We sequenced the genomes of two chikungunya virus isolates obtained from viremic patients who had traveled to Australia. The first patient acquired the infection in Bangladesh in 2017, and the second was infected in Thailand in 2019. Phylogenetic sequence analysis demonstrated that both isolates belonged to the East/Central/South African genotype.
ABSTRACT
We sequenced the genomes of two chikungunya virus isolates obtained from viremic patients who had traveled to Australia. The first patient acquired the infection in Bangladesh in 2017, and the second was infected in Thailand in 2019. Phylogenetic sequence analysis demonstrated that both isolates belonged to the East/Central/South African genotype.
ANNOUNCEMENT
Within the last 20 years, the reemergence and global spread of the arthropod-borne chikungunya virus (CHIKV) (genus Alphavirus, family Togaviridae) have resulted in explosive epidemics throughout the Pacific, Asia, and the Americas, affecting millions of people (1). Large-scale CHIKV outbreaks were recently recorded in Dhaka, Bangladesh, in 2017 (2) and Thailand in 2019 (3). Areas of nonendemicity, such as Queensland, Australia, which harbor populations of Aedes aegypti and Aedes albopictus mosquitoes, are at risk of CHIKV importation by viremic travelers and subsequent autochthonous transmission (1, 4).
In July 2017, a febrile male patient traveled to Brisbane, Australia, from Bangladesh. Similarly, in September 2019, a female patient with fever and joint pains arrived in Brisbane after recent travel in Thailand. Acute-phase sera were analyzed using a specific CHIKV reverse-transcription real-time PCR assay (4) which detected CHIKV RNA in each patient sample. The same patient sera were inoculated onto A. albopictus C6/36 cell monolayers. Two CHIKV isolates, namely, Bangladesh 2017 and Thail 2019, were recovered and subsequently used for whole-genome sequencing (WGS) and phylogenetic analysis.
To support ongoing and enhanced CHIKV surveillance within Australasia and provide contemporary sequences for continued scrutiny of existing molecular diagnostic assays, we performed WGS as previously described (5, 6). Briefly, total RNA was extracted from passage 1 C6/36 culture supernatants from each of the Bangladesh 2017 and Thail 2019 isolates using the QIAamp viral RNA extraction kit (Qiagen, Chadstone, Australia) without carrier RNA. Host and potentially contaminating microbial DNA was removed with DNase treatment (Heat&Run kit; ArcticZymes, Scientifix, South Yarra, Australia). RNA was converted to cDNA using the ProtoScript II first-strand cDNA kit (New England Biolabs), and second-strand cDNA synthesis was performed using an enzyme cocktail of Escherichia coli DNA ligase, DNA polymerase I, and RNase H (New England Biolabs). The Nextera XT kit was used for cDNA library construction, and paired-end (2 × 151 nucleotides [nt]) sequencing was performed using the V2 midoutput kit on a NextSeq 500 machine (Illumina, San Diego, CA).
Illumina sequencing yielded 10,299,664 and 16,336,098 reads for Bangladesh 2017 and Thail 2019, respectively, and raw sequence reads were processed using Geneious R10 version 10.2.6 software (7). Complete genome sequence assembly was achieved by mapping to a reference CHIKV genome sequence (strain CHIK31, GenBank accession number EU564335) that included 5′ and 3′ untranslated terminal sequences, using default parameters and the low-sensitivity setting. For Bangladesh 2017 (11,812 nt with 50.1% G+C content), a total of 4,908,657 reads with an average coverage depth of 45,504× were mapped to strain CHIK31. Similarly, a total of 8,240,564 reads with an average coverage depth of 78,217× were mapped to strain CHIK31 for Thail 2019 (11,812 nt with 50.1% G+C content).
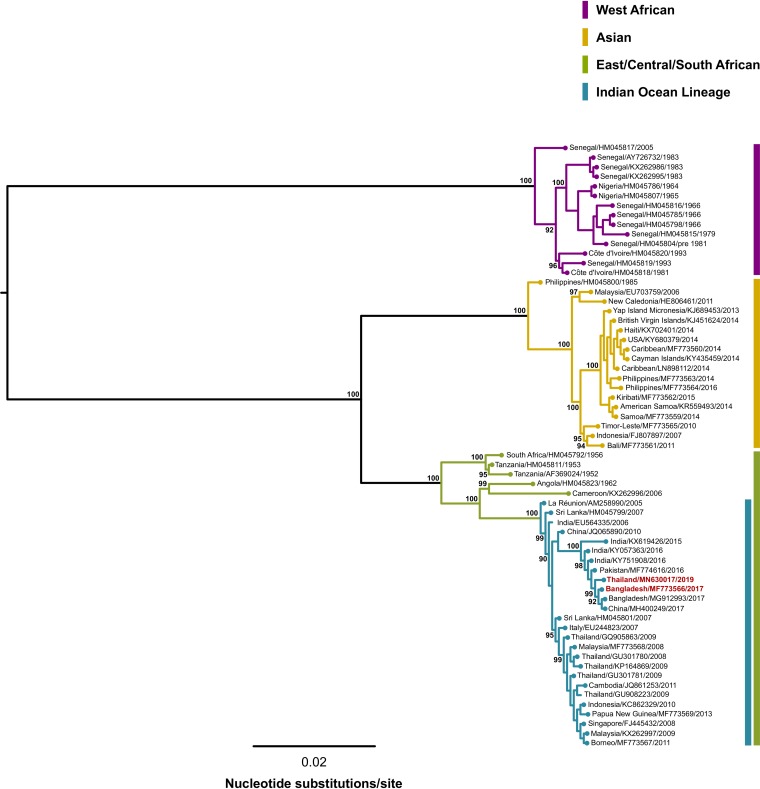
Genome sequence identities were determined using the online version of blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Bangladesh 2017 and Thail 2019 shared 99.83% sequence identity with and were most closely related to a Chinese 2017 isolate, QZ0823 (GenBank accession number MH400249), which shared 99.95% and 99.85% sequence identity with Bangladesh 2017 and Thail 2019, respectively. Further sequence and phylogenetic analysis (FastTree software version 2.1.11 [8] and the general time-reversible [GTR] nucleotide substitution model) of the region encompassing the putative nonstructural polyprotein (nsP) (nsP1-nsP2-nsP3 termination product or nsP1-nsP2-nsP3-nsP4 read-through product), the subgenomic 26S RNA promoter region, and the putative structural polyprotein (C-E3-E2-6K-E1) indicated that both Bangladesh 2017 and Thail 2019 grouped within the Indian Ocean lineage of the East/Central/South African (ECSA) genotype (Fig. 1). However, the E1-A226V mutation first detected during the 2005–2006 CHIKV epidemic on La Réunion Island (9) was not found in either strain.
