Abstract
Drugs of abuse can modify gene expression in brain reward and motivation centers, which contribute to the structural and functional remodeling of these circuits that impacts the emergence of a state of addiction. Our understanding of how addictive drugs induce transcriptomic plasticity in addiction-relevant brain regions, particularly in the striatum, has increased dramatically in recent years. Intracellular signaling machineries, transcription factors, chromatin modifications, and regulatory noncoding RNAs have all been implicated in the mechanisms through which addictive drugs act in the brain. Here, we briefly summarize some of the molecular mechanisms through which drugs of abuse can exert their transcriptional effects in the brain region, with an emphasis on the role for microRNAs in this process.
Keywords: cocaine, addiction, striatum, microRNA, MeCP2, BDNF, transcription factor
Abstract
Las drogas de abuso pueden modificar la expresión génica en los centros cerebrales de recompensa y motivación, lo que contribuye a la remodelación estructural y funcional de estos circuitos favoreciendo la aparición de una adicción. En los últimos años ha habido un importante aumento en la comprensión acerca del modo en que las drogas adictivas inducen la plasticidad transcriptómica en las regiones cerebrales relevantes para la adicción, particularmente en el cuerpo estriado. Las maquinarias de señalización intracelular, los factores de transcripción, las modificaciones de la cromatina y los ARNs reguladores no codificadores se han implicado en los mecanismos a través de los cuales las drogas adictivas actúan en el cerebro. En este artículo se resumen algunos de los mecanismos moleculares a través de los cuales las drogas de abuso pueden ejercer sus efectos transcripcionales en el cerebro, con énfasis en el papel de los microARNs en este proceso.
Abstract
Les stupéfiants peuvent modifier l’expression génique des centres cérébraux de récompense et de motivation, ce qui participe au remodelage structurel et fonctionnel de ces circuits influant sur l’apparition d’un état d’addiction. Notre compréhension du mécanisme selon lequel les substances addictives induisent une plasticité transcriptomique dans les régions cérébrales correspondant à l’addiction, surtout dans le striatum, a considérablement évolué au cours des dernières années. La signalisation intracellulaire, les facteurs de transcription, les modifications de la chromatine et les ARN de régulation non codants interviennent tous dans les mécanismes d’action des substances addictives sur le cerveau. Nous résumons brièvement ici certains des mécanismes moléculaires qui permettent aux stupéfiants d’exercer leurs effets transcriptionnels sur le cerveau, en insistant sur le rôle des microARN dans ce processus.
Introduction
Dorsal and ventral domains of the striatum receive extensive input from midbrain dopamine neurons, and this mesoaccumbens dopamine system has been heavily implicated in addiction-relevant behavioral abnormalities. Dopamine triggers plastic responses in the so-called direct pathway (striatonigral) and indirect pathway (striatopallidal) medium spiny neurons (dMSNs and iMSNs, respectively) of the striatum. While drugs of abuse differ in their pharmacological mechanisms of action, chronic exposure can often result in common molecular adaptations in MSNs, likely related to the fact that all major addictive drugs stimulate dopamine. It has been hypothesized that regulation of gene expression by transcriptional and post-transcriptional mechanisms plays a key role in the long lasting changes in brain function by drugs of abuse that precipitate the emergence of the behavioral abnormalities that define addiction. 1 , 2 To date, investigation of the transcriptional actions of drugs of abuse have focused on only a small fraction of the transcription factors that are likely to be involved. Beyond transcription, post-transcription mechanisms of gene regulation are increasingly recognized as important regulatory factors in addiction-relevant neuronal plasticity. Below, we briefly summarize findings on some of the most thoroughly explored transcription factors implicated in the transcriptional actions of addictive drugs in the striatum. In addition, we also summarize recent findings on the contribution of microRNAs, which are post-transcriptional regulators of gene expression, to the actions of addictive drugs.
FosB
ΔFosB is a member of the Fos family of transcription factors encoded by the fosB gene. ΔFosB can heterodimerize with the Jun family of proteins to form Activator Protein 1 (AP-1) complexes, which bind to AP-1 elements in the promoters of genes that contain AP-1 response elements to regulate their transcription. ΔFosB has a truncation on its C-terminal relative to other forms of FosB, which renders it less sensitive to protein degradation and hence increases its relative stability compared with other Fos family proteins. 3 , 4 Moreover, in vitro and in vivo studies have revealed that ΔFosB phosphorylation further stabilizes it. 5 , 6 This makes it an important candidate for transcriptional regulation in the context of regulating addiction-related behaviors, as its relative stabilization and long biological half-life provides a molecular mechanism by which changes in gene expression can persist for weeks or even longer after drug consumption.
