SUMMARY
Cell death is a vital and ubiquitous process that is tightly controlled in all organisms. However, the mechanisms underlying precise cell death control remain fragmented. As an important shared module in plant growth, development and immunity, Arabidopsis thaliana BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 (BAK1) and somatic embryogenesis receptor kinase 4 (SERK4) redundantly and negatively regulate plant cell death. By deploying an RNAi-based genetic screen for bak1/serk4 cell death suppressors, we revealed that cyclic nucleotide-gated channel 20 (CNGC20) functions as a hyperpolarization-activated Ca2+-permeable channel specifically regulating bak1/serk4 cell death. BAK1 directly interacts with and phosphorylates CNGC20 at specific sites in the C-terminal cytosolic domain, which in turn regulates CNGC20 stability. CNGC19, the closest homolog of CNGC20 with a low abundance compared with CNGC20, makes a quantitative genetic contribution to bak1/serk4 cell death only in the absence of CNGC20, supporting the biochemical data showing homo- and heteromeric assembly of the CNGC20 and CNGC19 channel complexes. Transcripts of CNGC20 and CNGC19 are elevated in bak1/serk4 compared with wild-type plants, further substantiating a critical role of homeostasis of CNGC20 and CNGC19 in cell death control. Our studies not only uncover a unique regulation of ion channel stability by cell surface-resident receptor kinase-mediated phosphorylation, but also provide evidence for fine-tuning Ca2+ channel functions in maintaining cellular homeostasis by the formation of homo-and heterotetrameric complexes.
Keywords: Plant cell death, cyclic nucleotide-gated channel, Ca2+-permeable channel, receptor kinase, cellular homeostasis, virus-induced gene silencing, phosphorylation
eTOC Summary:
Yu et al. show that BAK1/SERK4-mediated phosphorylation regulates Ca2+ channel CNGC20/CNGC19 stability for the precise control of plant cell death, revealing the critical function of CNGC20/CNGC19 in the regulation of cellular homeostasis for cell survival.
INTRODUCTION
Plants have evolved hundreds of cell surface-resident receptor-like kinases (RLKs) regulating plant growth, development, and defense [1, 2]. A large number of characterized RLKs contain an extracellular leucine-rich repeat (LRR) domain [1]. Several well-studied LRR-RLKs are bona fide receptors that perceive intrinsic and extrinsic factors derived from plant growth, development and environmental cues. These include FLAGELLIN-SENSING 2 (FLS2) perceiving bacterial flagellin in activating plant immune signaling, and BRASSINOSTEROID INSENSITIVE 1 (BRI1) perceiving plant growth hormone brassinosteroids (BRs) in regulating plant growth [3, 4]. BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (SERK3), together with other SERKs, function as coreceptors of FLS2, BRI1 and some other LRR-RLKs, and regulate a wide range of physiological responses ranging from male gametophyte development, plant growth, stomatal patterning, to plant immunity [5–7].
A unique yet largely uncharacterized function of BAK1 and its closest homolog SERK4 is the containment of cell death [8, 9]. The bak1/serk4 null mutant is postembryonic lethal associated with spontaneous cell death, constitutive H2O2 production, and pathogenesis-related (PR) gene induction [9]. The bak1–4/serk4–1 mutant shows severe seedling lethality and does not produce seeds preventing a classical suppressor screen. To understand the mechanisms underlying BAK1/SERK4-mediated cell death, we have designed a virus-induced gene silencing (VIGS), a type of RNA interference (RNAi), -based genetic screen for suppressors of bak1/serk4 cell death using a sequence-indexed library of Arabidopsis T-DNA insertion lines [10]. Silencing of BAK1 and SERK4 by Agrobacterium-mediated VIGS in wild type (WT) Col-0 plants triggers cell death as observed in the bak1–4/serk4–1 mutant. The screen has identified a series of mutants, including stt3a, which bears a mutation in the catalytic subunit of the oligosaccharyltransferase complex for protein N-glycosylation [10]. Members of cysteine-rich receptor-like kinases (CRKs), which are transcriptionally activated in bak1/serk4, are the client proteins of protein glycosylation regulating cell death process.
In this study, we report the involvement of cyclic nucleotide-gated channel 20 (CNGC20) and CNGC19 in regulating bak1/serk4 cell death. The RNAi-based screen identified CNGC20, but not other CNGCs, as a specific regulator of BAK1/SERK4 cell death. Although CNGC19 has no effect on its own, it contributes to bak1/serk4 cell death in the absence of CNGC20. Consistent with this, both CNGC20 and CNGC19 are hyperpolarization-activated Ca2+-permeable channels, and CNGC20 and CNGC19 have an additive effect on channel activity, suggesting that CNGC20 and CNGC19 form functional homo- and hetero-meric Ca2+ channels. BAK1 and SERK4 interact with and phosphorylate CNGC20, and likely CNGC19, which leads to the degradation of CNGC20 and CNGC19 proteins. In the bak1/serk4 mutant, both CNGC20/CNGC19 protein and transcript levels are increased, leading to mis-regulation of Ca2+ influx and signaling, as a consequence, cellular homeostasis cannot be maintained and plants undergo cell death. Our studies reveal the critical function of CNGC20 and CNGC19 in the precise control of cellular homeostasis for cell survival, and elucidate the regulation of CNGC20/CNGC19 protein stability by BAK1/SERK4-mediated phosphorylation.
RESULTS
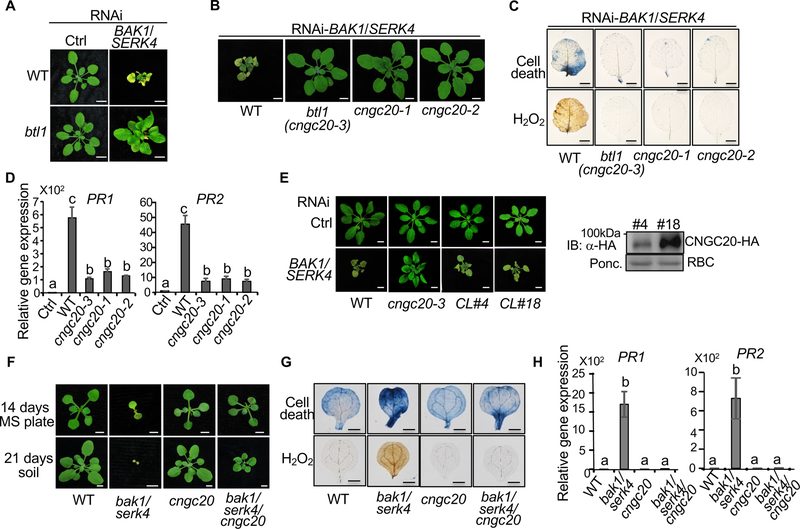
The cngc20 mutants suppress bak1/serk4 cell death
One mutant identified from VIGS-based genetic screen for suppressors of bak1/serk4 cell death, which was named bak to life 1 (btl1), is SALK_013823C. The btl1 mutant suppressed the dwarfism and leaf chlorosis triggered by RNAi-BAK1/SERK4 compared to WT plants (Figure 1A). Trypan blue staining confirmed that cell death was reduced in btl1 (Figure S1A). Staining with 3,3′-diaminobenzidine (DAB) showed that the elevated H2O2 accumulation caused by RNAi-BAK1/SERK4 was abolished in btl1 (Figure S1B). Compared to WT, btl1 showed reduced accumulation of PR1 and PR2 upon silencing of BAK1/SERK4 (Figure S1C). Furthermore, BTL1 specifically regulates BAK1/SERK4-mediated cell death and may not be involved in the cell death regulated by MEKK1, a MAP kinase (MAPK) kinase kinase downstream of BAK1/SERK4 in plant immunity [11], or BIR1, a BAK1-interacting LRR-RLK [12] (Figure S1D).
Figure 1. The cngc20 mutants suppress bak1/serk4 cell death.
(A) The btl1 mutant suppresses growth defects triggered by RNAi-BAK1/SERK4. Plant phenotypes are shown two weeks after VIGS of BAK1/SERK4 or a vector control (Ctrl). Bar=5 mm.
(B) The cngc20 mutants suppress growth defects triggered by RNAi-BAK1/SERK4. Plant phenotypes are shown for WT, btl1 (cngc20–3), cngc20–1, and cngc20–2 two weeks after VIGS of BAK1/SERK4. Bar=5 mm.
(C) The cngc20 mutants suppress cell death and H2O2 production triggered by RNAi-BAK1/SERK4. Plant true leaves after VIGS of BAK1/SERK4 were stained with trypan blue for cell death (top panel) and DAB for H2O2 accumulation (bottom panel). Bar=2 mm.
(D) The cngc20 mutants suppress PR1 and PR2 expression triggered by RNAi-BAK1/SERK4. The expression of PR1 and PR2 was normalized to the expression of UBQ10. The data are shown as mean ± SE from three independent repeats. The different letters denote statistically significant difference according to one-way ANOVA followed by Tukey test (p<0.05).
(E) Complementation of cngc20–3 with the CNGC20 genomic fragment (gCNGC20) under its native promoter restores growth defects by RNAi-BAK1/SERK4. CL#4 and CL#18 are two representative lines. Bar=5 mm. The HA epitope tagged CNGC20 proteins in transgenic plants were detected with an α-HA immunoblot (IB). Protein loading is shown by Ponceau S staining (Ponc.) for RuBisCo (RBC).
(F) The cngc20–3 mutant partially rescues the seedling lethality of bak1–4/serk4–1. Seedlings were grown on 1/2MS plate and photographed at 14 days post-germination (dpg) (top panel, Bar=2 mm.) or in soil and photographed at 21 dpg (bottom panel, Bar=5 mm). The bak1–4/serk4–1 mutant was photographed at 14 dpg and dead at 21 dpg.
(G) Alleviation of cell death and H2O2 accumulation in bak1–4/serk4–1/cngc20–3 compared to bak1–4/serk4–1. Bar=1 mm.
(H) Reduced PR1 and PR2 expression in bak1–4/serk4–1/cngc20–3 compared to bak1–4/serk4–1. The different letters denote statistically significant difference according to one-way ANOVA followed by Tukey test (p<0.05). The above experiments were repeated at least three times with similar results.
The btl1 mutant (SALK_013823C) was annotated to bear a T-DNA insertion at the 8th intron of AT1G60995 encoding an uncharacterized membralin domain-containing protein (Figure S1E). However, neither two additional mutant alleles of AT1G60995 (SALK_087793C and SALK_042821C) (Figure S1F–H), nor the complementation of AT1G60995 under either the 35S promoter or its native promoter (Figure S1I) supported that the annotated T-DNA insertion in AT1G60995 was responsible for the suppression of RNAi-BAK1/SERK4-induced cell death in btl1. To identify the causal mutation, we performed whole genome-sequencing analysis and revealed that btl1 carried two additional T-DNA insertions at AT1G11020, encoding a zinc finger superfamily protein, and AT3G17700, encoding CNGC20 (Figure S2A–C, Table S1 & S2). Neither mutant alleles of AT1G11020 (SALK_037558C and SAIL_302_A04) suppressed RNAi-BAK1/SERK4-induced cell death (Figure S1J).
Importantly, two additional alleles, including cngc20–1 (SALK_129133C) and cngc20–2 (SALK_074919C) (Figure S2B, D & E), suppressed RNAi-BAK1/SERK4-induced cell death (Figure 1B). Resembling btl1, which was renamed as cngc20–3, the cngc20–1 and cngc20–2 mutants abolished cell death (Figure 1C), elevated H2O2 (Figure 1C) and accumulation of PR1 and PR2 genes (Figure 1D) caused by RNAi-BAK1/SERK4. Transformation of the CNGC20 genomic fragment under the control of its native promoter into cngc20–3 restored RNAi-BAK1/SERK4-induced cell death (Figure 1E). In addition, we generated the bak1–4/serk4–1/cngc20–3 triple mutant by genetic crosses (Figure S2F). The bak1–4/serk4–1/cngc20–3 mutant overcame seedling lethality of bak1–4/serk4–1 and resembled WT plants at the two-week-old stage when grown on 1/2MS medium plates (Figure 1F). Cell death, H2O2 accumulation, and PR1 and PR2 expression were significantly ameliorated in bak1–4/serk4–1/cngc20–3 compared to those in bak1–4/serk4–1 (Figure 1G & H). When grown in soil, the bak1–4/serk4–1/cngc20–3 mutant developed true leaves whereas bak1–4/serk4–1 stopped development at the cotyledon stage (Figure 1F). Collectively, our genetic analyses demonstrate that the CNGC20 mutation leads to the suppression of bak1/serk4 cell death.
