Abstract
Early life adversity is associated with long-term effects on physical and mental health later in life, but the mechanisms are yet unclear. Epigenetic mechanisms program cell-type-specific gene expression during development, enabling one genome to be programmed in many ways, resulting in diverse stable profiles of gene expression in different cells and organs in the body. DNA methylation, an enzymatic covalent modification of DNA, has been one of the principal epigenetic mechanisms investigated. Emerging evidence is consistent with the idea that epigenetic processes are involved in embedding the impact of early-life experience in the genome and mediating between social environments and later behavioral phenotypes. Whereas there is evidence supporting this hypothesis in animal studies, human studies have been less conclusive. A major problem is the fact that the brain is inaccessible to epigenetic studies in humans and the relevance of DNA methylation in peripheral tissues to behavioral phenotypes has been questioned. In addition, human studies are usually confounded with genetic and environmental heterogeneity and it is very difficult to derive causality. The idea that epigenetic mechanisms mediate the life-long effects of perinatal adversity has attractive potential implications for early detection, prevention, and intervention in mental health disorders will be discussed.
Keywords: early life stress, perinatal stress, DNA methylation, epigenetics, early life adversity, NR3C1, glucocorticoid
Abstract
Si bien la adversidad en los inicios de la vida se asocia con efectos a largo plazo en la salud física y mental, los mecanismos aún no están claros. Los mecanismos epigenéticos programan durante el desarrollo la expresión génica específica de un tipo celular, lo que permite que un genoma se programe de muchas maneras, dando así como resultado diversos perfiles estables de expresión génica en diferentes células y órganos del cuerpo. Uno de los principales mecanismos epigenéticos investigados ha sido la metilación del ADN, una modificación enzimática covalente del ADN. La reciente evidencia es consistente con la idea que los procesos epigenéticos están involucrados en la incorporación del impacto de la experiencia de los inicios de la vida en el genoma y en la mediación entre ambientes sociales y fenotipos conductuales posteriores. Aunque la evidencia respalda esta hipótesis en estudios con animales, en humanos los resultados han sido menos concluyentes. Un problema importante es el hecho que el cerebro humano es inaccesible para los estudios epigenéticos y que se ha cuestionado.la relevancia de la metilación del ADN en los tejidos periféricos para los fenotipos conductuales. Además, los estudios en humanos suelen confundirse con la heterogeneidad genética y ambiental, y es muy difícil derivar la causalidad. La idea que los mecanismos epigenéticos median los efectos de la adversidad perinatal durante toda la vida tiene interesantes implicaciones potenciales para la detección temprana, la prevención y la intervención en los trastornos de salud mental que se discutirán.
Abstract
Les épreuves précoces de la vie sont associées à des effets à long terme sur la santé mentale et physique mais les mécanismes n’en sont toujours pas compris. Des mécanismes épigénétiques programment l’expression génique spécifique d’un type cellulaire au cours du développement permettant à un génome d’être programmé de diverses façons, ce qui entraîne divers profils stables d’expression génique dans différents organes et cellules du corps. La méthylation de l’ADN, une modification enzymatique covalente de l’ADN, est l’un des principaux mécanismes épigénétiques étudiés. D’après de nouvelles données, des processus épigénétiques jouent un rôle dans l’intégration de l’impact des expériences précoces de la vie dans le génome et dans la médiation entre l’environnement social et les phénotypes tardifs de comportement. Cette hypothèse est soutenue par des données issues d’études sur les animaux, mais les études sur les humains sont moins concluantes. L’inaccessibilité du cerveau aux études épigénétiques chez l’homme représente un problème majeur. Et la pertinence de la méthylation de l’ADN dans les tissus périphériques pour expliquer les phénotypes comportementaux a été remise en question. De plus, la causalité est très difficile à déduire car les études sur l’homme sont couramment faussées par une hétérogénéité génétique et environnementale. L’hypothèse que des mécanismes épigénétiques sous-tendent les effets à long terme du stress périnatal a des implications potentielles prometteuses dans la détection précoce, la prévention et le traitement des troubles mentaux. Elles seront discutées ici.
Introduction
Early life stress and social adversity have been associated with behavioral disorders later in life. 1 What are the mechanisms that mediate between experiences early in life and changes in stable phenotype later in life? Since these phenotypes are apparent a long time after the early experience, the changes in gene expression programming must be stable. It is postulated here that epigenetic mechanisms which evolved to confer cell-type-specific gene expression during embryonal and postnatal development are also involved in conferring experiential-specific gene expression profiles, mediating the phenotypic consequences of early life stress.
Epigenetic mechanisms
Epigenetic mechanisms explain how identical genes are differentially expressed in space and time in different cell types in the same individual. Epigenetic mechanisms establish cell-type-specific long-term states of gene expression during embryonal and postnatal development. 2 The focus in this review is on DNA methylation, which is a chemical covalent modification of the genetic “hardware” itself and has been the most studied epigenetic mechanism in relation to early-life stress.
