Abstract
The ribosomally synthesized and posttranslationally modified peptides (RiPPs), also called ribosomal peptide natural products (RPNPs), form a growing superfamily of natural products that are produced by many different organisms and particularly by bacteria. They are derived from precursor polypeptides whose modification by various dedicated enzymes helps to establish a vast array of chemical motifs. RiPPs have attracted much interest as a source of potential therapeutic agents, and in particular as alternatives to conventional antibiotics to address the bacterial resistance crisis. However, their ecological roles in nature are poorly understood and explored. The present review describes major RiPP actors in competition within microbial communities, the main ecological and physiological functions currently evidenced for RiPPs, and the microbial ecosystems that are the sites for these functions. We envision that the study of RiPPs may lead to discoveries of new biological functions and highlight that a better knowledge of how bacterial RiPPs mediate inter-/intraspecies and interkingdom interactions will hold promise for devising alternative strategies in antibiotic development.
Keywords: antimicrobial peptide (AMP), bacteria, bacteriocin, biofilm, natural product, bacterial communication, ecological strategies, microbial competition, ribosomal peptide natural product (RPNP), ribosomally synthesized and post-translationally modified peptide (RiPP)
Introduction
Microorganisms have a remarkable social life. They generally live in complex polymicrobial communities and diverse environments where interactions between individuals shape the composition of populations and communities as well as their functions (1–3). Microbial communities colonize niches at all levels of the biosphere, from soils and oceans to living organisms. Indeed, it is now firmly established that all organisms are living in tight association with microorganisms, forming the so-called holobionts that are found in all environments, marine or terrestrial, and all organisms from plants to vertebrates, including humans (4, 5). However, deciphering the mechanisms and interplays between the molecules and macromolecules from the host and microorganisms that make up the holobiont is still in its infancy.
Social interactions are widespread in microorganisms and play a major role in bacterial evolution and virulence. Either they are antagonistic, leading to the elimination of competitors, or they are cooperative or synergistic, which permits bacteria from obtaining benefits that could not be acquired by a single individual (2). These interactions lead to various behaviors, such as communication, toxin secretion, and acquisition of nutrients or metals that are essential for growth and development, such as iron for all bacteria (6, 7) or copper for methanotrophic bacteria (8). Adverse molecules, such as antimicrobials, have been considered for many years as weapons used by bacteria to inhibit or kill competitors in a given niche. But more recently, alternative hypotheses consider that antimicrobial compounds would act also as signaling molecules able to coordinate cooperative social interactions between bacteria (1, 9). Moreover, the production of antimicrobials would be influenced by social and competitive interactions between competing bacteria (10–13). Whereas the traditional model of bacterial life is that of cells swimming in a liquid environment, bacteria also commonly live in complex communities associated with various surfaces, either natural or human-engineered ones, which are called biofilms (14). Biofilms result from the coordinated action of several bacterial strains working together to acquire benefits. They often occur as a response to ecological competition from other strains and toxic stresses (15, 16). Antimicrobial compounds and communication or signaling molecules are involved in these processes through the so-called quorum-sensing (QS)3 mechanism (16). Currently, the ecological aspects of microbial research take particular importance both in environmental domains and in the field of bacterial pathogenesis (17), pointing to an acute need for understanding the underlying mechanisms.
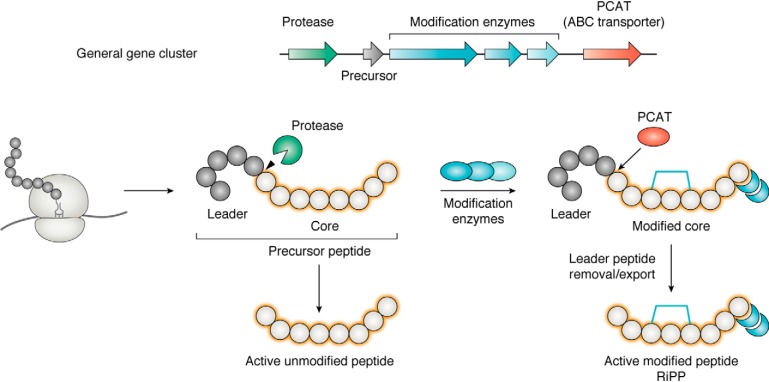
Bacteria can produce a broad range of metabolites that, among other functions, can mediate bacterial interactions and display antimicrobial activity. A large number of these natural products (NPs) originate from large multifunctional enzymatic complexes, nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs), or hybrid PKS/NRPS systems (18, 19). Another broad and diverse class of natural products consists of the superfamily of ribosomally derived molecules, the so-called ribosomally synthesized and posttranslationally modified peptides (RiPPs) (20), also termed ribosomal peptide natural products (RPNPs). Their biosynthesis starts with ribosomally synthesized linear peptides that are subject to enzymatic modifications. The peptide precursor is composed of a core peptide that is preceded by an N-terminal leader region, often involved in recognition events (21). In rare cases, the core peptide is followed by a C-terminal extension (i.e. a follower peptide). The core peptide is decorated with posttranslational modifications set up by dedicated enzymes (Fig. 1). The posttranslational modifications involved in RiPP biosynthesis are diverse, including for example dehydration, cyclodehydration, cyclization, glycosylation, and phosphorylation, resulting in a vast array of structures (20).
Figure 1.
General biosynthetic pathway leading to RiPP production, from the gene cluster to the mature active compound. The ribosomally synthesized precursor can be only cleaved by a protease (green) and exported by an ABC transporter (brown) as an unmodified peptide. For RiPP biosynthesis, the core peptide is fused to a leader peptide (gray) that contributes to the action of the posttranslational modification enzymes (blue). Cleavage of the leader and export of the RiPP are both generally ensured by a peptidase-containing ATP-binding transporter (PCAT) that accomplishes the two functions.
RiPPs are essentially produced by bacteria, but many eukaryotic cells, such as fungi, plants, or marine organisms, are also producers of such compounds (22). They are classified according to their structural features that rely on the types of posttranslational modifications. Among the most extensively studied classes, we can cite lanthipeptides and lantibiotics, thiopeptides, lasso peptides, cyanobactins, and thiazole/oxazole-modified microcins (TOMMs) (20). RiPPs are considered promising molecules to be developed for applications in environmental, medical, veterinary, and food industrial domains. They constitute a fast-expanding area of research due to increased genome-mining efforts and to the huge interest raised by their structures, biological properties, biosynthesis pathways, and posttranslational modification enzymes (for reviews, see Refs. 20 and 23–29). Metagenome analyses of various holobionts offer great promise for discovering novel RiPPs. The high abundance of RiPPs in the human microbiome has been reported based on genome-mining, metabolomics, and computational MS approaches (30–35). The development of novel and efficient software tools will allow expansion of the repertoire of RiPPs from diverse microbiomes, including those with unknown posttranslational modifications (36).
Despite the significant interest in RiPPs from an anthropocentric point of view, very little is known about the ecological roles of RiPPs in nature. Given that peptides mediate many biological processes across all domains of life, it is conceivable that RiPPs would play diverse roles in the social behavior and physiology of microorganisms. A number of RiPPs have been shown in vivo and/or in vitro to be involved in competition, communication, and various physiological functions such as biofilm formation and morphological development. The scope of this review will cover different classes of bacterial RiPPs and summarize current knowledge of their natural functions, particularly in the context of interactions within microbial communities and with their hosts.
RiPPs as actors of niche competitions
An overview of antibacterial RiPP families involved in competition
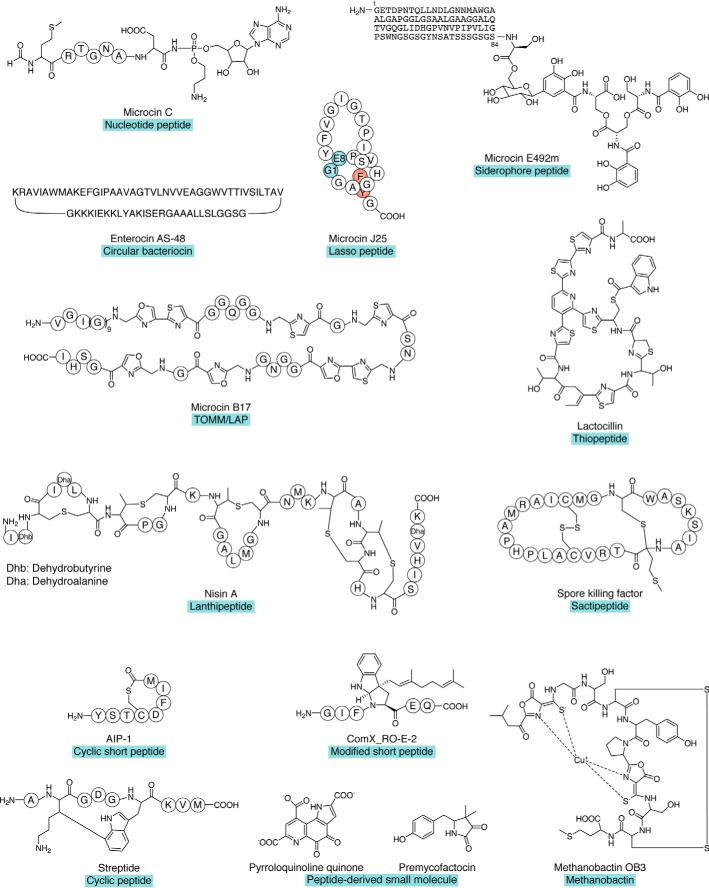
One obvious function for RiPPs is related to that of antimicrobial peptides (AMPs), which act as chemical weapons for defense and competition. Antibacterial RiPPs are posttranslationally modified bacteriocins or microcins produced by Gram-positive and Gram-negative bacteria (37, 38). Their biosynthetic gene clusters (BGCs) share common features. They encompass genes that encode at least one precursor peptide, one or several posttranslational modification enzymes, self-immunity proteins, and transporters that ensure the export of the RiPP and can be involved in immunity of the producer to the toxic RiPP in certain cases. The archetypes for each family of antibacterial RiPPs are described briefly below with regard to their structures (Fig. 2), biosynthesis pathways, and modes of action (Table 1). The roles in niche competition played by a number of such peptides have been evidenced. However for others, in particular the nucleotide peptide microcin C, there is not yet available information on their ecological roles in microbial communities, although it can be envisioned that they should fulfill similar functions.
Figure 2.
Representative structures of different classes of RiPPs cited in this review. Unmodified amino acids are circled. In the microcin J25 structure, the residues forming the macrolactam linkage and entrapping the tail within the ring by steric hindrance are highlighted in green and red, respectively.
Table 1.
Selected microbial RiPPs covering different chemical classes cited in this review for which an ecological function has been described
| RiPP class | RiPP name | Producing strains (examples) | Ecological role | Mode of antibacterial action (when applicable) | References |
|---|---|---|---|---|---|
| Circular bacteriocin | Enterocin AS-48/Bac-21 | E. faecalis AS-48, OG1RF | Interference competition | Pore-forming activity | 68 |
| Cyclic peptide | AIP | S. aureus, S. hominis | Quorum-sensing signala | 137, 152, 154 | |
| Cyclic peptide | Streptide | S. thermophilus | Quorum-sensing responseb | 175–177 | |
| Lanthipeptide (lantibiotic) | Nisinc | L. lactis | Autoinducing peptidea | Pore-forming activity; Blocking of peptidoglycan synthesis (binding to lipid II) | 83–86, 88 |
| Lanthipeptide (two-peptide lantibiotic) | Sh-lantibiotics-α/β | S. hominis A9 | Interference competition | Unknown | 135 |
| Lanthipeptide | SapB/SapT | S. coelicolor, S. tendae | Morphological development in Streptomyces | 194, 195 | |
| Lanthipeptide (lantibiotic) | Planosporicinc | Planomonospora alba | Autoinducing peptidea | Blocking of peptidoglycan biosynthesis (binding to lipid II) | 167, 238 |
| Lanthipeptide (lantibiotic) | Unnamed | S. pneumoniae | Quorum-sensing response; niche competition | Unknown | 170 |
| Lanthipeptide (two-peptide lantibiotics) | Cytolysin | E. faecalis | Autoinducing peptidea; virulence factor | Pore-forming activity | 169 |
| Lanthipeptide (two-peptide lantibiotics) | Lacticin 3147 (LtnA1/LtnA2) | L. lactis | Interference competition | Pore-forming activity (LtnA1/LtnA2/lipid II synergy) | 133, 134 |
| Lasso peptide | Microcin J25 | E. coli AY25 | Interference competition | Blocking of transcription (binding to RNA polymerase) | 55, 65 |
| Methanobactin | Methanobactin | Methylosinus trichosporium | Metallophore | 207 | |
| Modified short peptide | ComX | B. subtilis | Quorum-sensing signala | 158 | |
| Modified small peptide | RaxX | X. oryzae pv. oryzae | Host-bacteria interaction | 181 | |
| Peptide-derived small molecule | Pyrroloquinoline quinone | K. pneumoniae | Redox enzyme cofactor | 217 | |
| Peptide-derived small molecule | Mycofactocin | M. tuberculosis | Redox enzyme cofactor | 216, 225 | |
| Sactipeptide | Sporulation killing factor | B. subtilis 168 | Cannibalism | 202 | |
| Sactipeptide (sactibiotic) | Ruminococcin C | R. gnavus E1 | Interference competition | Perturbation of nucleic acid synthesis | 32, 33, 239 |
| Sactipeptide (two-peptide sactibiotic) | Thuricin CD (Trnα/Trnβ) | B. thuringiensis DPC 6431 | Interference competition | Pore-forming activity | 240 |
| Siderophore peptide | Microcin E492/E492m | K. pneumoniae RYC492 E. coli VCS257 with pJAM229 plasmid | Interference competition; exploitative competition (iron) | Pore-forming activity; Perturbation of mannose transport | 44–47 |
| Siderophore peptide | Microcin H47 Microcin M | E. coli H47, Nissle 1917, CA46, CA58 | Interference competition; exploitative competition (iron) | Perturbation of ATP synthesis | 42, 107 |
| Thiopeptide | Lactocillin | L. gasseri | Interference competition | Unknown | 31 |
| Thiopeptide | Thiocillinc | B. cereus | Stimulation of biofilm formation in Bacillus | Inhibition of ribosomal protein synthesis (binding to EF-Tu) | 191 |
| Thiopeptide | Thiostreptonc | S. laurentii | Morphological development in Streptomyces | Inhibition of ribosomal protein synthesis (binding to EF-Tu) | 200 |
| TOMM/LAP | Goadsporinc | Streptomyces sp. | Morphological development in Streptomyces | Unknown | 199 |
| TOMM/LAP | Listeriolysin S | L. monocytogenes | Interference competition (virulence factor) | Unknown | 139–141, 241 |
| TOMM/LAP | Streptolysin S | S. pyogenes | Virulence factor (cytotoxin) | 71 |
a The terms of autoinducing peptide and quorum-sensing signal are distinguished, the former describing peptides that only autoinduce their own production and the latter relating to peptides that also regulate transcription of other genes in addition to autoinduction.
b Other physiological functions have not been identified.
c Also an antibacterial peptide, but its function in niche competition has not yet been demonstrated.
Nucleotide peptides
The nucleotide peptide microcin C (Fig. 2) is synthesized by several strains of Escherichia coli as a leaderless precursor heptapeptide that has to undergo a two-step maturation in both the producer and the target bacterium for acquiring activity (for a review, see Ref. 39). First, posttranslational modifications of the precursor happen in the producing bacterium, leading to a formylated heptapeptide linked to a nucleotide moiety, which remains inactive. Second, after export outside of the producer, a double proteolytic cleavage occurs in the susceptible bacteria, providing the toxic entity, which is a nonhydrolyzable aspartyl-adenylate. This mimic of aspartyl adenylate is an inhibitor of aspartyl-tRNA synthetase, which therefore blocks protein synthesis at the translation step (40).
