Here, we report the detection of a novel alphavirus in Australian mosquitoes, provisionally named Yada Yada virus (YYV). Phylogenetic analysis indicated that YYV belongs to the mosquito-specific alphavirus complex. The assembled genome is 11,612 nucleotides in length and encodes two open reading frames.
ABSTRACT
Here, we report the detection of a novel alphavirus in Australian mosquitoes, provisionally named Yada Yada virus (YYV). Phylogenetic analysis indicated that YYV belongs to the mosquito-specific alphavirus complex. The assembled genome is 11,612 nucleotides in length and encodes two open reading frames.
ANNOUNCEMENT
Alphaviruses (genus Alphavirus, family Togaviridae) are small (10- to 12-kb) single-stranded positive-sense RNA viruses and include species important to human and animal health, such as Chikungunya virus and Eastern equine encephalitis virus (1). While these viruses are transmitted primarily by mosquitoes and pathogenic in their vertebrate hosts, there is a small complex of recently discovered alphaviruses that replicate only in mosquito cells (2–5). Here, we report the detection of an alphavirus belonging to this host-restricted complex in the Asia-Pacific region and provide the genome sequence for the novel virus, named Yada Yada virus (YYV).
Virus detection was performed using mosquitoes trapped as part of the Victorian Arbovirus Disease Control Program (6). Encephalitis virus surveillance (EVS) traps (7) were set up overnight each week in three locations in Victoria, Australia, for a total of 7 weeks in late 2016, resulting in 21 trap collections. Traps were sorted into 86 pools of up to 1,000 mosquitoes, which were homogenized in buffer AVL (Qiagen) and centrifuged. RNA was extracted from the supernatant with the QIAamp viral RNA minikit (Qiagen) and used for library preparation, which was performed using the Ovation universal transcriptome sequencing (RNA-Seq) system (NuGEN) with a customized mosquito rRNA depletion (8). The libraries were then treated with free adapter blocking reagent (Illumina) and sequenced on a HiSeq 3000 platform (Illumina) using 2 × 150 bp reads. A total of 909,467,304 paired reads were generated (mean, 10,575,201 per pool; range, 7,971,017 to 16,414,900).
Trinity v2.4.0 (9) was used to trim, normalize, and assemble the reads into contigs, which were taxonomically classified using DIAMOND BLASTx v0.9.22.123 (10) with the NCBI nonredundant (nr) database (acquired 2 September 2019) and an E value cutoff of 10−5. Reads were mapped to assembled contigs using BWA-MEM v0.7.17 r1188 (11). All analyses were performed using default parameters unless stated otherwise. Three of the 21 traps tested contained contigs that had the strongest BLASTx match to the mosquito-specific Eilat alphavirus (EILV). All three traps were collected in November 2016 in Mildura (latitude, 34.249617, longitude, 142.218261). The longest contig was 11,612 nucleotides (nt), with 21-fold average coverage depth and 75.7% amino acid identity to EILV. This contig represents the coding-complete YYV genome, with two open reading frames (ORFs), a 33-nt 3′ leader, a 470-nt 5′ trailer, and 53.4% G+C content. The two ORFs correspond to the structural (1,247 amino acids) and nonstructural (2,437 amino acids) proteins. Translation of the genome sequence was performed using the ExPASy Translate tool (12).
