. 27, 6962–6971 10.1128/MCB.02415-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ishikawa M., Numazawa S., and Yoshida T. (2005) Redox regulation of the transcriptional repressor Bach1. Free Radic. Biol. Med. 38, 1344–1352 10.1016/j.freeradbiomed.2005.01.021 [DOI] [PubMed] [Google Scholar]
- 48. Wiel C., Le Gal K., Ibrahim M. X., Jahangir C. A., Kashif M., Yao H., Ziegler D. V., Xu X., Ghosh T., Mondal T., Kanduri C., Lindahl P., Sayin V. I., and Bergo M. O. (2019) BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell 178, 330–345.e22 10.1016/j.cell.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 49. Lignitto L., LeBoeuf S. E., Homer H., Jiang S., Askenazi M., Karakousi T. R., Pass H. I., Bhutkar A. J., Tsirigos A., Ueberheide B., Sayin V. I., Papagiannakopoulos T., and Pagano M. (2019) Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e18 10.1016/j.cell.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marro S., Chiabrando D., Messana E., Stolte J., Turco E., Tolosano E., and Muckenthaler M. U. (2010) Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position −7007 of the FPN1 promoter. Haematologica 95, 1261–1268 10.3324/haematol.2009.020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., and Yamamoto M. (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275, 16023–16029 10.1074/jbc.275.21.16023 [DOI] [PubMed] [Google Scholar]
- 52. Bea F., Hudson F. N., Chait A., Kavanagh T. J., and Rosenfeld M. E. (2003) Induction of glutathione synthesis in macrophages by oxidized low-density lipoproteins is mediated by consensus antioxidant response elements. Circ. Res. 92, 386–393 10.1161/01.RES.0000059561.65545.16 [DOI] [PubMed] [Google Scholar]
- 53. Sekhar K. R., Crooks P. A., Sonar V. N., Friedman D. B., Chan J. Y., Meredith M. J., Starnes J. H., Kelton K. R., Summar S. R., Sasi S., and Freeman M. L. (2003) NADPH oxidase activity is essential for Keap1/Nrf2-mediated induction of GCLC in response to 2-indol-3-yl-methylenequinuclidin-3-ols. Cancer Res. 63, 5636–5645 [PubMed] [Google Scholar]
- 54. Wild A. C., Moinova H. R., and Mulcahy R. T. (1999) Regulation of γ-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274, 33627–33636 10.1074/jbc.274.47.33627 [DOI] [PubMed] [Google Scholar]
- 55. Sasaki H., Sato H., Kuriyama-Matsumura K., Sato K., Maebara K., Wang H., Tamba M., Itoh K., Yamamoto M., and Bannai S. (2002) Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 277, 44765–44771 10.1074/jbc.M208704200 [DOI] [PubMed] [Google Scholar]
- 56. Alam J., Stewart D., Touchard C., Boinapally S., Choi A. M., and Cook J. L. (1999) Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274, 26071–26078 10.1074/jbc.274.37.26071 [DOI] [PubMed] [Google Scholar]
- 57. Cao J. Y., Poddar A., Magtanong L., Lumb J. H., Mileur T. R., Reid M. A., Dovey C. M., Wang J., Locasale J. W., Stone E., Cole S. P. C., Carette J. E., and Dixon S. J. (2019) A genome-wide haploid genetic screen identifies regulators of glutathione abundance and ferroptosis sensitivity. Cell Rep. 26, 1544–1556.e8 10.1016/j.celrep.2019.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fan Z., Wirth A. K., Chen D., Wruck C. J., Rauh M., Buchfelder M., and Savaskan N. (2017) Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 6, e371 10.1038/oncsis.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roh J. L., Kim E. H., Jang H., and Shin D. (2017) Nrf2 inhibition reverses the resistance of cisplatin-resistant head and neck cancer cells to artesunate-induced ferroptosis. Redox Biol. 11, 254–262 10.1016/j.redox.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee P. J., Jiang B. H., Chin B. Y., Iyer N. V., Alam J., Semenza G. L., and Choi A. M. (1997) Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J. Biol. Chem. 272, 5375–5381 10.1074/jbc.272.9.5375 [DOI] [PubMed] [Google Scholar]