FIG 1.
Approximately maximum-likelihood phylogenetic tree (midpoint rooted for branch visibility only) inferred for 63 CHIKV genome sequences (excluding 5′ and 3′ untranslated regions) using FastTree version 2.1.11 software and the GTR nucleotide substitution model with the default setting of 20 for rate categories of sites (8). Percentage Shimodaira-Hasegawa-like local support values are shown for key nodes. Multiple sequence alignments were performed using the Multiple Alignment using Fast Fourier Transform (MAFFT) program version 7.450 and Geneious version 10.2.6 software. The three major CHIKV genotypes (West African, Asian, and ECSA) are shown, including grouping of the Bangladesh 2017 (GenBank accession number MF773566) and Thail 2019 (GenBank accession number MN630017) isolates (red font) within the ECSA-Indian Ocean lineage.
Data availability.
Raw sequencing reads were deposited in the Sequence Read Archive under the accession numbers SAMN13164600 (Bangladesh 2017) and SAMN13164601 (Thail 2019) and BioProject number PRJNA583228. The genome sequences have been deposited in GenBank under the accession numbers MF773566 (Bangladesh 2017) and MN630017 (Thail 2019).
ACKNOWLEDGMENTS
We are grateful to staff at the Royal Brisbane and Women’s Hospital, the Princess Alexandra Hospital, and the Public Health Virology Laboratory, Forensic and Scientific Services, who contributed to the collection of patient sera and clinical and travel histories.
This work received ethical clearance from the Forensic and Scientific Services Human Ethics Committee.
REFERENCES
- 1.Pyke AT, Moore PR, McMahon J. 2018. New insights into chikungunya virus emergence and spread from Southeast Asia. Emerg Microbes Infect 7:26. doi: 10.1038/s41426-018-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hossain MS, Hasan MM, Islam MS, Islam S, Mozaffor M, Khan MAS, Ahmed N, Akhtar W, Chowdhury S, Arafat SMY, Khaleque MA, Khan ZJ, Dipta TF, Asna S, Hossain MA, Aziz KS, Mosabbir AA, Raheem E. 2018. Chikungunya outbreak (2017) in Bangladesh: clinical profile, economic impact and quality of life during the acute phase of the disease. PLoS Negl Trop Dis 12:e0006561. doi: 10.1371/journal.pntd.0006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Intayot P, Phumee A, Boonserm R, Sor-Suwan S, Buathong R, Wacharapluesadee S, Brownell N, Poovorawan Y, Siriyasatien P. 2019. Genetic characterization of chikungunya virus in field-caught Aedes aegypti mosquitoes collected during the recent outbreaks in 2019, Thailand. Pathogens 8:121. doi: 10.3390/pathogens8030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Hurk AF, Hall-Mendelin S, Pyke AT, Smith GA, Mackenzie JS. 2010. Vector competence of Australian mosquitoes for chikungunya virus. Vector Borne Zoonotic Dis 10:489–495. doi: 10.1089/vbz.2009.0106. [DOI] [PubMed] [Google Scholar]
- 5.Huang B, Pyke AT, McMahon J, Warrilow D. 2017. Complete coding sequence of a case of chikungunya virus imported into Australia. Genome Announc 5:e00310-17. doi: 10.1128/genomeA.00310-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyke AT, Huang B, Warrilow D, Moore PR, McMahon J, Harrower B. 2017. Complete genome sequence of a highly divergent dengue virus type 2 strain, imported into Australia from Sabah, Malaysia. Genome Announc 5:e00546-17. doi: 10.1128/genomeA.00546-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequencing reads were deposited in the Sequence Read Archive under the accession numbers SAMN13164600 (Bangladesh 2017) and SAMN13164601 (Thail 2019) and BioProject number PRJNA583228. The genome sequences have been deposited in GenBank under the accession numbers MF773566 (Bangladesh 2017) and MN630017 (Thail 2019).