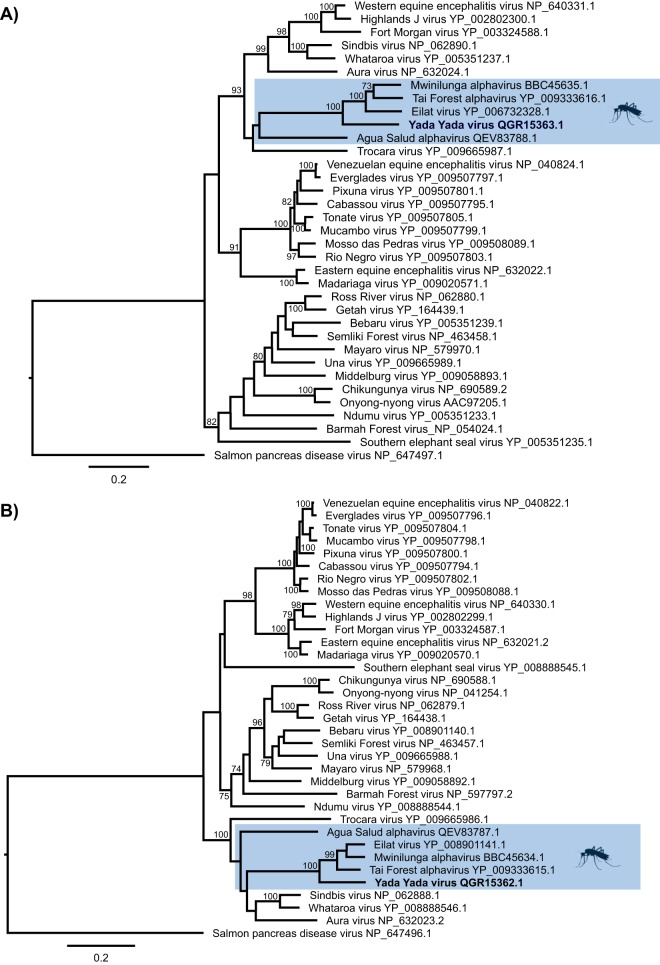
Phylogenetic analysis was performed by the creation of alignments of YYV and the structural and nonstructural protein sequences of other alphaviruses using MAFFT v7.429 (13), the removal of ambiguously aligned residues with TrimAl v1.4.1 (14), and maximum likelihood inference using PhyML v3.1 (15) employing the Le-Gascuel (LG) plus gamma distribution model of amino acid substitution and 1,000 bootstrap replicates. The resultant trees were then viewed in FigTree v1.4.4 (16). In both the structural (Fig. 1A) and nonstructural (Fig. 1B) protein trees, YYV was placed in the mosquito-specific alphavirus complex, suggesting that it might also have a restricted host range.
FIG 1.
Phylogenetic relationships of YYV and other alphaviruses based on alignments of structural proteins (850 amino acids) (A) and nonstructural proteins (1,287 amino acids) (B). Maximum likelihood trees were estimated using the LG plus gamma model of amino acid substitution in PhyML, with 1,000 bootstrap replicates, and rooted using salmon pancreas disease virus. Bootstrap values greater than 70% are shown beside the branches, and the GenBank accession numbers are shown with the virus names. The mosquito-specific complex is highlighted in blue with YYV in bold.
To investigate the vector of YYV, the assembled contigs were compared to a cytochrome oxidase I (COI) database of Australian mosquito species (17) using BLASTn v2.9.0+ (18) with an E value cutoff of 10−5, and the results were filtered for alignments >200 bp in length and matches of >95% identity. Only two mosquito species were present in all three traps, Anopheles annulipes and Culex australicus/Culex globocoxitus (these two Culex species are indistinguishable using COI), supporting previous studies that have detected mosquito-specific alphaviruses from only Anopheles and Culex species (2–5). Due to the homogenization of the traps, the YYV vector species cannot be definitively determined. However, read mapping showed that the abundance of A. annulipes was associated with YYV genome coverage, whereas the abundance of C. australicus/C. globocoxitus was not (data not shown).
The discovery of YYV expands the diversity and geographic range of the mosquito-specific alphavirus complex and in doing so will help reveal the virus origin and evolution of host switching (19). In addition, it is noteworthy that mosquito-specific viruses that are closely related to pathogenic vertebrate viruses have potential applications in vaccine development and as biocontrol agents (20).
Data availability.