The Fos family proteins are expressed in response to exposure to all known drugs of abuse. Levels of ΔFosB are increased in various reward-related regions of the brain, especially the striatum, 2 , 3 , 7 in response to repeated consumption of addictive drugs. Induction of ΔFosB in the ventral striatum (nucleus accumbens; NAc) and dorsal striatum by drugs of abuse is thought to occur exclusively in dMSNs, 1 , 3 with this action linked to addiction-related behavioral abnormalities. Indeed, dMSN-specific overexpression of ΔFosB in NAc of mice increases sensitivity to the stimulant and reward-related properties of cocaine. 8 ‑ 10 Virus-mediated manipulation of ΔFosB in the striatum suggests that it plays a key role in many of the transcriptional changes that are triggered by addictive drugs. In addition to addictive drugs, natural rewards can also induce ΔFosB in dMSNs in NAc. 3 , 10 ‑ 12 Notably, ΔFosB has a long half-life compared with other members of the Fos family of proteins. Consequently, levels of ΔFosB accumulate in the NAc and other addiction-related brain regions in response to repeated drug use, resulting in greater levels after prolonged drug use compared with acute use. This provides a transcriptional mechanism through which chronic drug exposure has enduring actions on the function of brain reward systems that persists after the initial pharmacological actions of the drug have waned. Thus, ΔFosB is hypothesized to regulate transcriptional responses to natural rewards, with maladaptive recruitment of this transcription factor for prolonged time periods by drugs of abuse driving transcriptional plasticity that leads to addiction.
Significant progress has been made in identifying transcriptional targets through which ΔFosB influences behavioral responses to drug of abuse. These include genes related to dendritic spine structure such as synaptotagmin, activity-regulated cytoskeleton-associated protein (ARC), microtubule associated proteins (MAPs), cyclin-dependent kinase-5 (CDK5), and kinesin. 13 ‑ 15 There have been reports implicating ΔFosB in glutamatergic signaling via modulating AMPA receptors (AMPARs). 9 , 16 This is consistent with the belief that ΔFosB mediates aspects of synaptic plasticity in MSNs after drug exposure. 17 , 18 ΔFosB also serves as an important epigenetic modulator. More specifically, it can serve as a molecular switch to enhance (CDK5 expression in response to cocaine) or repress (fos in response to amphetamine) gene expression by binding to specific promoters and recruiting epigenetic modulators. 2 , 19 , 20
Taking all these studies together, it has been postulated that by regulating a number of transcriptional as well as epigenetic regulatory proteins, this transcription factor may serve as a master regulator of drug-induced changes in both structural and synaptic plasticity. 17 , 21 ‑ 23
Cyclic AMP response element binding protein
The cyclic AMP response element (CRE)-binding protein (CREB) family of transcriptional activators plays important roles in the control of cellular metabolism, growth-factor-dependent cell survival, the function of brain circuits involved in the regulation of complex physiological processes such as learning and memory, and pathophysiological processes such as anxiety and drug addiction. Consistent with these diverse functions, CREB proteins are activated by phosphorylation at Ser133 in response to a number of signaling pathways, including mitogenic stimuli, cAMP, calcium and stress. Phosphorylation of CREB facilitates its nuclear translocation and subsequent binding to the scaffolding protein CREB-regulated transcription coactivator (CRTC), which plays a key role in recruiting other transcriptional components of the coactivator complex. This interaction between CREB and CRTC promotes expression of genes that express CRE-response elements through the recruitment of coactivators such as CREB-binding protein (CBP) and p300. CRTC can be deactivated by deacetylation by class III histone deacetylases (HDACs) such as sirtuin1 (SIRT1), which decreases CREB signaling. 24
Perturbations in CREB function in the striatum are thought to be involved in addiction-related transcriptional plasticity. Recurring exposure to drugs such as cocaine increases the activity of the cAMP–PKA pathway in the NAc, 25 which induces activation of CREB-mediated transcription of target genes. 14 , 26 Work from the Nestler group has shown that CREB signaling, and major components of this signaling cascade, are important regulators of cocaine reward in the NAc and the transition to compulsive cocaine use mediated by the dorsal striatum, reflected by escalating levels of drug intake and consumption that persists despite negative consequences. Infusion of cAMP analogs that activate PKA into the NAc of rats causes a time-delayed increase in intravenous self-administration (IVSA) behavior and shift the cocaine dose-response curve to the right, consistent with decreased sensitivity to the rewarding properties of the drug. Furthermore, inhibition of PKA signaling in the NAc can shift the dose-response curve for intravenous cocaine infusions to the left, suggesting that PKA regulates the rewarding property of the drug. 27
Evidence for a direct role for CREB came from their subsequent studies where viral mediated upregulation of CREB in rat accumbens resulted in reduction in the rewarding properties, while expression of a dominant negative CREB increased the rewarding properties of cocaine. 28 In addition further studies from their group showed similar effects of CREB modulation on the rewarding properties of morphine. 29 Similar observations were also observed in mouse models activating or inhibiting CREB function.