Siderophore peptides
Siderophore peptides are exemplified by microcins E492, M, and H47 (41, 42) (Fig. 2) (Table 1). Microcin E492 (MccE492) was initially characterized in Klebsiella pneumoniae RYC492 as an unmodified ribosomally synthesized peptide with a potent antibacterial activity directed essentially against Escherichia and Salmonella (43). This activity is associated with a pore-forming property (44, 45) and with interaction with the inner membrane components ManYZ of the mannose permease complex involved in mannose uptake (46). According to the growth conditions, this microcin is biosynthesized as the initially characterized unmodified form MccE492 (44) or as a posttranslationally modified form (MccE492m) that carries a linear catechol-type siderophore at the C terminus (47). The siderophore-microcin has a much higher antibacterial activity than the unmodified counterpart, which is reflected by an enlarged spectrum of activity, including Klebsiella and Enterobacter species. Actually, it has been shown that the unmodified microcin is an incompletely processed form and that the final mature compound (i.e. the siderophore microcin) is an RiPP (48). The MccE492/MccE492m biosynthesis requires four enzymes (MceC, MceD, MceI, and MceJ) that work with two precursors: the peptide precursor MceA and enterobactin, a catechol siderophore, which itself results from an NRPS pathway. Enterobactin is glucosylated and linearized by a C-glucosyl transferase (MceC) and an enterobactin esterase (MceD), successively, before an ester linkage is established between glucosylated enterobactin and the C-terminal serine of the peptide precursor. The resulting modified MceA is then processed at the membrane, and the mature modified peptide is exported after cleavage of the leader peptide (48, 49).
Lasso peptides
Lasso peptides are produced by Enterobacteriaceae (Proteobacteria), Actinobacteria, and a few Firmicutes. They are characterized by a [1]rotaxane knotted structure, which has been named after the shape adopted by the lasso of a cowboy (for reviews, see Refs. 50–54). The lasso topology is composed of a macrocycle closed by an isopeptide bond linkage from an Asp or Glu side chain to the N terminus of the core peptide. The resulting C-terminal tail is threaded through the ring and maintained in this entropically disfavored situation by bulky residues that firmly straddle the tail into the ring or, by disulfide bonds, or by an association of both means. Microcin J25 is the most intensively studied lasso peptide (Fig. 2 and Table 1). It remained for more than 10 years the archetype of this RiPP family. It is produced by E. coli AY25 (55) and deserved attention over the years due to its potent narrow-spectrum antibacterial activity, essentially directed against Salmonella and Escherichia. Its lasso structure was determined in the 2000s (56–59). Microcin J25 biosynthesis requires only four genes assembled on a plasmid (mcjABCD). It was shown that the lasso topology can be reconstituted in vitro from the precursor McjA in the presence of the two enzymes McjB and McjC and ATP only (60). McjC and McjB act as a lasso cyclase homologous to an asparagine synthetase and a leader peptidase, respectively, which presumably form a complex (lasso synthetase) (61). The mature lassoed microcin is then exported outside the producing cells by the ATP-binding cassette (ABC) exporter McjD, which ensures secretion of the toxic peptide into the environment. This both provides self-immunity to the producer and allows it to compete with other bacteria in the same niche thanks to the microcin (62, 63). Microcin J25 enters susceptible bacteria by hijacking the outer-membrane siderophore receptor FhuA/TonB-ExbB-ExbD–dependent pathway and the inner-membrane SbmA transporter to cross the double membrane of Gram-negative bacteria (for reviews, see Refs. 38 and 52) and subsequently inhibits gene transcription by inhibiting the RNA polymerase. Although many newly identified lasso peptides do not show significant antibacterial activities against tested strains, some of them are found to inhibit a range of bacterial RNA polymerases (64–66), indicating that the entry of the sensitive cells is the key determinant of the activity of lasso peptides.
Circular bacteriocins
Circular bacteriocins are characterized by a head-to-tail cyclization, which is actually the only posttranslational modification undergone by the ribosomal precursor. The BGCs of circular bacteriocins encode a membrane protein, which is hypothesized to ensure the N to C terminus cyclization and possibly the protease activity. About 14 circular bacteriocins have been characterized to date (67), among which is enterocin AS-48 (Fig. 2 and Table 1), the representative of the group, which has been characterized in Enterococcus faecalis strains from both food and clinical origins (68). Circular bacteriocins share a common, highly compact three-dimensional structural fold, which typically consists of an arrangement of 4–5 α-helices, packed into a helical bundle or a saposin-like fold (67, 69). Circular bacteriocins have been proposed to kill bacteria by interaction with the bacterial cell membrane, causing its permeabilization and leading to leakage of ions, dissipation of membrane potential, and cell death. The dimerization process of enterocin AS-48 at physiological pH was proposed to play a role in the mechanism of action. However, although it has been established for many circular bacteriocins that they did not require a receptor for antimicrobial activity, the maltose ABC transporter complex has been shown to be important for activity in the case of garvicin ML, leading to the hypothesis of a dual concentration-dependent mode of action, with membrane activity operating at higher concentrations than perturbation of maltose transport (70).
TOMMs-linear azol(in)e-containing peptides (LAPs)
LAPs exhibit various combinations of thiazole and oxazole or methyl-oxazole heterocycles. Although the term microcin is usually restricted to AMPs produced by Gram-negative bacteria, LAPs are also called thiazole/oxazole-modified microcins (TOMMs) (71) even when they are produced by Gram-positive bacteria. This nomenclature was adopted by homology to microcin B17 produced by E. coli (Fig. 2), the prototypic molecule endowed with such modifications (72) and the first RiPP to have its biosynthesis reconstituted in vitro (73). The thiazole/oxazole rings of microcin B17 are made from Cys and Ser/Thr residues by a three-component B17 synthetase that catalyzes dehydration and cyclization to form azolines, which are subsequently oxidized to azoles (74). Peptides from the TOMM family include several toxins from pathogenic bacteria (streptolysin S from Streptococcus pyogenes and other Streptococcus species, listeriolysin S from Listeria monocytogenes (Table 1), clostridiolysin S from Clostridium botulinum and Clostridium sporogenes, and stapholysin S from Staphylococcus aureus RF122, whose structures all remain elusive) and other TOMMs from nonpathogenic bacterial species (71, 74). Those have been shown to exert different biological activities among which are antibacterial properties (microcin B17 from E. coli, klebsazolicin from Klebsiella pneumoniae, goadsporin from soil Streptomyces sp. (Table 1), plantazolicin from the saprophyte Bacillus amyloliquefaciens FZB42 (75), or trifolitoxin and phazolicin from the legume symbionts Rhizobium leguminosarum bv. trifolii T24 (76) and Rhizobium sp. Pop5, respectively (77)). It has been firmly established that microcin B17 blocks DNA gyrase (74, 78) and that phazolicin and klebsazolicin inhibit the ribosome (77), but the modes of action of other TOMM peptides remain largely unknown.
Thiopeptides
Thiopeptides are macrocyclic peptides containing a characteristic six-membered nitrogen-containing ring, oxazole/thiazol(in)e moieties, and/or dehydroamino acids (Fig. 2). They are produced by and show potent activity against Gram-positive pathogens, such as methicillin-resistant S. aureus and Clostridium difficile (79). Their mode of action consists of inhibition of protein synthesis, interfering either with the 50S ribosomal subunit (80) or the elongation factor Tu (81). Biosynthesis of the oxazole/thiazole motifs is similar to that of TOMMs. The central nitrogen-containing ring is synthesized by a dedicated enzyme catalyzing a [4 + 2]-cycloaddition reaction. Thorough genome mining studies have revealed that the human microbiome harbors abundant thiopeptide BGCs (31) and that Actinobacteria or Bacilli species are the dominant producers of thiopeptides (82), raising the questions of their ecological roles in the related complex ecosystems (e.g. soil environment and human microbiota).
Lanthipeptides and lantibiotics
Lanthipeptides, including those that have antimicrobial properties, called lantibiotics, are produced by and active against Gram-positive bacteria. They are generally inactive against Enterobacteriaceae and other Gram-negative bacteria. The major producers are lactic acid bacteria (e.g. Lactococcus and Streptococcus) and certain Staphylococcus strains. Lanthipeptides are characterized by the presence of the unusual residues lanthionine and 3-methyllanthionine (for reviews, see Refs. 83–86). These residues result from dehydration of serine and threonine in the core peptide to give dehydroamino acids that cyclize with cysteines via the formation of thioether linkages between β-carbons. Nisin, produced by Lactococcus lactis, is the first studied lantibiotic (Fig. 2 and Table 1). It has been used as a food preservative (Nisaplin® and Niprosin®) in more than 80 countries for over 50 years. Its mechanism of action involves binding to and sequestration of lipid II, the essential peptidoglycan precursor for cell wall biosynthesis. This interaction both blocks peptidoglycan biosynthesis and causes the formation of lipid II–nisin heteromolecular pores in the membrane bilayer, leading to leakage of ions and essential metabolites. Other lantibiotics called “two-peptide lantibiotics” (or two-component lantibiotics) are produced as two distinct peptide entities, which function in a synergistic fashion to give optimal activity, whereas separately they are not active or are weakly active (87). Each component of two-peptide lantibiotics fulfills one of the two roles involved in the mechanism of action of nisin (i.e. binding to lipid II and pore formation) (88). They are exemplified by lacticin 3147 (consisting of peptides LtnA1/LtnA2) (Table 1) produced by L. lactis and haloduracin (peptides Halα/Halβ) from Bacillus halodurans, the most studied of the group (52). The structure and mechanism of action of lacticin 3147 have been completely deciphered, showing the specific role of each of the two peptide components LtnA1 and LtnA2 in the synergistic mechanism (89, 90). LtnA2 can form pores in membrane bilayers without the help of LtnA1 or lipid II. LtnA1 cannot form pores in membrane bilayers but has the ability to bind lipid II with its C terminus. The synergistic mechanism of action of the two lacticin 3147 components appears flexible, as lipid II binding enhances the membrane activity, which, however, can happen, although to a lesser extent, independently of lipid II.
Sactipeptides and sactibiotics
Similar to lanthipeptides and lantibiotics, the term sactipeptides includes the subclass of antimicrobial sactibiotics (91, 92). They are characterized by an intramolecular sulfur to α-carbon linkage between a cysteine and another residue (Fig. 2) and, as such, are distinguished from lanthipeptides/lantibiotics (thioether linkages between β-carbons) and from sactipeptide-like peptides that contain sulfur to β- or γ-carbon linkages (93). The sactionine motif is established by a radical-based mechanism ensured by a radical SAM enzyme called sactisynthase. Only few sactipeptides have been characterized to date. They are produced mainly by the Bacillus genus (subtilosin A from B. subtilis 168, the first reported sactipeptide, the sporulation-killing factor (SKF) from B. subtilis 168 (Fig. 2) (Table 1), thurincin H from Bacillus thuringiensis SF361, the two-peptide thuricin CD from the human fecal isolate B. thuringiensis DPC 6431, and huazacin or thuricin Z from B. thuringiensis serovar Huazhongensis) (25, 32, 93, 94). They have been found recently in Ruminococcus (ruminococcins from Ruminococcus gnavus isolated from the human microbiota) (32, 33) and presumably in Staphylococcus (hyicin from Staphylococcus hyicus 4244). The structure of ruminococcin C was recently established, showing that this sactipeptide is stabilized by four thioether bonds that generate a double hairpin fold (32, 33).
RiPP-mediated competition mechanisms
Competition has long been recognized as a major regulatory process in populations and community dynamics, which structures the ecological systems. It involves one organism decreasing the survival, development, or reproduction of others. Competition is categorized into two major modes identified in ecosystems, including microbial communities either hosted in holobionts or living in open environments: (i) exploitative competition, which is an indirect process and occurs via resource consumption when organisms compete for common nutrients and one organism depletes its surroundings of nutrients, and (ii) interference competition or direct competition, which happens when one individual directly attacks another, generally by the secretion of harmful molecules (9, 95, 96). The battle for iron is a typical example of exploitative competition (97), whereas the production of antimicrobial molecules and toxins (bacteriocins, microcins, and other NPs) exemplifies interference competition. In some cases, interference competition can be driven through an indirect action of a metabolite that triggers at sublevel concentration the production of an antibiotic molecule. This process, which involves a cross-talk between antibiotic biosynthesis pathways, is exemplified by jadomycin B, an angucyclin antibiotic produced by Streptomyces venezuaelae, which at subinhibitory concentration regulates the production of prodigiosin (an NRPS-derived antibiotic) by Streptomyces coelicolor and concomitantly its morphological development (98).
It has been proposed that bacteria have evolved to detect and respond to ecological competition by modifying regulatory networks and drive responses to defend them and counterattack. This physiological response that detects harms caused by other cells but not from abiotic stresses has been called competition sensing (9). Engagement in interference and exploitative mechanisms of competition contributes to colonization resistance, the process by which in healthy conditions and in the absence of harmful molecules, microbiota can efficiently inhibit colonization and overgrowth by invading nonindigenous microorganisms, including pathogens (99, 100).
Among the different roles played (or supposed to be played) by RiPPs in nature, their involvement in microbial competition is the most documented one. The gut microbiota is probably the most extensively studied and the better known natural microbial ecosystem due to its extremely high importance for human health (101). Indeed, perturbation of its equilibrium (dysbiosis) is correlated to many diseases. Therefore, it has been analyzed from both a strict microbiological and a biochemical aspect to gain a better understanding of the interplay between molecules from the microorganisms and the host. Moreover, the therapeutic potential of the molecules secreted by the gut microbiota has increased in the context of bacterial resistance and its implications for human, animal, and environmental health (102). Currently, other mammal-associated microbiota are increasingly being studied, and among them, skin microbiota is pretty well-documented (103, 104). In this context, it has been evidenced both in vivo and in vitro that certain RiPPs are actors in microbiota and particularly in the gut microbiota. To describe these aspects, we selected several classes of RiPPs produced by Gram-negative and Gram-positive bacteria, including modified microcins and bacteriocins, which illustrate different contexts in the field.
Interference competition driven by RPNPs in Gram-negative bacteria
The siderophore peptides microcins M and H47 use two complementary strategies to serve as efficient actors in colonization of the gut and competition. They are produced by E. coli Nissle 1917 (also called Mutaflor and abbreviated as EcN), a probiotic strain originally isolated in 1917 by the army surgeon Alfred Nissle from the feces of a soldier who resisted a severe outbreak of shigellosis during the First World War (105). Today it is one of the most investigated bacterial strains, but despite its commercialization and numerous applications, the mechanisms underlining its positive properties to regulate intestinal disorders of infectious or inflammatory origin remain still obscure (106). Microcins M and H47 initially identified in EcN (107) have been shown further to be siderophore peptides (42). Although the potent bacterial inhibitory properties of microcins in vitro were known for a long time (108), their action and interplay with competitors in vivo remained elusive (109). However, microcins M and H47 were recently demonstrated for the first time to mediate competition among Enterobacteriaceae exclusively in the inflamed gut, which represents highly competitive conditions for resources, and particularly for iron (110). Indeed, in such conditions, EcN colonizes the inflamed gut, where it uses its microcins for niche competition against related Enterobacteriaceae, without modifying substantially the gut microbiota. Furthermore, in these conditions, EcN microcins impair the growth of enterobacterial pathogens, such as Salmonella enterica subsp. enterica serovar Typhimurium or adherent invasive E. coli. Moreover, in a recent study, evidence that the posttranslational modification of EcN microcins with a siderophore is required for antibacterial activity in vivo has been obtained (111). This study also shows that the biosynthesis pathway of siderophore microcins tightly depends on that of the genotoxin colibactin (112), which is also produced by EcN and is correlated with the frequency and severity of colorectal cancer in humans (113), making the use of this commercialized probiotic problematic.