DNA methylation and hydroxymethylation
Both adenine and cytosine bases in DNA could be enzymatically modified by DNA methylation. The methyl donor for the reaction is S-adenosyl methionine (SAMe). 3 The role of cytosine methylation in gene regulation has been extensively studied in mammalian DNA 4 while the presence of N(6)-methyladenine in vertebrates and human DNA was discovered recently and its role in gene regulation and cellular differentiation is yet unclear. 5 In vertebrates, DNA methylation is prevalent in the dinucleotide CG, which is a palindromic sequence (5’CG3’/5’GC3’). 6 This enables semiconservative replication of DNA methylation from the template DNA (5’CG3’) to the nascent DNA (5’GC3’) by the DNA methyltransferase 1 (DNMT1) enzyme, a maintenance DNMT1. 7 The maintenance of DNA methylation in dividing cells implies that a new methylation introduced into a CG sequence in DNA will be inherited through many cell divisions. A different set of enzymes, de novo methyltransferases DNMT3a and DNMT3b, introduce methyl groups to DNA irrespective of the DNA methylation state of the template, 8 , 9 which will be maintained in following cell divisions by the maintenance DNMT1. Thus, a change in DNA methylation in response to a transient signal introduced by early life stress can be “memorized” in the genome and serve as a genomic memory of the functional state of the gene. Cell-specific DNA methylation patterns are formed during development, are involved in programming cell-type-specific gene expression profiles, 3 and are essential for vertebrate development. 10 , 11 Examination of methylation patterns of genes in different tissues revealed an inverse correlation between DNA methylation of promoter regions and gene activity. 11 , 12 A recent survey of the state of methylation of all promoters in the brain that were physically engaged in active transcription revealed that all active promoters are unmethylated without exception. 13 In contrast, methylation in gene bodies containing the coding regions of the gene, the exons, and the introns seems to positively correlate with gene expression. 14 Interestingly, recently discovered N(6)-Methyladenine is poised at exons of transcribed genes and positively correlates with gene expression. 5
The methyl moiety on cytosine is further modified by oxidation to 5 hydroxymethyl-cytosine, 5-formylcytosine and 5-carboxylcytosine by ten-eleven translocation (TET 1-3) monooxygenases. 15 The role that these modifications play in controlling gene expression is unclear; however, it is plausible that these oxidized modifications of the methyl moiety provide further fine tuning of epigenetic regulation of genes and are implicated in gene activation during development. 16
DNA methylation plays a causal role in regulation of gene expression
The inverse correlation between DNA methylation of promoters and transcription suggested that DNA methylation in promoters silences gene expression. 11 The main evolutionary lesson derived from studying bacterial restriction-modification systems was that DNA methylation interfered with the interaction of proteins and their recognition elements in DNA; bacterial restriction enzymes did not cleave their recognition sequence when it was methylated. 17 Similarly, it was shown that DNA methylation interferes with binding of transcription factors to their methylated recognition sequence, resulting in inhibition of transcription initiation. 18 DNA methylation also attracts a specific class of proteins to methylated DNA; methylated DNA binding proteins. These proteins such as MeCP2 recruit chromatin inactivation complexes to promoter resulting in a silencing chromatin configuration. 19 Three lines of evidence support a causal role for DNA methylation in gene expression. First, in vitro methylation of promoters silences reporter gene expression when transfected into cells. 20 Second, treating cells with DNA methylation inhibitor 5-azacytidine activates gene expression. 21 Third, knockdown of DNMT1 in cells 22 or genetic knockout in mice results in changes in gene expression. 10 There is still no evidence that demethylation of a specific set of sites can induce gene expression.
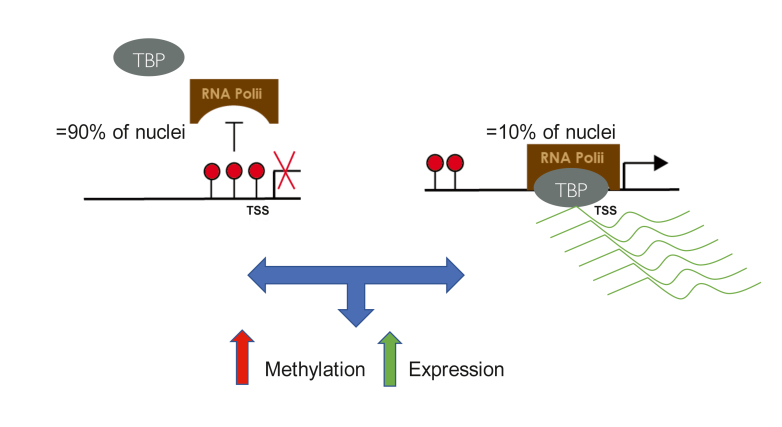
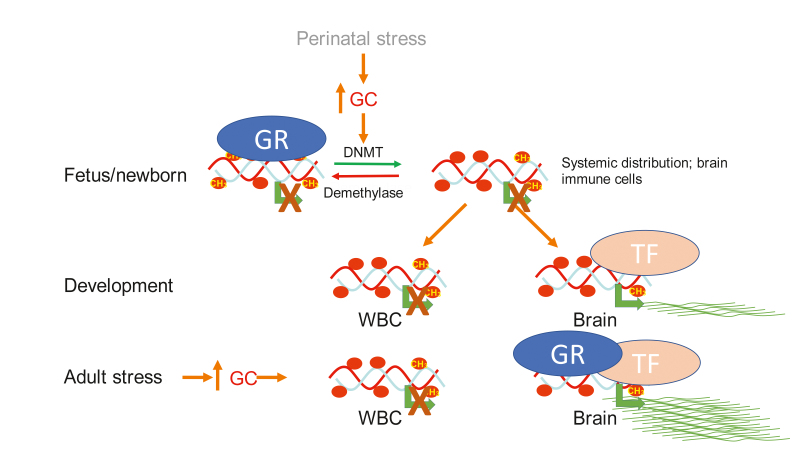
However, recent genome-wide correlations between promoter methylation and gene expression reveal a modest inverse correlation between gene expression and DNA methylation of promoters. 13 It should be noted that the simplistic idea that DNA methylation in promoters is just a mere inverse image of steady-state mRNA levels that dominates the literature is inconsistent with the data and the complexity of epigenetic regulation. First, DNA methylation controls transcription turn-on while most gene expression studies measure steady-state mRNA. Second, expression and DNA methylation are heterogeneous in almost any population of cells. High expression might be occurring in a small fraction of cells that are hypomethylated, while the majority of cells are methylated and silenced. In such a case overlaying methylation and expression, as is commonly done, provides a misleading picture of a highly methylated gene that is highly expressed. 13 RNApolII that is phosphorylated at serine 5 (PS5) is the form of RNApolII present on promoters that is turning on transcription. 23 Bisulfite mapping of PS5 bound DNA and input DNA revealed that promoters engaged in transcription were invariably unmethylated. Even when the overall methylation of a promoter at a brain sample was high, PS5 bound DNA was invariably unmethylated ( Figure 1 ). 13 Thus, the methylation level of a promoter provides a digital count of the fraction of cells in a population where this promoter is silenced. Third, although loss of methylation in a promoter is necessary for expression, it is not sufficient. Demethylation removes a barrier for expression, but expression might be realized at the right time or context when the needed factors or signals are present. 24 For example, prenatal demethylation of an enhancer of the tyrosine aminotransferase gene in the liver programs the gene to respond to glucocorticoids stimulus postnatally. 25 This is important for our discussion of how early life stress triggers phenotypes in adulthood. Gene expression is responsive to time- and context-dependent signals such as neurotransmitters or hormones. DNA methylation anticipates future transcriptional response to triggers; comparing steady-state expression with DNA methylation does not capture the full meaning and scope of the regulatory roles of differential methylation ( Figure 2 ). 13