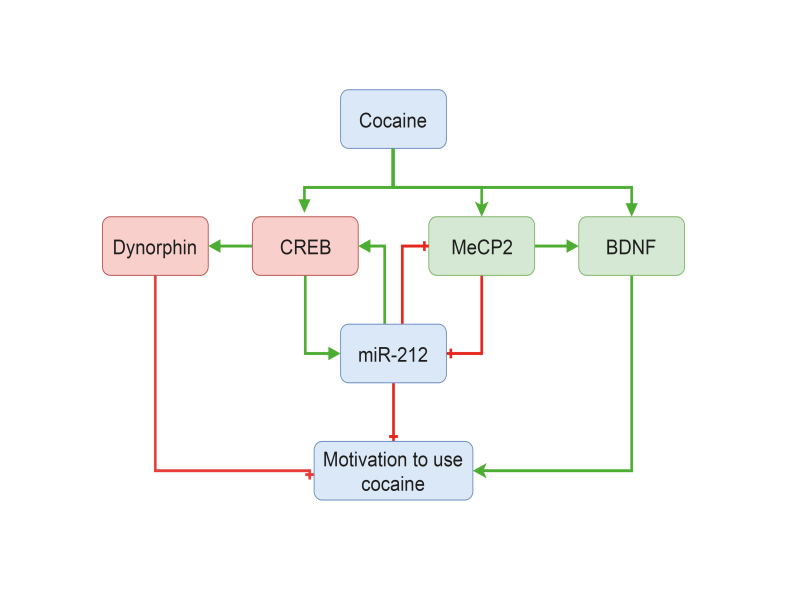
Extended (6 h) daily access to intravenous cocaine self-administration can precipitate a compulsive-like increase in consumption, a process termed escalation of intake. Rats that demonstrate escalation of cocaine intake show increased levels of phosphorylated CREB in the dorsal striatum . It has been shown that overexpression of the CREB coactivator CRTC1 in dorsal striatum blocks the emergence of escalated levels of cocaine intake in rats with extended daily access to the drug but does not alter the stable intake seen in rats with restricted (1 h) daily access. 30 This may suggest that engagement of CREB/CRTC-mediated transcription in the dorsal striatum of rats with extended access to cocaine may serve as a “protective” homeostatic response that counters the rewarding and motivational actions of cocaine. This interpretation is consistent the fact that CREB signaling in the NAc has also been shown to oppose the rewarding and motivational effects of cocaine 28 which likely reflects CREB-induced increases in the transcription of the anti-reward endogenous opioid dynorphin and related genes. In addition to CREB signaling, extended daily access to cocaine also increases levels of brain-derived neurotrophic factor (BDNF) and the transcriptional repressor methyl CpG-binding protein 2 (MeCP2) in the dorsal striatum. 31 Moreover, virus-mediated knockdown of BDNF or MeCP2 in the striatum reduced cocaine intake in rats with extended but not restricted daily access to the drug. 31 Since both BDNF and Mecp2 are CREB-responsive genes, these observations suggest that CREB, MeCP2, and BDNF may represent components of a larger transcriptional mechanism in the striatum that modulates the addiction-relevant actions of cocaine. These findings reflect the complexity of CREB signaling in the striatum, where it can stimulate the transcription of dynorphin and other anti-reward genes yet also promote the expression of BDNF and other genes that can enhance reward processes. Hence, the balance between CREB-induced increases in the expression of pro- and anti-addiction transcriptional programs likely plays a key role in determining vulnerability to, and the emergence of, drug addiction ( Figure 1 ).
In addition to MeCP2 and BDNF, CREB regulates the expression of many other genes in the striatum that are involved in synaptic plasticity, neuronal growth, and cell adhesion. These include genes involved in transmission (syntaxin1A, dynorphin), intracellular signaling (adenylyl cyclase VI), and cell growth (BDNF), as well as other gene modulators such as c-Fos and Mef2. 14 Many of these genes are very likely involved in regulating addiction-related plasticity in the striatum. For example, the genes encoding the subunits of NMDA receptors (GluN1 and GluN2B) contain a CREB-binding site, and it has been reported that in accumbens slice cultures, activation of CREB increases protein levels of both GluN1 and GluN2B subunits, but not GluN2A subunits. 32 , 33 As additional molecular targets of CREB in the striatum and other addiction-related brain areas are identified, characterization of their contribution to drug-related behaviors will further help us by providing information on how different drugs of abuse may reshape addiction-related circuits. Indeed, much work remains to be done to fully understand how CREB and related transcriptional regulators influence the emergence of drug addiction and their role in vulnerability versus resilience to this disorder.
Figure 1. Interactions between cAMP response element-binding protein (CREB), methyl CpG-binding protein 2 (MeCP2), brain-derived neurotrophic factor (BDNF) and miR-212 in the control of cocaine intake. Cocaine increases the activity or expression of CREB, MeCP2, BDNF, and miR-212 in the striatum. In turn, CREB stimulates the expression of dynorphin and other genes that serve to decrease the motivational properties of cocaine and protect against addiction. Conversely, MeCP2 and BDNF increase the motivational properties of cocaine and increase vulnerability to addiction. CREB, MeCP2, and BDNF all share reciprocal interactions with miR-212, with miR-212 modulating the activity or expression to protect against the development of compulsive cocaine use. Red lines indicate an inhibitory action on activity or expression. Green lines indicated a stimulatory action.