Specific function of CNGC20 in plant cell death control
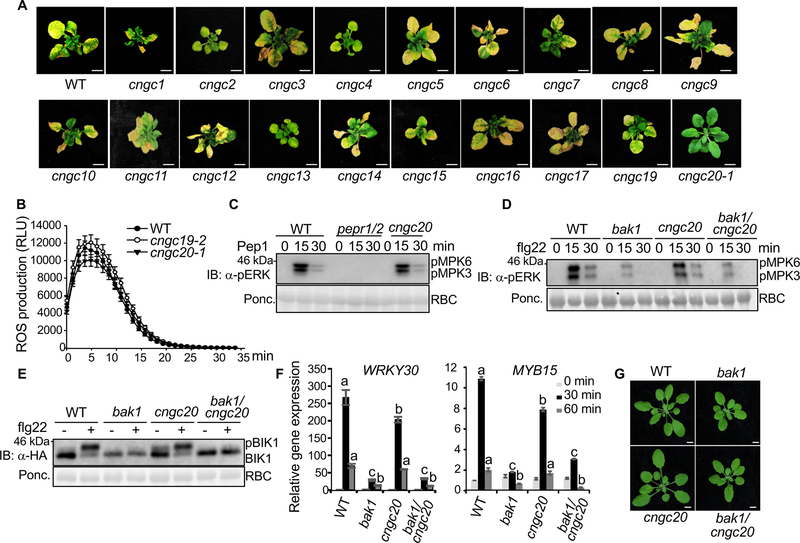
The Arabidopsis genome encodes 20 CNGCs mediating abiotic and biotic stresses and developmental processes [13, 14]. To assess the involvement of additional CNGCs in bak1/serk4 cell death, we systemically characterized all the available cngc mutants for suppression of RNAi-BAK1/SERK4-induced cell death (Figure 2A & Table S3). The cngc18 mutant is male sterile and could not set homozygous seeds [15]. The cngc2 and cngc4 mutants, also known as defense, no death 1 (dnd1–1) and dnd2–1 respectively, are derived from ethyl methanesulfonate (EMS) mutagenesis and produce truncated proteins [16–18]. Genotyping and RT-PCR analyses of cngc1, 3, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 and 19 mutants confirmed that all these mutants, except cngc7 showing a WT genotyping, are homozygous for the T-DNA insertion and showed reduced CNGC transcripts accordingly (Figure S3A & B). However, none of these cngc mutants, including cngc19, the gene of which is the closest homolog of CNGC20, suppressed RNAi-BAK1/SERK4-induced cell death (Figure 2A), indicating a specific function of CNGC20 in bak1/serk4 cell death.
Figure 2. Specific function of CNGC20 in bak1/serk4 cell death.
(A) Unlike cngc20–1, other cngc mutants do not suppress growth defects by RNAi-BAK1/SERK4. Plant phenotypes are shown two weeks after VIGS of BAK1/SERK4. Bar=1 cm.
(B) The cngc19 and cngc20 mutants exhibit similar flg22-induced ROS production as WT plants. Leave discs from four-week-old WT, cngc19–2 and cngc20–1 plants were treated with 100 nM flg22 for 35 min. The data are shown as means ± SE from 24 leaf discs.
(C) The cngc20–3 mutant exhibits similar Pep1-induced MAPK activation as WT. Ten-day-old seedlings were treated without or with 100 nM Pep1 for 15 and 30 min. The MAPK activation was analyzed by immunoblot with α-pERK antibody (top panel), and the protein loading is shown by Ponceau S staining for RBC (bottom panel). The pepr1/2 is the Pep1 receptor PEPR1 and PEPR2 double mutant.
(D) The cngc20–1 mutant does not interfere with the compromised flg22-induced MAPK activation in bak1–4. Ten-day-old seedlings were treated without or with 100 nM flg22 for 15 and 30 min. MAPK activation was analyzed by immunoblotting with α-pERK antibody (top panel), and protein loading is shown by Ponceau S staining for RBC (bottom panel).
(E) The cngc20–1 mutant does not affect the compromised flg22-induced BIK1 phosphorylation in bak1–4. Protoplasts from different plants were transfected with HA-tagged BIK1 and treated with 100 nM flg22 for 10 min. BIK1-HA proteins were detected by immunoblotting using α-HA antibody (top panel), and protein loading is shown by Ponceau S staining for RBC (bottom panel).
(F) The cngc20–1 mutant did not affect the compromised flg22-induced gene expression in bak1–4. Ten-day-old seedlings were treated without or with 100 nM flg22 for 30 or 60 min for qRT-PCR analysis. The data are shown as mean ± SE from three independent repeats. The different letters indicate statistically significant differences from WT within the same time point according to two-way ANOVA followed by Tukey test (p<0.05).
(G) The cngc20–1 mutant does not affect the growth phenotype of bak1–4. Four-week-old soil-grown plants are shown. The cngc20–1/bak1–4 mutant exhibited similar growth phenotypes, including rounder leaves and shorter petioles, as bak1–4 compared to WT plants. Bar=5 mm.
The above experiments were repeated at least three times with similar results.
CNGC20 and CNGC19 belong to group IV of Arabidopsis CNGCs together with CNGC2 and CNGC4, which have been shown to regulate plant immunity [16–21]. CNGC19, not CNGC20, has been recently shown to regulate Arabidopsis defense against Spodoptera herbivory [22]. Plants have developed a two-tiered immune system: effector-triggered immunity (ETI) and microbe/danger-associated molecular pattern (MAMP/DAMP)-triggered immunity (PTI) to ward off pathogen invasions [23–25]. CNGC11 and CNGC12 are involved in plant ETI [26]. We assessed the involvement of CNGC20 in plant ETI and/or PTI. The cngc20 mutants showed similar pathogen resistance as WT, as shown in bacterial growth and symptom development to Pseudomonas syringae pv. tomato (Pst) DC3000 carrying avrRpt2 (Figure S4A) or avrRps4 (Figure S4B). ETI is often associated with localized cell death, referred as the hypersensitive response (HR). The progression of Pst avrRpt2- or Pst avrRpm1-triggered HR was similar in the cngc20 mutants as that in WT plants (Figure S4C & D). In addition, the cngc20 mutants did not affect plant resistance to the virulent P. syringae pv. maculicola (Psm) ES4326 in terms of bacterial growth and disease symptom development (Figure S4E). Furthermore, the cngc20 mutants did not affect MAMP flg22-, a 22-amino acid synthetic peptide corresponding to bacterial flagellin, or DAMP Pep1-induced early PTI signaling events such as ROS production, MAPK activation, and receptor-like cytoplasmic kinase (RLCK) BIK1 phosphorylation (Figure 2B–E). These data suggest that unlike CNGC2, 4, 11 and 12, CNGC20 is not involved in plant ETI and PTI.
BAK1, the shared co-receptor of multiple LRR-RLKs, positively regulates plant immunity and development [5, 6]. The bak1 mutant is compromised in flg22-triggered immunity and BR-mediated development with rounder leaves and shorter petioles. Since the mutation in cngc20 suppressed bak1/serk4 cell death, we tested whether cngc20 interferes with the bak1–4 deficiency in plant immunity and growth using the bak1–4/cngc20–1 double mutant. The compromised flg22-induced MAPK activation and BIK1 phosphorylation remained similar in bak1–4/cngc20–1 as those in bak1–4 (Figure 2D & E). Similar to bak1–4, bak1–4/cngc20–1 showed compromised expression of flg22-induced genes, WRKY30 and MYB15, compared with WT or cngc20–1 (Figure 2F). In addition, the cngc20–1/bak1–4 mutant resembles bak1–4 in its growth morphology (Figure 2G). Collectively, the evidence suggests a specific involvement of CNGC20 in BAK1/SERK4-mediated cell death containment, uncoupled from BAK1/SERK4 functions in plant immunity and growth.
BAK1 interacts with and phosphorylates CNGC20
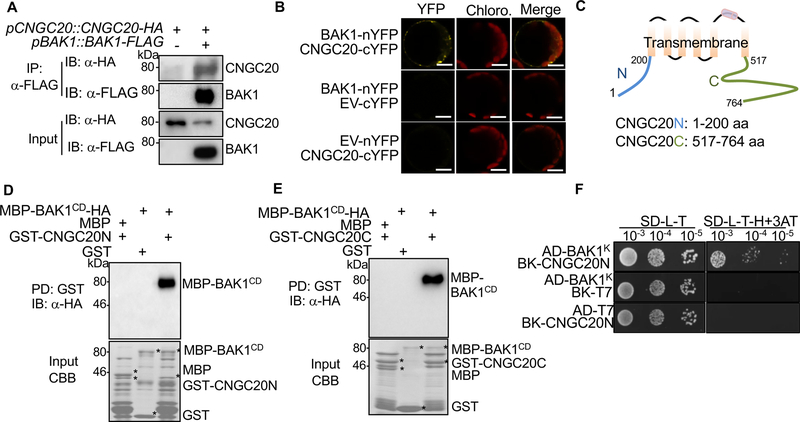
Both CNGC20 and BAK1 exhibit plasma membrane-localization [27, 28]. We examined whether CNGC20 associates with BAK1. A co-immunoprecipitation (Co-IP) assay from co-expression of FLAG-tagged CNGC20 and HA-tagged BAK1 in Arabidopsis protoplasts indicated that BAK1 co-immunoprecipitated CNGC20 (Figure S5A). When transiently expressed in Nicotiana benthamiana, BAK1-FLAG immunoprecipitated CNGC20-HA (Figure S5B). We further transformed pCNGC20::CNGC20-HA into pBAK1::BAK1-FLAG transgenic plants. BAK1 immunoprecipitated CNGC20 when both were expressed under their native promoters, indicating that BAK1 associates with CNGC20 in vivo (Figure 3A). The association between BAK1 and CNGC20 at the plasma membrane was confirmed by a bimolecular fluorescence complementation (BiFC) assay (Figure 3B). The observed puncta of BAK1-nYFP and CNGC20-cYFP close to plasma membrane suggest that they may also localize to endosomal compartments [28]. CNGC20 is a transmembrane protein consisting of N-terminal and C-terminal cytosolic domains (CNGC20N and CNGC20C) and six transmembrane domains (Figure 3C). The BAK1 cytosolic domain (BAK1CD), including juxtamembrane and kinase domains, immunoprecipitated both CNGC20N and CNGC20C (Figure S5C). We further tested whether CNGC20N or CNGC20C directly interacted with BAK1 in an in vitro pull-down assay. The maltose-binding protein (MBP)-tagged BAK1CD (MBP-BAK1CD) was pulled down by glutathione S-transferase (GST)-tagged CNGC20N or CNGC20C but not GST itself (Figure 3D & E). Moreover, the interaction between the BAK1 kinase domain (BAK1K) and CNGC20N was confirmed by a yeast two-hybrid assay (Figure 3F). Interestingly, the BAK1 kinase inactive mutant (BAK1CDKM) showed a reduced interaction with CNGC20C, indicating a requirement of BAK1 kinase activity for a full interaction (Figure S5D). Together, the data elucidate that CNGC20 interacts with BAK1 via its N- and C-terminal cytosolic domains.
Figure 3. BAK1 interacts with CNGC20.
(A) CNGC20 associates with BAK1 in transgenic plants. Protein extracts from transgenic plants carrying pBAK1::BAK1-FLAG and pCNGC20::CNGC20-HA were immunoprecipitated with α-FLAG agarose beads (IP: α-FLAG) and immunoblotted with α-HA (IB: α-HA) or α-FLAG (IB: α-FLAG) (top two panels). Protein inputs are shown with immunoblotting before immunoprecipitation (bottom two panels).
(B) Interaction between CNGC20 and BAK1 by BiFC assay in Arabidopsis protoplasts. BAK1-nYFP (BAK1 fused with N-terminal YFP) and CNGC20-cYFP (CNGC20 fused with C-terminal YFP) proteins were transiently co-expressed in protoplasts. YFP signals were observed using a confocal microscopy. EV indicates the empty vector control. Bar=10 μm.
(C) Schematic diagram of CNGC20 protein structure indicating the transmembrane and cytosolic N-terminal and C-terminal domains (hereafter, CNGC20N and CNGC20C).
(D) CNGC20N interacts with the BAK1 cytosolic domain (BAK1CD) in an in vitro pull-down (PD) assay. GST or GST-CNGC20N immobilized on glutathione sepharose beads was incubated with MBP or MBP-BAK1CD-HA proteins. The beads were washed and pelleted for immunoblotting with α-HA antibody (PD: GST; IB: α-HA) (top panel). Coomassie blue staining (CBB) of input proteins is shown on the bottom.
(E) CNGC20C interacts with BAK1CD in vitro. A similar assay using CNGC20C was performed as in (D).