Although this review focuses on DNA methylation and no other epigenetic modifications, it should be noted that DNA methylation and chromatin modifications are interrelated. For example, histone acetylation at H3K9 is often correlated with regions of DNA hypomethylation and both are associated with gene expression (for a review see ref 26). Histone methylation at H3K4, which is a marker of active promoters, inhibits binding of DNA methylating enzymes DNMT3A and DNMT3B, thus keeping these active regions hypomethylated. On the other hand, histone methylation at H3K36 which occurs at actively transcribed gene bodies serves to recruit DNA methylating enzymes DNMT3A and 3B to methylated gene bodies of actively transcribed genes (for a review see ref 27).
Figure 1. The same gene could be methylated and silenced in most neurons and unmethylated and highly transcriptionally active in few neurons. Measuring DNA methylation and mRNA expression from such a sample will show high methylation and high expression. However, truly all the expression that we measure comes from a few cells, while in most cells in the sample the gene promoter is methylated and expression is silenced. RNApolII, RNA polII transcription initiation complex; horizontal arrow, transcription; red balloons, methylated sites; horizonal curved lines, mRNA molecules.
Figure 2. Experience triggered, systemic epigenetic alterations anticipates response to future signals. A model for epigenetic programming by early life stress. Perinatal stress perceived by the brain triggers release of glucocorticoids (GC) from the adrenal in the mother prenatally or the newborn postnatally. GC activate nuclear glucocorticoid receptors across the body, which epigenetically program (demethylate) genes that are targets of GR in brain and white blood cells (WBC). The demethylation events are insufficient for activation of these genes. A-brain specific factor (TF) is required for expression and will activate low expression of the gene in the brain but not in blood. During adulthood a stressful event transiently triggers a very high level of expression of the GR regulated gene specifically in the brain. Horizontal arrow, transcription; circles, CpG sites; CH3 in circles, methylated sites; empty circles, unmethylated CpG sites; horizonal curved lines, mRNA.
Evidence for epigenetic programming by early life social environment; the role of the mother
The first evidence that DNA methylation might be mediating long-term programming of DNA function in response to time-limited social exposure came from studies of the effects of differences in maternal care on life-long stress responsivity in rats. Animals that were reared by a high licking and grooming mother exhibited a more measured stress responsiveness than animals reared by a low licking and grooming mother. 28 The differences in stress behavior between offspring of high and low maternal care were not genetically determined, since this phenotype was transferred by a foster mother as well as the biological (genetic) mother. 28 Differences in maternal care are associated with differences in DNA methylation, histone acetylation, transcription factor binding at the glucocorticoid hormone receptor ( NR3C1 ) gene promoter, and expression in the hippocampus of the adult offspring. 29 Animals that received high maternal care exhibited increased H3K9 acetylation, reduced DNA methylation, and increased expression of the glucocorticoid receptor gene in the hippocampus in comparison with the low-care animals. H3K9 acetylation is a hallmark of active genes and there is a close bilateral relation between increased histone acetylation and reduced DNA methylation. 26
Epigenetic marks are laid down and maintained by enzymes that either add or remove epigenetic modifications and are therefore potentially reversible in contrast to genetic changes. There is therefore a potential therapeutic angle to the discovery of a causal epigenetic driver of behavioral phenotypes. 30 Indeed, treatment of adult rats with methionine, an upstream source of methyl moieties for methylation reactions including DNA methylation, reversed the behavior of high maternal care offspring to a behavior characteristic of low maternal care offspring, while trichostatin A (TSA), a histone deacetylase inhibitor (HDACi), which elevated histone acetylation, triggered loss of DNA demethylation 31 and activation of the glucocorticoid receptor reversed the behavior of offspring of low maternal care, which became similar to offspring of high maternal care mothers. 29 , 32 These studies support a causal role for epigenetic programming by maternal behavior and point to potential epigenetic therapeutics for behavioral/psychiatry disorders. 30
These early studies also provided a plausible mechanism and a working paradigm for further exploration of the molecular links between experience and epigenetic reprogramming. Stimulation of a serotoninergic cAMP-dependent signaling pathway in the hippocampus of offspring by maternal care leads to activation of the transcription factor nerve growth induced factor A (NGFIA), 33 which in turn recruits epigenetic factors such as the histone acetyl transferase (CREB binding protein) CBP 33 and the methylated DNA binding domain protein MBD2 34 to the Nr3c1 gene resulting in epigenetic programming of the gene.