Taken altogether, these studies show on one side that siderophore microcins M and H47 efficiently act in microbial competition through both exploitative competition (i.e. in the present case, competition for iron) and interference competition (contact-dependent killing) (96) (Fig. 3A), because they both trap catechol siderophores to anchor them at the peptide C terminus and hijack siderophore receptors for import. On the other side, they highlight the versatility of the biosynthetic assembly lines to produce both virulence factors and competition molecules and the consequential fine margin between pathogenicity and probiotic activity for a given strain.
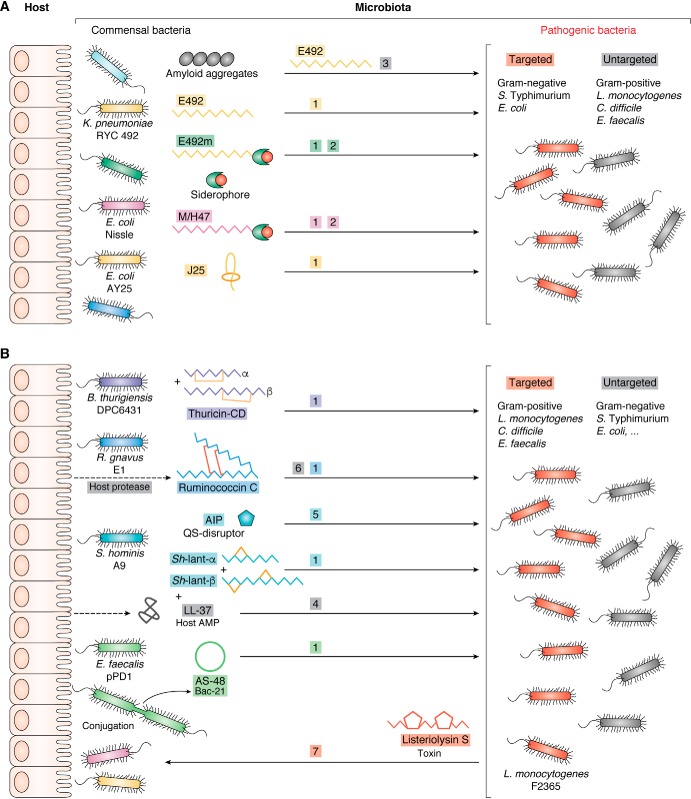
Figure 3.
Schematic of the main strategies of Gram-negative (A) and -positive (B) bacteria for colonization resistance and competition involving RPNPs. 1, interference competition using RPNPs as contact-dependent killer molecules (several examples from both Gram-negative and -positive bacteria, among which are the lasso peptide microcin J25 and the two-peptide sactibiotic thuricin CD, respectively); 2, exploitative competition exemplified by the battle for iron involving competition between siderophores and siderophore-modified peptides for the same high affinity receptors (MccE492m from K. pneumoniae RYC 492 and MccH47 from E. coli Nissle 1917); 3, reservoir of killing RPNPs using functional amyloids for delivering killers (MccE492 from K. pneumoniae RYC 492); 4, synergy between RPNPs and host AMPs (Sh-lantibiotic-α and Sh-lantibiotic-β from S. hominis A9, each synergizing with the host cathelicidin LL-37); 5, quorum-sensing interference (an autoinducing RPNP from S. hominis A9 acts as a disruptor of quorum sensing that inhibits the secretion of the virulence factor PSMα in S. aureus; 6, cooperation between host and RPNP producing bacterium in the gut for the final processing step leading to mature RPNP acting by interference competition (ruminococcin C from R. gnavus E1); 7, manipulation of commensals by the pathogen to occupy the niche and increase virulence (the pathogenic strain L. monocytogenes F2365 produces listeriolysin S, a TOMM virulence factor, which targets commensal bacteria occupying the same niche and devoid of self-immunity to the toxin, to increase nutrient level and favor invasion).
Microcin J25 initially produced by a newborn infant fecal isolate of E. coli AY25 has been known for about 25 years for exerting a potent and narrow-spectrum antibacterial activity directed essentially against Escherichia, Salmonella, and Shigella at concentrations in the nanomolar to micromolar range (55). The activity of microcin J25 in complex matrices and in vivo in a mouse model of Salmonella infection was evidenced (114). Competitive exclusion of Salmonella in poultry under field conditions has been applied since the 1990s in several countries, using diverse preparations, including complex consortia of bacterial strains (115). But it was not until recent years that evidence of the involvement of microcin J25 in these properties was obtained. In that way, the ability of microcin J25 to compete the pathogen Salmonella in the human or animal gut conditions, as well as its stability in the different GI tract compartments, were examined using gut simulators. The degradome of microcin J25 was examined in both dynamic (TIM1 dynamic simulator) and static models of digestion, using antibacterial assays, LC-MS/MS, and molecular networking analysis (116). The stability and activity are quite good in the stomach acidic conditions, but the lassoed microcin partly degrades in the compartment mimicking the duodenum conditions, in particular upon the action of elastase, one of the main enzymes in this GI compartment, showing that protection of the microcin will be required for further applications. Our current work indicates that microcin J25 has a potent antagonistic activity against the pathogen Salmonella enterica subsp. enterica serovar Newport without strong perturbation of the microbiota composition in an in vitro model of colon.4 The efficacy of microcin J25 in the GI tract was also evaluated in vivo as a strategy to control the negative effects of postweaning stress in pig husbandry (117). Microcin J25 used as a feed additive was shown to reduce inflammation, attenuate diarrhea, and improve growth performance for weaned pigs, which are particularly susceptible to pathogen infections. Otherwise, the EcN strain was engineered to express and secrete microcin J25 (strain E. coli Nissle(J25)) (118). When administered to turkeys challenged with pathogenic Salmonella, the engineered probiotic E. coli Nissle(J25) significantly reduced the S. enterica Enteritidis counts in turkey caeca compared with nonengineered EcN, indicating that the lassoed microcin is responsible for this competitive effect. Therefore, rigorous scientific bases obtained thanks to the numerous studies on the structure, mechanisms of antibacterial properties, and activity in gut (both in in vitro models and in vivo) of microcin J25 testify to the role of the lasso peptide as a competition molecule in the gut microbial community (Fig. 3A). These data also support the possibility of translation of the ecological properties of an RiPP to society via potential applications to the animal feed industry.
Functional amyloids as reservoirs of toxic compounds and modulators of antibacterial activity
Siderophore microcins develop another strategy to exert their dominance in microbial communities by using their ability to form or not form functional amyloids (depending on the environmental conditions), which serve as reservoirs of toxic compounds. Amyloid aggregates have been studied initially as associated with misfolded proteins involved in cytotoxicity and pathologies, including prion diseases, diabetes type II, or Alzheimer's and Parkinson's diseases (119, 120). However, in the last 2 decades, it has been shown that amyloids have critical functions in all domains of life and that their production can be beneficial for the organism (121). Particularly in the bacterial world, the amyloid state represents a functional structure, which can ensure important functions in bacterial fitness and social behavior (122, 123). Functional amyloids take part in virulence, cytotoxicity, adhesion to surfaces, biofilm formation as well as protein/peptide reservoirs, and their role continues to expand.
Posttranslational modification was shown to play a particular role in both the formation and function of amyloids, which ensures a subtle role in microbial competition. Indeed, microcin E492 (MccE492) has the capacity to form amyloid fibrils in vitro and in vivo in the extracellular space, and this property was correlated with a loss of antibacterial activity of the peptide (124, 125). Similar to the extracellular toxin inactivation process, intracellular amyloid accumulation does not have a toxic effect (126). Indeed, amyloid fiber nucleation by the MccE492m modified form of the microcin appears less efficient than by the unmodified peptide, although both forms can be incorporated into preformed fibers (125). It was shown that in vitro amyloid formation by MccE492 is a dynamic and reversible process and that amyloids could represent a reservoir of toxic molecules (127, 128). Although such a process has not been shown in vivo, it can be envisaged that the toxic or antibacterial compounds would be stocked when the concentration reaches a high level and released when required upon modification of the environmental conditions (Fig. 3A). Furthermore, bacteria generally use dedicated immunity proteins or ABC transporters encoded in the bacteriocin gene cluster to inactivate or expel the toxin they produce and make them resistant. Thus, amyloidogenesis would be a strategy used by bacteria to modulate the release of potent weapons against niche-occupying competitors and simultaneously contribute to self-immunity.
Interference competition driven by RPNPs in Gram-positive bacteria
Enterococci are Gram-positive bacteria belonging to the commensal gut microbiota, but they are also opportunistic pathogens that can be responsible for significant diseases in immune-compromised people. An E. faecalis strain, OG1RF, harboring the sex pheromone–responsive conjugative plasmid pPD1 (129) has been shown to replace indigenous enterococci and outcompete E. faecalis strains lacking the pPD1 plasmid in a model of colonization of the mouse gut with E. faecalis (130, 131). This model allowed the establishment of long-term colonization of the gut by a marked strain without the need for antibiotics by delivering the strain in drinking water, therefore modeling commensal colonization. The plasmid pPD1 encodes bacteriocin 21 (Bac-21), which is actually identical (129) to the well-characterized 70-amino acid circular bacteriocin AS-48. In this mammalian GI tract model, Bac-21 was shown to provide colonization advantage in the highly competitive environment of the gut to outcompete indigenous enterococci and drive the competition between closely related bacterial species. Moreover, pPD1 is actively transferred to other E. faecalis strains by conjugation in the GI tract, showing that this strategy can be used to enhance the number of Bac-21 producers in the niche and eliminate more efficiently and rapidly the susceptible population. Thus, the circular bacteriocin Bac-21/enterocin AS-48 serves as a killing factor that facilitates competition between the E. faecalis producer and other enterococci that are not able to synthesize this RiPP (Fig. 3B). Therefore, the production of circular bacteriocin by commensal bacteria, such as E. faecalis, drives niche competition in the GI tract by the interference competition strategy boosted by a conjugation transfer strategy of the encoding plasmid to enhance the level of production of the killer molecule.
Thuricin CD is a two-component sactibiotic produced by B. thuringiensis DPC 6431 isolated from a human fecal isolate (132) (Table 1). The two distinct 30-amino acid sactipeptides, Trn-α and Trn-β, forming thuricin CD act synergistically (optimal 1:2 ratio) to kill the pathogens Clostridium difficile, the causative agent of hospital-acquired infections, and L. monocytogenes at nanomolar concentrations. But they have a moderate impact on most other genera, indicating a very restricted spectrum of activity. In an in vitro distal colon model, thuricin CD killed highly efficiently a wide range of clinical C. difficile isolates while having a low impact on other genera and in particular on gastrointestinal commensal strains, such as Lactobacillus casei and Bacillus lactis, known to contribute to microbiota health. Thuricin CD is thus significantly stable in the gut conditions, where it can selectively outcompete a pathogen by interference competition (Fig. 3B) without perturbing the commensal microbiota.
Recently, ruminococcin C purified from caecal contents of rats associated with the human symbiont R. gnavus E1 has been shown to kill pathogenic Clostridia and Gram-positive multidrug-resistant bacteria without showing toxicity to eukaryotic cells (33). It was proposed to inhibit nucleic acid synthesis in a way similar to that of the commercial antibiotic metronidazole. Moreover, ruminococcin C maturation from the inactive precursor into its active form was shown to involve two successive steps. The first one occurs in the R. gnavus producer thanks to a specific zinc-metallopeptidase, whereas the second one is ensured by the human pancreatic trypsin (Fig. 3B). This is the first description in RiPPs of a two-step maturation process involving enzymes from both the symbiont and the host, which affords an example of the tight cooperation between host and associated bacteria for the production of a competition molecule identified as a RiPP.
By contrast, the broad-spectrum two-peptide lantibiotic lacticin 3147 produced by the WT strain L. lactis DPC 3147, which requires the synergistic action of its two components LtnA1 and LtnA2 (Table), inhibits beneficial commensal Gram-positive bacteria as well as the pathogen C. difficile in an anaerobic fecal-based fermentation protocol (133). Moreover, it is not stable, both in vivo in pig and in vitro and ex vivo in conditions simulating the mammalian GI tract (134). Degradation of the two peptide components LtnA1 and LtnA2 and concomitant significant loss of antibacterial activity were shown to be mainly due to the α-chymotrypsin activity in in vitro experiments, with the LtnA1 peptide being more susceptible to digestion than LtnA2. This two-peptide lantibiotic, while having a good activity against C. difficile in vitro, thus appears to be a poor competitor in the intestinal context. This series of examples points to the necessity of examining the antibacterial properties of a given RiPP in its ecological context.
If the gut microbiota is the most largely studied ecological niche in terms of bacterial competition or communication mediated by diverse natural compounds, the skin microbiota is also the site for such complex interactions. The ability of two lantibiotics produced by Staphylococcus hominis A9 to compete S. aureus, a pathogen closely related to the commensal producer, has been evidenced in the context of atopic dermatitis, where the commensal bacterial community is deficient (135). This led to the isolation and characterization of two lantibiotics termed Sh-lantibiotic-α and Sh-lantibiotic-β (Table 1) that were shown to participate in the host defense. Furthermore, they potentiate LL-37, a human AMP of the cathelicidin family, which protects the skin from infections by invasive bacteria (136). Thus, it is shown that a synergy between a commensal bacterial RiPP and a host AMP leads to an improved protection of the host (Fig. 3B). Moreover, a second competition strategy was identified in S. hominis. A small peptide was isolated and identified as a branched-cyclic nonapeptide (Fig. 2) close to other Staphylococcus autoinducing peptides (AIPs) involved in the QS mechanism (137) (also see “Functions in communication and QS”). It was shown to inhibit the S. aureus system regulating the secretion of a virulence factor (phenol-soluble modulin α (PSMα)) that promotes inflammation in mice hosts (Fig. 3B). Therefore, a complex array of interactions using RiPPs as communication and competition molecules is established between commensals and pathogens. It includes synergistic effects between RiPPs and AMPs and the production of an AIP by commensals (Fig. 3B). This highlights the importance of a complex and well-balanced chemical dialog in maintaining a healthy symbiosis system.
Another example of competition in the skin microbiota is illustrated by the interaction between Propionibacterium acnes and Staphylococcus epidermidis. A group of P. acnes strains isolated from both healthy and acne-affected skin exhibited higher antibacterial activity toward S. epidermidis. Comparative genomics showed that they differ in other groups of P. acnes strains by harboring a genomic island encoding a thiopeptide BGC, which is likely to be responsible for the anti-S. epidermidis activity. This study highlights a likely role of mediating interspecies interactions in the microbiota played by thiopeptides (138). In further support of this assumption, thiopeptide BGCs have been identified as the dominant RiPP clusters in genomes and metagenomes of the human microbiota (31, 35). A thiopeptide lactocillin has been identified from a vaginal isolate of Lactobacillus gasseri (Fig. 2 and Table 1). It shows activity to Gram-positive pathogens, including S. aureus, E. faecalis, Gardnerella vaginalis, and Corynebacterium aurimucosum, implicating its role in protecting the vaginal microbiota against invading pathogens. Remarkably, it has no activities against vaginal commensal bacteria, which was hypothesized to be related to an evolved resistance by the related microbiota to a compound produced by their community (31).