(F) The BAK1 kinase domain (BAK1K) interacts with CNGC20N in a yeast two-hybrid assay. The interaction between BAK1K and CNGC20N was tested on synthetic defined (SD) medium without leucine, tryptophan and histidine (SD-L-T-H) supplemented with 1 mM 3-amino-1, 2, 4-triazole (3AT). pGADT7 and pGBKT7 are empty vectors. Serial dilutions of the yeast colonies were plated.
The above experiments were repeated three times with similar results.
See also Figure S5.
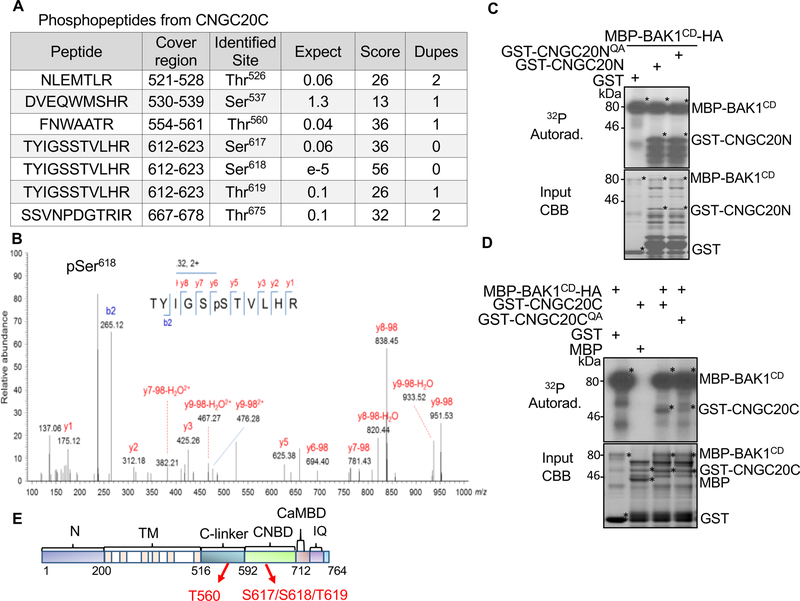
We next tested whether BAK1 could phosphorylate CNGC20. An in vitro kinase assay indicated that BAK1CD, but not BAK1CDKM, directly phosphorylated both CNGC20N and CNGC20C (Figure S5E & F). The BAK1CD-phosphorylated CNGC20N and CNGC20C proteins were subjected to trypsin digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. We identified Ser184 as a confident and Ser156 as a less confident BAK1CD phosphorylation site on CNGC20N (Figure S6A), and multiple confident BAK1CD phosphorylation sites on CNGC20C, including Thr560, Ser617, and Ser618 (Figure 4A, B, S6B & C). In addition, Thr619 was identified as a potential phosphorylation site on CNGC20C (Figure 4A). Both Ser156 and Ser184 have potential phosphorylatable residues next to them, Thr155 and Ser185 on CNGC20N. We mutated all four of these residues on CNGC20N to Ala, and the Thr155ASer156ASer184ASer185A quadruple mutant (CNGC20NQA) did not affect the phosphorylation by BAK1 (Figure 4C). CNGC20C Thr560A also did not affect the phosphorylation by BAK1 (Figure S6D). However, the Thr560ASer617ASer618AThr619A quadruple mutant (CNGC20CQA) of CNGC20C compromised its phosphorylation by BAK1 (Figure 4D), implying an important role for CNGC20 C-terminal phosphorylation by BAK1. These phosphorylation sites locate to the C-linker and cyclic nucleotide-binding domain (CNBD) (Figure 4E & S6E). This re-elaborates a postulated regulatory role of the C-terminus for CNGC functions. For example, several residues in the C-terminus of human hyperpolarization-activated cyclic nucleotide-gated (HCN) channels and Arabidopsis CNGC11/CNGC12 are critical for inter-subunit interactions [29, 30].
Figure 4. BAK1 phosphorylates CNGC20.
(A) Phosphorylation sites and the corresponding phosphopeptides from CNGC20C region identified from LC-MS/MS analysis. Expect indicates the number of times the peptide is matched by chance (the smaller this value is, the more significant the peptide identification is). Score is a measure of how well the experimental MS/MS spectrum matches to the stated peptide based on calculated probability (P) that the observed match is random. The reported score is: −10Log(P) (the higher the value is, the more confident the peptide identification is). Dupes is the number of additional matches to the same peptide with the same modifications and charge.
(B) LC-MS/MS analysis reveals that Ser618 of CNGC20C is phosphorylated by BAK1CD.
(C) The Thr155A/Ser156A/Ser183A/Ser184A quadruple mutant of CNGC20N (CNGC20NQA) does not affect its phosphorylation by BAK1 (top panel). The kinase assay was performed by incubating MBP-BAK1CD with GST, GST-CNGC20N or GST-CNGC20NQA as a substrate. Phosphorylation of CNGC20N by BAK1CD is shown with autoradiography (top panel). Protein loading control is shown by CBB (bottom panel).
(D) The Thr560A/Ser617A/Ser618A/Thr619A quadruple mutant of CNGC20C (CNGC20CQA) compromises its phosphorylation by BAK1CD (top panel). A similar assay using GST-CNGC20C or GST-CNGC20CQA was performed as in (C).
(E) Schematic diagram of the CNGC20 protein motifs with identified phosphorylation sites. Cytoplasmic N-terminus (N, 1–200); transmembrane region (TM, 201–516); C-linker (517–592); cyclic nucleotide binding domain (CNBD, 593–712); calmodulin binding domain (CaMBD, 713–732); IQ domain (737–752). Numbers indicate amino acid positions. The two red arrows indicate locations of phosphorylation residues.
The above experiments, except LC-MS/MS, were repeated three times with similar results.
BAK1 regulates CNGC20 stability
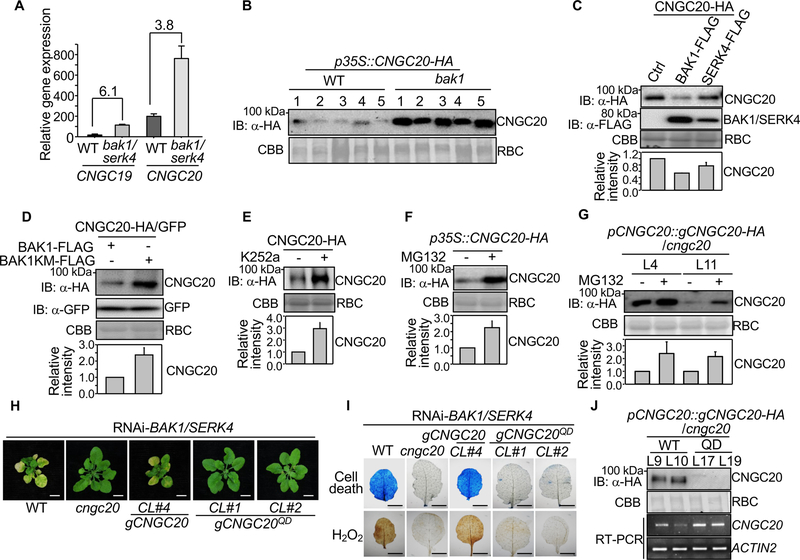
Since loss of CNGC20 suppressed bak1/serk4 cell death and BAK1 phosphorylates CNGC20, it is tempting to speculate that abundance and/or activity of CNGC20 is regulated by BAK1-mediated phosphorylation. Notably, the Lotus japonicus CNGC20 homolog makes a quantitative contribution to root development and infection by nitrogen-fixing rhizobia and its expression level is a key for its function [31]. In addition to tissue expression specificity, transcript levels of some CNGCs are regulated upon pathogen infections [32]. We observed an elevated level (about 4–6 fold) of CNGC20 and CNGC19 transcripts in bak1–4/serk4–1 compared with WT (Figure 5A) based on RNA-Seq analysis [10]. When expressed under the control of the 35S promoter, the protein expression level of CNGC20-HA in bak1–4 was much higher than that in WT (Figure 5B). These data indicate that both CNGC20 transcripts and proteins are regulated by BAK1 and SERK4. Conversely, when co-expressing BAK1 or SERK4 with CNGC20 in Arabidopsis protoplasts, the protein level of CNGC20, but not the GFP control, was reduced (Figure 5C & S6F). The CNGC20 protein level was higher when coexpressed with the BAK1 kinase inactivate mutant than that with WT BAK1 (Figure 5D), implying the importance of BAK1 phosphorylation on CNGC20 abundance. Similarly, the kinase inhibitor K252a increased the protein level of CNGC20-HA (Figure 5E). Further, pretreatment with MG132, a protein degradation inhibitor, increased CNGC20 protein accumulation when transiently expressed in N. benthamiana (Figure 5F) or in the pCNGC20::gCNGC20-HA/cngc20 transgenic plants (Figure 5G). Therefore, the stability of CNGC20 proteins is likely regulated by BAK1 phosphorylation.
Figure 5. BAK1 destabilizes CNGC20 through phosphorylation.
(A) Transcripts of CNGC19 and CNGC20 are upregulated in bak1–4/serk4–1. Mean gene expression levels of CNGC19 and CNGC20 in bak1–4/serk4–1 and WT were obtained from RNA-Seq data [10]. The data are shown as mean ± SD from two independent repeats. The numbers indicate fold changes of gene expression (bak1–4/serk4–1 vs. WT).
(B) BAK1 is required for CNGC20 destabilization. The CNGC20 cDNA fragment with a C-terminal HA tag under the 35S promoter was introduced into WT or bak1–4. About 20 independent T1 transgenic lines were obtained and five representative lines from each background are shown for immunoblot analysis with α-HA.
(C) CNGC20 proteins are destabilized by co-expression with BAK1 or SERK4. CNGC20-HA was co-expressed with empty vector (Ctrl), BAK1-FLAG or SERK4-FLAG in Arabidopsis protoplasts for 12 hr. Protein expression was analyzed with α-HA or α-FLAG immunoblot.
(D) BAK1, not the BAK1 kinase mutant (BAK1KM), destabilizes CNGC20 proteins. CNGC20-HA or GFP proteins were co-expressed with BAK1-FLAG or BAK1KM-FLAG in Arabidopsis protoplasts for 12 hr. Protein expression was analyzed with α-HA or α-GFP immunoblot.
(E) Pretreatment of kinase inhibitor K252a stabilizes CNGC20-HA in Arabidopsis protoplasts. Protoplasts expressing CNGC20-HA were treated with 0.05% DMSO (−) or 1 μM K252a for 12 hr. Protein expression was analyzed with α-HA immunoblot.
(F) The protein degradation inhibitor MG132 stabilizes CNGC20-HA proteins in N. benthamiana. CNGC20-HA was expressed in N. benthamiana by Agrobacterium-mediated transient assay. 0.1% DMSO (−) or 2 μM MG132 was infiltrated 3 hr before the samples were collected at two days post-inoculation (dpi). Total proteins were analyzed by immunoblot with α-HA.
(G) Stabilization of CNGC20-HA proteins by MG132 in transgenic plants. Ten-day-old pCNGC20::gCNGC20-HA/cngc20–1 seedlings were pretreated with 0.1% DMSO (−) or 2 μM MG132 for 3 hr before total proteins were isolated for immunoblot analysis with α-HA.
Quantifications of CNGC20-HA relative intensity from the immunoblots (IB) are shown on the bottom in (C), (D), (E), (F) and (G) as mean ± SE from three independent repeats. The intensity of CNGC20-HA band normalized to RBC by CBB staining in the first lane in (C) and (D), and the non-treatment in (E), (F) and (G) was set as 1.0.
(H) Complementation of cngc20–1 with the Thr560D/Ser617D/Ser618D/Thr619D quadruple mutant of CNGC20 genomic fragment (gCNGC20QD) under its native promoter does not restore growth defects by RNAi-BAK1/SERK4. CL#1 and CL#2 are two representative lines. Bar=5 mm. CL#4 is the complementation line with WT CNGC20.
(I) The CNGC20QD mutant does not restore cell death and H2O2 production triggered by RNAi-BAK1/SERK4. True leaves of WT, cngc20–1, CNGC20 (CL#4) and CNGC20QD (CL#1 and CL#2) after RNAi-BAK1/SERK4 were stained with trypan blue for cell death (top panel) and DAB for H2O2 accumulation (bottom panel). Bar=2 mm.
(J) CNGC20-HA proteins accumulate more than CNGC20QD-HA proteins in transgenic plants. Plants from two independent homozygous T3 lines of pCNGC20::gCNGC20-HA/cngc20–1 and pCNGC20::gCNGC20QD-HA/cngc20–1 were subjected to immunoblot with α-HA antibody. RT-PCR was performed with CNGC20 specific primers to detect the transcript level of CNGC20 and CNGC20QD. ACTIN2 was used as an internal control.