- 61. Gong P., Hu B., Stewart D., Ellerbe M., Figueroa Y. G., Blank V., Beckman B. S., and Alam J. (2001) Cobalt induces heme oxygenase-1 expression by a hypoxia-inducible factor-independent mechanism in Chinese hamster ovary cells: regulation by Nrf2 and MafG transcription factors. J. Biol. Chem. 276, 27018–27025 10.1074/jbc.M103658200 [DOI] [PubMed] [Google Scholar]
- 62. Jiang Y., Mao C., Yang R., Yan B., Shi Y., Liu X., Lai W., Liu Y., Wang X., Xiao D., Zhou H., Cheng Y., Yu F., Cao Y., Liu S., Yan Q., and Tao Y. (2017) EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics 7, 3293–3305 10.7150/thno.19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adedoyin O., Boddu R., Traylor A., Lever J. M., Bolisetty S., George J. F., and Agarwal A. (2018) Heme oxygenase-1 mitigates ferroptosis in renal proximal tubule cells. Am. J. Physiol. Renal Physiol. 314, F702–F714 10.1152/ajprenal.00044.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith C. C., Davidson S. M., Lim S. Y., Simpkin J. C., Hothersall J. S., and Yellon D. M. (2007) Necrostatin: a potentially novel cardioprotective agent? Cardiovasc. Drugs Ther. 21, 227–233 10.1007/s10557-007-6035-1 [DOI] [PubMed] [Google Scholar]
- 65. Oerlemans M. I., Liu J., Arslan F., den Ouden K., van Middelaar B. J., Doevendans P. A., and Sluijter J. P. (2012) Inhibition of RIP1-dependent necrosis prevents adverse cardiac remodeling after myocardial ischemia-reperfusion in vivo. Basic Res. Cardiol. 107, 270 10.1007/s00395-012-0270-8 [DOI] [PubMed] [Google Scholar]
- 66. Nakanome A., Brydun A., Matsumoto M., Ota K., Funayama R., Nakayama K., Ono M., Shiga K., Kobayashi T., and Igarashi K. (2013) Bach1 is critical for the transformation of mouse embryonic fibroblasts by Ras(V12) and maintains ERK signaling. Oncogene 32, 3231–3245 10.1038/onc.2012.336 [DOI] [PubMed] [Google Scholar]
- 67. Lee J., Yesilkanal A. E., Wynne J. P., Frankenberger C., Liu J., Yan J., Elbaz M., Rabe D. C., Rustandy F. D., Tiwari P., Grossman E. A., Hart P. C., Kang C., Sanderson S. M., Andrade J., et al. (2019) Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature 568, 254–258 10.1038/s41586-019-1005-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mansoori B., Mohammadi A., Asadzadeh Z., Shirjang S., Minouei M., Abedi Gaballu F., Shajari N., Kazemi T., Gjerstorff M. F., Duijf P. H. G., and Baradaran B. (2019) HMGA2 and Bach-1 cooperate to promote breast cancer cell malignancy. J. Cell. Physiol. 234, 17714–17726 [DOI] [PubMed] [Google Scholar]
- 69. Han W., Zhang Y., Niu C., Guo J., Li J., Wei X., Jia M., Zhi X., Yao L., and Meng D. (2019) BTB and CNC homology 1 (Bach1) promotes human ovarian cancer cell metastasis by HMGA2-mediated epithelial-mesenchymal transition. Cancer Lett. 445, 45–56 10.1016/j.canlet.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 70. Lee J., Lee J., Farquhar K. S., Yun J., Frankenberger C. A., Bevilacqua E., Yeung K., Kim E. J., Balázsi G., and Rosner M. R. (2014) Network of mutually repressive metastasis regulators can promote cell heterogeneity and metastatic transitions. Proc. Natl. Acad. Sci. U.S.A. 111, E364–E373 10.1073/pnas.1304840111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saigusa D., Okamura Y., Motoike I. N., Katoh Y., Kurosawa Y., Saijyo R., Koshiba S., Yasuda J., Motohashi H., Sugawara J., Tanabe O., Kinoshita K., and Yamamoto M. (2016) Establishment of protocols for global metabolomics by LC-MS for biomarker discovery. PloS One 11, e0160555 10.1371/journal.pone.0160555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rasband W. S. (1997–2012) ImageJ, U.S. National Institutes of Health, Bethesda, MD [Google Scholar]
- 73. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.