Myocyte enhancing factor-2
The myocyte enhancing factor-2 (MEF2) family of transcription factors is comprised of four nuclear proteins, named MEF2A-D, that were originally identified in muscle tissue. 34 , 35 Later, these transcription factors were shown to be expressed in neurons of various regions of the brain, including MSNs in the striatum. MEF2 regulates the transcription of a wide range of genes. Flavell and colleagues used a genome-wide targeting strategy to identify activity-dependent MEF2 targets, including genes that regulate synapse development and function. These genes contribute to synapse weakening (eg, Homer 1a, kcna1, and kcna4 potassium channels) as well as synapse strengthening (eg, BDNF and adenylyl cyclase VIII), suggesting that MEF2 exerts a complex action in synaptic plasticity. 36 The members of this family can form either homodimers or heterodimers to modulate gene expression. Depending on the protein recruited by these complexes, they can act either as activators or repressors. 2 , 37
In a study by Pulipparachauvil and colleagues, cocaine was shown to suppress striatal MEF2 activity, with this action related to cocaine-induced activation of a D1 receptor–cAMP-dependent signaling event that resulted in inhibition of calcineurin, a Ca 2+- dependent protein phosphatase. This reduction in MEF2 activity played a key role in cocaine-induced increases in MSN dendritic spine number, but appeared to oppose sensitization to the locomotor stimulatory actions of the drug. 2 , 37 This finding also speaks to the current uncertainly about the role for drug-induced increases in the structural complexity of MSNs in response to drugs of abuse, with some studies suggesting that such plasticity contributes to addiction-related behavioral abnormalities and other studies suggesting that such plasticity protects against addiction. Cocaine increases MEF2C expression in rat cortex and striatum. Cocaine is thought to activate SILK1, which in turn phosphorylates and thereby inactivates HDAC5, which then de-represses MEF2 transcription. 38 As MEF2 is known to play an important role in regulating structural and synaptic plasticity, cocaine-induced increases in MEF2 activity are likely involved in addiction-relevant transcriptional plasticity in response to cocaine exposure.
Other transcription factors
The transcription factors discussed above are those that have been most extensively investigated in the context of drug addiction. Others transcription factors, including NF-κB, 39 , 40 glucocorticoid receptor (GR), nucleus accumbens 1 transcription factor (NAC1), and signal transducers and activators of transcription (STATs), have also been implicated in transcriptional responses to cocaine and other drugs of abuse. The role for GR in stress and psychiatric disorders is well characterized. 41 - 44 Levels of the mRNA of NAC-1 are increased in the rat forebrain weeks after cocaine exposure. 45 , 46 Virus-mediated overexpression of NAC-1 in the NAc of rats prevented the development (but not the expression) of locomotor sensitization in response to repeated administration of cocaine, suggesting that it may have a homeostatic, compensatory role similar to that described above for CREB. Thus, it has been suggested that elevated levels of NAC1 in the accumbens may influence expression of behaviors sometimes seen in drug addiction, particularly in those dependent on psychomotor stimulants, such as paranoia, by regulating gene transcription. 47 More recently, a study by the Nestler group has implicated E2F3a in regulating cocaine action in accumbens. 48 Some of the other transcription factors associated with addiction-like behaviors include Npas4, 49 PGC1-α 50 SMAD3, 51 Egr3, 52 and BRG1, 53 to name but a few. With the advent of new sequencing technologies the list of transcriptional processes shown to be impacted by addictive drugs such as cocaine is likely to grow and further highlight the key importance of gene regulatory processes in addiction. In particular single-cell sequencing technologies are likely to reveal gene expression programs that are recruited by drugs of abuse in a cell type-specific manner to drive the emergence of addiction-related behavioral abnormalities.
MicroRNAs
MicroRNAs (miRNAs), which in their mature form are ~22 nucleotides long, are perhaps the best characterized class of functional non-protein coding regulatory RNA. miRNAs are important regulators of gene expression at the post-transcriptional level, and hundreds of miRNAs have been identified across mammalian species. The latest miRbase entry includes over 2500 human miRNAs, 54 with more than 60% of human protein-coding genes thought to be under miRNA control. 55 Therefore, miRNAs participate in the control of virtually all important physiological processes. The canonical mode of miRNA action is believed to be via binding of the miRNA through its so-called “seed sequence” to complementary sequence in the 3’ untranslated region (3’UTR) of gene transcripts, leading to mRNA degradation or translational repression. 56 , 57 Most of the target prediction algorithms that predict transcripts regulated by miRNAs rely on conservation of putative target sequences to identify functional miRNA binding sites. A large number of such algorithms have been developed, including Targetscan, MiRanda, and TarBase. These algorithms can facilitate the identification of miRNA binding sites in the 3’UTRs of target genes and enable the discovery of functional miRNA-mRNA interactions. Importantly, other poorly characterized factors are likely to influence miRNA-mRNA interactions and search algorithms are likely to improve as these factors become better understood. As described below, miRNAs are emerging as important regulators of drug-induced plasticity in the brain.