Listeriolysin S is the first bacteriocin to be reported from the Gram-positive genus Listeria (139) (Table 1). It belongs to the TOMM family of RiPPs that contain thiazole and oxazole rings as posttranslational modifications. Contrary to most microcins and bacteriocins that act as competitors due to their antimicrobial properties, listeriolysin S is a virulence factor produced by epidemic strains of the food-borne pathogen Listeria, L. monocytogenes F2365, responsible for listeriosis outbreaks. Indeed, with the cytotoxin streptolysin S from S. pyogenes, listeriolysin S belongs to a class of TOMM virulence peptides from pathogens (71). Listeriolysin S has been shown to manipulate the host microbiota, targeting selectively direct competitors, L. lactis, S. aureus, and L. monocytogenes, lacking the listeriolysin S operon (which confers self-immunity), to provide a more favorable niche for the pathogenic Listeria strains in a mouse oral infection model (140). Moreover, listeriolysin S does not contribute to injuries caused to host tissues by L. monocytogenes infection or to virulence in host organs, but it is specifically produced in the gastrointestinal tract and targets exclusively bacteria in the gut, leading to altered gut microbiota (140, 141). The listeriolysin S TOMM can thus act as a virulence factor that only and selectively targets bacterial cells in vivo without detrimental effects on the host cells (Fig. 3B). Importantly, the dehydratase/dehydrogenase LlsB, which is encoded in the TOMM operon and is putatively involved in establishment of the lysteriolysin S posttranslational modification, was shown to be required for the mice gut colonization, whereas lysteriolysin S itself is not. LlsB thus plays an additional role for virulence, which is hypothesized as participating in the posttranslational modification of another molecule. This shows here again the subtle and essential roles played by posttranslational modifications in the competition strategies of bacteria.
Functions in communication and QS
Microorganisms are now widely appreciated for their ability to communicate and coordinate social traits. To do so, they employ a QS strategy, which involves the production, diffusion, and perception of an extracellular signaling molecule that regulates subsequently gene expression at the community level (142). QS allows the microorganisms to alter behavior collectively in response to changes in the biotic or abiotic environment, thus granting them an adaptive advantage (143). Autoinduction of the production of signaling molecules is a hallmark of QS. In contrast to Gram-negative bacteria that use frequently small molecules, such as homoserine lactones, as autoinducers, Gram-positive bacteria employ commonly ribosomal peptide–mediated QS mechanisms (144, 145). For the focus of this review, we only include examples of QS-related peptides with post-translational modifications, although many of them are linear, resulting from the peptidase cleavage from a precursor. The involvement of RiPPs in QS can be categorized into three types. (i) the RiPP itself is a QS signaling molecule, termed peptide pheromone, and the process regulates other physiological aspects; (ii) the RiPP is an AIP, but it only controls its own production; and (iii) the RiPP biosynthesis is tightly regulated by QS that relies on other signals.
RiPPs as QS signals
The most prominent example is the AIPs, derived from the agrABCD system, which controls the production of virulence factors and biofilm formation in several pathogens, including S. aureus, L. monocytogenes, and Clostridium species (146–150) (Table 1). These are short peptides (7–12 amino acids) with a C-terminal five-member thiolactone ring (Fig. 2). Except for the conserved Cys required for thiolactone formation, amino acid composition in AIPs is highly variable, with a high frequency of hydrophobic residues (151, 152). AIP maturation and regulation have been extensively studied in S. aureus. AIP is made from the precursor AgrD by a membrane-bound endopeptidase AgrB. Matured in and released to the extracellular environment, AIP binds to the sensor histidine kinase AgrC. This induces signal transduction via phospho-relay to the response regulator, AgrA, which in turn activates the expression of the agrABCD operon and an effector RNA gene that elicits QS responses. AgrC and AgrA form a canonical two-component system (TCS), as frequently seen in peptide-related QS mechanisms. AIPs activate their cognate AgrC receptor and can modulate the activity of AgrC receptors from other related species (152, 153). Thus, it can be envisioned that AIP-mediated interspecies cross-talks play a role in maintaining homeostasis of related microbiota in a complex environment. Indeed, as mentioned above, it has been evidenced that commensal, non-S. aureus staphylococcal strains in the human skin microbiota can block the agr QS system of S. aureus by secreting different AIPs, hence preventing its damage to the skin (137, 154). Such a phenomenon has implications for the development of therapeutic strategies using AIP variants (153, 155, 156). Similarly, E. faecalis harbors a cyclic peptide-based fsrABCD QS system that regulates virulence. It differs from the agr system in the autoinducer, termed gelatinase biosynthesis-activating pheromone, that contains a 9-residue lactone ring at the C terminus.
ComX pheromones derived from the comQXPA system of B. subtilis regulate genetic competence, surfactin production, and biofilm formation (Table 1). They are short peptides (4–13 amino acids) with a conserved, modified Trp residue that is formed upon isoprenylation and cyclization (157, 158) (Fig. 2). Regulation by ComX via a dedicated TCS (ComP/A) parallels that of the agr system. Great variation in terms of amino acid sequence and the nature of the isoprenyl modification on ComX has been observed among Bacillus strains, both contributing to their activation specificity of the TCS (159–161). Although some QS interferences were detected among Bacillus strains isolated from the same location, the lack of consistency in the interference pattern suggested that ComX diversification is driven by gain-of-function, such as genetic competence upon TCS activation, instead of competition advantages (159). This is in contrast with AIPs in staphylococcal species.
RiPP autoinducer that only autoregulates its own biosynthesis
This phenomenon is frequently observed for lantibiotic production, including nisin, subtilin, mersacidin (162, 163), cytolysin (164), microbisporicin (165, 166), and planosporicin (167), although the regulation mechanisms differ. In this case, a trigger signal is required to initiate the RiPP production. Autoinduction of cytolysin, a two-peptide lantibiotic and a virulence factor associated with hemolysis of E. faecalis, is remarkably triggered by the presence of eukaryotic cells (168, 169). As for microbisporicin and planosporicin produced by Actinobacteria, their initial autoinduction is induced by nutrient limitation. It is suggested that this would allow the coordination of antibiotic production across all mycelium cells to achieve an ecologically effective concentration (166). However, the advantages of autoinduction of antibacterial peptides in niche competition remain to be firmly demonstrated.
RiPPs regulated by QS
QS-controlled production of RiPPs, in particular modified bacteriocins, has been observed and intensively studied in streptococci. Placing antibacterial synthesis under QS regulation would allow these bacteria to fine-adjust interaction with the host and other species according to environmental and cell population conditions. For example, the human pathogen Streptococcus pneumoniae produces a lantibiotic as a QS response triggered by the presence of galactose via an internalized pheromone-based QS system (170) (Table 1). As galactose is a major sugar of the human nasopharynx, lantibiotic production at high cell density thus would be a strategy to gain competition advantages for S. pneumoniae during colonization of this niche. Moreover, as seen in S. pneumoniae and Streptococcus mutans, bacteriocin production is tightly regulated by the QS system involved in the regulation of competence (171, 172). This would provide new DNA materials from the lysis of competitors to be taken up by the pathogen and to stimulate biofilm formation, which has been demonstrated to confer an adaptive advantage in S. pneumoniae when grown on the mucosal surface (173). This phenomenon has also been shown to have a huge impact on the outcome of mixed-species biofilms, using dual-biofilm formed by S. mutans and Candida albicans as a model (174).
As another example, Streptococcus thermophilus produces a short cyclic peptide, named streptide, having a C–C linkage between a Lys and a Trp residue (Fig. 2 and Table 1). The cyclization is introduced on the precursor peptide by a radical SAM enzyme. Streptide biosynthesis is under the control of a QS system involving a small hydrophobic peptide (SHP) as signaling molecule and a transcriptional regulator (i.e. SHP/Rgg system) (175–177). Genome-mining efforts identified a large panel of streptide-like biosynthetic systems in streptococci, all likely within the context of QS control (178). Biochemical characterization of these radical SAM enzymes led to the discovery of novel cyclization motifs such as β-thioether and α-ether linkages (179, 180). Such unprecedented chemical diversity would reflect functional divergence. However, the roles of streptide-like molecules remain elusive.
Host-bacteria interaction
Few studies exist in probing the roles of microbial RiPPs in host-bacteria interaction. Recently, it was discovered that the plant pathogen Xanthomonas oryzae pv. oryzae utilizes a tyrosine-sulfonated small peptide (RaxX) as a mimic of plant peptide hormones to activate the immune receptor XA21 in rice, triggering immune responses (Table 1). RaxX thus functions as an immunogen and has a role in the virulence (181). Although RaxX resembles phosphorylated peptides that are commonly involved in the regulation of various biological processes, the mode of biosynthesis of RaxX that requires pathway-encoded modification enzymes and a precursor peptide parallels the canonical RiPP pathways. RaxX apparently belongs to a large family of bacterial and plant RiPPs with a sulfated tyrosine, suggesting that such plant-bacteria interaction is prevalent in nature. Interestingly, sulfonation can potentially occur on other RiPPs, such as lasso peptides, as their BGCs from proteobacteria frequently encode sulfotransferases (182). Sulfonated molecules are rather unusual in prokaryotes; however, known examples are frequently involved in the communication between eukaryotic hosts and bacterial symbionts or pathogens. Those include the factor Nod from the nitrogen-fixing rhizobium Sinorhizobium meliloti (183) and sulfolipid-1 from Mycobacterium tuberculosis (184). It thus remains a promising direction to study sulfonated RiPPs in host-bacteria interaction. In addition, there are several remarkable RiPPs isolated from animal symbionts, such as polytheonamides produced by a sponge symbiont (185, 186) and cyanobactins by the Prochloron cyanobacteria symbionts of tunicates (187). It would be very interesting to decipher whether these peptides play a role in the symbiosis.
Effects on microbial physiology
Biofilm formation and morphological development
Being unicellular organisms, many bacteria have evolved to adopt multicellular lifestyles, manifested by, for example, the formation of biofilms, fruiting bodies, or morphological differentiation (188). It is now largely appreciated that some natural antibiotics function as signaling molecules at subinhibitory concentrations in these processes, playing a role in modulating the physiology of the producing bacteria or the interacting ones (189, 190). Consistent with this view, a thiopeptide produced by Bacillus cereus, thiocillin, was identified to stimulate biofilm formation in B. subtilis using a coculture approach (Table 1). Other Bacillus strains with thiopeptide BGCs in the genome produced the same effect (191, 192). Similarly, another thiopeptide, thiostrepton, was found to stimulate the biofilm formation in Pseudomonas aeruginosa at a concentration that does not inhibit the pathogen's growth (193) (Table 1). Although the molecular mechanisms underlying the biofilm induction effect remain unknown, these studies highlight a role of thiopeptides in modulating bacterial physiology. Given the dominant prevalence of thiopeptide BGCs in metagenomes of the human microbiota, future studies should focus on the function of these molecules in microbial competition and interaction (31, 35).
The Streptomyces genus is extensively studied for its extraordinary specialized metabolism. This is intimately linked to its complex life cycle encompassing vegetative mycelia, aerial hyphae, and spores. Some RiPPs are identified to mediate the morphological development in Streptomyces. The lanthipeptides SapB and SapT from S. coelicolor and Streptomyces tendae, respectively, are required for the emergence of aerial hyphae from vegetative cells (194, 195) (Table 1). They function as surfactants by reducing the surface tension for aerial growth (196). This developmental stage can be interfered with by the surfactants produced by other bacteria, adding a new strategy of interspecies interaction. It has been shown that the lipopeptide surfactin produced by B. subtilis inhibits Streptomyces aerial development, whereas it is required for its own aerial structure formation toward spore genesis (197, 198). Furthermore, the TOMM peptide goadsporin produced by Streptomyces sp. induces spore formation and/or pigment production in other Streptomyces species (199) (Table 1). Hence, goadsporin has been used as an elicitor at subinhibitory concentration for triggering specialized metabolite production in Streptomyces. Similarly, thiostrepton produced by Streptomyces laurentii was identified to stimulate pellet formation accompanied by a decrease in pigment synthesis in S. coelicolor (200).
Cannibalism
Microbial antimicrobial peptides are commonly considered as part of the chemical weapons for the microorganisms to eliminate competitors from other species. On the other side, they can be used against a distinct subpopulation of the same species, in analogy to cannibalism, for the benefit of the whole community. In filamentous fungi, this function is hypothesized to be fulfilled by Cys-rich antifungal peptides (201). In B. subtilis, under nutrient limitation conditions, cells produce a disulfide-containing sactipeptide, termed sporulation killing factor (SKF) (202) (Fig. 2 and Table 1), and a sporulation-delaying protein to lyse nonsporulation sibling cells (203). This allows the release of nutrients to sustain the growth of sporulating cells, hence delaying the time- and energy-consuming process of sporulation. The ecological significance of such cannibalism in B. subtilis would be to avoid the growth disadvantage when the nutrients become available again. It is more difficult to resume growth from spores compared with vegetative cells (204). Moreover, the cannibalism has been shown to stimulate biofilm formation via the increased extracellular matrix production in B. subtilis, and both processes were triggered by the NRPS-derived lipopeptide surfactin (205). Interestingly, synthesis of an extracellular matrix and sporulation have been recently shown to be implicated in the interaction of B. subtilis and Pseudomonas chlororaphis that modulates plant co-colonization (206). This highlights that peptide-mediated cannibalism in sporulating bacteria is not only a mechanism of intraspecies interaction, but also should have a wider implication in interspecies interactions.
Metal acquisition
Acquisition of nutrients such as transition metals from the environment is an essential trait for bacteria to survive. Iron acquisition via siderophores is a well-studied phenomenon that involves uniquely nonribosomal peptide products. Remarkably, some methanotrophic bacteria secrete methanobactin RiPPs as chalkophores (i.e. compounds with high binding affinity for copper) to acquire copper from the environment (207) (Fig. 2 and Table 1). The majority of these bacteria utilize a copper-dependent, membrane-bound particulate, methane monooxygenase, which catalyzes the first step of methane metabolization to form methanol. An elaborate copper acquisition system thus helps to meet the high needs of copper in methanotrophs (208). Methanobactins were first discovered in cultures of Methylosinus trichosporium OB3b under copper starvation conditions and later from several other methanotrophic species (8, 207). They bind copper ions with high affinity (Kd at 10−21 to 10−19 m for Cu+ and 10−14 to 10−11 for Cu2+). Copper chelation occurs via two bidentate ligands, each composed of a nitrogen-containing heterocycle, such as oxazolone with an adjacent thioamide group (Fig. 2). Structural variations in amino acid composition were found to influence the copper affinity (209). The copper-methanobactin complex is transported into the cell by an active transport process, involving a TonB-dependent transporter encoded in the pathway and a periplasmic binding protein (210, 211), although the function of the latter in transport is not clearly known. Such an import mechanism is consistent with that of other metallophores (212). It has been shown that the transporter displays relaxed substrate promiscuity (210), suggesting a possible pirating of methanobactins from other species. These features have important implications in the ecology of methanotrophs in nature. Interestingly, methanobactin-like BGCs have been identified in the genomes of many nonmethanotrophic bacteria (213). It remains to be seen whether the derived peptides have a similar function for the producers as known methanobactins. Interestingly, some RiPP BGCs, such as certain lasso peptide pathways in proteobacteria, encode dedicated TonB-dependent receptors (214, 215), suggesting a possible role in transporting cargo molecules, in analogy to metallophores. This direction deserves further investigations.
