Summary
Viral diseases severely affect crop yield and quality, thereby threatening global food security. Genetic improvement of plant virus resistance is essential for sustainable agriculture. In the last decades, several modern technologies were applied in plant antiviral engineering. Here we summarized breakthroughs of the two major antiviral strategies, RNA silencing and genome editing. RNA silencing strategy has been used in antiviral breeding for more than thirty years, and many crops engineered to stably express small RNAs targeting various viruses have been approved for commercial release. Genome editing technology has emerged in the past decade, especially CRISPR/Cas, which provides new methods for genetic improvement of plant virus resistance and accelerates resistance breeding. Finally, we discuss the potential of these technologies for breeding crops, and the challenges and solutions they may face in the future.
Keywords: Virus, plant engineering, resistance, RNA silencing, genome editing, CRISPR/Cas
Introduction
The global population has increased by over 25% in the last 20 years and is projected to increase from 7.7 billion in 2019 to around 10 billion by 2050 (Tilman et al., 2011). As the global population continues to grow, the provision of adequate food has emerged as one of the major challenges at present (Cheeseman, 2016; Legg et al., 2014). However, the growth rate of grain production does not seem to have kept pace with population growth: the global per capita grain production has declined over the last 20 years (Legg et al., 2014; Suweis et al., 2015).
In addition to restricted arable land and water resources, the growth of global food production is further limited by pests and diseases. The yield of cultivated plants is threatened by pests which cause yield loss of 20–40%, while bacterial and fungal pathogens reduce crop yields by about 15% and viruses reduce yields by 3–7% (Oerke and Dehne, 2004). Compared with pests or other diseases, losses caused by viruses are not so great, but the outbreak of viral diseases can cause serious problems. Economic losses caused by viral diseases can reach USD 60–80 billion annually. This is due to the fact that chemical pesticides can be used for bacterial or fungal diseases and insect pests, but there is currently no traditional chemical pesticide that directly targets viral diseases. At present, the main strategy to control viral diseases in the field is to use pesticides or natural predators to control the vectors, or use physical barriers, such as reflective mulches and insect‐proof nets (Legg et al., 2014). However, complex epidemiological factors associated with viral disease outbreaks, such as rapid evolution of viruses, vector migration dynamics and unpredictable expansion of viral host range, make it very difficult to develop effective long‐term disease management strategies (Zaidi et al., 2016).
The use of virus‐resistant varieties in agricultural production is the most economical and effective way to reduce losses caused by viral diseases, thus the current situation requires the development of highly effective and durable virus‐resistant/immune crop varieties to combat increasingly serious viral diseases. Conventional antiviral breeding plays an essential role in crop improvement but usually requires large growing populations of crops over multiple generations, which is a rather time‐consuming and laborious process. The emergence of genetic engineering, which directly alters the organism's genetic information using modern biotechnology, has significantly accelerated the process and efficiency of breeding (Christou, 2013).
Increasing knowledge about the molecular mechanism of plant–virus interactions and the advancement of biotechnology provides new opportunities for engineering plant resistance to viruses (Duan et al., 2012; Mahas and Mahfouz, 2018; Yin and Qiu, 2019). This review summarizes current antiviral biotechnology strategies, compares their advantages and disadvantages and discusses their application prospects and challenges.
Engineering RNA silencing‐based resistance against viruses
As early as 1985, Sanford and Johnston put forward the elegant concept of pathogen‐derived resistance (PDR; Figure 1), whose core theory was that expressing the pathogen genetic elements in plants will destroy the pathogenicity of the parasitic pathogens (Sanford and Johnston, 1985). The Beachy lab conducted pioneering work in 1986 which induced Tobacco mosaic virus (TMV) resistance in tobacco through the introduction of gene constructs expressing the viral coat protein (CP) (Figure 1; Abel et al., 1986). Subsequently, there have been numerous attempts to generate virus resistance by transforming plants with various viral genes or genome fragments, leading to successful development of virus‐resistant crops for commercial application (Baulcombe, 1994; Beachy, 1993; Lomonossoff, 1995; Wilson, 1993), although the mechanisms of PDR were still unclear at that time (Baulcombe, 1996).
Figure 1.