MicroRNAs in addiction
One of the first studies to investigate the role for miRNAs in the addiction-related actions of cocaine was published by Schaefer and coworkers, who established that cocaine can induce robust alterations in the expression of a large number of miRNAs in the striatum. A subset of these cocaine-responsive miRNAs were shown to regulate the expression levels of genes implicated in addiction, including BDNF, FosB, and Cdk5r1. They showed that genetic ablation of Argonaute 2 (Ago2), which is the major catalytic unit of the RNA-induced silencing complex (RISC), in iMSNs of the striatum dramatically reduced rewarding effects of cocaine in mice. This is important because the RISC complex plays an important role in regulating the suppressive effects of miRNAs on target transcripts and, in some cases, also regulates the maturation of miRNAs. The Ago2-iMSN mutant mice also demonstrated reduced levels of cocaine self-administration compared with their wild-type littermates across the entire dose-response curve, suggesting that the motivation to consume cocaine was decreased in the mutant animals. These findings suggest that Ago2, and the miRNAs that require Ago2 to function, play an important role in controlling the behavioral actions of cocaine. 58
Subsequent studies have identified a large number of miRNAs whose expression is altered by drugs of abuse and extensive information from the last decade of work has established their critical role in regulating different aspects of substance-use disorders. Some of the key miRNAs include miR-181, miR-212, miR-124, miR-9, and Let-7, and the list continues to grow. Although predominantly found intracellularly, significant pools of miRNAs have also been observed in the extracellular environment, including blood. Recent studies have shown that some miRNAs can be encapsulated into exosomes and released, and are capable of transmitting information between cells. There has been good correlation of serum miRNA expression and certain cancers, suggesting their potential as putative biomarkers for disease. Furthermore, it has been demonstrated that miRNAs can be packaged and, upon depolarization, 59 , 60 can be released from neurons. A few studies have assessed serum miRNA profiles in response to drugs like nicotine, alcohol, and amphetamine. 61 ‑ 64 Interpretation of results from studies measuring serum miRNAs requires careful consideration, as circulating miRNAs are generally expressed at relatively low levels and their functional importance and relationship with miRNA signaling in the brain is unclear. Hence, more studies are needed to determine whether circulating miRNAs are also detected in patients diagnosed with substance use disorder for other psychostimulants. However, this opens new avenues for research on mechanisms of drug dependence as well as development of biomarkers for drug dependence.
miR-212/132 cluster in addiction
Previous studies from our group have shown that miR-212 plays a critical role in regulating compulsive cocaine intake in rats. 30 , 31 We showed that miR-212 (and also miR-132) was upregulated in the dorsal striatum of rats with a history of cocaine consumption under extended (6 h) daily access conditions. By contrast, miR-212 levels were unaltered in rats with restricted daily access to cocaine and in rats that received non-volitional cocaine infusions time-locked to rats that volitionally consumed cocaine during extended access conditions, compared with cocaine-naïve control rats. Importantly, it is the extended access rats that show compulsive-like cocaine-taking behaviors. Virus-mediated overexpression of miR-212 in the dorsal striatum dramatically decreased cocaine intake in rats with extended daily access but had no effects in rats with restricted daily drug access. In addition, we detected a dramatic downward shift in the dose-response curve of cocaine infusions in the extended access rats in which miR-212 was overexpressed, suggesting that their motivation to consume cocaine was markedly decreased. Conversely, antisense oligonucleotide-mediated inhibition of miR-212 signaling in the striatum, increased cocaine intake in rats with extended but not in those with restricted daily access to the drug. These findings suggest that striatal miR-212 signaling may protect against the actions of cocaine that drive the emergence of compulsive consumption of the drug in much the same way that striatal CREB signaling also protects against the addiction-related actions of cocaine (see above), 30 ( Figure 1 ).
As CREB opposes the motivational properties of cocaine, 9 , 28 and CREB can stimulate increases in miR-212 expression 30 ( Figure 1 ), we tested the hypothesis that miR-212 signaling in the dorsal striatum may inhibit cocaine consumption under extended access conditions by recruiting and enhancing striatal CREB activity through a positive feedback loop. Indeed, we found that miR-212 dramatically enhances CREB signaling both in vitro and in vivo in the striatum. 30 This suggests that miR-212 expression is induced in the striatum by cocaine, then subsequently sensitizes CREB signaling, which attenuates the motivational properties of the drug through expression of other CREB-responsive genes. Precisely how does miR-212 regulate CREB signaling? We found that miR-212 increases the activity of Raf1 kinase by repressing the Raf1 inhibitor SPREAD1 (Sprouty-related, EVH1 domain-containing protein 1). This results in enhanced Raf1 activity, which in turn phosphorylates and sensitizes the activity of adenylyl cyclases in the striatum. 30 These observations suggest that miR-212 controls cocaine intake in part by amplifying the CREB activity.