Enzyme cofactors
Pyrroloquinoline quinone (PQQ) and mycofactocin are two examples of RiPPs functioning as enzyme cofactors (Table 1). They share parallel biosynthesis: both are derived from the cross-linking of a Tyr and another residue (Glu in PQQ and Val in mycofactocin) in the precursor, followed by peptide cleavage and further modifications on the aromatic ring (for a review on biosynthesis, see Refs. 216 and 217) (Fig. 2). However, the structure of mycofactocin is not yet completely elucidated. PQQ is produced mainly by Gram-negative bacteria (218). It serves as a redox cofactor of dehydrogenases, including alcohol and glucose dehydrogenases (219, 220), which allows the producer to use these substances as carbon sources. It is conceivable that this confers on PQQ-producing bacteria a competitive growth advantage in certain environments. As for mycofactocin, its BGC is present in many Actinobacteria and particularly in Mycobacterium species. Bioinformatic analysis revealed an association of mycofactocin BGCs with the presence of genes encoding nicotinoproteins (i.e. enzymes with bound NAD(P) as cofactor) (221), such as short-chain, iron-dependent, and zinc-dependent dehydrogenases (222). This indicates that these proteins may utilize mycofactocin as a redox cofactor. Recent experimental data indeed showed that the mycofactocin pathway is essential for primary alcohol metabolism in Mycobacteria (223). Precisely, it was demonstrated that the methanol dehydrogenase in Mycobacterium smegmatis uses mycofactocin as an in vivo electron acceptor (224). Very recently, the biosynthesis, structure, and redox potential have been thoroughly characterized for premycofactocin (225) (Fig. 2). This study has demonstrated for the first time that premycofactocin is a biologically active redox cofactor used by M. smegmatis carveol dehydrogenase in the oxidation of carveol. Therefore, the physiological function of premycofactocin and, by extension, mycofactocin, resembles that of PQQ as peptide-derived redox cofactors.
Concluding remarks and future directions
Microbial RiPPs are the most documented among known RiPPs, with a huge number of studies on their structures, structure/activity relationships, mechanisms of action, and biosynthesis, concerning in particular the posttranslational modifications and their dedicated enzymes. Although RiPPs' antibiotic properties and biosynthesis pathways are currently well-studied with the purpose of identifying alternative strategies to the use of conventional antibiotics in the context of the microbial resistance crisis and the One Health context approved by the World Health Organization, RiPPs' ecological roles remain poorly documented. For instance, cyanobactins (including the well-known patellamides), which are ribosomal peptide macrocycles produced by several cyanobacteria species associated with marine organisms (sponges and ascidians) showing important biomedical activities, have been intensively studied with regard to their biosynthesis mechanisms and their biological properties (226–228), but their ecological functions have not been approached.
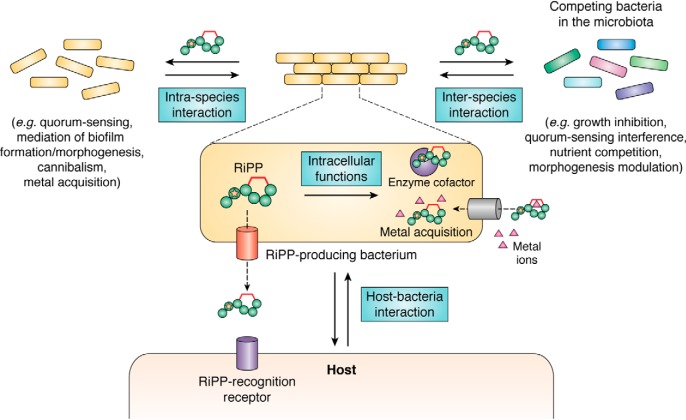
This review provides for the first time an overview of the various roles that are currently known for microbial RiPPs in nature (Fig. 4), particularly in the context of microbial communities and from the perspective of multicellular development. At the present time, competition is the ecological function that has been most clearly demonstrated for some microbial RiPPs, but it appears that the domain is still in its infancy, and the field is very likely to witness more and more active developments in the next few years. This will be made possible in the hologenome (i.e. the collective genomic content of all individuals within a holobiont) concept of evolution (4, 5), thanks to an increasing number of studies deciphering the roles played and the interactions of various microbiota that are progressively evidenced to be associated with all organisms and form holobionts (marine organisms, insects, birds, plants, mammals …). Certainly, current technological advances, including genome mining tools and gene manipulation methods, MS imaging, analytical and spectrometric, and single-cell imaging techniques, will also favor the discovery of novel roles played by RiPPs in microbial ecology and in a large diversity of both organisms and ecosystems. This allows us to presume that ecological roles will be evidenced more and more rapidly and will concern an increasing number of RiPPs and microbiota. As of now, we can speculate that ecosystems where presently RiPPs have not been evidenced as actors in bacterial competitions or other ecological functions, whereas unmodified antimicrobial peptides and bacteriocins or NRPS-derived NPs are, will appear as prolific niches to study. They can be exemplified by the rumen microbiota (229, 230) or the microbiota of honeybees (231) or lepidopterans (232), where the secreted RiPP antibacterial compounds remain to be identified and to be evidenced in competition. Indeed, the production of antimicrobial metabolites by social bee gut symbionts has been shown to contribute to augmenting their resistance to pathogens, in addition to stimulating host-immune responses or competing for space and nutrients, thus participating in reduction of pathogen-related bee declines (233), but their identification still remains to be achieved.
Figure 4.
Schematic summary of known microbial RiPP functions in the context of interkingdom, interspecies, and intraspecies interactions.
Hence, the future of RiPPs as actors in ecosystems will presumably pass not only through a fast development of the incipient directions described in this review, but also a deep exploration of the arrays of tight connections established between them that will open new tracks for discovering novel functions ensured by these NPs. In this sense, deciphering more deeply the fine-tuning regulations managing RiPP production, their effects at low doses, their pheromone function, and their roles in QS and biofilm formation as well as in interbacterial predation is expected to expand our knowledge of their native function. Furthermore, as mentioned above, elucidating the tight relationships between the aforementioned functions and DNA acquisition by naturally competent bacteria (234, 235) will afford rich perspectives. Indeed, natural transformation is a driving force for adaptation and evolution through horizontal gene transfer (236), and involvement of bacteriocins in the process has already been suggested (171, 237), prefiguring a putative role that could be played by RiPPs in these processes. Overall, it is a much greater challenge to decipher the native roles of microbial RiPPs, and multidisciplinary collaboration from chemists, microbiologists, and microbial ecologists is definitely required.
Implementing such directions, in addition to the already developed studies on RiPP biosynthetic machineries and on their structures or mechanisms of action, will increase the panel of novel molecules and strategies to fight against bacterial resistance to antimicrobials while preserving commensal bacteria in microbiota, thus affording new opportunities for agricultural or farming practices as well as human, animal, and environmental health in the One Health context for the benefit of society.
The authors declare that they have no conflicts of interest with the contents of this article.
S. Rebuffat and I. Fliss, unpublished results.
- QS
- quorum sensing
- NP
- natural product
- NRPS
- nonribosomal peptide synthetase
- PKS
- polyketide synthase
- RiPP
- ribosomally synthesized and posttranslationally modified peptide
- RPNP
- ribosomal peptide natural product
- TOMM
- thiazole/oxazole-modified microcin
- AMP
- antimicrobial peptide
- BGC
- biosynthetic gene cluster
- LAP
- linear azol(in)e-containing peptide
- SKF
- sporulation killing factor
- EcN
- E. coli Nissle 1917
- GI
- gastrointestinal
- MccE492
- microcin E492
- AIP
- autoinducing peptide
- TCS
- two-component system
- SHP
- small hydrophobic peptide
- PQQ
- pyrroloquinoline quinone.
References
- 1. Hibbing M. E., Fuqua C., Parsek M. R., and Peterson S. B. (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Inglis R. F., Brown S. P., and Buckling A. (2012) Spite versus cheats: competition among social strategies shapes virulence in Pseudomonas aeruginosa. Evolution 66, 3472–3484 10.1111/j.1558-5646.2012.01706.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Inglis R. F., West S., and Buckling A. (2014) An experimental study of strong reciprocity in bacteria. Biol. Lett. 10, 20131069 10.1098/rsbl.2013.1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris J. J. (2018) What is the hologenome concept of evolution? F1000Res. 7, F1000 Faculty Rev–1664 10.12688/f1000research.14385.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenberg E., and Zilber-Rosenberg I. (2018) The hologenome concept of evolution after 10 years. Microbiome 6, 78 10.1186/s40168-018-0457-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hider R. C., and Kong X. (2010) Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657 10.1039/b906679a [DOI] [PubMed] [Google Scholar]
- 7. McRose D. L., Seyedsayamdost M. R., and Morel F. M. M. (2018) Multiple siderophores: bug or feature? J. Biol. Inorg. Chem. 23, 983–993 10.1007/s00775-018-1617-x [DOI] [PubMed] [Google Scholar]
- 8. Dassama L. M., Kenney G. E., and Rosenzweig A. C. (2017) Methanobactins: from genome to function. Metallomics 9, 7–20 10.1039/C6MT00208K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cornforth D. M., and Foster K. R. (2013) Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293 10.1038/nrmicro2977 [DOI] [PubMed] [Google Scholar]
- 10. Abrudan M. I., Smakman F., Grimbergen A. J., Westhoff S., Miller E. L., van Wezel G. P., and Rozen D. E. (2015) Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl. Acad. Sci. U.S.A. 112, 11054–11059 10.1073/pnas.1504076112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cornforth D. M., and Foster K. R. (2015) Antibiotics and the art of bacterial war. Proc. Natl. Acad. Sci. U.S.A. 112, 10827–10828 10.1073/pnas.1513608112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Netzker T., Flak M., Krespach M. K., Stroe M. C., Weber J., Schroeckh V., and Brakhage A. A. (2018) Microbial interactions trigger the production of antibiotics. Curr. Opin. Microbiol. 45, 117–123 10.1016/j.mib.2018.04.002 [DOI] [PubMed] [Google Scholar]
- 13. Molloy E. M., and Hertweck C. (2017) Antimicrobial discovery inspired by ecological interactions. Curr. Opin. Microbiol. 39, 121–127 10.1016/j.mib.2017.09.006 [DOI] [PubMed] [Google Scholar]
- 14. Hall-Stoodley L., Costerton J. W., and Stoodley P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 15. Oliveira N. M., Martinez-Garcia E., Xavier J., Durham W. M., Kolter R., Kim W., and Foster K. R. (2015) Biofilm formation as a response to ecological competition. PLoS Biol. 13, e1002191 10.1371/journal.pbio.1002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma D., Misba L., and Khan A. U. (2019) Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8, 76 10.1186/s13756-019-0533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korgaonkar A., Trivedi U., Rumbaugh K. P., and Whiteley M. (2013) Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 110, 1059–1064 10.1073/pnas.1214550110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fischbach M. A., and Walsh C. T. (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- 19. Marahiel M. A. (2016) A structural model for multimodular NRPS assembly lines. Nat. Prod. Rep. 33, 136–140 10.1039/C5NP00082C [DOI] [PubMed] [Google Scholar]
- 20. Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., et al. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 10.1039/C2NP20085F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burkhart B. J., Hudson G. A., Dunbar K. L., and Mitchell D. A. (2015) A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 10.1038/nchembio.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo S., and Dong S. H. (2019) Recent advances in the discovery and biosynthetic study of eukaryotic RiPP natural products. Molecules 24, E1541 10.3390/molecules24081541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hetrick K. J., and van der Donk W. A. (2017) Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 38, 36–44 10.1016/j.cbpa.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu W., and Schmidt E. W. (2017) Three principles of diversity-generating biosynthesis. Acc. Chem. Res. 50, 2569–2576 10.1021/acs.accounts.7b00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hudson G. A., and Mitchell D. A. (2018) RiPP antibiotics: biosynthesis and engineering potential. Curr. Opin. Microbiol. 45, 61–69 10.1016/j.mib.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burkhart B. J., Schwalen C. J., Mann G., Naismith J. H., and Mitchell D. A. (2017) YcaO-dependent posttranslational amide activation: biosynthesis, structure, and function. Chem. Rev. 117, 5389–5456 10.1021/acs.chemrev.6b00623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Repka L. M., Chekan J. R., Nair S. K., and van der Donk W. A. (2017) Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 117, 5457–5520 10.1021/acs.chemrev.6b00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu W., Dong S.-H., Sarkar S., Nair S. K., and Schmidt E. W. (2018) The biochemistry and structural biology of cyanobactin pathways: enabling combinatorial biosynthesis. Methods Enzymol. 604, 113–163 10.1016/bs.mie.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sikandar A., and Koehnke J. (2019) The role of protein–protein interactions in the biosynthesis of ribosomally synthesized and post-translationally modified peptides. Nat. Prod. Rep. 36, 1576–1588 10.1039/C8NP00064F [DOI] [PubMed] [Google Scholar]
- 30. Liu L., Hao T., Xie Z., Horsman G. P., and Chen Y. (2016) Genome mining unveils widespread natural product biosynthetic capacity in human oral microbe Streptococcus mutans. Sci. Rep. 6, 37479 10.1038/srep37479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donia M. S., Cimermancic P., Schulze C. J., Wieland Brown L. C., Martin J., Mitreva M., Clardy J., Linington R. G., and Fischbach M. A. (2014) A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 10.1016/j.cell.2014.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balty C., Guillot A., Fradale L., Brewee C., Boulay M., Kubiak X., Benjdia A., and Berteau O. (2019) Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J. Biol. Chem. 294, 14512–14525 10.1074/jbc.RA119.009416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chiumento S., Roblin C., Kieffer-Jaquinod S., Tachon S., Leprètre C., Basset C., Aditiyarini D., Olleik H., Nicoletti C., Bornet O., Iranzo O., Maresca M., Hardré R., Fons M., Giardina T., et al. (2019) Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci. Adv. 5, eaaw9969 10.1126/sciadv.aaw9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huo L., Zhao X., Acedo J. Z., Estrada P., Nair S. K., and Van der Donk W. A. (2019) Characterization of a dehydratase and methyltransferase in the biosynthesis of a ribosomally-synthesized and post-translationally modified peptide in Lachnospiraceae. Chembiochem 20, 1–11 10.1002/cbic.201900483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aleti G., Baker J. L., Tang X., Alvarez R., Dinis M., Tran N. C., Melnik A. V., Zhong C., Ernst M., Dorrestein P. C., and Edlund A. (2019) Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio 10, e00321–19 10.1128/mBio.00321-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao L., Gurevich A., Alexander K. L., Naman C. B., Leão T., Glukhov E., Luzzatto-Knaan T., Vargas F., Quinn R., Bouslimani A., Nothias L. F., Singh N. K., Sanders J. G., Benitez R. A. S., Thompson L. R., et al. (2019) MetaMiner: a scalable peptidogenomics approach for discovery of ribosomal peptide natural products with blind modifications from microbial communities. Cell Syst. S2405-4712(19)30312-6 10.1016/j.cels.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duquesne S., Destoumieux-Garzón D., Peduzzi J., and Rebuffat S. (2007) Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 24, 708–734 10.1039/b516237h [DOI] [PubMed] [Google Scholar]