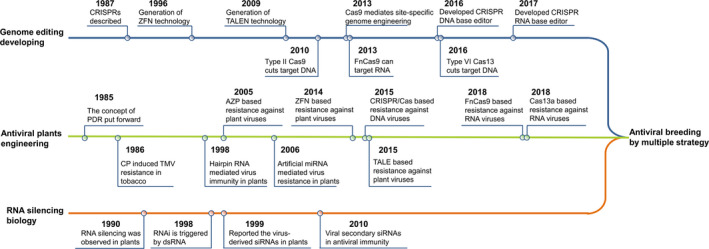
Timeline of antiviral plant engineering, genome editing and RNA silencing technology developing research fields. Key developments in all three fields are shown. In future, these fields will merged together, and multiple strategies will combined to server for antiviral breeding.
Meanwhile, the RNA silencing phenomenon in plants was first discovered in 1990 (Napoli et al., 1990), and has since been widely characterized in many eukaryotic organisms such as fungi, animals and plants (Figure 1) (Baulcombe, 2004; Fire et al., 1998; Guo and Kemphues, 1995; Hannon, 2002; Romano and Macino, 1992). RNA silencing, also referred to as RNA interference (RNAi), is activated by the presence of double‐stranded RNA molecules (dsRNAs) and induces gene expression inhibition or suppression in a nucleotide sequence‐specific manner (Hannon, 2002; Voinnet, 2005). In plants, several key protein families are involved in RNA silencing, including Dicer‐like (DCL), Argonautes (AGO), RNA‐dependent RNA Polymerase (RDR) and Suppressor of Gene Silencing (SGS). As type III RNases, DCL proteins process dsRNA or miRNA precursors into siRNA or miRNA, respectively, of 20‐ to 24‐nt long with a two‐base overhang at the 3′ end. These siRNAs or miRNAs are incorporated into the endonuclease AGO proteins to form RNA‐induced silencing complex (RISC). Directed by its containing siRNA/miRNA, RISC can bind to target mRNA or noncoding RNA and then silence the target gene expression by cleaving target RNA and rendering its degradation, or recruiting DNA and histone modifiers and inhibiting the transcription of the target gene. The cleaved target RNA may be recognized by RDR proteins which amplify the dsRNA to enhance the silence effect. SGS proteins stabilize the dsRNA substrate for DCLs to produce secondary siRNAs and reinforce the RNA silencing process (Figure 2; Ding, 2010; Ipsaro and Joshua‐Tor, 2015; Voinnet, 2005).
Figure 2.
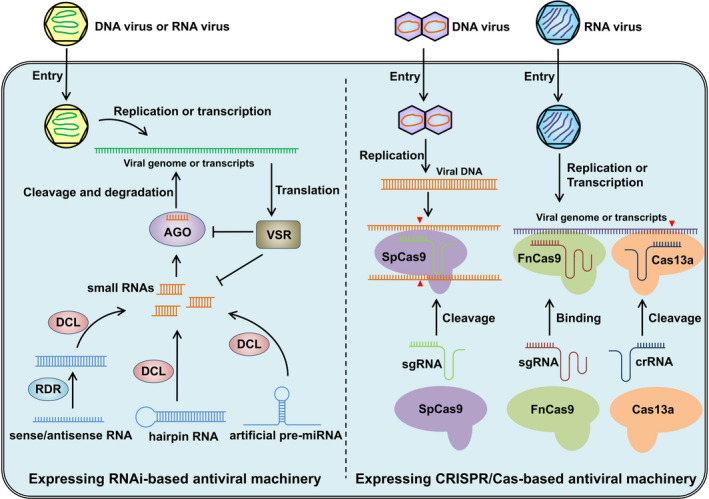
Schematic diagram depicting RNA silencing and CRISPR/Cas strategies to target plant viruses. The diagram on the left shows the mechanism of RNA silencing‐based antiviral engineering. The plant cells are transgenic expressing or exogenous applied of virus‐derived sense/antisense RNA, hairpin RNA, or artificial pre‐miRNA to produce the small RNAs targeting viral genome or transcripts. The small RNAs are loaded into the AGO protein to guide the cleavage of the viral RNA, which induces the degradation of the viral genomic RNA or mRNA. Plant viruses can encode VSR to counter the RNA silencing based resistance by targeting the AGO protein or small RNAs. The diagram on the right shows the mechanism of CRISPR/Cas‐based antiviral engineering. The CRISPR system consists of sgRNA and Cas protein. The transgenic or transit expression of the virus targeting sgRNA and its cognate Cas protein can effectively inhibit the virus infection. Upon DNA virus entry into the plant cell, for example the geminivirus, the viral genome is converted to a doublestranded DNA intermediate, which could be targeted and cleaved by the Cas9 protein from Streptococcus pyogenes (SpCas9). For RNA viruses, Cas9 from Francisella novicida (FnCas9) and Cas13a has been proved to confer virus resistance effectively. Guided by their cognate sgRNA or crRNA, the FnCas9 and Cas13a can bind or cleave the viral genome or transcripts, respectively. Arrowheads in red indicate cleave sites in the viral.