In a follow-up study, we investigated the mechanisms through which cocaine regulates miR-212 levels in the striatum. 31 The miR-212 gene is located in a CpG-enriched region of the genome, suggesting that DNA methylation may play a role in its expression. MeCP2 binds to methylated DNA and can serve as a repressor of gene expression by recruiting transcriptional repressors. 65 Therefore, we hypothesized that MeCP2 plays a role in cocaine-induced regulation of miR-212 expression in striatum. Consistent with this hypothesis, we found that knockdown of MeCP2 increased miR-212 expression in cultured cells. 31 Similarly, RNA interference-mediated knockdown of MeCP2 transcripts in the striatum increased the stimulatory effects of cocaine on miR-212 expression and markedly reduced cocaine intake in rats with extended access to the drug. These data suggest that MeCP2 exerts an inhibitory effect on miR-212 expression and, as such, may promote the development of addiction by limiting the protective actions of miR-212 against cocaine-induced plasticity in the striatum 31 ( Figure 1 ). The miRNA miR-132 is closely related to miR-212, and they both share the same “seed” sequence, thought to be critical for target specification. Importantly, miR-132 was shown to inhibit MeCP2 expression by direct binding with the 3′UTR 66 of MeCP2 transcripts in brain. Based on this observation, we hypothesized that miR-212 may also inhibit MeCP2. Consistent with this hypothesis, miR-212 was shown to exert a negative influence over MeCP2 expression in vitro and in the striatum in vivo. 31 These findings suggest that a negative feedback loop exists between miR-212 and MeCP2 in which each inhibits the expression of the other. Based on this finding, we hypothesize that homeostatic interactions between miR-212 and MeCP2 may play a critical role in determining vulnerability to compulsive cocaine use addiction. Currently, little is known about the role for miR-212 in regulating other behavioral abnormalities associated with cocaine use, such as sensitization of locomotor activity, withdrawal-related symptoms, or vulnerability to relapse-like drug-seeking during periods of abstinence.
Summary
Over recent years, our understanding of how drugs of abuse recruit transcription factors to trigger plasticity-relevant gene expression in the striatum and other addiction-relevant brain sites has increased dramatically. We have also gained considerable insights into how noncoding RNAs and other post-transcriptional regulatory mechanisms control gene expression and thereby regulate the actions of drugs of abuse in the striatum. Emerging technologies, such as single-cell sequencing, are likely to further revolutionize our understanding of how addictive drugs remodel brain reward and motivation circuitries. Critical will be identification of approaches to leverage this information into the development of new therapeutics that can modulate or even reverse the transcriptional actions of abuse drugs in the striatum and elsewhere in the brain to facilitate cessation effects and combat addiction. New approaches to modulating transcription factors, RNAs, and the protein machineries through which they act, in a safe and effective manner, will be required to translate these important insights into new therapeutics to combat drug addiction.
Acknowledgments
Paul Kenny is cofounder of Eolas Therapeutics Inc, is a consultant for Takeda Pharmaceuticals and Alkermes, and has a research award from Eli Lilly. The authors have no conflict of interest to declare
Contributor Information
Purva Bali, Nash Family Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Paul J. Kenny, Nash Family Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
REFERENCES
- 1.Nestler EJ. Transcriptional mechanisms of drug addiction. Clin Psychopharmacol Neurosci. 2012;10(3):136–143. doi: 10.9758/cpn.2012.10.3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12(11):623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nestler EJ. Transcriptional mechanisms of addiction: role of ΔFosB. Philos Trans R Soc B Biol Sci. 2008;363(1507):3245–3255. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H-F, Zhou W-H, Zhu H-Q, Lai M-J, Chen W-S. Microinjection of M(5) muscarinic receptor antisense oligonucleotide into VTA inhibits FosB expression in the NAc and the hippocampus of heroin sensitized rats. Neurosci Bull. 2007;23(1):1–8. doi: 10.1007/s12264-007-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulery-Reynolds PG, Castillo MA, Vialou V, Russo SJ, Nestler EJ. Phosphorylation of ΔFosB mediates its stability in vivo. Neuroscience. 2009;158(2):369–372. doi: 10.1016/j.neuroscience.2008.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulery PG, Rudenko G, Nestler EJ. Regulation of ΔFosB stability by phosphorylation. J Neurosci. 2006;26(19):5131–5142. doi: 10.1523/JNEUROSCI.4970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrotti LI, Weaver RR, Robison B, et al Distinct patterns of ΔFosB induction in brain by drugs of abuse. Synapse. 2008;62(5):358–369. doi: 10.1002/syn.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby CR, Whisler K, Steffen C, Nestler EJ, Self DW. Striatal cell type-specific overexpression of ΔFosB enhances incentive for cocaine. J Neurosci. 