Early life adversity and DNA methylation in animal models
Animal studies confirmed DNA methylation programming by early life experience. For example, methylation of brain-derived nerve growth factor (BDNF) is altered in response to an abusive caretaker in the prefrontal cortex, 35 and arginine vasopressin ( Avp ) is demethylated in the paraventricular nucleus in response to early life stress. 36 Maternal separation in two strains of mice triggered sex and strain specific anxiety and heightened stress responsivity later in life and DNA methylation differences in Nr3C1 , Avp , and Nrda4 promoter regions. 37 Avp, Nr3c1, and Nr4a1 are involved in regulation of the HPA axis: the observed changes in methylation potentially affect regulation of expression of these genes and as a consequence the HPA axis. Prenatal stress in rats was shown to affect DNA methylation and expression in the hippocampus and prefrontal cortex of the Glycoprotein M6A ( Gpm6a ) gene, which is involved in filopodium motility and possibly synaptic extension. 38 Animal models were also used to link prenatal stress and schizophrenia-like phenotypes later in life in mice such as deficits in social interaction, prepulse inhibition, and fear conditioning. 39 In these animals DNMT1 and 3a and hydroxy-methylation and methylation in the reelin and Gad67 promoters are elevated in GABAergic neurons in the frontal cortex. 39 The effects of reelin and GAD 67 would be consistent with impaired GABA-ergic inhibition, which is seen in models of schizophrenia.
Early life stress impacts a broad transcriptome and DNA methylation landscape in brain and other tissues; alterations in DNA methylation follow a developmental trajectory and are sex-specific
Maternal care affects a wide transcriptomic landscape in the hippocampus 40 and broad genomic regions including the entire protocadherin α, β, and γ gene families. 41 Several of the protocadherin genes are associated with neurodevelopment and neuropsychiatric disorders 42 consistent with functional implications for these epigenetic alterations. The response to early life stress and maternal behavior is also not limited to the brain and involves at least the immune system as well. Studies of rhesus macaques revealed differentially methylated regions in both T cells and prefrontal cortex in monkeys who were separated from their mothers after birth. 43 There was a small overlap between differentially methylated regions in brain and immune cells; however, there were notably many tissue specific differentially methylated sites. 43 The immune system has been long known to be impacted by early life stress in both animal models and in humans. 44 , 45 The response to behavioral experiences, stress, and adversity early in life is system-wide and is not limited to the brain, while most probably driven by sensing mechanisms in the brain. Several lines of study suggest a bilateral dialogue between immune and neuroendocrine functions in humans and animals and a crosstalk which affects behavior immunity and inflammation. 46
The placenta is also impacted by maternal social experience and early life stress. Maternal social rank differences in nonhuman primates were associated with broad changes in DNA methylation in placentae. 47 The changes in DNA methylation that were associated with maternal rank found in placentae overlapped with gene expression alterations associated with social rank in adult rhesus monkeys discovered in a different, independent study. 48 These cross-sectional analyses are consistent with the hypothesis that epigenetic markers of early social experience in placentae are predictive of changes in expression later in life. The placenta is potentially a unique resource for early life biomarkers of psychiatric and immunological risks. This hypothesis was recently tested in a genetic model of glucocorticoid depletion in mice. Nr3c1-/+ mice exhibit sex-specific differences in DNA methylation in the placentae, which predicted anxiety behavior in adulthood. 49
In rhesus macaques, DNA methylation profiles dynamically evolve from birth to adolescence and are sex-dependent. 50 Maternal deprivation alters the normal evolution of DNA methylation profiles. Early DNA methylation differences in maternally deprived macaques anticipate later changes in methylation. 50 This might explain how early life experiences impact phenotypes at a later time point in life.
Evidence for epigenetic programming by early life stress in humans
In comparison with animal studies, it is more difficult to obtain evidence in humans for associations of DNA methylation with early life stress. Heterogeneity of human populations results in confounding genetic and environmental factors. Moreover, since the brain is inaccessible for DNA methylation analysis in living humans, most studies are limited to peripheral tissues such as saliva and white blood cells, and the relevance to brain physiology and pathology is uncertain. Obtaining any causal evidence or mechanistic insights linking DNA methylation alterations with physiological function is difficult in human studies, and even temporal relationships between exposure, DNA methylation, and phenotypic outcome mostly rely on cross-sectional design. The low absolute differences in methylation seen in most human behavioral EWAS raise questions about their biological significance.