The above experiments, except A, were repeated three times with similar results.
To elucidate the role of BAK1 phosphorylation on CNGC20, we generated the phosphomimetic form of CNGC20 (CNGC20QD), in which four critical phosphorylation sites (Thr560/Ser617/Ser618/Thr619) in CNGC20C were mutated to Asp, and transformed into cngc20–1. Unlike CNGC20, CNGC20QD did not restore cell death, H2O2 accumulation and PR gene expression caused by silencing of BAK1/SERK4 (Figure 5H, 5I & S6G), suggesting that BAK1 phosphorylation of CNGC20 at Thr560/Ser617/Ser618/Thr619 suppresses bak1/serk4 cell death. Consistent with the notion that BAK1 phosphorylation negatively regulates CNGC20 protein stability, the CNGC20QD-HA protein level was substantially lower than CNGC20-HA and barely detectable in the pCNGC20::gCNGC20QD-HA complementation lines compared with the pCNGC20::gCNGC20-HA plants (Figure 5J). Notably, the reduced abundance of CNGC20QD-HA proteins was not due to the reduction of CNGC20 transcripts (Figure 5J). Thus, the data suggest that BAK1-mediated phosphorylation at Thr560/Ser617/Ser618/Thr619 negatively regulates CNGC20 abundance and the homeostasis of CNGC20 is vital in bak1/serk4 cell death.
CNGC19 and CNGC20 are hyperpolarization-activated Ca2+ permeable channels
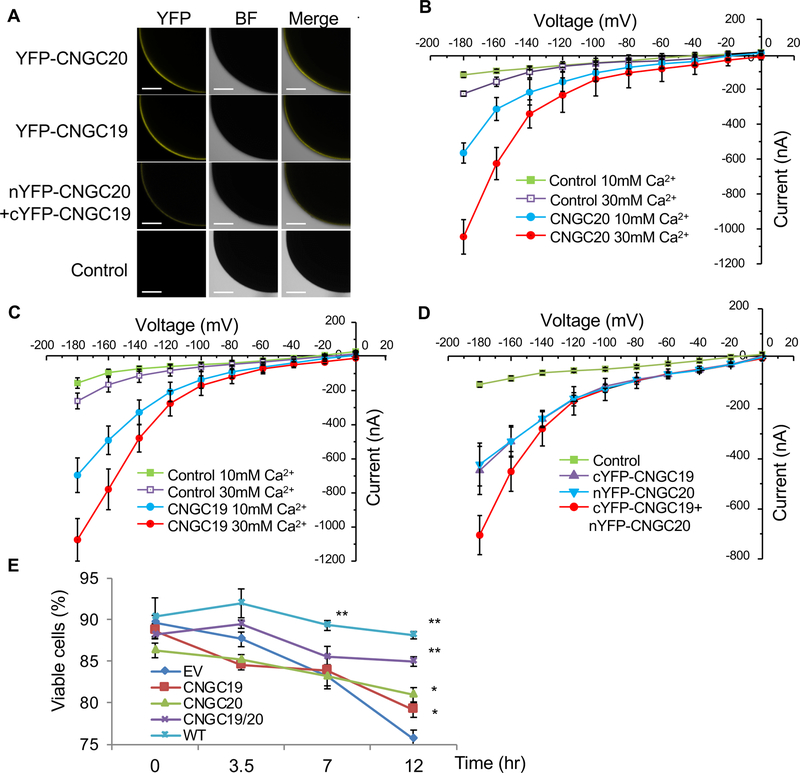
CNGCs function as non-selective cation channels. Evidence from electrophysiological and physiological studies supports that plant CNGCs are inward rectifying Ca2+ channels [21, 31, 33]. To determine whether CNGC20 possesses permeability to Ca2+, we injected YFP-CNGC20 capped RNAs into Xenopus oocytes for two-electrode voltage clamping (Figure 6A). Oocytes expressing YFP-CNGC20, but not the water-injected controls, exhibited significant inward currents at negative voltages in the presence of 10 or 30 mM Ca2+ (Figure 6B), indicating CNGC20 possesses Ca2+ channel activity. The CNGC20 channel activity appeared higher with 30 mM Ca2+ than that with 10 mM Ca2+ (Figure 6B), suggesting a Ca2+ dose-dependent activity. Of 20 CNGCs in Arabidopsis, CNGC19 and CNGC20 share the highest homology and are tandemly located on Chromosome 3 [13, 14]. Expression of YFP-CNGC19 in Xenopus oocytes also led to a Ca2+ dose-dependent channel activity (Figure 6A & C). Apparently, when CNGC19 and CNGC20 were co-expressed together in Xenopus oocytes, the channel activity was increased compared to CNGC19 or CNGC20 alone (Figure 6A & D). Likewise, when expressed in the yeast mutant strain K927 (cchl::TRP1), which shows a lethality phenotype upon treatment with α-mating factor due to the lack of the CCH1 Ca2+ channel [34], CNGC20 or CNGC19 alone weakly, but significantly, complemented the phenotype while the combination of CNGC20 and CNGC19 showed a strong complementation (Figure 6E). These results suggest that both CNGC19 and CNGC20 confer hyperpolarization-activated Ca2+ permeable channel activity. Ca2+ is an essential second messenger in nearly every aspect of cellular signaling programs [35]. Therefore, we speculate that mis-regulation of Ca2+ influx through CNGCs in bak1/serk4 leads to the uncontrolled activation of downstream signaling events, contributing to cell death, ROS production and defense gene induction.
Figure 6. CNGC20 and CNGC19 are Ca2+-permeable channels.
(A) Confocal fluorescence images of oocytes expressing YFP-CNGC20, YFP-CNGC19 or co-expressing nYFP-CNGC20 (N-terminal YFP fused with CNGC20) and cYFP-CNGC19 (C-terminal YFP fused with CNGC19). Water-injected control is showed on the bottom. YFP signals were observed using a confocal microscopy. BF indicates bright field. Bar=200 μm.
(B) CNGC20 exhibits Ca2+-permeable channel activity in Xenopus oocyte. Current-voltage relationship was recorded in oocytes expressing YFP-CNGC20 in the presence of CaCl2. Voltage steps of 0 to −180 mV in 20 mV decrements. Data shown are means ± SE, Control in 10 mM CaCl2 (n = 8) or 30 mM CaCl2 (n=5), CNGC20 in 10 mM CaCl2 (n = 8) or 30 mM CaCl2 (n = 8).
(C) CNGC19 exhibits Ca2+-permeable channel activity when expressed in Xenopus oocytes. Current-voltage relationship was recorded in oocytes expressing YFP-CNGC19 in the presence of CaCl2. Voltage steps of 0 to −180 mV in 20 mV decrements. Data shown are means ± SE, Control in 10 mM CaCl2 (n = 10) or 30 mM CaCl2 (n=8), CNGC19 in 10 mM CaCl2 (n=15) or 30 mM CaCl2 (n = 9).
(D) CNGC19 and CNGC20 additively enhance channel activity in Xenopus oocyte. Current-voltage relationship was recorded in oocytes injected with water control (n=6), cYFP-CNGC19 (n=7), nYFP-CNGC20 (n=7), or cYFP-CNGC19+nYFP-CNGC20 (n=12) in the presence of 10 mM CaCl2.
(E) Enhanced Ca2+-permeable channel activity of CNGC19/CNGC20 in yeast complementation analysis. CNGC19/CNGC20 complemented the Ca2+-uptake deficient mutant K927 (cchl::TRP1). Time course after addition of 20 μM α-mating factor is shown. Each data point is average of three independent samples (=independent yeast transformation colony). 200 cells were scored and the percentage of viable cells was calculated.
The above experiments were repeated three times with similar results.
CNGC19 contributes to bak1/serk4 cell death in the absence of CNGC20 and dimerizes with CNGC20
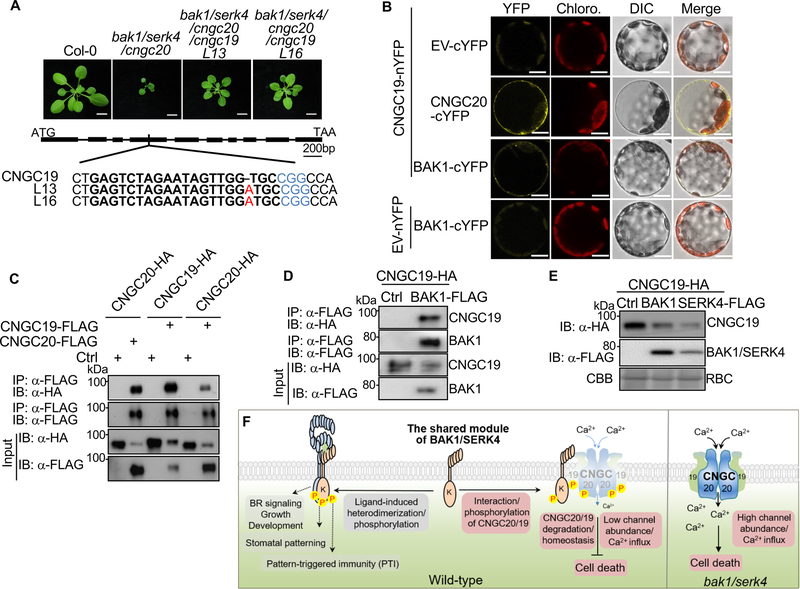
Although the cngc19 single mutant did not affect bak1/serk4 cell death, the additive effects of CNGC19 and CNGC20 in channel activity and complementing yeast mutant growth (Figure 6D & E) prompted us to test whether CNGC19 and CNGC20 additively regulate bak1/serk4 cell death. We generated the bak1–4/serk4–1/cngc20–1/cngc19 quadruple mutants using CRISPR/Cas9 genome editing to introduce lesions in CNGC19 in the bak1–4/serk4–1/cngc20–1 background. The homozygous bak1–4/serk4–1/cngc20–1/cngc19 quadruple mutants contain an adenine insertion in the fourth exon of CNGC19, which caused a frameshift mutation leading to a premature stop codon (Figure 7A & S7A). When grown on soil, the bak1/serk4/cngc20 mutant showed the growth retardation three weeks after germination and could not set seeds at 23°C (Figure 1F). However, the bak1/serk4/cncg19/cngc20 quadruple mutants almost fully rescued the cell death phenotype of bak1/serk4 and were able to set seeds normally (Figure 7A). Thus, CNGC19 makes undetectable contributions to bak1/serk4 cell death in the presence of CNGC20, however, it contributes to bak1/serk4 cell death when CNGC20 is absent. Notably, the CNGC20 transcripts were about ~10 fold higher than CNGC19 in WT plants (Figure 5A), reinforcing the primary role of CNGC20 in regulating bak1/serk4 cell death. Taken together, CNGC20 plays a major role with a quantitative effect of CNGC19 in regulating bak1/serk4 cell death.
Figure 7.
CNGC20 and CNGC19 heteromerize and contribute additively to bak1/serk4 cell death (A) The CRISPR/Cas9 cngc19 mutants further rescue the growth defect of bak1–4/serk4–1/cngc20–1. Plants grown in soil were photographed at 21 dpg. Bar=1 cm. Scheme of CNGC19 with the gene editing site is shown below. Solid bars indicate exons and lines indicate introns. Both line13 and line16 of the quadruple mutants carry an adenine insertion (red) in the fourth exon of CNGC19. Selected target sequences (20 base pairs) are shown in bold and protospacer adjacent motif (PAM) sequences are shown in blue.
(B) CNGC19 associates with CNGC20 and BAK1 in a BiFC assay. CNGC19-nYFP was co-expressed with EV-cYFP, CNGC20-cYFP, or BAK1-cYFP in Arabidopsis protoplasts. Reconstructed YFP signals were observed using a confocal microscopy 12 hr after transfection. EV indicates the empty vector. Bar=10 μm.
(C) Homo- and heteromerization of CNGC19 and CNGC20 in Arabidopsis protoplasts. Proteins were transiently co-expressed in Arabidopsis protoplasts and subjected for immunoprecipitation assays with α-FLAG agarose beads for IP and with α-HA or α-FLAG for IB (top two panels). The input control is shown on the bottom two panels.
(D) CNGC19 associates with BAK1 in Arabidopsis protoplasts. BAK1-FLAG and CNGC19-HA proteins were transiently co-expressed in Arabidopsis protoplasts. Co-IP was performed as in (C).
(E) CNGC19 proteins are destabilized by co-expression with BAK1 or SERK4. CNGC19-HA was co-expressed with Ctrl, BAK1-FLAG or SERK4-FLAG in Arabidopsis protoplasts for 12 hr. Protein expression was analyzed with α-HA or α-FLAG immunoblot.