- 38. Drider D., and Rebuffat S. (2011) Prokaryotic Antimicrobial Peptides: From Genes to Applications, 1st Ed., Springer-Verlag, New York [Google Scholar]
- 39. Severinov K., and Nair S. K. (2012) Microcin C: biosynthesis and mechanisms of bacterial resistance. Fut. Microbiol. 7, 281–289 10.2217/fmb.11.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Metlitskaya A., Kazakov T., Kommer A., Pavlova O., Praetorius-Ibba M., Ibba M., Krasheninnikov I., Kolb V., Khmel I., and Severinov K. (2006) Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic Microcin C. J. Biol. Chem. 281, 18033–18042 10.1074/jbc.M513174200 [DOI] [PubMed] [Google Scholar]
- 41. Azpiroz M. F., and Laviña M. (2004) Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 48, 1235–1241 10.1128/AAC.48.4.1235-1241.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vassiliadis G., Destoumieux-Garzón D., Lombard C., Rebuffat S., and Peduzzi J. (2010) Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob. Agents Chemother. 54, 288–297 10.1128/AAC.00744-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Lorenzo V. (1984) Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch. Microbiol. 139, 72–75 10.1007/BF00692715 [DOI] [PubMed] [Google Scholar]
- 44. de Lorenzo V., and Pugsley A. P. (1985) Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27, 666–669 10.1128/AAC.27.4.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lagos R., Wilkens M., Vergara C., Cecchi X., and Monasterio O. (1993) Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett. 321, 145–148 10.1016/0014-5793(93)80096-D [DOI] [PubMed] [Google Scholar]
- 46. Bieler S., Silva F., Soto C., and Belin D. (2006) Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 188, 7049–7061 10.1128/JB.00688-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas X., Destoumieux-Garzón D., Peduzzi J., Afonso C., Blond A., Birlirakis N., Goulard C., Dubost L., Thai R., Tabet J. C., and Rebuffat S. (2004) Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279, 28233–28242 10.1074/jbc.M400228200 [DOI] [PubMed] [Google Scholar]
- 48. Vassiliadis G., Peduzzi J., Zirah S., Thomas X., Rebuffat S., and Destoumieux-Garzón D. (2007) Insight into siderophore-carrying peptide biosynthesis: enterobactin is a precursor for microcin E492 posttranslational modification. Antimicrob. Agents Chemother. 51, 3546–3553 10.1128/AAC.00261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nolan E. M., Fischbach M. A., Koglin A., and Walsh C. T. (2007) Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J. Am. Chem. Soc. 129, 14336–14347 10.1021/ja074650f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hegemann J. D., Zimmermann M., Xie X., and Marahiel M. A. (2015) Lasso peptides: an intriguing class of bacterial natural products. Acc. Chem. Res. 48, 1909–1919 10.1021/acs.accounts.5b00156 [DOI] [PubMed] [Google Scholar]
- 51. Maksimov M. O., Pan S. J., and James Link A. (2012) Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 29, 996–1006 10.1039/c2np20070h [DOI] [PubMed] [Google Scholar]
- 52. Li Y., Zirah S., and Rebuffat S. (2015) Lasso Peptides: Bacterial Strategies to Make and Maintain Bioactive Entangled Scaffolds, 1st Ed., Springer-Verlag, New York [Google Scholar]
- 53. Cheung-Lee W. L., and Link A. J. (2019) Genome mining for lasso peptides: past, present, and future. J. Ind. Microbiol. Biotechnol. 46, 1371–1379 10.1007/s10295-019-02197-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan S., Moore G., and Nodwell J. (2019) Put a bow on it: knotted antibiotics take center stage. Antibiotics 8, E117 10.3390/antibiotics8030117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Salomón R. A., and Farías R. N. (1992) Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174, 7428–7435 10.1128/jb.174.22.7428-7435.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bayro M. J., Mukhopadhyay J., Swapna G. V., Huang J. Y., Ma L. C., Sineva E., Dawson P. E., Montelione G. T., and Ebright R. H. (2003) Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J. Am. Chem. Soc. 125, 12382–12383 10.1021/ja036677e [DOI] [PubMed] [Google Scholar]
- 57. Rosengren K. J., Clark R. J., Daly N. L., Göransson U., Jones A., and Craik D. J. (2003) Microcin J25 has a threaded sidechain-to-backbone ring structure and not a head-to-tail cyclized backbone. J. Am. Chem. Soc. 125, 12464–12474 10.1021/ja0367703 [DOI] [PubMed] [Google Scholar]
- 58. Wilson K. A., Kalkum M., Ottesen J., Yuzenkova J., Chait B. T., Landick R., Muir T., Severinov K., and Darst S. A. (2003) Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail. J. Am. Chem. Soc. 125, 12475–12483 10.1021/ja036756q [DOI] [PubMed] [Google Scholar]
- 59. Rebuffat S., Blond A., Destoumieux-Garzon D., Goulard C., and Peduzzi J. (2004) Microcin J25, from the macrocyclic to the lasso structure: implications for biosynthetic, evolutionary and biotechnological perspectives. Curr. Prot. Pept. Sci. 5, 383–391 10.2174/1389203043379611 [DOI] [PubMed] [Google Scholar]
- 60. Duquesne S., Destoumieux-Garzón D., Zirah S., Goulard C., Peduzzi J., and Rebuffat S. (2007) Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem. Biol. 14, 793–803 10.1016/j.chembiol.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 61. Yan K. P., Li Y., Zirah S., Goulard C., Knappe T. A., Marahiel M. A., and Rebuffat S. (2012) Dissecting the maturation steps of the lasso peptide microcin J25 in vitro. Chembiochem 13, 1046–1052 10.1002/cbic.201200016 [DOI] [PubMed] [Google Scholar]
- 62. Choudhury H. G., Tong Z., Mathavan I., Li Y., Iwata S., Zirah S., Rebuffat S., van Veen H. W., and Beis K. (2014) Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. U.S.A. 111, 9145–9150 10.1073/pnas.1320506111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bountra K., Hagelueken G., Choudhury H. G., Corradi V., El Omari K., Wagner A., Mathavan I., Zirah S., Yuan Wahlgren W., Tieleman D. P., Schiemann O., Rebuffat S., and Beis K. (2017) Structural basis for antibacterial peptide self-immunity by the bacterial ABC transporter McjD. EMBO J. 36, 3062–3079 10.15252/embj.201797278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Metelev M., Arseniev A., Bushin L. B., Kuznedelov K., Artamonova T. O., Kondratenko R., Khodorkovskii M., Seyedsayamdost M. R., and Severinov K. (2017) Acinetodin and klebsidin, RNA polymerase targeting lasso peptides produced by human isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem. Biol. 12, 814–824 10.1021/acschembio.6b01154 [DOI] [PubMed] [Google Scholar]
- 65. Kuznedelov K., Semenova E., Knappe T. A., Mukhamedyarov D., Srivastava A., Chatterjee S., Ebright R. H., Marahiel M. A., and Severinov K. (2011) The antibacterial threaded-lasso peptide capistruin inhibits bacterial RNA polymerase. J. Mol. Biol. 412, 842–848 10.1016/j.jmb.2011.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheung-Lee W. L., Parry M. E., Jaramillo Cartagena A., Darst S. A., and Link A. J. (2019) Discovery and structure of the antimicrobial lasso peptide citrocin. J. Biol. Chem. 294, 6822–6830 10.1074/jbc.RA118.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perez R. H., Zendo T., and Sonomoto K. (2018) Circular and leaderless bacteriocins: biosynthesis, mode of action, applications, and prospects. Front. Microbiol. 9, 2085 10.3389/fmicb.2018.02085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grande Burgos M. J., Pulido R. P., Del Carmen López Aguayo M., Gálvez A., and Lucas R. (2014) The cyclic antibacterial peptide enterocin AS-48: isolation, mode of action, and possible food applications. Int. J. Mol. Sci. 15, 22706–22727 10.3390/ijms151222706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Towle K. M., and Vederas J. C. (2017) Structural features of many circular and leaderless bacteriocins are similar to those in saposins and saposin-like peptides. MedChemComm 8, 276–285 10.1039/C6MD00607H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gabrielsen C., Brede D. A., Hernández P. E., Nes I. F., and Diep D. B. (2012) The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother. 56, 2908–2915 10.1128/AAC.00314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Molloy E. M., Cotter P. D., Hill C., Mitchell D. A., and Ross R. P. (2011) Streptolysin S-like virulence factors: the continuing sagA. Nat. Rev. Microbiol. 9, 670–681 10.1038/nrmicro2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yorgey P., Lee J., Kördel J., Vivas E., Warner P., Jebaratnam D., and Kolter R. (1994) Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. U.S.A. 91, 4519–4523 10.1073/pnas.91.10.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li Y. M., Milne J. C., Madison L. L., Kolter R., and Walsh C. T. (1996) From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274, 1188–1193 10.1126/science.274.5290.1188 [DOI] [PubMed] [Google Scholar]
- 74. Collin F., and Maxwell A. (2019) The microbial toxin microcin B17: prospects for the development of new antibacterial agents. J. Mol. Biol. 431, 3400–3426 10.1016/j.jmb.2019.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Molohon K. J., Blair P. M., Park S., Doroghazi J. R., Maxson T., Hershfield J. R., Flatt K. M., Schroeder N. E., Ha T., and Mitchell D. A. (2016) Plantazolicin is an ultranarrow-spectrum antibiotic that targets the Bacillus anthracis membrane. ACS Infect. Dis. 2, 207–220 10.1021/acsinfecdis.5b00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Triplett E. W., and Barta T. M. (1987) Trifolitoxin production and nodulation are necessary for the expression of superior nodulation competitiveness by Rhizobium leguminosarum bv. trifolii strain T24 on clover. Plant Physiol. 85, 335–342 10.1104/pp.85.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Travin D. Y., Watson Z. L., Metelev M., Ward F. R., Osterman I. A., Khven I. M., Khabibullina N. F., Serebryakova M., Mergaert P., Polikanov Y. S., Cate J. H. D., and Severinov K. (2019) Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition. Nat. Commun. 10, 4563 10.1038/s41467-019-12589-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pierrat O. A., and Maxwell A. (2003) The action of the bacterial toxin microcin B17: insight into the cleavage-religation reaction of DNA gyrase. J. Biol. Chem. 278, 35016–35023 10.1074/jbc.M304516200 [DOI] [PubMed] [Google Scholar]
- 79. Bagley M. C., Dale J. W., Merritt E. A., and Xiong X. (2005) Thiopeptide antibiotics. Chem. Rev. 105, 685–714 10.1021/cr0300441 [DOI] [PubMed] [Google Scholar]
- 80. Reusser F. (1969) Mode of action of berninamycin: an inhibitor of protein biosynthesis. Biochemistry 8, 3303–3308 10.1021/bi00836a026 [DOI] [PubMed] [Google Scholar]
- 81. Heffron S. E., and Jurnak F. (2000) Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 Å resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry 39, 37–45 10.1021/bi9913597 [DOI] [PubMed] [Google Scholar]
- 82. Schwalen C. J., Hudson G. A., Kille B., and Mitchell D. A. (2018) Bioinformatic expansion and discovery of thiopeptide antibiotics. J. Am. Chem. Soc. 140, 9494–9501 10.1021/jacs.8b03896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lubelski J., Rink R., Khusainov R., Moll G. N., and Kuipers O. P. (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65, 455–476 10.1007/s00018-007-7171-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Asaduzzaman S. M., and Sonomoto K. (2009) Lantibiotics: diverse activities and unique modes of action. J. Biosci. Bioeng. 107, 475–487 10.1016/j.jbiosc.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 85. Knerr P. J., and van der Donk W. A. (2012) Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81, 479–505 10.1146/annurev-biochem-060110-113521 [DOI] [PubMed] [Google Scholar]
- 86. Ortega M. A., and van der Donk W. A. (2016) New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 23, 31–44 10.1016/j.chembiol.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lawton E. M., Ross R. P., Hill C., and Cotter P. D. (2007) Two-peptide lantibiotics: a medical perspective. Mini Rev. Med. Chem. 7, 1236–1247 10.2174/138955707782795638 [DOI] [PubMed] [Google Scholar]
- 88. Islam M. R., Nagao J., Zendo T., and Sonomoto K. (2012) Antimicrobial mechanism of lantibiotics. Biochem. Soc. Trans. 40, 1528–1533 10.1042/BST20120190 [DOI] [PubMed] [Google Scholar]
- 89. Wiedemann I., Böttiger T., Bonelli R. R., Wiese A., Hagge S. O., Gutsmann T., Seydel U., Deegan L., Hill C., Ross P., and Sahl H.-G. (2006) The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61, 285–296 10.1111/j.1365-2958.2006.05223.x [DOI] [PubMed] [Google Scholar]
- 90. Bakhtiary A., Cochrane S. A., Mercier P., McKay R. T., Miskolzie M., Sit C. S., and Vederas J. C. (2017) Insights into the mechanism of action of the two-peptide lantibiotic lacticin 3147. J. Am. Chem. Soc. 139, 17803–17810 10.1021/jacs.7b04728 [DOI] [PubMed] [Google Scholar]
- 91. Flühe L., and Marahiel M. A. (2013) Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis. Curr. Opin. Chem. Biol. 17, 605–612 10.1016/j.cbpa.2013.06.031 [DOI] [PubMed] [Google Scholar]
- 92. Mathur H., Rea M. C., Cotter P. D., Hill C., and Ross R. P. (2015) The sactibiotic subclass of bacteriocins: an update. Curr. Protein Pept. Sci. 16, 549–558 10.2174/1389203716666150515124831 [DOI] [PubMed] [Google Scholar]
- 93. Hudson G. A., Burkhart B. J., DiCaprio A. J., Schwalen C. J., Kille B., Pogorelov T. V., and Mitchell D. A. (2019) Bioinformatic mapping of radical S-adenosylmethionine-dependent ribosomally synthesized and post-translationally modified peptides identifies new Cα, Cβ, and Cγ-linked thioether-containing peptides. J. Am. Chem. Soc. 141, 8228–8238 10.1021/jacs.9b01519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mo T., Ji X., Yuan W., Mandalapu D., Wang F., Zhong Y., Li F., Chen Q., Ding W., Deng Z., Yu S., and Zhang Q. (2019) Thuricin Z, a narrow-spectrum sactibiotic that targets cell membrane. Angew. Chem. Int. Ed. Engl. 10.1002/anie.201908490 10.1002/anie.201908490 [DOI] [PubMed] [Google Scholar]
- 95. Stubbendieck R. M., and Straight P. D. (2016) Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155 10.1128/JB.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Raffatellu M. (2018) Learning from bacterial competition in the host to develop antimicrobials. Nat. Med. 24, 1097–1103 10.1038/s41591-018-0145-0 [DOI] [PubMed] [Google Scholar]
- 97. Wilson B. R., Bogdan A. R., Miyazawa M., Hashimoto K., and Tsuji Y. (2016) Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol. Med. 22, 1077–1090 10.1016/j.molmed.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang W., Ji J., Li X., Wang J., Li S., Pan G., Fan K., and Yang K. (2014) Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111, 5688–5693 10.1073/pnas.1324253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Buffie C. G., and Pamer E. G. (2013) Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lawley T. D., and Walker A. W. (2013) Intestinal colonization resistance. Immunology 138, 1–11 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Thursby E., and Juge N. (2017) Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Donia M. S., and Fischbach M. A. (2015) Small molecules from the human microbiota. Science 349, 1254766 10.1126/science.1254766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schommer N. N., and Gallo R. L. (2013) Structure and function of the human skin microbiome. Trends Microbiol. 21, 660–668 10.1016/j.tim.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yamazaki Y., Nakamura Y., and Núñez G. (2017) Role of the microbiota in skin immunity and atopic dermatitis. Allergol. Int. 66, 539–544 10.1016/j.alit.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 105. Nissle A. (1918) Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Med. Klin. 2, 29–30 [Google Scholar]
- 106. Jacobi C. A., and Malfertheiner P. (2011) Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig. Dis. 29, 600–607 10.1159/000333307 [DOI] [PubMed] [Google Scholar]
- 107. Patzer S. I., Baquero M. R., Bravo D., Moreno F., and Hantke K. (2003) The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149, 2557–2570 10.1099/mic.0.26396-0 [DOI] [PubMed] [Google Scholar]
- 108. Baquero F., and Moreno F. (1984) The microcins. FEMS Microbiol. Lett. 23, 117–124 10.1111/j.1574-6968.1984.tb01046.x [DOI] [Google Scholar]