Several approaches were used to overcome some but not all these inherent difficulties. First, studies used an evolutionary approach and examined whether epigenetic alterations in human post-mortem brain samples replicated observations in rodents and nonhuman primate experiments. For example, the region of the Nr3c1 gene that was differentially methylated by maternal care in rat hippocampus was also differentially methylated by child abuse in post-mortem hippocampi of humans. 51 This overlap extended well beyond the Nr3c1 gene, and a broad evolutionary conserved response was observed across a syntenic locus that covers the Nr3c1 gene and the protocadherin alpha, beta, and gamma gene families. 41
Although post-mortem studies examine epigenetic programming in physiologically relevant tissues, they represent only a final and single stage that does not capture the dynamic evolution of environments and epigenetic programming in living humans. This is possible only in peripheral tissues such as blood and saliva. Studies have examined in the last decade alterations in DNA methylation in candidate genes in response to maternal stress, early life stress, and early life trauma including the NR3C1 52 , 53 exon 1f region, which corresponds to the region that is altered in hippocampi of rats, the proximal regulator of glucocorticoid receptor FKBP5 , 54 BDNF , 55 OXTR , 56 and the serotonin transporter SLC6A4 . 57 Low socioeconomic status in childhood was associated with increased methylation in AVP , FKBP5 , and OXTR , and two inflammation-related genes CCL1 and CD1D . 58 DNA methylation differences in CD1D and FKBP5 were associated with expression differences and these changes in expression were negatively correlated with changes in DNA methylation. 58
A third approach was to combine a human blood study with cross-species and cross-tissue (brain/blood) comparisons at multiple time points. Using this approach, the Morc1 gene was found to be differentially methylated in CD34+ cells from cord bloods from children who were exposed to prenatal stress compared with controls, in CD3+ T cells from newborn and adolescent monkeys and in the prefrontal cortex of adult rats. 59 Morc1 was also associated with major depressive disorder (MDD) in a gene-set-based analysis of data from a genome-wide association study, 59 possibly pointing to a link between epigenetic programming by early life stress and psychiatric disorders later in life.
A fourth approach was to combine blood/saliva DNA methylation with brain imaging. Methylation of the serotonin transporter gene Slc6a4 correlated with serotonin synthesis in the orbitofrontal cortex measured with positron emission tomography (PET) and childhood physical aggression. 60
Broad signature of early life experience in DNA methylation
The epigenetic response associated with early life stress has a broad footprint in DNA methylation in blood 61 , 62 and brain. 63 DNA methylation measured in 40 adults enrolled in the 1958 British Birth Cohort showed association with early life socioeconomic status in multiple loci 64 in blood. Socioeconomic status differences were associated with DNA methylation differences in nonhuman primates 47 and humans in placentae at birth. 65 Childhood abuse was associated with methylation in multiple loci in DNA 66 and multiple changes in DNA methylation were observed in a study that compared people who suffered from post-traumatic stress disorder (PTSD) who were also exposed to childhood adversity with those exposed only to adult trauma. 62 Prenatal maternal stress 59 was associated with differential methylation in multiple loci in CD34+ cells from cord blood. However, the functional meaning of small epigenetic changes in methylation in blood in multiple loci is yet unclear.
In contrast to this line of studies that showed consistent but nevertheless small differences in DNA methylation in candidate genes and genome wide analyses, a recent study shed doubt on the association between childhood trauma and DNA methylation changes in blood. 67 The study analyzed the largest number of samples to date and found no evidence for association of DNA methylation in blood with childhood trauma, even in candidate genes that were shown in multiple studies to associate with early life adversity, after correction for confounding factors such as cigarette smoking. The study compels us to replicate and revisit this question. 67
Natural disasters, a quasi-experimental design for examining causal relations between stress and epigenetic programming
The Quebec 1988 ice storm provided an opportunity to examine the impact of randomized objective stress of mothers on children who were born around the time of the storm. An epigenome-wide association study of T cells obtained from these children 13 years later revealed many changes in DNA methylation that correlated with maternal objective stress, 68 while mothers’ cognitive appraisal of the stress associated with a different set of DNA methylation alterations. 69 Analysis of the functional gene networks that were altered with perinatal maternal stress showed predominantly genes involved in immunity and inflammation, as well as the insulin pathway. 68
Consistent with the functional network analysis, DNA methylation was found to mediate the effects of prenatal maternal objective stress 70 and mother’s cognitive appraisal 69 on BMI and obesity in the children, the effect of cognitive appraisal on child’s C-peptide in adolescence (a measure of endogenous insulin secretion), 71 as well as the effect of objective prenatal stress on cytokine production in the children. 72 These data are consistent with a causal link between prenatal stress, DNA methylation in immune cells, and metabolic and immune system phenotypes.
What are the mechanisms that translate prenatal stress into DNA methylation alterations in multiple tissues?
A likely suspect to mediate a system-wide epigenetic response to stress is the glucocorticoid stress hormone. Glucocorticoids are released in response to social stress, are distributed systemically, have nuclear receptors that are distributed across the body, and are epigenetic modulators. Other endocrine molecules are also candidates.