Besides the regulatory roles in plant growth and development, the dsRNA‐mediated RNA silencing also serves as a host antiviral defense mechanism (Ding, 2010). Since the progressive understanding of the RNA silencing mechanism and its role in antiviral immunity, RNA silencing has been deployed in crop improvement for viral resistance (Ding and Voinnet, 2007). Generation of virus‐derived dsRNA is a general feature when successful resistance is achieved in plants. Many approaches have been developed for engineering virus‐resistant transgenic plants, mostly based on different precursor RNA for siRNA production, including sense/antisense RNA, hairpin RNA (hpRNA) and artificial miRNA precursors (Figure 2; Duan et al., 2012).
Thus far, RNA silencing technology has been successfully applied to target over 60 species of economically important plant viruses, including Papaya ringspot virus (PRSV) (Bau et al., 2003; Fitch et al., 1992; Ye and Li, 2010), Banana bunchy top virus (BBTV) (Elayabalan et al., 2013; Shekhawat et al., 2012), Citrus tristeza virus (CTV) (Soler et al., 2012), Plum pox virus (PPV) (Guo et al., 1998; Hily et al., 2007; Ravelonandro et al., 2014; Scorza et al., 2001; Wittner et al., 1998), Maize streak virus (MSV) (Shepherd et al., 2007), Maize dwarf mosaic virus (MDMV) (Zhang et al., 2010, 2013), Soybean mosaic virus (SMV) (Gao et al., 2015; Wang et al., 2001) and Tomato yellow leaf curl virus (TYLCV; Antignus et al., 2004; Fuentes et al., 2006). Nearly 30 crop species have been engineered to stably express small RNAs targeting various viruses, dozens of which have been approved for commercial release in several countries, such as papaya resistant to PRSV (Gonsalves, 2006; Ye and Li, 2010), potato resistant to Potato leafroll virus (PLRV) or Potato virus Y (PVY) and squash resistant to Cucumber mosaic virus (CMV) or Zucchini yellow mosaic virus (ZYMV) (Table 2).
These successful examples were all obtained by genetically modified methods; however, these transgenic approaches are not only time‐consuming and expensive, but also suffer significant regulation and public acceptance issues. To address these limitations and public concerns, several approaches that involve exogenous application of naked dsRNA proved to successfully trigger the RNA silencing pathway against pathogenic viruses (Gan et al., 2010; Kaldis et al., 2018; Lau et al., 2014; Namgial et al., 2019; Robinson et al., 2014; Tenllado et al., 2003; Worrall et al., 2019). However, the obvious shortcoming of this strategy is that it has a very short virus protection window of 5–7 days post‐application (Mitter et al., 2017b).Recently, a research group used a novel approach of delivering dsRNA using layered double hydroxide nanosheets as carriers and successfully established CMV resistance in tobacco plants (Mitter et al., 2017a). This approach not only increases the stability of dsRNA in plants, but also provides a sustained release of dsRNA to extend the virus protection period.
Engineering ZFN‐ or TALEN‐based resistance against viruses
A decade ago, a new approach, referred to as genome editing, emerged that makes it possible to manipulate the genetic information in different cell types and organisms. Zinc finger nucleases (ZFNs) and transcription activator‐like effector nucleases (TALENs) were the first‐generation tools of genome editing technology (Figure 1) (Boch et al., 2009; Kim et al., 1996; Moscou and Bogdanove, 2009). Both ZFNs and TALENs are chimeric proteins created by fusing a DNA‐binding domain (DBD) from a zinc finger protein or transcription activator‐like effector, respectively, to the non‐specific cleavage domain of the enzyme FokI. The DBD determines a specific nucleotide recognition in the DNA target and the cleavage domain cleaves DNA to produce the double‐strand breaks (DSB) in the targeted site (Boch et al., 2009; Kim et al., 1996; Moscou and Bogdanove, 2009; Urnov et al., 2010). In eukaryotes, the DSBs are repaired by non‐homologous end joining (NHEJ) or homologous recombination, and both repairing mechanisms may induce mutations in the particular genomic location (Wyman and Kanaar, 2006).