2003;23(6):2488–2493. doi: 10.1523/JNEUROSCI.23-06-02488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelz MB, Chen J, Carlezon Jr WA, et al Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DL, Vialou V, Rios L, et al The influence of δfosb in the nucleus accumbens on natural reward-related behavior. J Neurosci. 2008;28(41):10272–10277. doi: 10.1523/JNEUROSCI.1531-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitchers KK, Frohmader KS, Vialou V, et al DeltaFosB in the nucleus accumbens is critical for reinforcing effects of sexual reward. Genes Brain Behav. 2010;9(7):831–840. doi: 10.1111/j.1601-183X.2010.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werme M, Messer C, Olson L, et al ΔFosB regulates wheel running. J Neurosci. 2002;22(18):8133–8138. doi: 10.1523/JNEUROSCI.22-18-08133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibb JA, Chen J, Taylor JR, et al Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- 14.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and ΔFosB. Nat Neurosci. 2003;6:1208. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 15.Renthal W, Kumar A, Xiao G, et al Genome wide analysis of chromatin regulation by cocaine reveals a novel role for sirtuins. Neuron. 2009;62(3):335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vialou V, Robison AJ, LaPlant QC, et al ΔFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat Neurosci. 2010;13(6):745–752. doi: 10.1038/nn.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo SJ, Dietz DM, Dumitriu D, Malenka RC, Nestler EJ. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010;33(6):267–276. doi: 10.1016/j.tins.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf ME, Ferrario CR. AMPA receptor plasticity in the nucleus accumbens after repeated exposure to cocaine. Neurosci Biobehav Rev. 2010 doi: 10.1016/j.neubiorev.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Choi K-H, Renthal W, et al Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Renthal W, Carle TL, Maze I, et al ΔFosB mediates epigenetic desensitization of the fos gene after chronic amphetamine exposure. J Neurosci. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10:561. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- 22.Maze I, Covington HE, Dietz DM, et al Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47:33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Dentin R, Chen D, et al A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger RZ, Beitner-Johnson D, Sevarino KA, Crain SM, Nestler EJ. A general role for adaptations in G-proteins and the cyclic AMP system in mediating the chronic actions of morphine and cocaine on neuronal function. Brain Res. 1991;548(1):100–110. doi: 10.1016/0006-8993(91)91111-d. [DOI] [PubMed] [Google Scholar]
- 26.Shaw-Lutchman TZ, Barrot M, Wallace T, et al Regional and cellular mapping of cAMP response element-mediated transcription during naltrexone-precipitated morphine withdrawal. J Neurosci. 2002;22(9):3663–3672. doi: 10.1523/JNEUROSCI.22-09-03663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Self DW, Genova LM, Hope BT, Barnhart WJ, Spencer JJ, Nestler EJ. Involvement of cAMP-dependent protein kinase in the nucleus accumbens in cocaine self-administration and relapse of cocaine-seeking behavior. J Neurosci. 1998;18(5):1848–1859. doi: 10.1523/JNEUROSCI.18-05-01848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlezon WA, Thome J, Olson VG, et al Regulation of cocaine reward by CREB. Science. 1998;282(5397):2272–2275. doi: 10.1126/science.282.5397.2272. [DOI] [PubMed] [Google Scholar]
- 29.Barrot M, Olivier JDA, Perrotti LI, et al CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci U S A. 2002;99(17):11435–11440. doi: 10.1073/pnas.172091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollander JA, Im H-I, Amelio AL, et al Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010 doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown TE, Lee BR, Mu P, et al A silent synapse-based mechanism for cocaine-induced locomotor sensitization. J Neurosci. 2011;31(22):8163–8174. doi: 10.1523/JNEUROSCI.0016-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein M, Pieri I, Uhlmann F, Pfizenmaier K, Eisel U. Cloning and characterization of promoter and 5′-UTR of the NMDA receptor subunit ϵ2: evidence for alternative splicing of 5′-non-coding exon. Gene. 1998;208(2):259–269. doi: 10.1016/s0378-1119(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 34.Dietrich J-B. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 2013;352(2):179–190. doi: 10.1007/s00441-013-1565-2. [DOI] [PubMed] [Google Scholar]
- 35.Brand NJ. Myocyte enhancer factor 2 (MEF2) Int J Biochem Cell Biol. 1997;29(12):1467–1470. doi: 10.1016/s1357-2725(97)00084-8. [DOI] [PubMed] [Google Scholar]
- 36.Flavell SW, Kim T-K, Gray JM, et al Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pulipparacharuvil S, Renthal W, Hale CF, et al Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59(4):621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jean-Bernard D, Hiroshi T, Sylvie G, Alejandro B, Jean Z. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse. 2011;66(1):61–70. doi: 10.1002/syn.20988. [DOI] [PubMed] [Google Scholar]