In support of this hypothesis, prenatal synthetic glucocorticoid exposure in the guinea pig triggered broad alterations in DNA methylation, histone acetylation, and gene expression in the fetal hippocampus. 73 , 74 Further evidence for the involvement of glucocorticoid pathway came from examining the impact of fetal hemizygous depletion of Nr3c1 on DNA methylation in placenta at birth in mice. Nr3c1 heterozygosity leads to altered DNA methylation in a sex-specific manner across multiple sites in the placenta 49 ; DNA methylation of several genes in the placenta correlated with anxiety-like behavior in adults, suggesting a temporal relationship between DNA methylation alterations and development of behavioral phenotypes. 49
A recent study provided evidence for this mechanism in human cells. Exposure of human hippocampal neuron progenitors to glucocorticoids resulted in long-lasting broad changes in DNA methylation, which were not associated with steady-state gene expression. However, re-exposure of the neurons to glucocorticoids resulted in an enhanced response. 75 This is consistent with the idea that methylation defines the potential responsivity of the gene to future triggers. Differential methylation in glucocorticoid-treated hippocampal neurons overlapped with transcriptional response to glucocorticoids in peripheral blood and a polyepigenetic score computed from differentially methylated sites in neurons predicted exposure to prenatal glucocorticoids in newborn cord blood. 75 These data further confirm correspondence of brain blood DNA methylation responses to glucocorticoids.
Summary and perspectives
Animal studies have established plausible molecular pathways between perinatal social exposures, DNA methylation changes, and phenotype. Human studies have been criticized, however, for the small differences in DNA methylation, the small sample sizes, and confounding genetic heterogeneity and other common environmental exposures such as age, smoking, and alcohol. Indeed, a recent large epigenome-wide methylation study failed to confirm previously documented associations with early life trauma, casting doubt on previously described associations in peripheral tissues. 67
One of the most criticized facets of studies of human early adversity epigenetics is the use of blood or white blood cells for studying behavior, a brain function. Inaccessibility of the brain in living humans precludes, except for post-mortem studies, examination of “functionally relevant” epigenetic profiles. Studies therefore examined blood or saliva DNA as a surrogate tissue. The assumption is that certain DNA methylation profiles show the same interindividual differences in blood and brain or saliva and brain. Several studies have delineated such sites, 76 , 77 and some studies use these comparisons as a guide. Methylation differences in “brain-specific genes” were reported in blood in several human studies. However, it is unclear whether corresponding changes exist in the brain.
Animal studies and experiments in human hippocampal neurons provide a plausible mechanism linking perinatal stress and changes in DNA methylation across several tissues. Involvement of the stress hormone in triggering these changes could explain how a stress sensed by the brain can have a systemic effect across several tissues and target overlapping sites in different tissues, since glucocorticoids have nuclear receptors in many tissues and overlapping glucocorticoid recognition elements in different tissues. It is possible then that DNA methylation changes only poise genes for expression, 24 while the presence of a brain-specific transcription factor activates the gene in the brain. Therefore, although changes in methylation in brain specific genes in response to early life stress happen in several tissues, they will be functionally relevant only in the brain. Thus, absence of correlation between DNA methylation alterations and steady-state expression in peripheral tissues does not necessarily imply that the changes in DNA methylation are functionally irrelevant. In addition, even in the brain, DNA methylation alterations might be programming genes for response to future triggers such as glucocorticoids or neuronal activation, and this might not be reflected in steady-state transcription ( Figure 2 ).
Questions remain on the functional meaning of the changes in DNA methylation for either brain function or peripheral physiological functions. These could be addressed by linking the genes affected to central phenotypes using imaging and mediation analyses in humans, as was described above, as well as pharmacology, targeted gene depletion, or epigenetic editing in animals or primary human hippocampal cultures. Studies of natural disasters provide quasi-experimental designs to draw a causal link between stress and changes in epigenetic states and mediation analyses define a causal link between early life stress, DNA methylation changes, and the phenotype. Although the Quebec ice storm study provides preliminary support for a causal link between perinatal stress, DNA methylation, and phenotypic changes in adults, further similar studies are needed. One of the main difficulties in relating DNA methylation alterations to phenotype is how to link DNA methylation changes in multiple genes with a specific phenotype, since current methods in genetics and pharmacology focus on specific targets.
Although the focus in the field is mainly on examining “brain relevant” DNA methylation marks in peripheral tissue, it is suggested here that the response to stress is systemic and that the immune system and brain are interactive partners in a coordinated developmental and physiological response. Moreover, early life stress is known to impact immune, 78 cardiovascular, and metabolic disorders. 79 DNA methylation alterations in the immune system are therefore an important component of the system-wide response to perinatal stress. Examining how peripheral changes in DNA methylation mediate changes in immunity and inflammatory systems and how they interact with the brain are critical for a comprehensive understanding of epigenetic programming by early life stress and its functional consequences.
Acknowledgments
MS is funded by the Canadian Institute of Health Research PJT-159583. The author has no conflicts of interest to disclose.