(F) A model for cell death regulation by BAK1/SERK4-mediated phosphorylation of CNGC20/CNGC19. As a shared module, BAK1/SERK4 regulate plant growth, development, and immunity via ligand-induced heterodimerization and transphosphorylation with their cognate receptor kinases. Unique to cell death regulation, BAK1/SERK4 associate with and phosphorylate the plasma membrane-localized Ca2+ permeable channels CNGC20/CNGC19. Specific phosphorylation at the C-terminal cytosolic domain modulates the degradation of the CNGC20/CNGC19 complex and cellular homeostasis. In the bak1/serk4 mutant, without BAK1/SERK4 phosphorylation, CNGC20/CNGC19 are overproduced, leading to mis-regulation of Ca2+ influx and downstream signaling, eventually causing cell death. The abundance of CNGC20 is likely more than CNGC19.
The above experiments were repeated three times with similar results.
See also Figure S7.
Conventional animal CNGC channels exist as heterotetramers composed of A and B subunits [29]. Plant CNGCs also form both homomeric and heteromeric tetramers [15, 36]. To examine whether CNGC19 and CNGC20 also assemble a multi-unit protein complex, we tested the association between CNGC19 and CNGC20 with BiFC and Co-IP assays. CNGC20 associated with CNGC19 in Arabidopsis protoplasts (Figure 7B & C). In addition, both CNGC20 and CNGC19 were self-associated, suggesting that CNGC20 and CNGC19 likely function as homo- and heteromeric protein complexes (Figure 7C). Human CNGCs assemble via the C-terminal cytosolic domain [29]. Similarly, CNGC20C self-associated in Arabidopsis protoplasts (Figure S7B). Similar to CNGC20, CNGC19 also associated with BAK1 (Figure 7B & D). The four critical BAK1 phosphorylation sites (Thr560/Ser617/Ser618/Thr619) in CNGC20 are all conserved in CNGC19 (Figure S6E). In accordance, the CNGC19 protein level was regulated by BAK1/SERK4 (Figure 7E), likely through phosphorylation on those conserved sites. Thus, the data indicate functional homo- and heteromeric channel complexes of CNGC20 and CNGC19 regulated by BAK1 and SERK4.
The additive function of CNGC19 and CNGC20 in bak1/serk4 cell death led us to examine whether they regulate immune signaling in a similar manner. The cngc19/cngc20 double mutants were generated by introducing the same CRISPR/Cas9 CNGC19 construct into cngc20–1. flg22-induced MAPK activation remained similar to WT in two independent cngc19/cngc20 lines (Figure S7C). The cngc19/cngc20 mutant also did not affect flg22-induced Ca2+ influx, another early PTI event, which was measured by the single-wavelength fluorescent Ca2+ indicator GCaMP3 (Figure S7D). Neither cngc20 nor cngc19/cngc20 affected Pep1-mediated seedling growth inhibition (Figure S7E). Additionally, disease resistance to Pst DC3000 or Pst DC3000 avrRpt2 in cngc19/cngc20 plants was similar to that of WT (Figure S7F & G). Therefore, CNGC19 and CNGC20 are unlikely involved in plant PTI and ETI.
DISCUSSION
As a shared coreceptor, BAK1 interacts with and trans-phosphorylates cognate receptors [5, 6]. Few BAK1 substrates in transducing intracellular signaling have been identified. We show here that BAK1 directly interacts with and phosphorylates a functional Ca2+-permeable channel CNGC20, and likely CNGC19, at C-terminal specific sites (Figure 7F). The C-terminal cytosolic domain of mammalian CNGCs has been recognized to be essential for subunit interactions and cyclic nucleotide gating [29]. Intriguingly, BAK1-mediated phosphorylation regulates CNGC20 and CNGC19 stability and homeostasis. Although the regulatory mechanism remains unclear, BAK1 also interacts with CNGC17, which may form a functional cation-translocating unit with H+-ATPases for Phytosulfokine (PSK) receptor-mediated root growth [37]. Additionally, BAK1 phosphorylates sugar transport protein 13 (STP13) to enhance its monosaccharide uptake activity, thereby competing with bacteria for extracellular sugars likely as a defense mechanism [38]. Apparently, in addition to signaling molecules, ion channels and transporters are also regulated by cell surface-resident RLKs for their homeostasis and activation.
Two recent reports show that plant RLCKs phosphorylate and activate CNGCs mediating MAMP-induced Ca2+ influx in plant PTI [21, 33]. Arabidopsis RLCK BIK1 interacts and activates CNGC2 and CNGC4 by phosphorylation, leading to an increase of cytosolic Ca2+ [21]. Rice OsRLCK185 directly interacts and phosphorylates OsCNGC9 to activate its channel activity and regulate rice resistance to blast disease [33]. Notably, phosphorylation by BIK1, a direct substrate of BAK1 [39], contributes to the activation of CNGC2/CNGC4 channel activity in PTI signaling, whereas phosphorylation by BAK1 diminishes CNGC19/CNGC20-mediated signaling in controlling cell survival. In addition, CNGC2 and CNGC4 likely assemble into a functional heterotetrameric Ca2+ channel and neither CNGC2 nor CNGC4 alone is a functional channel [21], which is different from that CNGC20 and CNGC19 form functional homo- and hetero-meric Ca2+ channels. These parallel but distinct regulatory mechanisms further emphasize the importance of BAK1 in bifurcating the signaling specificity of various biological processes, as well as the diverse functions and regulations of CNGC family proteins.
It has been shown previously that protein glycosylation is involved in regulating bak1/serk4 cell death [10]. There are six and eight potential glycosylation sites in CNGC20 and CNGC19 respectively. It will be interesting to test in the future whether CNGC19/CNGC20 are glycosylated and whether glycosylation is important for their function. In addition, cytoplasmic export of mRNAs plays a role in bak1/serk4 cell death [40]. It is possible that the nucleocytoplasmic trafficking facilitates the export of CNGC20 and CNGC19 transcripts from nucleus, which further contributes to the cell death in bak1/serk4. Alternatively, it has been hypothesized that BAK1/SERK4 may play a role in keeping in check the specific cellular perturbation and safeguarding cellular homeostasis for cell survival [8]. This notion is supported by our current results that BAK1/SERK4 keep CNGC19/CNGC20 proteins at a low level under normal growth conditions. When this regulation is absent (i.e. in the bak1/serk4 mutant,) cellular homeostasis or cell survival cannot be maintained, thus, as a consequence, the cell undergoes cell death. In summary, our results pinpoint a regulatory mechanism where the homeostasis of CNGC20 and CNGC19 modulated by receptor-like kinase BAK1-mediated phosphorylation is the key for precise control of plant cell death, which expands the portfolio of CNGC functions and regulations.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Libo Shan (lshan@tamu.edu). we will distribute the plasmids and transgenic plants freely to the scientific community upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis thaliana and growth conditions
All Arabidopsis plants used in this study were in the Columbia-0 (Col-0) background. The various mutants and transgenic lines used in this study were described in the Key Resources Table. Arabidopsis lines were grown in soil (Metro Mix 366) in a growth room at 23°C, 45% humidity and 85μE m−2s−1 light with a 12-hr light/12-hr dark photoperiod for two-weeks before VIGS assays or 30 days for protoplast isolation, ROS production and pathogen assays. Seedlings were germinated on plates containing half-strength Murashige and Skoog medium (1/2MS) with 0.5% sucrose, 0.8% agar and 2.5 mM MES at pH 5.7, and grown under the same growth condition as above for 10 days. The seedlings were transferred to a 6-well tissue culture plate with 2 ml H2O for overnight, and then used for indicated assays.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA-Peroxidase | Roche | Cat # 12013819001; RRID:AB_439705 |
| Anti-FLAG-Peroxidase | Sigma-Aldrich | Cat # A8592; RRID:AB_259529 |
| Anti-GFP | Roche | Cat # 11814460001; RRID:AB_390913 |
| Anti-Mouse IgG HRP-linked antibody | Cell Signaling | Cat # 7076; RRID:AB_330924 |
| Phospho-p44/42 MAPK (Erk1/2) Antibody | Cell Signaling | Cat #9101; RRID:AB_331646 |
| Anti-rabbit IgG HRP-linked antibody | Cell Signaling | Cat #7074; RRID:AB_2099233 |
| Anti-FLAG M2 Affinity gel | Sigma-Aldrich | Cat # 2220; RRID:AB_10063035 |
| Protein G Agarose | Roche | Cat # 05015952001 |
| Bacterial and Virus Strains | ||
| Agrobacterium tumefaciens GV3101 | [10] | N/A |
| E. coli BL21 | [42] | N/A |
| Pseudomonas syringae pv. tomato DC3000 (Pst) | [41] | N/A |
| P. syringae pv. maculicola ES4326 (Psm) | [44] | N/A |
| Pst avrRpt2 | [42] | N/A |
| Pst avrRpm1 | [43] | N/A |
| Pst avrRps4 | [43] | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| flg22 | [48] | N/A |
| Pep1 | [48] | N/A |
| MG132 | AG Scientific | Cat #99533-80-9 |
| K252a | AG Scientific | Cat #133407-82-6 |
| RiboZol™ RNA Extraction Reagent | AMRESCO | Cat # N580 |
| IPTG | Sigma-Aldrich | Cat# I6758 |
| Pierce glutathione agarose | Thermo Scientific | Cat# 16101 |
| amylose resin | New England BioLabs | Cat# E8021L |
| Luminol | Sigma-Aldrich | Cat#A8511 |
| Peroxidase from horseradish | Sigma-Aldrich | Cat#P6782 |
| RNase-free DNase I | New England BioLabs | Cat # M0303L |
| Critical Commercial Assays | ||
| M-MuLV Reverse Transcriptase | New England BioLabs | Cat # M0253L |
| iTaq SYBR green Supermix | Bio-Rad | Cat # 1725124 |
| mMESSAGE mMACHINE T7 high yield RNA Transcription Kit | Ambion | Cat # AM1344 |
| Phusion U Hot Start DNA polymerase | Thermo Fisher | Cat # F555 |
| USER enzyme mix | New England Biolabs | M550 |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis thaliana Col-0 wild-type | [10] | N/A |
| bak1–4 | [10] | N/A |
| btl1 (cngc20–3) | ABRC | SALK _013823C |
| cngc1 | ABRC | SAIL_443_B11 |
| cngc2 | [16] | dnd1–1 |
| cngc3 | ABRC | SALK_056832C |
| cngc4 | [17] | dnd2–1 |
| cngc5 | ABRC | SALK_149893C |
| cngc6 | ABRC | SALK_042207 |
| cngc7 | ABRC | SALK_060871C |
| cngc8 | ABRC | GABI_101C03 |
| cngc9 | ABRC | SALK_026086 |
| cngc10 | ABRC | SALK_015952C |
| cngc11 | ABRC | SALK_026568C |
| cngc12 | ABRC | SALK_092657 |
| cngc13 | ABRC | SALK_060826 |
| cngc14 | ABRC | WiscDsLox437E09 |
| cngc15 | ABRC | CS93507 |
| cngc16 | ABRC | SAIL_726_B04 |
| cngc17 | ABRC | SALK_041923 |
| cngc19 | ABRC | SALK_007105 |
| cngc20–1 | ABRC | SALK_129133C |
| cngc20–2 | ABRC | SALK_074919C |
| bak1–4 | [10] | N/A |
| bak1–4/serk4–1 | [10] | N/A |
| pepr1/2 | [19] | N/A |
| bak1–4/cngc20–1 | This paper | N/A |
| bak1–4/serk4–1/cngc20–3 | This paper | N/A |
| bak1–4/serk4–1/cngc20–1 | This paper | N/A |
| bak1–4/serk4–1/cngc20–1/cngc19 | This paper | N/A |
| pCNGC20::gCNGC20-HA/cngc20–3 | This paper | N/A |
| pCNGC20::gCNGC20T560/S617/S618/T619D-HA/cngc20–1 | This paper | N/A |
| p35S::CNGC20-HA/Col-0 | This paper | N/A |
| p35S::CNGC20-HA/bak1–4 | This paper | N/A |
| p35S::AT1G60995-HA/btl1 | This paper | N/A |
| pAT1G60995::AT1G60995-HA/btl1 | This paper | N/A |
| Xenopus laevis | [49] | N/A |