The YYV genome sequence has been deposited in GenBank under the accession number MN733821. The sequencing reads are available in the SRA database via BioProject accession number PRJNA594295.
ACKNOWLEDGMENTS
We thank Dale Hutchinson for conducting the trapping and Karen Brown for her assistance in processing the mosquitoes.
This work was funded by the Biosciences Research Innovation Fund Program provided by the Victorian Department of Jobs, Precincts, and Regions. The Victorian Arbovirus Disease Control Program is funded by the Department of Health and Human Services. J.B. is supported by an Australian Government research training program scholarship. E.C.H. is supported by an Australian Research Council Australian Laureate fellowship (FL170100022).
REFERENCES
- 1.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, Powers A, Weaver SC, ICTV Report Consortium . 2018. ICTV virus taxonomy profile: Togaviridae. J Gen Virol 99:761–762. doi: 10.1099/jgv.0.001072. [DOI] [PubMed] [Google Scholar]
- 2.Hermanns K, Zirkel F, Kopp A, Marklewitz M, Rwego IB, Estrada A, Gillespie TR, Drosten C, Junglen S. 2017. Discovery of a novel alphavirus related to Eilat virus. J Gen Virol 98:43–49. doi: 10.1099/jgv.0.000694. [DOI] [PubMed] [Google Scholar]
- 3.Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa APT, Savji N, Popov VL, Sherman MB, Lipkin WI, Tesh RB, Weaver SC. 2012. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci U S A 109:14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermanns K, Marklewitz M, Zirkel F, Overheul GJ, Page RA, Loaiza JR, Drosten C, van Rij RP, Junglen S. 2019. Agua Salud alphavirus defines a novel lineage of insect-specific alphaviruses discovered in the New World. J Gen Virol. doi: 10.1099/jgv.0.001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torii S, Orba Y, Hang'ombe BM, Mweene AS, Wada Y, Anindita PD, Phongphaew W, Qiu Y, Kajihara M, Mori-Kajihara A, Eto Y, Harima H, Sasaki M, Carr M, Hall WW, Eshita Y, Abe T, Sawa H. 2018. Discovery of Mwinilunga alphavirus: a novel alphavirus in Culex mosquitoes in Zambia. Virus Res 250:31–36. doi: 10.1016/j.virusres.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Wong S, Brown K, Crowder J, Chea S, Mee P, Batovska J, Lynch S. 2017. Victorian Arbovirus Disease Control Program annual report 2016–2017. Agriculture Victoria, Victoria, Australia. [Google Scholar]
- 7.Ritchie SA, Kline DL. 1995. Comparison of CDC and EVS light traps baited with carbon dioxide and octenol for trapping mosquitoes in Brisbane, Queensland (Diptera: Culicidae). Aust J Entomol 34:215–218. doi: 10.1111/j.1440-6055.1995.tb01322.x. [DOI] [Google Scholar]
- 8.Batovska J, Mee PT, Lynch SE, Sawbridge TI, Rodoni BC. 2019. Sensitivity and specificity of metatranscriptomics as an arbovirus surveillance tool. Sci Rep 9:19398. doi: 10.1038/s41598-019-55741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 11.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 [q-bio.GN]. https://arxiv.org/abs/1303.3997.
- 12.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 16.Rambaut A. 2019. FigTree v1.4: tree figure drawing tool. GitHub. https://github.com/rambaut/figtree/releases.
- 17.Batovska J, Blacket MJ, Brown K, Lynch SE. 2016. Molecular identification of mosquitoes (Diptera: Culicidae) in southeastern Australia. Ecol Evol 6:3001–3011. doi: 10.1002/ece3.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall RA, Hobson-Peters J. 2018. Newly discovered mosquito viruses help control vector-borne viral diseases. Microbiol Aust 39:72–75. doi: 10.1071/MA18020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The YYV genome sequence has been deposited in GenBank under the accession number MN733821. The sequencing reads are available in the SRA database via BioProject accession number PRJNA594295.