- 39.Ang E, Chen J, Zagouras P, et al Induction of nuclear factor-κB in nucleus accumbens by chronic cocaine administration. J Neurochem. 2008;79(1):221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- 40.Russo SJ, Wilkinson MB, Mazei-Robison MS, et al Nuclear factor B signaling regulates neuronal morphology and cocaine reward. J Neurosci. 2009;29(11):3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christoffel DJ, Golden SA, Russo SJ. Structural and synaptic plasticity in stress-related disorders. Rev Neurosci. 2011;22(5):535–549. doi: 10.1515/RNS.2011.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michela M, Vincenzo PP. Interaction between glucocorticoid hormones, stress and psychostimulant drugs. Eur J Neurosci. 2002;16(3):387–394. doi: 10.1046/j.1460-9568.2002.02089.x. [DOI] [PubMed] [Google Scholar]
- 43.Ambroggi F, Turiault M, Milet A, et al Stress and addiction: glucocorticoid receptor in dopaminoceptive neurons facilitates cocaine seeking. Nat Neurosci. 2009;12:247. doi: 10.1038/nn.2282. [DOI] [PubMed] [Google Scholar]
- 44.Rose AK, Shaw SG, Prendergast MA, Little HJ. The importance of glucocorticoids in alcohol dependence and neurotoxicity. Alcohol Clin Exp Res. 2010;34(12):2011–2018. doi: 10.1111/j.1530-0277.2010.01298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackler SA, Homan YX, Korutla L, Conti AC, Blendy JA. The mouse nac1 gene, encoding a cocaine-regulated bric-a-brac tramtrac broad complex/pox virus and zinc finger protein, is regulated by ap1. Neuroscience. 2003;121(2):355–361. doi: 10.1016/s0306-4522(03)00376-2. [DOI] [PubMed] [Google Scholar]
- 46.Korutla L, Wang PJ, Lewis DM, Neustadter JH, Stromberg MF, Mackler SA. Differences in expression, actions and cocaine regulation of two isoforms for the brain transcriptional regulator NAC1. Neuroscience. 2002;110(3):421–429. doi: 10.1016/s0306-4522(01)00518-8. [DOI] [PubMed] [Google Scholar]
- 47.Chandrasekar V, Dreyer J-L. The brain-specific neural zinc finger transcription factor 2b (NZF-2b/7ZFMyt1) suppresses cocaine self-administration in rats. Front Behav Neurosci. 2010;4:14. doi: 10.3389/fnbeh.2010.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cates HM, Heller EA, Lardner CK, et al Transcription Factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol Psychiatry. 2018;84(3):167–179. doi: 10.1016/j.biopsych.2017.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi M, Carreira MB, Cooper YA, et al HDAC5 and its target gene, Npas4, function in the nucleus accumbens to regulate cocaine-conditioned behaviors. Neuron. 2017;96(1):130–144. doi: 10.1016/j.neuron.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra R, Engeln M, Francis TC, Konkalmatt P, Patel D, Lobo MK. A role for PGC1-alpha in nucleus accumbens neuron subtypes in cocaine action. Biol Psychiatry. 2017;81(7):564–572. doi: 10.1016/j.biopsych.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gancarz AM, Wang Z-J, Schroeder GL, et al Activin-receptor signaling regulates cocaine-primed behavioral and morphological plasticity. Nat Neurosci. 2015;18(7):959–961. doi: 10.1038/nn.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandra R, Francis TC, Konkalmatt P, et al Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci. 2015;35(20):7927–7937. doi: 10.1523/JNEUROSCI.0548-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z-J, Martin JA, Mueller LE, et al BRG1 in the nucleus accumbens regulates cocaine-seeking behavior. Biol Psychiatry. 2016;80(9):652–660. doi: 10.1016/j.biopsych.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 58.Schaefer A, Im H-I, Venø MT, et al Argonaute 2 in dopamine 2 receptor–expressing neurons regulates cocaine addiction. J Exp Med. 2010;207(9):1843–1851. doi: 10.1084/jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldie BJ, Dun MD, Lin M, et al Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014;42(14):9195–9208. doi: 10.1093/nar/gku594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Li S, Li L, et al Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi K, Yokota S, Tatsumi N, Fukami T, Yokoi T, Nakajima M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol Appl Pharmacol. 2013;272(1):154–160. doi: 10.1016/j.taap.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 62.Li H, Li C, Zhou Y, et al Expression of microRNAs in the serum exosomes of methamphetamine-dependent rats vs. ketamine-dependent rats. Exp Ther Med. 2018;15(4):3369–3375. doi: 10.3892/etm.2018.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee A, Waters D, Camacho OM, Minet E. Quantification of plasma microRNAs in a group of healthy smokers, ex-smokers and non-smokers and correlation to biomarkers of tobacco exposure. Biomarkers. 2015;20(2):123–131. doi: 10.3109/1354750X.2014.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Y, Zhang K, Jiang H, et al Decreased expression of plasma MicroRNA in patients with methamphetamine (MA) use disorder. J Neuroimmune Pharmacol. 2016;11(3):542–548. doi: 10.1007/s11481-016-9671-z. [DOI] [PubMed] [Google Scholar]
- 65.Guy J, Cheval H, Selfridge J, Bird A. The role of meCP2 in the brain. Annu Rev Cell Dev Biol. 2011;27(1):631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 66.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]