| Saccharomyces cerevisiae strain W303–1A | H Iida laboratory (Tokyo Gakugei University) | N/A |
| Saccharomyces cerevisiae strain K927 | H Iida (Tokyo Gakugei University). | N/A |
| Saccharomyces cerevisiae strain AH109 | [41] | N/A |
| Oligonucleotides | ||
| Primers for VIGS, cloning and point mutation, see Table S4 | This paper | N/A |
| Primers for genotyping, see Table S4 | This paper | N/A |
| Primers for qRT-PCR and RT-PCR, see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| pYL156 (pTRV-RNA2) | [10] | N/A |
| pTRV-RNA1 | [10] | N/A |
| pYL156-GFP | [10] | N/A |
| pYL156-BAK1/SERK4 | [10] | N/A |
| pYL156-MEKK1 | [10] | N/A |
| pYL156-BIR1 | [10] | N/A |
| pHBT | [41] | N/A |
| pGST | [45] | N/A |
| pMAL-c2 | [45] | N/A |
| pCB302 | [10] | N/A |
| pGADT7 | [41] | N/A |
| pGBKT7 | [41] | N/A |
| pNB1 | [49] | N/A |
| pHEE401E | [47] | N/A |
| pCBC-DT1T2 | [47] | N/A |
| pHBT-p35S::GCaMP3 | This paper | N/A |
| pYES2 | [34] | N/A |
| pHBT-BAK1-HA | [45] | N/A |
| pHBT-BIK1-HA | [45] | N/A |
| pHBT-BAK1-FLAG | [45] | N/A |
| pHBT-SERK4-FLAG | [45] | N/A |
| pMAL-BAK1CD-HA | [45] | N/A |
| pMAL-BAK1CDKM-HA | [45] | N/A |
| pHBT-CNGC20-HA | This paper | N/A |
| pHBT-CNGC20-FLAG | This paper | N/A |
| pHBT-CNGC20-cYFP | This paper | N/A |
| pHBT-CNGC19-HA | This paper | N/A |
| pHBT-CNGC19-nYFP | This paper | N/A |
| pHBT-CNGC20C-HA | This paper | N/A |
| pHBT-CNGC20N-FLAG | This paper | N/A |
| pHBT-CNGC20NT145/S146/S183/S184A-HA | This paper | N/A |
| pHBT-CNGC20CT560A-HA | This paper | N/A |
| pHBT-CNGC20CT560/S617/S618/T619A-HA | This paper | N/A |
| pHBT-CNGC20CT560/S617/S618/T619D-HA | This paper | N/A |
| pHBT-p35S::gCNGC20-HA | This paper | N/A |
| pHBT-pCNGC20::gCNGC20-HA | This paper | N/A |
| pGST-CNGC20N | This paper | N/A |
| pGST-CNGC20C | This paper | N/A |
| pGST-CNGC20NT145/S146/S183/S184A | This paper | N/A |
| pGST-CNGC20CT560A | This paper | N/A |
| pGST-CNGC20CT560/S617/S618/T619A | This paper | N/A |
| pGADT7-BAK1K | This paper | N/A |
| pGBKT7-CNGC20N | This paper | N/A |
| pCAMBIA1300-pCNGC20::gCNGC20-HA | This paper | N/A |
| pCAMBIA1300-pCNGC20::gCNGC20T560/S617/S618/T619D-HA | This paper | N/A |
| pNB1YFP-CNGC19 | This paper | N/A |
| pNB1YFP-CNGC20 | This paper | N/A |
| pNB1YC-CNGC19 | This paper | N/A |
| pNB1YN-CNGC20 | This paper | N/A |
| pHEE401E-CNGC19 | This paper | N/A |
| pCB302–35S::CNGC20-HA | This paper | N/A |
| pCB302-p35S::AT1G60995-HA | This paper | N/A |
| pCB302-pAT1G60995::AT1G60995-HA | This paper | N/A |
| pCB302-pBAK1::BAK1-FLAG | This paper | N/A |
| pYES2-CNGC19 | This paper | N/A |
| pYES2-CNGC20 | This paper | N/A |
| Software and Algorithms | ||
| Bowtie aligner software | [50] | http://bowtie-bio.sf.net |
| CLC Genomics Workbench 6.0.1 software | QIAGEN | http://www.clcbio.com |
| LTQ Orbitrap XL LC-MS/MS system | Thermo Scientific | N/A |
| Mascot | Matrix Science | Version 2.2.2 |
| Olympus Fluoview Viewer | Olympus | Version 3.0 |
| ZEN | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
| LAS-X | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Nicotiana benthamiana and growth conditions
Nicotiana benthamiana was grown in greenhouses in soil under a 12-hr light/12-hr dark photoperiod at 23°C.
Bacterial strains
The various bacteria strains used in this study were described in the Key Resources Table. Pseudomonas syringae pv. tomato (Pst) DC3000 were grown on the King’s B medium plates with 50 μg/ml rifampicin. Pst DC3000 carrying avrRpt2, avrRps4 or avrRpm1 was grown with 50 μg/ml kanamycin and 50 μg/ml rifampicin [41–43]. P. syringae pv. maculicola ES4326 (Psm) was grown with 50 μg/ml Streptomycin [44]. All the Pseudomonas strains were grown on plates at 28°C for 2 days for further inoculum preparation.
METHOD DETAILS
Plasmid construction and generation of transgenic plants
The VIGS of BAK1/SERK4, MEKK1 and BIR1 constructs and the pHBT-BIK1-HA, pHBT-BAK1-HA, pHBT-BAK1-FLAG, pMAL-BAK1CD-HA, and pMAL-BAK1CDKM-HA constructs were reported previously [10, 45]. The CNGC19 and CNGC20 genes were amplified from Col-0 cDNA with primers containing NcoI at N-terminus and StuI at C-terminus (Supplemental Table S4), and ligated into a plant protoplast expression vector pHBT under the control of a CaMV 35S promoter with an HA, FLAG, cYFP or nYFP tag at C-terminus. N-terminus and C-terminus of CNGC20 (CNGC20N and CNGC20C) were cloned using the above constructs as the templates and primers as listed in the Supplemental Table S4. The point mutations of CNGC20NT145/S146/S183/S184A, CNGC20CT560A, CNGC20CT560/S617/S618/T619A, CNGC20T560/S617/S618/T619D were generated by site-directed mutagenesis with primers listed in Supplemental Table S4. To construct the E.coli expression vectors, CNGC20N, CNGC20C, CNGC20NT145/S146/S183/S184A, CNGC20CT560A and CNGC20CT560/S617/S618/T619A were subcloned into a modified GST fusion protein expression vector pGEX4T-1 (Pharmacia) using BamHI (the enzyme site located in front of NcoI on the pHBT vector) and StuI digestion. The CNGC20N was subcloned into a modified pGBKT7 vector (Clontech) for yeast two-hybrid assays using BamHI and StuI digestion. To introduce CNGC20 into the binary vectors, the CNGC20 fragment was released from the pHBT vector using BamHI and StuI digestion and ligated into the pCB302 (35S promoter) or pMDC (2×35S promoter) binary vectors.
To construct the pCAMBIA1300 binary vector containing the native promoter driven gCNGC20 for Agrobacterium-mediated transformation, the genomic fragment of the CNGC20 gene (gCNGC20) was amplified from Col-0 genomic DNA using primers containing NcoI at N-terminus and StuI at C-terminus (Supplemental Table S4), and ligated into pHBT under the control of the 35S promoter with an HA epitope-tag at its C-terminus to obtain the pHBT-p35S::gCNGC20-HA vector. The CNGC20 promoter (2 kb upstream of the start codon) was amplified from Col-0 genomic DNA using primers containing XhoI at N-terminus and NcoI at C-terminus (Supplemental Table S4), and ligated into pHBT-p35S::gCNGC20-HA to obtain the pHBT-pCNGC20::gCNGC20-HA vector. The native promoter driven gCNGC20 together with the HA epitope and the NOS terminator were released from pHBT-pCNGC20::gCNGC20-HA using XhoI/EcoRI digestion and ligated into pCAMBIA1300 to obtain the pCAMBIA1300-pCNGC20::gCNGC20-HA binary vector. The point mutations of gCNGC20T560/S617/S618/T619D were generated by site-directed mutagenesis in the corresponding vector.
To construct the oocyte expression vectors for electrophysiology studies, CNGC19 and CNGC20 were cloned into the pNB1 serial vectors using the USER method to obtain pNB1YFP-CNGC19, pNB1YFP-CNGC20, pNB1YC-CNGC19 and pNB1YN-CNGC20 expression vectors [46]. Uracil-containing forward primer was designed as 5’-GGCTTAAU + sequence complementary to the target gene-3’, and reverse primer as 5’-GGTTTAAU + sequence complementary to the target gene-3’. PCR was performed with Phusion U Hot Start DNA polymerase (Thermo Fisher, F555) according to manufacturer’s instruction. The reaction mixture containing PCR product, USER enzyme mix (New England Biolabs, M550), and PacI/Nt.BbvCI digested pNB1 vector was incubated 20 min at 37°C followed by 20 min at 25°C, and then transformed into chemically competent E. coli cells. PCR and Sanger sequencing were performed to confirm the positive clones.
To construct the CRISPR/Cas9 CNGC19 vector, two suitable guide RNAs (gRNAs) without predicted off-targets were designed via the website http://chopchop.cbu.uib.no/. gRNA1, located in the 4th exon of CNGC19 (5’-GAGTCTAGAATAGTTGGTGC-3’), and gRNA2, located in the 5th exon of CNGC19 (5’-CGAAGTCACAACGAGATCTG-3’), were incorporated into the gRNA expression cassette by the first round PCR using pCBC-DT1T2 [47] as a template. The resulting PCR fragments were further used as the template to incorporate the BsaI restriction enzyme site through the second round PCR. The final PCR fragments were inserted into the BsaI site of the CRISPR/Cas9 pHEE401E vector [47].
The AT1G60995 gene was amplified from Col-0 cDNA with primers containing XbaI and NcoI at N-terminus and StuI at C-terminus (Supplemental Table S4), and ligated into the binary vector pCB302 under the control of the 35S promoter with an HA tag at C-terminus to generate pCB302-p35S::AT1G60995-HA. The promoter of AT1G60995 (520 bp upstream of the start codon) was amplified from Col-0 genomic DNA using primers containing SacI at N-terminus and XbaI at C-terminus (Supplemental Table S4), and ligated into pCB302-p35S::AT1G60995-HA to generate pCB302-pAT1G60995::AT1G60995-HA vector.
The promoter of BAK1 (1.5kb upstream of the start codon) was amplified from Col-0 genomic DNA using primers containing XhoI and XbaI at N-terminus and BamHI at C-terminus (Supplemental Table S4), and ligated into pHBT-p35S::BAK1-FLAG to obtain pHBT-pBAK1::BAK1-FLAG. The native promoter driven BAK1 was released using XbaI/StuI digestion and ligated into pCB302 with a FLAG tag at the C terminus to obtain the pCB302-pBAK1::BAK1-FLAG binary vector.
The sequences of all genes, promoters or gRNAs were verified by the Sanger-sequencing. These binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis using the floral dipping method.
Agrobacterium-mediated floral dipping
Agrobacterium tumefaciens GV3101 containing the binary vector was cultured at 28°C in LB liquid medium with 50 μg/ml Kanamycin and 25 μg/ml Gentamicin. Bacteria were harvested by centrifugation at 3000 rpm for 15 min and the pellet was suspended with buffer containing 50 mM MES (pH 5.5–5.7), 5% sucrose and 200 μl/L silwetL-77 at the density of OD600 =0.8. Arabidopsis flower buds were dipped thoroughly to the bacteria solution and then the dipped plants were covered with a dome for 24 hr to maintain high humidity. After that, plants were placed in the greenhouses under 12-hr light/12-hr dark light period and seeds were harvested for transgenic plants selection.
Elicitor and chemical inhibitor treatments
The elicitors flg22 and Pep1 were reported previously [48]. MG132 (AG Scientific #99533-80-9) and K252a (AG Scientific #133407-82-6) were diluted from DMSO stock solutions (2 mM for MG132 and 2 mM for K252a).
Trypan blue and DAB staining
Trypan blue staining and 3, 3′-diaminobenzidine (DAB) staining were performed according to procedures described previously with modifications. Briefly, the excised plant leaves were immersed in trypan blue staining solution (2.5 mg/mL trypan blue in lactophenol [lactic acid: glycerol: liquid phenol: H2O = 1:1:1:1]) or DAB solution (1 mg/mL DAB in 10 mM Na2HPO4 and 0.05% Tween 20). Samples were vacuum-infiltrated for 30 min and then incubated for 8 hr at 25°C with gentle shaking at 75 rpm. Subsequently, samples were transferred to trypan blue destaining solution (ethanol: lactophenol = 2:1) or DAB destaining solution (ethanol: acetic acid: glycerol = 3:1:1) and incubated at 65°C for 30 min. The samples were then incubated in fresh destaining solution at room temperature until complete destaining. Pictures were taken under a dissecting microscope with samples in 10% glycerol.