- 109. Altenhoefer A., Oswald S., Sonnenborn U., Enders C., Schulze J., Hacker J., and Oelschlaeger T. A. (2004) The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 40, 223–229 10.1016/S0928-8244(03)00368-7 [DOI] [PubMed] [Google Scholar]
- 110. Sassone-Corsi M., Nuccio S. P., Liu H., Hernandez D., Vu C. T., Takahashi A. A., Edwards R. A., and Raffatellu M. (2016) Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 10.1038/nature20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Massip C., Branchu P., Bossuet-Greif N., Chagneau C. V., Gaillard D., Martin P., Boury M., Sécher T., Dubois D., Nougayrède J.-P., and Oswald E. (2019) Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 15, e1008029 10.1371/journal.ppat.1008029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Xue M., Kim C. S., Healy A. R., Wernke K. M., Wang Z., Frischling M. C., Shine E. E., Wang W., Herzon S. B., and Crawford J. M. (2019) Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685 10.1126/science.aax2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Nougayrède J. P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., Buchrieser C., Hacker J., Dobrindt U., and Oswald E. (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- 114. Lopez F. E., Vincent P. A., Zenoff A. M., Salomón R. A., and Farías R. N. (2007) Efficacy of microcin J25 in biomatrices and in a mouse model of Salmonella infection. J. Antimicrob. Chemother. 59, 676–680 10.1093/jac/dkm009 [DOI] [PubMed] [Google Scholar]
- 115. Stavric S., and D'Aoust J. Y. (1993) Undefined and defined bacterial preparations for the competitive exclusion of Salmonella in poultry: a review. J. Food Prot. 56, 173–180 10.4315/0362-028X-56.2.173 [DOI] [PubMed] [Google Scholar]
- 116. Naimi S., Zirah S., Hammami R., Fernandez B., Rebuffat S., and Fliss I. (2018) Fate and biological activity of the antimicrobial lasso peptide microcin J25 under gastrointestinal tract conditions. Front. Microbiol. 9, 1764 10.3389/fmicb.2018.01764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yu H. T., Ding X. L., Li N., Zhang X. Y., Zeng X. F., Wang S., Liu H. B., Wang Y. M., Jia H. M., and Qiao S. Y. (2017) Dietary supplemented antimicrobial peptide microcin J25 improves the growth performance, apparent total tract digestibility, fecal microbiota, and intestinal barrier function of weaned pigs. J. Anim. Sci. 95, 5064–5076 10.2527/jas2017.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Forkus B., Ritter S., Vlysidis M., Geldart K., and Kaznessis Y. N. (2017) Antimicrobial probiotics reduce Salmonella enterica in turkey gastrointestinal tracts. Sci. Rep. 7, 40695 10.1038/srep40695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Eisenberg D. S., and Sawaya M. R. (2017) Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem. 86, 69–95 10.1146/annurev-biochem-061516-045104 [DOI] [PubMed] [Google Scholar]
- 120. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 121. Pham C. L., Kwan A. H., and Sunde M. (2014) Functional amyloid: widespread in Nature, diverse in purpose. Essays Biochem. 56, 207–219 10.1042/bse0560207 [DOI] [PubMed] [Google Scholar]
- 122. Cámara-Almirón J., Caro-Astorga J., de Vicente A., and Romero D. (2018) Beyond the expected: the structural and functional diversity of bacterial amyloids. Crit. Rev. Microbiol. 44, 653–666 10.1080/1040841X.2018.1491527 [DOI] [PubMed] [Google Scholar]
- 123. Shanmugam N., Baker M. O. D. G., Ball S. R., Steain M., Pham C. L. L., and Sunde M. (2019) Microbial functional amyloids serve diverse purposes for structure, adhesion and defence. Biophys. Rev. 11, 287–302 10.1007/s12551-019-00526-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bieler S., Estrada L., Lagos R., Baeza M., Castilla J., and Soto C. (2005) Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 280, 26880–26885 10.1074/jbc.M502031200 [DOI] [PubMed] [Google Scholar]
- 125. Marcoleta A., Marín M., Mercado G., Valpuesta J. M., Monasterio O., and Lagos R. (2013) Microcin E492 amyloid formation is retarded by posttranslational modification. J. Bacteriol. 195, 3995–4004 10.1128/JB.00564-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Aguilera P., Marcoleta A., Lobos-Ruiz P., Arranz R., Valpuesta J. M., Monasterio O., and Lagos R. (2016) Identification of key amino acid residues modulating intracellular and in vitro microcin E492 amyloid formation. Front. Microbiol. 7, 35 10.3389/fmicb.2016.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Shahnawaz M., and Soto C. (2012) Microcin amyloid fibrils A are reservoir of toxic oligomeric species. J. Biol. Chem. 287, 11665–11676 10.1074/jbc.M111.282533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Shahnawaz M., Park K. W., Mukherjee A., Diaz-Espinoza R., and Soto C. (2017) Prion-like characteristics of the bacterial protein Microcin E492. Sci. Rep. 7, 45720 10.1038/srep45720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tomita H., Fujimoto S., Tanimoto K., and Ike Y. (1996) Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178, 3585–3593 10.1128/jb.178.12.3585-3593.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kommineni S., Bretl D. J., Lam V., Chakraborty R., Hayward M., Simpson P., Cao Y., Bousounis P., Kristich C. J., and Salzman N. H. (2015) Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 10.1038/nature15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kommineni S., Kristich C. J., and Salzman N. H. (2016) Harnessing bacteriocin biology as targeted therapy in the GI tract. Gut Microbes 7, 512–517 10.1080/19490976.2016.1233089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Rea M. C., Sit C. S., Clayton E., O'Connor P. M., Whittal R. M., Zheng J., Vederas J. C., Ross R. P., and Hill C. (2010) Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107, 9352–9357 10.1073/pnas.0913554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Rea M. C., Clayton E., O'Connor P. M., Shanahan F., Kiely B., Ross R. P., and Hill C. (2007) Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56, 940–946 10.1099/jmm.0.47085-0 [DOI] [PubMed] [Google Scholar]
- 134. Gardiner G. E., Rea M. C., O'Riordan B., O'Connor P., Morgan S. M., Lawlor P. G., Lynch P. B., Cronin M., Ross R. P., and Hill C. (2007) Fate of the two-component lantibiotic lacticin 3147 in the gastrointestinal tract. Appl. Environ. Microbiol. 73, 7103–7109 10.1128/AEM.01117-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nakatsuji T., Chen T. H., Narala S., Chun K. A., Two A. M., Yun T., Shafiq F., Kotol P. F., Bouslimani A., Melnik A. V., Latif H., Kim J. N., Lockhart A., Artis K., David G., et al. (2017) Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Nizet V., Ohtake T., Lauth X., Trowbridge J., Rudisill J., Dorschner R. A., Pestonjamasp V., Piraino J., Huttner K., and Gallo R. L. (2001) Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414, 454–457 10.1038/35106587 [DOI] [PubMed] [Google Scholar]
- 137. Williams M. R., Costa S. K., Zaramela L. S., Khalil S., Todd D. A., Winter H. L., Sanford J. A., O'Neill A. M., Liggins M. C., Nakatsuji T., Cech N. B., Cheung A. L., Zengler K., Horswill A. R., and Gallo R. L. (2019) Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl. Med. 11, eaat8329 10.1126/scitranslmed.aat8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Christensen G. J., Scholz C. F., Enghild J., Rohde H., Kilian M., Thürmer A., Brzuszkiewicz E., Lomholt H. B., and Brüggemann H. (2016) Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152 10.1186/s12864-016-2489-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cotter P. D., Draper L. A., Lawton E. M., Daly K. M., Groeger D. S., Casey P. G., Ross R. P., and Hill C. (2008) Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4, e1000144 10.1371/journal.ppat.1000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Quereda J. J., Meza-Torres J., Cossart P., and Pizarro-Cerdá J. (2017) Listeriolysin S: a bacteriocin from epidemic Listeria monocytogenes strains that targets the gut microbiota. Gut Microbes 8, 384–391 10.1080/19490976.2017.1290759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Quereda J. J., Dussurget O., Nahori M. A., Ghozlane A., Volant S., Dillies M. A., Regnault B., Kennedy S., Mondot S., Villoing B., Cossart P., and Pizarro-Cerda J. (2016) Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5706–5711 10.1073/pnas.1523899113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Papenfort K., and Bassler B. L. (2016) Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mukherjee S., and Bassler B. L. (2019) Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 10.1038/s41579-019-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Sturme M. H. J., Kleerebezem M., Nakayama J., Akkermans A. D. L., Vaughan E. E., and de Vos W. M. (2002) Cell to cell communication by autoinducing peptides in Gram-positive bacteria. Antonie Van Leeuwenhoek 81, 233–243 10.1023/A:1020522919555 [DOI] [PubMed] [Google Scholar]
- 145. Thoendel M., and Horswill A. R. (2010) Biosynthesis of peptide signals in Gram-positive bacteria. Adv. Appl. Microbiol. 71, 91–112 10.1016/S0065-2164(10)71004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Lyon G. J., and Novick R. P. (2004) Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25, 1389–1403 10.1016/j.peptides.2003.11.026 [DOI] [PubMed] [Google Scholar]
- 147. Martin M. J., Clare S., Goulding D., Faulds-Pain A., Barquist L., Browne H. P., Pettit L., Dougan G., Lawley T. D., and Wren B. W. (2013) The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J. Bacteriol. 195, 3672–3681 10.1128/JB.00473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yu Q., Lepp D., Mehdizadeh Gohari I., Wu T., Zhou H., Yin X., Yu H., Prescott J. F., Nie S. P., Xie M. Y., and Gong J. (2017) The Agr-Like quorum sensing system is required for pathogenesis of necrotic enteritis caused by Clostridium perfringens in poultry. Infect. Immun. 85, e00975–16 10.1128/IAI.00975-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Cooksley C. M., Davis I. J., Winzer K., Chan W. C., Peck M. W., and Minton N. P. (2010) Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76, 4448–4460 10.1128/AEM.03038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zetzmann M., Sánchez-Kopper A., Waidmann M. S., Blombach B., and Riedel C. U. (2016) Identification of the agr peptide of Listeria monocytogenes. Front. Microbiol. 7, 989 10.3389/fmicb.2016.00989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rémy B., Mion S., Plener L., Elias M., Chabrière E., and Daudé D. (2018) Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front. Pharmacol. 9, 203 10.3389/fphar.2018.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Gless B. H., Bojer M. S., Peng P., Baldry M., Ingmer H., and Olsen C. A. (2019) Identification of autoinducing thiodepsipeptides from staphylococci enabled by native chemical ligation. Nat. Chem. 11, 463–469 10.1038/s41557-019-0256-3 [DOI] [PubMed] [Google Scholar]
- 153. Canovas J., Baldry M., Bojer M. S., Andersen P. S., Gless B. H., Grzeskowiak P. K., Stegger M., Damborg P., Olsen C. A., and Ingmer H. (2016) Cross-talk between Staphylococcus aureus and other Staphylococcal species via the agr quorum sensing system. Front. Microbiol. 7, 1733 10.3389/fmicb.2016.01733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Paharik A. E., Parlet C. P., Chung N., Todd D. A., Rodriguez E. I., Van Dyke M. J., Cech N. B., and Horswill A. R. (2017) Coagulase-negative Staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22, 746–756.e5 10.1016/j.chom.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gless B. H., Peng P., Pedersen K. D., Gotfredsen C. H., Ingmer H., and Olsen C. A. (2017) Structure–activity relationship study based on autoinducing peptide (AIP) from dog pathogen S. schleiferi. Org. Lett. 19, 5276–5279 10.1021/acs.orglett.7b02550 [DOI] [PubMed] [Google Scholar]
- 156. Yang T., Tal-Gan Y., Paharik A. E., Horswill A. R., and Blackwell H. E. (2016) Structure–function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol. 11, 1982–1991 10.1021/acschembio.6b00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Esmaeilishirazifard E., De Vizio D., Moschos S. A., and Keshavarz T. (2017) Genomic and molecular characterization of a novel quorum sensing molecule in Bacillus licheniformis. AMB Express 7, 78 10.1186/s13568-017-0381-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Okada M., Sato I., Cho S. J., Iwata H., Nishio T., Dubnau D., and Sakagami Y. (2005) Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1, 23–24 10.1038/nchembio709 [DOI] [PubMed] [Google Scholar]
- 159. Ansaldi M., Marolt D., Stebe T., Mandic-Mulec I., and Dubnau D. (2002) Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol. Microbiol. 44, 1561–1573 10.1046/j.1365-2958.2002.02977.x [DOI] [PubMed] [Google Scholar]
- 160. Okada M., Sugita T., and Abe I. (2017) Posttranslational isoprenylation of tryptophan in bacteria. Beilstein J. Org. Chem. 13, 338–346 10.3762/bjoc.13.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ansaldi M., and Dubnau D. (2004) Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J. Bacteriol. 186, 15–21 10.1128/JB.186.1.15-21.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Schmitz S., Hoffmann A., Szekat C., Rudd B., and Bierbaum G. (2006) The lantibiotic mersacidin is an autoinducing peptide. Appl. Environ. Microbiol. 72, 7270–7277 10.1128/AEM.00723-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kleerebezem M. (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25, 1405–1414 10.1016/j.peptides.2003.10.021 [DOI] [PubMed] [Google Scholar]
- 164. Haas W., Shepard B. D., and Gilmore M. S. (2002) Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415, 84–87 10.1038/415084a [DOI] [PubMed] [Google Scholar]
- 165. Foulston L. C., and Bibb M. J. (2010) Microbisporicin gene cluster reveals unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 107, 13461–13466 10.1073/pnas.1008285107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Fernández-Martínez L. T., Gomez-Escribano J. P., and Bibb M. J. (2015) A relA-dependent regulatory cascade for auto-induction of microbisporicin production in Microbispora corallina. Mol. Microbiol. 97, 502–514 10.1111/mmi.13046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Sherwood E. J., and Bibb M. J. (2013) The antibiotic planosporicin coordinates its own production in the actinomycete Planomonospora alba. Proc. Natl. Acad. Sci. U.S.A. 110, E2500–E2509 10.1073/pnas.1305392110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Coburn P. S., Pillar C. M., Jett B. D., Haas W., and Gilmore M. S. (2004) Enterococcus faecalis senses target cells and in response expresses cytolysin. Science 306, 2270–2272 10.1126/science.1103996 [DOI] [PubMed] [Google Scholar]
- 169. Van Tyne D., Martin M. J., and Gilmore M. S. (2013) Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 5, 895–911 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Hoover S. E., Perez A. J., Tsui H. C., Sinha D., Smiley D. L., DiMarchi R. D., Winkler M. E., and Lazazzera B. A. (2015) A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol. Microbiol. 97, 229–243 10.1111/mmi.13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. van der Ploeg J. R. (2005) Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187, 3980–3989 10.1128/JB.187.12.3980-3989.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wang C. Y., Patel N., Wholey W. Y., and Dawid S. (2018) ABC transporter content diversity in Streptococcus pneumoniae impacts competence regulation and bacteriocin production. Proc. Natl. Acad. Sci. U.S.A. 115, E5776–E5785 10.1073/pnas.1804668115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Wholey W. Y., Kochan T. J., Storck D. N., and Dawid S. (2016) Coordinated bacteriocin expression and competence in Streptococcus pneumoniae contributes to genetic adaptation through neighbor predation. PLoS Pathog. 12, e1005413 10.1371/journal.ppat.1005413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Sztajer H., Szafranski S. P., Tomasch J., Reck M., Nimtz M., Rohde M., and Wagner-Döbler I. (2014) Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271 10.1038/ismej.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Ibrahim M., Guillot A., Wessner F., Algaron F., Besset C., Courtin P., Gardan R., and Monnet V. (2007) Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189, 8844–8854 10.1128/JB.01057-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Schramma K. R., Bushin L. B., and Seyedsayamdost M. R. (2015) Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink. Nat. Chem. 7, 431–437 10.1038/nchem.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fleuchot B., Guillot A., Mézange C., Besset C., Chambellon E., Monnet V., and Gardan R. (2013) Rgg-associated SHP signaling peptides mediate cross-talk in streptococci. PLoS One 8, e66042 10.1371/journal.pone.0066042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Bushin L. B., Clark K. A., Pelczer I., and Seyedsayamdost M. R. (2018) Charting an unexplored streptococcal biosynthetic landscape reveals a unique peptide cyclization motif. J. Am. Chem. Soc. 140, 17674–17684 10.1021/jacs.8b10266 [DOI] [PubMed] [Google Scholar]