These genome editing techniques not only integrate, delete and/or mutate genes of interest, but also provide a new weapon in the arsenal against plant viruses. As early as in 2005, Sera developed an artificial zinc finger protein (AZP), which lacks the cleavage domain compared to ZFN, targeting the intergenic region (IR) of Beet severe curly top virus (BSCTV, family Geminiviridae) in Arabidopsis (Figure 1) (Sera, 2005). The IR of geminiviruses contains a stem‐loop structure which is essential for virus replication by viral replication initiator protein (Rep) binding (Hanley‐Bowdoin et al., 2013). The transgenically expressed AZP efficiently binds the IR of BSCTV, thus blocking the Rep binding and subsequently suppressing the infection of the virus (Sera, 2005). Similar work has also been applied to reduce replication of Rice tungro bacilliform virus (RTBV) in Arabidopsis by expressing an AZP which was able to recognize and block the viral promoter sequences (Ordiz et al., 2010). ZFN technology, unlike AZP, involves both DBD and DNA cleavage domains, and was also applied to target the Rep gene of two begomoviruses, Tomato yellow leaf curl China virus (TYLCCNV) and Tobacco curly shoot virus (TbCSV), in tobacco plants, and showed a significant inhibition of viral replication (Figure 1) (Chen et al., 2014). TALEs, which lack the nucleases domain compared to TALEN, were developed to combat these begomoviruses using a similar approach. TALEs were engineered to target conserved motifs among begomoviruses. Tobacco plants expressing the TALEs displayed resistance to TbCSV and TYLCCNV, while resistance to Tomato leaf curl Yunnan virus (TLCYnV) was partial (Figure 1) (Cheng et al., 2015). TALEN technologies, a nuclease domain fused to TALE, has not been reported against plant viruses, although it has been explored for potential antiviral applications in some human viruses, such as hepatitis B virus (HBV), hepatitis C virus (HCV) and human immunodeficiency virus (HIV; Bloom et al., 2015).
Although the use of these genome editing platforms has led to important advances, each has unique limitations, and their use in plants is far from routine. Thus, applications of ZFNs and TALENs have rapidly been surpassed by a new emerging genome editing system in various organisms, including plants.
Engineering CRISPR/Cas‐based resistance against viruses
The clustered, regularly interspaced short palindromic repeats/CRISPR‐associated protein (CRISPR/Cas) system is based on an adaptive immune system that eliminates invasion of foreign plasmids or viral DNA via cleavage in bacteria and archaea (Bhaya et al., 2011). CRISPR/Cas genome editing systems consist of an endonuclease Cas protein and a single‐guide RNA (sgRNA) which directs the Cas protein to the DNA or RNA target. Moreover, sgRNA contains a scaffold for Cas protein binding and a user‐defined approximately 20‐nt long spacer sequence for genome targeting (Figure 1; Cong et al., 2013; Mali et al., 2013). Owing to its simplicity, high efficiency and affordability compared with precedent ZFN or TALEN, many labs working in different fields have turned to this technology. Many efforts are under way to reveal the potential application of CRISPR/Cas9 to control human viruses such as HIV, HBV, Epstein‐Barr and plant viruses (Price et al., 2016).
The original CRISPR/Cas system from Streptococcus pyogenes, used for genome editing, targets the DNA. Thus, the CRISPR/Cas9 machinery was first exploited to combat the geminivirus by targeting its viral genomic DNA during the replication stage (Mahas and Mahfouz, 2018; Yin and Qiu, 2019). Three groups reported the successful use of CRISPR/Cas9 to generate geminivirus resistance in tobacco and Arabidopsis (Figure 1, Table 1). They designed sgRNAs to target the IR, Rep or CP loci, and significantly reduced or abolished disease symptoms of several geminiviruses (Figure 2; Ali et al., 2015; Baltes et al., 2015; Ji et al., 2015). In addition to its application to model plants, the same system was recently also used in barley and established highly efficient resistance against Wheat dwarf virus (WDV) (Kis et al., 2019). For double‐stranded DNA viruses, CRISPR/Cas9 has also been shown to be effective in inhibiting the virulence of Cauliflower mosaic virus (CaMV) in Arabidopsis (Table 1; Liu et al., 2018).
Table 1.