Agrobacterium-mediated virus-induced gene silencing (VIGS) assay
Plasmids containing binary TRV vectors pTRV-RNA1 and pTRV-RNA2 derivatives, pYL156-BAK1/SERK4, pYL156-MEKK1, pYL156-BIR1, pYL156-GFP (the vector control) were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Bacterial cultures were first grown in LB medium containing 50 μg/ml kanamycin and 25 μg/ml gentamicin for overnight and then sub-cultured in fresh LB medium containing 50 μg/ml kanamycin and 25 μg/ml gentamicin supplemented with 10 mM MES and 20 μM acetosyringone for overnight at 28°C in a roller drum. Cells were pelleted by 4200 rpm centrifugation, re-suspended in a solution containing 10 mM MgCl2, 10 mM MES and 200 μM acetosyringone, adjusted to OD600 of 1.5 and incubated at 25°C for at least 3 hr. Bacterial cultures containing pTRV-RNA1 and pTRV-RNA2 derivatives were mixed at a 1:1 ratio and inoculated into the first pair of true leaves of two-week-old soil-grown plants using a needleless syringe.
Electrophysiological Studies in Xenopus laevis Oocytes
Capped RNAs (cRNAs) were in vitro transcribed from the linearized pNB1 vectors using the mMESSAGE mMACHINE T7 high yield RNA Transcription Kit following the manufacturer’s protocol (Ambion). The quality of cRNAs was checked by Nanodrop and concentration was adjusted to the same level and stored at −80°C until injection. The expression and Two-Electrode Voltage-Clamp Recordings Xenopus oocytes were performed as described previously [49]. Xenopus oocytes were harvested at the stage V to VI and kept in a ND96 perfusion solution (96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, 10 mM sorbitol, pH was adjusted to 7.4 with NaOH) for overnight prior to injections. Each oocyte was injected with 25 ng cRNAs or the same amount of water. Injected oocytes were incubated in perfusion solution at 18 °C for two days, and the protein expression was detected by YFP fluorescence signals (excited at 514 nm) using the Leica SP8 Laser confocal microscope. The currents were recorded with hyperpolarized pulses of a 0.1-s pre-pulse at −40 mV, followed by voltage steps of 0 to −180 mV (step at −20 mV, 1.5-s duration) and a 0.5-s deactivation at −40 mV using Axon Axoclamp 900A Microelectrode Amplifier. The bath solution for current recording contained 10 mM or 30 mM CaCl2, 10 mM MES-Tris pH7.4, and osmolality was adjusted to 220 mOsm/L with mannitol. The pipette solution contained 3 M KCl.
Analysis of mutations at CRISPR-Cas9 target sites
Genomic DNAs were extracted from individual CRISPR/Cas9 transgenic T1 plants and WT plants. About 1100-bp fragments covering the targeted locus of the gRNAs were amplified by PCR using primers gCNGC19-CRISPR listed in Supplemental Table S4 and subjected for enzyme digestions. NlaIV was used to detect the mutation in the first target site and BglII was used for the second target site. Sanger sequencing of the PCR products using both the forward and reverse primers of gCNGC19-CRISPR was performed to verify the presence of mutations. Transgenic plants carrying mutations, especially in the gRNA1 target locus, were kept to harvest seeds. PCR products obtained using DNAs from individual T2 plants were analyzed by enzyme digestion and Sanger sequencing as mentioned above to confirm the mutations in T2 plants, and T3 homozygous mutants carrying mutation in the gRNA1 target locus were kept for seeds.
Next generation sequence analysis of cngc20–3 (SALK_013823C) to map T-DNA insertions
The genomic DNAs of cngc20–3 were isolated for 100 nt paired-end sequencing on an Illumina HiSeq 2000 platform at Texas AgriLife Genomics and Bioinformatics Service (TAGS) (College Station, TX, USA). The Illumina reads of cngc20–3 were first aligned to TAIR10 release of the Col-0 genome using the Bowtie aligner software with no more than three mis-matches [50]. A total of 9,527,475 (account for 90.6%) reads have been aligned. The unaligned reads were recovered and mapped to the T-DNA vector (pROK2) sequence with no more than three mismatches, and 2,182 reads have been mapped. The remaining unaligned reads (988,418), which were neither aligned to the Col-0 genome sequence nor the T-DNA vector sequence, were of interest to map over the T-DNA insertion breakpoint and reveal their locations. These reads were first blasted against the T-DNA vector sequence to retrieve any reads hitting to the T-DNA vector sequence and then blasted against the Col-0 genome sequence. In total, 20 reads aligned to both the T-DNA boarder sequence and the Col-0 genome sequence were retrieved and verified by the CLC Genomics Workbench 6.0.1 software (http://www.clcbio.com). Three major breakpoints in AT1G60995, AT1G11020 and AT3G17700 (CNGC20) were detected and confirmed by Sanger-sequencing using primers upstream and downstream of each breakpoint.
RT-PCR and qRT-PCR analysis
Total RNA was isolated from ten-day-old seedlings grown on 1/2MS plates or leaves of soil-grown plants two weeks after Agrobacterial inoculation for VIGS assay with TRIzol reagent. RNA was reverse transcribed to synthesize first strand cDNA with M-MuLV reverse transcriptase and oligo (dT) primer following RNase-free DNase I treatment. RT-PCR analysis was carried out using Taq DNA polymerase. Fragments of target genes were amplified using the primers listed in Table S4. UBQ1 or ACTIN2 was used as an internal control. Fragments were separated in 1.5% agarose gel and revealed by ethidium bromide staining and UV light exposure. Quantitative RT-PCR (qRT-PCR) analysis was carried out using iTaq SYBR green Supermix supplemented with ROX in an ABI GeneAmp® PCR System 9700. The expression of genes was normalized to the expression of UBQ10 or ACTIN2.
MAPK assay
Ten-day-old seedlings grown on 1/2MS plates were transferred to water for overnight-recovery and then treated with 100 nM flg22 for indicated times. Each sample containing three seedlings was grounded in 40 ul of extraction buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 5 mM EDTA, 1% Triton X-100, 1 mM Na3VO4, 1 mM NaF, 1 mM DTT, 1:200 complete protease inhibitor cocktail from Sigma). Supernatant was collected after 12,000 rpm centrifugation for 5 min at 4°C and protein samples with 1 x SDS buffer were loaded on 10% SDS-PAGE gel to detect pMPK3, pMPK4 and pMPK6 by immunoblot with α-pERK1/2 antibody.
BIK1 phosphorylation assay
Protoplasts were isolated from four-week-old WT, bak1–4, cngc20–1 and bak1–4/cngc20–1 plants according to the protocol described previously [51]. 200 μl of protoplasts at the density of 2 × 105/ml for each genotype were transfected with 20 μg of plasmid DNA carrying HA-tagged BIK1 (pHBT-p35S::BIK1-HA) at the concentration of 1.8 μg/μl. Protoplasts were incubated at 25°C for 12 hr and treated with or without 100 nM flg22 for 10 min. Samples were subjected for 10% SDS-PAGE and immunoblotting with α-HA-HRP antibody. The flg22-induced BIK1 phosphorylation was evidenced as a mobility shift from immunoblot.
Bacterial infection assay
Pseudomonas. syringae pv. maculicola ES4326 (Psm), P. syringae pv. tomato (Pst) DC3000 (avrRpt2), Pst DC3000 (avrRpm1) and Pst DC3000 (avrRps4) were cultured for overnight at 28°C in the King’s B medium with the appropriate antibiotics (50 μg/ml streptomycin, rifampicin or kanamycin). Bacteria were harvested by centrifugation at 3500 rpm, washed with ddH2O, and adjusted to the desired density with 10 mM MgCl2. Leaves of four-week-old plants were hand-infiltrated with bacterial suspension using a 1-ml needleless syringe and collected at the indicated time for HR or bacterial growth assays. To measure bacterial growth, two leaf discs were ground in 100 μl H2O and serial dilutions were plated on TSA medium (1% Bacto tryptone, 1% sucrose, 0.1% glutamic acid, 1.5% agar) with appropriate antibiotics. Bacterial colony forming units (cfu) were counted 2 days and 4 days after inoculation. Each data point is shown as triplicates.
In vivo co-immunoprecipitation (Co-IP) assay
Arabidopsis protoplasts were transfected with a pair of constructs tested (the empty vector as the negative control) and incubated for 12 hr. Samples were collected by centrifugation and lysed with Co-IP buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% Triton X-100 and protease inhibitor cocktail from Roche) by vertexing. For Co-IP in Nicotiana benthamiana, leaves of three-week-old soil-grown plants were hand-infiltrated with different pairs of Agrobacterium tumefaciens carrying indicated vectors. Overnight cultured bacteria were harvested by centrifugation and re-suspended in buffer (10 mM MES, pH5.7, 10 mM MgCl2, 200 μM acetosyringone) at OD600=1.5. Leaf samples were harvested two days post-inoculation and subjected to homogenization with Co-IP buffer. Protein extract was pre-incubated with protein-G-agarose beads for 1 hr at 4°C with gentle shaking on a rocker. Immunoprecipitation was carried out with α-FLAG agarose for 3 hr at 4°C. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100). The immunoprecipitated proteins and input proteins were analyzed by immunoblotting with indicated antibodies.
In vitro pull-down and kinase assays
Fusion proteins were expressed in E. coli BL21 strain using LB medium supplemented with 0.25 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) [42]. Glutathione-S-transferase (GST), GST-CNGC20N, GST-CNGC20NT145/S146/S183/S184A, GST-CNGC20C, GST-CNGC20CT560A and GST-CNGC20CT560/S617/S618/T619A were purified with Pierce glutathione agarose, and maltose binding protein (MBP), MBP-BAK1CD and MBP-BAK1CDKM proteins were purified using amylose resin according to standard protocols. MBP fusion proteins (tagged with HA) were pre-incubated with prewashed glutathione agarose in 300 μL incubation buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 0.1mM EDTA and 0.5% Triton X-100) for 0.5 hr at 4°C. After centrifugation, the supernatant was collected and incubated with prewashed GST, GST-CNGC20N or GST-CNGC20C beads for another 1 hr. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH7.5, 300 mM NaCl, 0.1mM EDTA and 0.1% Triton X-100). Proteins were detected with an α-HA antibody by immunoblotting. For in vitro kinase assay, The 1 μg MBP, MBP-BAK1CD or MBP-BAK1CDKM proteins were incubated with 5 μg GST, GST-CNGC20N, GST-CNGC20NT145/S146/S183/S184A, GST-CNGC20C, GST-CNGC20CT560A or GST-CNGC20CT560/S617/S618/T619A in the kinase reaction buffer (20 mM Tris-HCl, pH7.5, 20 mM MgCl2, 5 mM EDTA, 1 mM DTT and 100 μM ATP) in the presence of 5 μCi [32P]-γ-ATP for 2 hr at room temperature. The reactions were stopped by adding SDS sample buffer, and protein phosphorylation was visualized by autoradiography in 10% SDS-PAGE.
Mass spectrometry analysis
The in vitro phosphorylation for MS analysis was performed in a 20 μl reaction for 2 hr at RT. The reaction buffer contains 20 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 5 mM EDTA, 1 mM DTT, 5 mM ATP, 10 μg GST-CNGC20N or GST-CNGC20C and 1 μg MBP-BAK1CD. The phosphorylated GST-CNGC20N or GST-CNGC20C proteins were resolved by 10% SDS-PAGE gel. The gel was stained with Thermo GelCode Blue Safe Protein Stain and destained with ddH2O. The corresponding bands were sliced and subjected for in-gel digestion with trypsin. The phosphopeptides were enriched and analyzed using a LTQ Orbitrap XL LC-MS/MS system (Thermo Scientific) as previously described [43]. The MS/MS spectra were analyzed with Mascot (Matrix Science; version 2.2.2), and the identified phosphorylated peptides were manually inspected to ensure confidence in phosphorylation site assignment.
Yeast two-hybrid assay
The plasmids of pGADT7 (empty vector) or pGADT7-BAK1K were introduced into the yeast strain AH109. The plasmids of pGBKT7 or pGBKT7-CNGC20N were introduced into AH109 containing pGADT7 or pGADT7-BAK1K. Polyethylene glycol/LiAc-mediated yeast transformation was performed according to the protocol of Yeastmaker Yeast Transformation System 2 (Clontech). The yeast colonies containing both pGADT7 and pGBKT7 were selected on the synthetic defined (SD) medium without leucine and tryptophan (SD-L-T), and the interaction was tested on the SD medium without histidine, leucine and tryptophan (SD-H-L-T), and supplemented with 1 mM 3-amino-1, 2, 4-triazole (3-AT).