- 179. Caruso A., Bushin L. B., Clark K. A., Martinie R. J., and Seyedsayamdost M. R. (2019) Radical approach to enzymatic β-thioether bond formation. J. Am. Chem. Soc. 141, 990–997 10.1021/jacs.8b11060 [DOI] [PubMed] [Google Scholar]
- 180. Clark K. A., Bushin L. B., and Seyedsayamdost M. R. (2019) Aliphatic ether bond formation expands the scope of radical SAM enzymes in natural product biosynthesis. J. Am. Chem. Soc. 141, 10610–10615 10.1021/jacs.9b05151 [DOI] [PubMed] [Google Scholar]
- 181. Luu D. D., Joe A., Chen Y., Parys K., Bahar O., Pruitt R., Chan L. J. G., Petzold C. J., Long K., Adamchak C., Stewart V., Belkhadir Y., and Ronald P. C. (2019) Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. U.S.A. 116, 8525–8534 10.1073/pnas.1818275116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hegemann J. D., Zimmermann M., Zhu S., Klug D., and Marahiel M. A. (2013) Lasso peptides from proteobacteria: genome mining employing heterologous expression and mass spectrometry. Biopolymers 100, 527–542 10.1002/bip.22326 [DOI] [PubMed] [Google Scholar]
- 183. Gibson K. E., Kobayashi H., and Walker G. C. (2008) Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42, 413–441 10.1146/annurev.genet.42.110807.091427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Gilmore S. A., Schelle M. W., Holsclaw C. M., Leigh C. D., Jain M., Cox J. S., Leary J. A., and Bertozzi C. R. (2012) Sulfolipid-1 biosynthesis restricts Mycobacterium tuberculosis growth in human macrophages. ACS Chem. Biol. 7, 863–870 10.1021/cb200311s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Freeman M. F., Gurgui C., Helf M. J., Morinaka B. I., Uria A. R., Oldham N. J., Sahl H.-G., Matsunaga S., and Piel J. (2012) Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 338, 387–390 10.1126/science.1226121 [DOI] [PubMed] [Google Scholar]
- 186. Wilson M. C., Mori T., Rückert C., Uria A. R., Helf M. J., Takada K., Gernert C., Steffens U. A. E., Heycke N., Schmitt S., Rinke C., Helfrich E. J. N., Brachmann A. O., Gurgui C., Wakimoto T., et al. (2014) An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 10.1038/nature12959 [DOI] [PubMed] [Google Scholar]
- 187. Lin Z., Torres J. P., Tianero M. D., Kwan J. C., and Schmidt E. W. (2016) Origin of chemical diversity in prochloron-tunicate symbiosis. Appl. Environ. Microbiol. 82, 3450–3460 10.1128/AEM.00860-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Claessen D., Rozen D. E., Kuipers O. P., Søgaard-Andersen L., and van Wezel G. P. (2014) Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115–124 10.1038/nrmicro3178 [DOI] [PubMed] [Google Scholar]
- 189. Romero D., Traxler M. F., López D., and Kolter R. (2011) Antibiotics as signal molecules. Chem. Rev. 111, 5492–5505 10.1021/cr2000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. López D., Fischbach M. A., Chu F., Losick R., and Kolter R. (2009) Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 106, 280–285 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Bleich R., Watrous J. D., Dorrestein P. C., Bowers A. A., and Shank E. A. (2015) Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 112, 3086–3091 10.1073/pnas.1414272112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Townsley L., and Shank E. A. (2017) Natural-product antibiotics: cues for modulating bacterial biofilm formation. Trends Microbiol. 25, 1016–1026 10.1016/j.tim.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ranieri M. R. M., Chan D. C. K., Yaeger L. N., Rudolph M., Karabelas-Pittman S., Abdo H., Chee J., Harvey H., Nguyen U., and Burrows L. L. (2019) Thiostrepton hijacks pyoverdine receptors to inhibit growth of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, e00472–19 10.1128/AAC.00472-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kodani S., Hudson M. E., Durrant M. C., Buttner M. J., Nodwell J. R., and Willey J. M. (2004) The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 101, 11448–11453 10.1073/pnas.0404220101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Kodani S., Lodato M. A., Durrant M. C., Picart F., and Willey J. M. (2005) SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol. Microbiol. 58, 1368–1380 10.1111/j.1365-2958.2005.04921.x [DOI] [PubMed] [Google Scholar]
- 196. Willey J. M., Willems A., Kodani S., and Nodwell J. R. (2006) Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 59, 731–742 10.1111/j.1365-2958.2005.05018.x [DOI] [PubMed] [Google Scholar]
- 197. Straight P. D., Willey J. M., and Kolter R. (2006) Interactions between Streptomyces coelicolor and Bacillus subtilis: role of surfactants in raising aerial structures. J. Bacteriol. 188, 4918–4925 10.1128/JB.00162-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gaskell A. A., Giovinazzo J. A., Fonte V., and Willey J. M. (2012) Multi-tier regulation of the streptomycete morphogenetic peptide SapB. Mol. Microbiol. 84, 501–515 10.1111/j.1365-2958.2012.08041.x [DOI] [PubMed] [Google Scholar]
- 199. Onaka H., Tabata H., Igarashi Y., Sato Y., and Furumai T. (2001) Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot. 54, 1036–1044 10.7164/antibiotics.54.1036 [DOI] [PubMed] [Google Scholar]
- 200. Wang H., Zhao G., and Ding X. (2017) Morphology engineering of Streptomyces coelicolor M145 by sub-inhibitory concentrations of antibiotics. Sci. Rep. 7, 13226 10.1038/s41598-017-13493-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Meyer V., and Jung S. (2018) Antifungal peptides of the AFP family revisited: are these cannibal toxins? Microorganisms 6, E50 10.3390/microorganisms6020050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Liu W. T., Yang Y. L., Xu Y., Lamsa A., Haste N. M., Yang J. Y., Ng J., Gonzalez D., Ellermeier C. D., Straight P. D., Pevzner P. A., Pogliano J., Nizet V., Pogliano K., and Dorrestein P. C. (2010) Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 16286–16290 10.1073/pnas.1008368107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Allenby N. E., Watts C. A., Homuth G., Prágai Z., Wipat A., Ward A. C., and Harwood C. R. (2006) Phosphate starvation induces the sporulation killing factor of Bacillus subtilis. J. Bacteriol. 188, 5299–5303 10.1128/JB.00084-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. González-Pastor J. E. (2011) Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 35, 415–424 10.1111/j.1574-6976.2010.00253.x [DOI] [PubMed] [Google Scholar]
- 205. López D., Vlamakis H., Losick R., and Kolter R. (2009) Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74, 609–618 10.1111/j.1365-2958.2009.06882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Molina-Santiago C., Pearson J. R., Navarro Y., Berlanga-Clavero M. V., Caraballo-Rodriguez A. M., Petras D., García-Martín M. L., Lamon G., Haberstein B., Cazorla F. M., de Vicente A., Loquet A., Dorrestein P. C., and Romero D. (2019) The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat. Commun. 10, 1919 10.1038/s41467-019-09944-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Kenney G. E., and Rosenzweig A. C. (2018) Methanobactins: maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem. 293, 4606–4615 10.1074/jbc.TM117.000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. DiSpirito A. A., Semrau J. D., Murrell J. C., Gallagher W. H., Dennison C., and Vuilleumier S. (2016) Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev. 80, 387–409 10.1128/MMBR.00058-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. El Ghazouani A., Baslé A., Gray J., Graham D. W., Firbank S. J., and Dennison C. (2012) Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 8400–8404 10.1073/pnas.1112921109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Dassama L. M., Kenney G. E., Ro S. Y., Zielazinski E. L., and Rosenzweig A. C. (2016) Methanobactin transport machinery. Proc. Natl. Acad. Sci. U.S.A. 113, 13027–13032 10.1073/pnas.1603578113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Balasubramanian R., Kenney G. E., and Rosenzweig A. C. (2011) Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 286, 37313–37319 10.1074/jbc.M111.284984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Ghssein G., Brutesco C., Ouerdane L., Fojcik C., Izaute A., Wang S., Hajjar C., Lobinski R., Lemaire D., Richaud P., Voulhoux R., Espaillat A., Cava F., Pignol D., Borezée-Durant E., and Arnoux P. (2016) Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352, 1105–1109 10.1126/science.aaf1018 [DOI] [PubMed] [Google Scholar]
- 213. Kenney G. E., and Rosenzweig A. C. (2013) Genome mining for methanobactins. BMC Biol. 11, 17 10.1186/1741-7007-11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Maksimov M. O., Pelczer I., and Link A. J. (2012) Precursor-centric genome-mining approach for lasso peptide discovery. Proc. Natl. Acad. Sci. U.S.A. 109, 15223–15228 10.1073/pnas.1208978109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Zimmermann M., Hegemann J. D., Xie X., and Marahiel M. A. (2013) The astexin-1 lasso peptides: biosynthesis, stability, and structural studies. Chem. Biol. 20, 558–569 10.1016/j.chembiol.2013.03.013 [DOI] [PubMed] [Google Scholar]
- 216. Ayikpoe R., Govindarajan V., and Latham J. A. (2019) Occurrence, function, and biosynthesis of mycofactocin. Appl. Microbiol. Biotechnol. 103, 2903–2912 10.1007/s00253-019-09684-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Klinman J. P., and Bonnot F. (2014) Intrigues and intricacies of the biosynthetic pathways for the enzymatic quinocofactors: PQQ, TTQ, CTQ, TPQ, and LTQ. Chem. Rev. 114, 4343–4365 10.1021/cr400475g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Shen Y. Q., Bonnot F., Imsand E. M., RoseFigura J. M., Sjölander K., and Klinman J. P. (2012) Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51, 2265–2275 10.1021/bi201763d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Yakushi T., and Matsushita K. (2010) Alcohol dehydrogenase of acetic acid bacteria: structure, mode of action, and applications in biotechnology. Appl. Microbiol. Biotechnol. 86, 1257–1265 10.1007/s00253-010-2529-z [DOI] [PubMed] [Google Scholar]
- 220. Anthony C. (2001) Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxid. Redox Signal. 3, 757–774 10.1089/15230860152664966 [DOI] [PubMed] [Google Scholar]
- 221. Piersma S. R., de Vries S., and Duine J. A. (1996) Nicotinoprotein alcohol/aldehyde oxidoreductases. in Enzymology and Molecular Biology of Carbonyl Metabolism 6: Advances in Experimental Medicine and Biology (Weiner H., Lindahl R., Crabb D. W., and Flynn T. G., eds) pp. 425–434, Springer, Boston [Google Scholar]
- 222. Haft D. H., and Basu M. K. (2011) Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755 10.1128/JB.00040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Krishnamoorthy G., Kaiser P., Lozza L., Hahnke K., Mollenkopf H. J., and Kaufmann S. H. E. (2019) Mycofactocin is associated with ethanol metabolism in Mycobacteria. mBio 10, e00190–19 10.1128/mBio.00190-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Dubey A. A., and Jain V. (2019) Mycofactocin is essential for the establishment of methylotrophy in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 516, 1073–1077 10.1016/j.bbrc.2019.07.008 [DOI] [PubMed] [Google Scholar]
- 225. Ayikpoe R. S., and Latham J. A. (2019) MftD catalyzes the formation of a biologically active redox center in the biosynthesis of the ribosomally synthesized and post-translationally modified redox cofactor mycofactocin. J. Am. Chem. Soc. 141, 13582–13591 10.1021/jacs.9b06102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Jaspars M. (2014) The origins of cyanobactin chemistry and biology. Chem. Commun. (Camb.) 50, 10174–10176 10.1039/C3CC49252D [DOI] [PubMed] [Google Scholar]
- 227. Ruffner D. E., Schmidt E. W., and Heemstra J. R. (2015) Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth. Biol. 4, 482–492 10.1021/sb500267d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Koehnke J., Bent A. F., Houssen W. E., Mann G., Jaspars M., and Naismith J. H. (2014) The structural biology of patellamide biosynthesis. Curr. Opin. Struct. Biol. 29, 112–121 10.1016/j.sbi.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Odenyo A. A., Mackie R. I., Stahl D. A., and White B. A. (1994) The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: development of probes for Ruminococcus species and evidence for bacteriocin production. Appl. Environ. Microbiol. 60, 3688–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Zhao X., and van der Donk W. A. (2016) Structural characterization and bioactivity analysis of the two-component lantibiotic Flv system from a ruminant bacterium. Cell Chem. Biol. 23, 246–256 10.1016/j.chembiol.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Vásquez A., Forsgren E., Fries I., Paxton R. J., Flaberg E., Szekely L., and Olofsson T. C. (2012) Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7, e33188 10.1371/journal.pone.0033188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Shao Y., Chen B., Sun C., Ishida K., Hertweck C., and Boland W. (2017) Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75 10.1016/j.chembiol.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 233. Palmer-Young E. C., Raffel T. R., and McFrederick Q. S. (2019) pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitology 146, 380–388 10.1017/S0031182018001555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Attaiech L., and Charpentier X. (2017) Silently transformable: the many ways bacteria conceal their built-in capacity of genetic exchange. Curr. Genet. 63, 451–455 10.1007/s00294-016-0663-6 [DOI] [PubMed] [Google Scholar]
- 235. Sun D. (2018) Pull in and push out: mechanisms of horizontal gene transfer in bacteria. Front. Microbiol. 9, 2154 10.3389/fmicb.2018.02154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Veening J. W., and Blokesch M. (2017) Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat. Rev. Microbiol. 15, 621–629 10.1038/nrmicro.2017.66 [DOI] [PubMed] [Google Scholar]
- 237. Zaccaria E., Wels M., van Baarlen P., and Wells J. M. (2016) Temporal regulation of the transformasome and competence development in Streptococcus suis. Front. Microbiol. 7, 1922 10.3389/fmicb.2016.01922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Castiglione F., Cavaletti L., Losi D., Lazzarini A., Carrano L., Feroggio M., Ciciliato I., Corti E., Candiani G., Marinelli F., and Selva E. (2007) A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon Actinomycete Planomonospora sp. Biochemistry 46, 5884–5895 10.1021/bi700131x [DOI] [PubMed] [Google Scholar]
- 239. Crost E. H., Ajandouz E. H., Villard C., Geraert P. A., Puigserver A., and Fons M. (2011) Ruminococcin C, a new anti-Clostridium perfringens bacteriocin produced in the gut by the commensal bacterium Ruminococcus gnavus E1. Biochimie 93, 1487–1494 10.1016/j.biochi.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 240. Mathur H., Fallico V., O'Connor P. M., Rea M. C., Cotter P. D., Hill C., and Ross R. P. (2017) Insights into the mode of action of the sactibiotic thuricin CD. Front. Microbiol. 8, 696 10.3389/fmicb.2017.00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Quereda J. J., Nahori M. A., Meza-Torres J., Sachse M., Titos-Jiménez P., Gomez-Laguna J., Dussurget O., Cossart P., and Pizarro-Cerdá J. (2017) Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively pokaryotic cells in vivo. mBio 8, e00259–17 10.1128/mBio.00259-17 [DOI] [PMC free article] [PubMed] [Google Scholar]