REFERENCES
- 1.Syed SA, Nemeroff CB. Early life stress, mood, and anxiety disorders. Chronic Stress (Thousand Oaks) 2017 doi: 10.1177/2470547017694461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. Embo J. 1998;17(17):4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 4.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–778. doi: 10.1016/S0140-6736(18)31268-6. [DOI] [PubMed] [Google Scholar]
- 5.Xiao CL, Zhu S, He M. et al N(6)-methyladenine DNA modification in the human genome. Mol Cell. 2018;71(2):306–318. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Gruenbaum Y, Stein R, Cedar H, Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- 7.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh CL. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol Cell Biol. 1999;19(12):8211–8218. doi: 10.1128/mcb.19.12.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases [letter] Nat Genet. 1998;19(3):219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 10.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 11.Szyf M, Avraham-Haetzni K, Reifman A, et al DNA methylation pattern is determined by the intracellular level of the methylase. Proc Natl Acad Sci U S A. 1984;81(11):3278–3282. doi: 10.1073/pnas.81.11.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, Pelizzola M, Dowen RH, et al Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massart R, Suderman M, Mongrain V, Szyf M. DNA methylation and transcription onset in the brain. Epigenomics. 2017;9(6):797–809. doi: 10.2217/epi-2016-0184. [DOI] [PubMed] [Google Scholar]
- 14.Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829(11):1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Ito S, Shen L, Dai Q, et al Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szyf M. The elusive role of 5’-hydroxymethylcytosine. Epigenomics. 2016;8(11):1539–1551. doi: 10.2217/epi-2016-0076. [DOI] [PubMed] [Google Scholar]
- 17.Arber W, Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 18.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18(13):3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan X, Ng HH, Johnson CA, et al Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex [see comments] Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 20.Vardimon L, Kressmann A, Cedar H, Maechler M, Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc Natl Acad Sci U S A. 1982;79(4):1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PA, Taylor SM, Mohandas T, Shapiro LJ. Cell cycle-specific reactivation of an inactive X-chromosome locus by 5- azadeoxycytidine. Proc Natl Acad Sci U S A. 1982;79(4):1215–1219. doi: 10.1073/pnas.79.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szyf M, Rouleau J, Theberge J, Bozovic V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. J Biol Chem. 1992;267(18):12831–12836. [PubMed] [Google Scholar]
- 23.Sogaard TM, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282(19):14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 24.Cheishvili D, Christiansen S, Stochinsky R, et al DNA methylation controls unmethylated transcription start sites in the genome in trans. Epigenomics. 2017;9(5):611–633. doi: 10.2217/epi-2016-0141. [DOI] [PubMed] [Google Scholar]
- 25.Thomassin H, Flavin M, Espinas ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20(8):1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Alessio AC, Szyf M. Epigenetic tete-a-tete: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol. 2006;84(4):463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 28.Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286(5442):1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 29.Weaver IC, Cervoni N, Champagne FA, et al Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 30.Szyf M. Prospects for the development of epigenetic drugs for CNS conditions. Nat Rev. 2015;14(7):461–474. doi: 10.1038/nrd4580. [DOI] [PubMed] [Google Scholar]
- 31.Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276(44):40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 32.Weaver IC, Champagne FA, Brown SE, et al Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25(47):11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver IC, D’Alessio AC, Brown SE, et al The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27(7):1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver IC, Hellstrom IC, Brown SE, et al The methylated-DNA binding protein MBD2 enhances NGFI-A (egr-1)-mediated transcriptional activation of the glucocorticoid receptor. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652) doi: 10.1098/rstb.2013.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murgatroyd C, Patchev AV, Wu Y, et al Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 37.Kember RL, Dempster EL, Lee TH, Schalkwyk LC, Mill J, Fernandes C. Maternal separation is associated with strain-specific responses to stress and epigenetic alterations to Nr3c1, Avp, and Nr4a1 in mouse. Brain Behav. 2012;2(4):455–467. doi: 10.1002/brb3.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monteleone MC, Adrover E, Pallares ME, Antonelli MC, Frasch AC, Brocco MA. Prenatal stress changes the glycoprotein GPM6A gene expression and induces epigenetic changes in rat offspring brain. Epigenetics. 2014;9(1):152–160. doi: 10.4161/epi.25925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matrisciano F, Tueting P, Dalal I, et al Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–194. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103(9):3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suderman M, McGowan PO, Sasaki A, et al , Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus Proc Natl Acad Sci U S A. 2012;109(suppl 2):17266. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14(5):557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 43.Provencal N, Suderman MJ, Guillemin C, et al The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T Cells. J Neurosci. 2012;32(44):15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lubach GR, Coe CL, Ershler WB. Effects of early rearing environment on immune responses of infant rhesus monkeys. Brain Behav Immun. 1995;9(1):31–46. doi: 10.1006/brbi.1995.1004. [DOI] [PubMed] [Google Scholar]
- 45.Schlewinski E. Studies on the influence of psychological factors on the immune system: the result of infantile stimulation on the disease course in neonatal mice with bacterial infection. Z Psychosom Med Psychoanal. 1975;21(4):390–399. [PubMed] [Google Scholar]
- 46.Bottaccioli AG, Bottaccioli F, Minelli A. Stress and the psyche-brain-immune network in psychiatric diseases based on psychoneuroendocrineimmunology: a concise review. Ann N Y Acad Sci. 2019;1437(1):31–42. doi: 10.1111/nyas.13728. [DOI] [PubMed] [Google Scholar]