Bimolecular fluorescence complementation and subcellular localization assay
Protoplasts from four-week-old WT were transfected with different pairs of BiFC constructs as shown in the figures. Fluorescence signals in the protoplasts were examined 12 hr after transfection using the Zeiss LSM 780 NLO multiphoton confocal system. YFP and chlorophyll fluorescence signals were excited at 514 and 633 nm respectively. Images were captured in multichannel mode with bright field, and processed with Zeiss ZEN microscope software.
Growth inhibition assay
Four days after germination on 1/2MS plates, Arabidopsis seedlings with uniform root lengths were transferred to 24-well culture plates containing 500 μl liquid 1/2MS supplemented without or with 1 μM Pep1. Two seedlings were placed in one well and four repeats were performed. Seedlings were photographed at 7 days after transfer and the root length of individual seedlings were measured.
ROS assay
Around 25 leaves of four-week-old soil-grown Arabidopsis plants for each genotype were excised into leaf discs (5-mm diameter) and then cut into leaf strips, followed by an overnight incubation with water in 96-well plates to eliminate the wounding effect. ROS burst was determined by a luminol-based assay. Leaf strips were soaked with solution containing 50 μM luminol and 10 μg/mL horseradish peroxidase supplemented with 100 nM flg22. The measurement was performed immediately after adding the solution with a Multilabel Plate Reader (Perkin-Elmer; Victor X3) for a period of ~35 min. The values for ROS production from each line were indicated as means of relative light units.
Semi-quantification of Ca2+ signals
Protoplasts were isolated from four-week-old WT and CRISPR/Cas9 cngc19/cngc20–1 mutants and transfected with pHBT-p35S::GCaMP3 plasmid DNAs [52, 53]. Protoplasts were incubated at 25°C for 12 hr and transferred to a Greiner 96-well black plate followed by treatment with ddH2O (Ctrl) or 100 nM flg22. GCaMP3 fluorescence was measured immediately upon treatment using a Perkin Elmer VICTOR X3 Multilabel Plate Reader with an excitation at 485 nm and emission detection at 535 nm. Measurements were recorded for each well at 0.2s intervals, for total duration as indicated in the figures. The difference of the absolute fluorescence value with the control value for each experiment was normalized to the control value as (F-Feq)/Feq (where F was the measured fluorescence at a given time point and Feq was the averaged measurement for the samples at the final equilibrated time points measured). Plotted values were averages of five replicate wells, with the SE represented by error bars.
Ca2+ yeast mutant complementation
Saccharomyces cerevisiae strains W303–1A (WT) and K927 (cch1::TRP1 null mutant) were provided by Dr. H Iida (Tokyo Gakugei University). The Ca2+ channel mutant strain K927 was transformed with pYES2 empty vector, pYES2-CNGC19, pYES2-CNGC20, or both. To test for complementation of the cch1 mutation, yeast transformants were grown to logarithmic phase in synthetic minimal media and were diluted to 106 cells/ml and exposed to 20 μM α-mating factor in modified synthetic minimal media containing 100 μM CaCl2 [34, 54]. 100 μl aliquots of cells were harvested by centrifugation and resuspended in 10 mg/ml Trypan blue solution at various time points. Yeast viability was measured by assessing the ratio of stained to unstained cells under a bright field microscope. A minimum of 200 cells were counted for each transformant.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data for quantification analyses are presented as mean ± standard error (SE) or standard deviation (SD). The statistical analyses were performed by Student’s t-test or one-way analysis of variance (ANOVA) test (* P < 0.05, ** P < 0.01, *** P < 0.001). Number of replicates is shown in the figure legends.
DATA AND CODE AVAILABILITY
This manuscript did not generate new datasets or code.
Supplementary Material
TABLE S4. PRIMERS USED IN THIS STUDY. RELATED TO STAR METHODS.
Highlights:
CNGC20 specifically regulates bak1/serk4 cell death.
BAK1 phosphorylates CNGC20 and regulates CNGC20 stability.
CNGC19 contributes to bak1/serk4 cell death in the absence of CNGC20.
CNGC20 and CNGC19 form complexes and are Ca2+ permeable channels.
ACKNOWLEDGEMENTS
We thank the Arabidopsis Biological Resource Center (ABRC) for Arabidopsis T-DNA insertion library and various mutant seeds, Dr. Qijun Chen (China Agricultural University, China) for the CRISPR/Cas9 system, Dr. Tim Devarenne for critical reading of the manuscript, and members of the laboratories of L.S. and P.H. for discussions and comments of the experiments. The work was supported by National Institutes of Health (NIH) (R01GM092893) and National Science Foundation (NSF) (IOS-1252539) to P.H, and NIH (R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S. G.X. and W.S. were partially supported by China Scholarship Council (CSC). B.R. was partially supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPS), Brazil.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
References:
- 1.Belkhadir Y, Yang L, Hetzel J, Dangl JL, and Chory J (2014). The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci 39, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohmann U, Lau K, and Hothorn M (2017). The Structural Basis of Ligand Perception and Signal Activation by Receptor Kinases. Annual Review of Plant Biology, Vol 68 68, 109–137. [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Gomez L, and Boller T (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- 4.Li J, and Chory J (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- 5.Li J (2010). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr Opin Plant Biol 13, 509–514. [DOI] [PubMed] [Google Scholar]
- 6.Ma X, Xu G, He P, and Shan L (2016). SERKing Coreceptors for Receptors. Trends Plant Sci 21, 1017–1033. [DOI] [PubMed] [Google Scholar]
- 7.Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, and Shan L (2015). Differential Function of Arabidopsis SERK Family Receptor-like Kinases in Stomatal Patterning. Curr Biol 25, 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17, 1116–1122. [DOI] [PubMed] [Google Scholar]
- 9.He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, Russell SD, and Li J (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol 17, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 10.de Oliveira MVV, Xu G, Li B, de Souza Vespoli L, Meng X, Chen X, Yu X, de Souza SA, Intorne AC, de A. Manhães AME, et al. (2016). Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nature Plants 2, 15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, and Zhang Y (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18, 1190–1198. [DOI] [PubMed] [Google Scholar]
- 12.Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. [DOI] [PubMed] [Google Scholar]
- 13.Talke IN, Blaudez D, Maathuis FJ, and Sanders D (2003). CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci 8, 286–293. [DOI] [PubMed] [Google Scholar]
- 14.DeFalco TA, Moeder W, and Yoshioka K (2016). Opening the Gates: Insights into Cyclic Nucleotide-Gated Channel-Mediated Signaling. Trends Plant Sci 21, 903–906. [DOI] [PubMed] [Google Scholar]
- 15.Pan Y, Chai X, Gao Q, Zhou L, Zhang S, Li L, and Luan S (2019). Dynamic Interactions of Plant CNGC Subunits and Calmodulins Drive Oscillatory Ca(2+) Channel Activities. Dev Cell 48, 710–725 e715. [DOI] [PubMed] [Google Scholar]
- 16.Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK Jr., and Bent AF (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci U S A 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurkowski GI, Smith RK Jr., Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, and Bent AF (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact 17, 511–520. [DOI] [PubMed] [Google Scholar]
- 18.Balague C, Lin B, Alcon C, Flottes G, Malmstrom S, Kohler C, Neuhaus G, Pelletier G, Gaymard F, and Roby D (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Walker RK, Zhao YC, and Berkowitz GA (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proceedings of the National Academy of Sciences of the United States of America 109, 19852–19857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, and Berkowitz GA (2007). Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19, 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. [DOI] [PubMed] [Google Scholar]
- 22.Meena MK, Prajapati R, Krishna D, Divakaran K, Pandey Y, Reichelt M, Mathew MK, Boland W, Mithofer A, and Vadassery J (2019). The Ca(2+) Channel CNGC19 Regulates Arabidopsis Defense Against Spodoptera Herbivory. Plant Cell 31, 1539–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones JD, and Dangl JL (2006). The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- 24.Cui H, Tsuda K, and Parker JE (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66, 487–511. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Feng B, He P, and Shan L (2017). From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu Rev Phytopathol 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, and Klessig DF (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer C, Kugler A, Hoth S, and Dietrich P (2013). An IQ domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol 54, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, and de Vries SC (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee CH, and MacKinnon R (2017). Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell 168, 111–120 e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Hamid H, Chin K, Moeder W, Shahinas D, Gupta D, and Yoshioka K (2013). A suppressor screen of the chimeric AtCNGC11/12 reveals residues important for intersubunit interactions of cyclic nucleotide-gated ion channels. Plant Physiol 162, 1681–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiasson DM, Haage K, Sollweck K, Brachmann A, Dietrich P, and Parniske M (2017). A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moeder W, Urquhart W, Ung H, and Yoshioka K (2011). The role of cyclic nucleotide-gated ion channels in plant immunity. Mol Plant 4, 442–452. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Liu X, Zhang A, Ren Y, Wu F, Wang G, Xu Y, Lei C, Zhu S, Pan T, et al. (2019). A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin K, Moeder W, Abdel-Hamid H, Shahinas D, Gupta D, and Yoshioka K (2010). Importance of the alphaC-helix in the cyclic nucleotide binding domain for the stable channel regulation and function of cyclic nucleotide gated ion channels in Arabidopsis. J Exp Bot 61, 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boudsocq M, and Sheen J (2010). Stress Signaling II: Calcium Sensing and Signaling. Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation, 75–90. [Google Scholar]
- 36.Chin K, DeFalco TA, Moeder W, and Yoshioka K (2013). The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol 163, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladwig F, Dahlke RI, Stuhrwohldt N, Hartmann J, Harter K, and Sauter M (2015). Phytosulfokine Regulates Growth in Arabidopsis through a Response Module at the Plasma Membrane That Includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27, 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada K, Saijo Y, Nakagami H, and Takano Y (2016). Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 354, 1427–1430. [DOI] [PubMed] [Google Scholar]
- 39.Lin W, Li B, Lu D, Chen S, Zhu N, He P, and Shan L (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci U S A 111, 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du JB, Gao Y, Zhan YY, Zhang SS, Wu YJ, Xiao Y, Zou B, He K, Gou XP, Li GJ, et al. (2016). Nucleocytoplasmic trafficking is essential for BAK1-and BKK1-mediated cell-death control. Plant Journal 85, 520–531. [DOI] [PubMed] [Google Scholar]
- 41.Shan LB, He P, Li JM, Heese A, Peck SC, Nurnberger T, Martin GB, and Sheen J (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host & Microbe 4, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, and Shan L (2015). Phosphorylation of Trihelix Transcriptional Repressor ASR3 by MAP KINASE4 Negatively Regulates Arabidopsis Immunity. The Plant cell 27, 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)-dependent protein kinases. PLoS Pathog 9, e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Cheng C, Cui F, de Oliveira MV, Yu X, Meng X, Intorne AC, Babilonia K, Li M, Li B, et al. (2014). Modulation of RNA Polymerase II Phosphorylation Downstream of Pathogen Perception Orchestrates Plant Immunity. Cell Host Microbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nour-Eldin HH, Hansen BG, Nørholm MHH, Jensen JK, and Halkier BA (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic acids research 34, e122–e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, and Chen QJ (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mang H, Feng B, Hu Z, Boisson-Dernier A, Franck CM, Meng X, Huang Y, Zhou J, Xu G, Wang T, et al. (2017). Differential Regulation of Two-Tiered Plant Immunity and Sexual Reproduction by ANXUR Receptor-Like Kinases. The Plant cell 29, 3140–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao X, Horie T, Xue S, Leung HY, Katsuhara M, Brodsky DE, Wu Y, and Schroeder JI (2009). Differential Sodium and Potassium Transport Selectivities of the Rice OsHKT2;1 and OsHKT2;2 Transporters in Plant Cells. Plant physiology 152, 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–U354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He P, Shan L, and Sheen J (2007). The use of protoplasts to study innate immune responses. Methods Mol Biol 354, 1–9. [DOI] [PubMed] [Google Scholar]
- 52.DeFalco TA, Toyota M, Phan V, Karia P, Moeder W, Gilroy S, and Yoshioka K (2017). Using GCaMP3 to Study Ca2+ Signaling in Nicotiana Species. Plant Cell Physiol 58, 1173–1184. [DOI] [PubMed] [Google Scholar]
- 53.Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, and Gilroy S (2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115. [DOI] [PubMed] [Google Scholar]
- 54.Muller EM, Locke EG, and Cunningham KW (2001). Differential regulation of two Ca(2+) influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 159, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S4. PRIMERS USED IN THIS STUDY. RELATED TO STAR METHODS.
Data Availability Statement
This manuscript did not generate new datasets or code.