- 47.Massart R, Suderman MJ, Nemoda Z, et al The signature of maternal social rank in placenta deoxyribonucleic acid methylation profiles in rhesus monkeys. Child Dev. 2017;88(3):900–918. doi: 10.1111/cdev.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tung J, Barreiro LB, Johnson ZP, et al Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci U S A. 2012;109(17):6490–6495. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt M, Lax E, Zhou R, et al Fetal glucocorticoid receptor (Nr3c1) deficiency alters the landscape of DNA methylation of murine placenta in a sex-dependent manner and is associated to anxiety-like behavior in adulthood. Transl Psychiatry. 2019;9(1):23. doi: 10.1038/s41398-018-0348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massart R, Nemoda Z, Suderman MJ, et al Early life adversity alters normal sex-dependent developmental dynamics of DNA methylation. Dev Psychopathol. 2016;28(4pt2):1259–1272. doi: 10.1017/S0954579416000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGowan PO, Sasaki A, D’Alessio AC, et al Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perroud N, Paoloni-Giacobino A, Prada P, et al Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cicchetti D, Handley ED. Methylation of the glucocorticoid receptor gene, nuclear receptor subfamily 3, group C, member 1 (NR3C1), in maltreated and nonmaltreated children: Associations with behavioral undercontrol, emotional lability/negativity, and externalizing and internalizing symptoms. Dev Psychopathol. 2017;29(5):1795–1806. doi: 10.1017/S0954579417001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klengel T, Mehta D, Anacker C, et al Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Unternaehrer E, Luers P, Mill J, et al Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF ) after acute psychosocial stress. Transl Psychiatry. 2012;2:e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unternaehrer E, Meyer AH, Burkhardt SC, et al Childhood maternal care is associated with DNA methylation of the genes for brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) in peripheral blood cells in adult men and women. Stress. 2015;18(4):451–461. doi: 10.3109/10253890.2015.1038992. [DOI] [PubMed] [Google Scholar]
- 57.Provenzi L, Giorda R, Beri S, Montirosso R. SLC6A4 methylation as an epigenetic marker of life adversity exposures in humans: A systematic review of literature. Neurosci Biobehav Rev. 2016;71:7–20. doi: 10.1016/j.neubiorev.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 58.Needham BL, Smith JA, Zhao W, et al Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics. 2015;10(10):958–969. doi: 10.1080/15592294.2015.1085139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nieratschker V, Massart R, Gilles M, et al MORC1 exhibits cross-species differential methylation in association with early life stress as well as genome-wide association with MDD. Transl Psychiatry. 2014;4:e429. doi: 10.1038/tp.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang D, Szyf M, Benkelfat C, et al Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PLoS One. 2012;7(6):e39501. doi: 10.1371/journal.pone.0039501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Houtepen LC, Vinkers CH, Carrillo-Roa T, et al Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nat Commun. 2016;7:10967. doi: 10.1038/ncomms10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mehta D, Klengel T, Conneely KN, et al Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110(20):8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suderman MM, Sasaki PO, Huang A, et al Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci U S A. 2012;109(suppl 2):17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borghol N, Suderman M, McArdle W, et al Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41(1):62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santos HP, Bhattacharya A, Martin EM, et al Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics. 2019 doi: 10.1080/15592294.2019.1614743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suderman M, Borghol N, Pappas JJ, et al Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Med Genomics. 2014;7:13. doi: 10.1186/1755-8794-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marzi SJ, Sugden K, Arseneault L, et al Analysis of DNA methylation in young people: limited evidence for an association between victimization stress and epigenetic variation in blood. Am J Psychiatry. 2018;175(6):517–529. doi: 10.1176/appi.ajp.2017.17060693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao-Lei L, Massart R, Suderman MJ, et al DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: project ice storm. PLoS One. 2014;9(9):e107653. doi: 10.1371/journal.pone.0107653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao-Lei L, Dancause KN, Elgbeili G, Laplante DP, Szyf M, King S. Pregnant women’s cognitive appraisal of a natural disaster affects their children’s BMI and central adiposity via DNA methylation: Project Ice Storm. Early Hum Devel. 2016;103:189–192. doi: 10.1016/j.earlhumdev.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Cao-Lei L, Dancause KN, Elgbeili G, et al DNA methylation mediates the impact of exposure to prenatal maternal stress on BMI and central adiposity in children at age 13(1/2) years: Project Ice Storm. Epigenetics. 2015;10(8):749–761. doi: 10.1080/15592294.2015.1063771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao-Lei L, Dancause KN, Elgbeili G, Laplante DP, Szyf M, King S. DNA methylation mediates the effect of maternal cognitive appraisal of a disaster in pregnancy on the child’s C-peptide secretion in adolescence: Project Ice Storm. PLoS One. 2018;13(2):e0192199. doi: 10.1371/journal.pone.0192199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao-Lei L, Veru F, Elgbeili G, Szyf M, Laplante DP, King S. DNA methylation mediates the effect of exposure to prenatal maternal stress on cytokine production in children at age 13(1/2) years: Project Ice Storm. Clin Epigenetics. 2016;8:54. doi: 10.1186/s13148-016-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crudo A, Suderman M, Moisiadis VG, et al Glucocorticoid programming of the fetal male hippocampal epigenome. Endocrinology. 2013;154(3):1168–1180. doi: 10.1210/en.2012-1980. [DOI] [PubMed] [Google Scholar]
- 74.Crudo A, Petropoulos S, Suderman M, et al Effects of antenatal synthetic glucocorticoid on glucocorticoid receptor binding, DNA methylation, and genome-wide mRNA levels in the fetal male hippocampus. Endocrinology. 2013;154(11):4170–4181. doi: 10.1210/en.2013-1484. [DOI] [PubMed] [Google Scholar]
- 75.Provencal N, Arloth J, Cattaneo A, et al Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1820842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walton E, Hass J, Liu J, et al Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull. 2015;42(2):406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farre P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenetics Chromatin. 2015;8:19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slopen N, Loucks EB, Appleton AA, et al Early origins of inflammation: An examination of prenatal and childhood social adversity in a prospective cohort study. Psychoneuroendocrinology. 2015;51:403–413. doi: 10.1016/j.psyneuen.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCrory C, Dooley C, Layte R, Kenny RA. The lasting legacy of childhood adversity for disease risk in later life. Health Psychol. 2015;34(7):687–696. doi: 10.1037/hea0000147. [DOI] [PubMed] [Google Scholar]