CRISPR/Cas technologies developed for viral pathogen resistance in plants. Cas9 was obtained from Streptococcus pyogenes (SpCas9), Cas9 from Francisella novicida (FnCas9) Cas13a from Leptotrichia shahii (LshCas13a)
| Target | CRISPR system | Host plants | Virus | Genus | Family | Viral genome type | References |
|---|---|---|---|---|---|---|---|
| Viral genome | SpCas9 | Tobacco | Tomato yellow leaf curl virus | Begomovirus | Geminiviridae | ssDNA | Ali et al. (2015, 2016) |
| Cotton leaf curl Kokhran virus | |||||||
| Merremia mosaic virus | |||||||
| Tobacco and Arabidopsis | Beet severe curly top virus | Curtovirus | Ali et al. (2015), Ji et al. (2015) | ||||
| Beet curly top virus | |||||||
| Tobacco | Bean yellow dwarf virus | Mastrevirus | Baltes et al. (2015) | ||||
| Barley | Wheat dwarf virus | Kis et al. (2019), Liu et al. (2018) | |||||
| Arabidopsis | Cauliflower mosaic virus | ||||||
| Caulimovirus | Caulimoviridae | dsDNA | |||||
| FnCas9 | Tobacco | Tobacco mosaic virus | Tobamovirus | Virgaviridae | +ssRNA | Zhang et al. (2018) | |
| Tobacco and Arabidopsis | Cucumber mosaic virus | Cucumovirus | Bromoviridae | Zhang et al. (2018) | |||
| LshCas13a | Tobacco | Turnip mosaic virus | Potyvirus | Potyviridae | +ssRNA | Aman et al. (2018) | |
| Tobacco | Tobacco mosaic virus | Tobamovirus | Virgaviridae | +ssRNA | Zhang et al. (2019) | ||
| Rice | Southern rice black‐streaked dwarf virus | Fijivirus | Reoviridae | dsRNA | Zhang et al. (2019) | ||
| Rice stripe mosaic virus | Cytorhabdovirus | Rhabdoviridae | ‐ssRNA | Zhang et al. (2019) | |||
| Potato | Potato virus Y | Potyvirus | Potyviridae | +ssRNA | Zhan et al. (2019) | ||
| Host factor | SpCas9 | Cucumber | Cucumber vein yellowing virus | Ipomovirus | Potyviridae | +ssRNA | Chandrasekaran et al. (2016) |
| Cassava | Cassava brown streak virus | Gomez et al. (2019) | |||||
| Cucumber and Arabidopsis | Zucchini yellow mosaic virus | Potyvirus | Chandrasekaran et al. (2016), Pyott et al. (2016) | ||||
| Papaya ringspot virus | |||||||
| Turnip mosaic virus |
Off‐target effect is the major issue of genome editing, which occurs due to tolerance of sgRNA sequence mismatches and extended expression of Cas9 nuclease (Tsai and Joung, 2016). Ji et al. (2015, 2018) tested for the off‐target effects in their Arabidopsis line expressing the virus‐targeting CRISPR/Cas9 construct. In order to overcome the off‐target effect, they used virus‐induced promoters instead of constitutive promoter, to drive the Cas9 expression. Thus, the CRISPR/Cas9 antiviral system will be only expressed when the virus invades the plant cells. Ingeniously, no off‐target effect was detected by deep sequencing in candidate sites of the virus‐inducible genome editing Arabidopsis (Ji et al., 2018). This kind of virus‐inducible genome editing system could be widely applicable for generating virus‐resistant plants without off‐target costs (Table 2).
Table 2.
Comparison of the strategies used in engineering antiviral plants
| Strategies | Advantages | Disadvantages | Applications |
|---|---|---|---|
| RNA silencing | High efficiency and have several successful commercialized transgenic antiviral crops | Many viruses can encode VSRs to counter the defense of RNA silencing | Engineering resistance for the viruses without strong VSR, better effect on RNA viruses than DNA viruses |
| CRISPR/Cas targeting DNA | The eukaryotic plant viruses have not evolved to possess the ability to counter this immune defense coming from prokaryote | The targeted DNA viruses might be repaired and escape the engineered resistance. Off‐target effect may produce some heritable mutations in the host genome | Suites for the viruses having dsDNA genome or dsDNA replication intermediate. A virus‐induced promoter could reduce the off‐target effect |
| CRISPR/Cas targeting RNA | The targeted RNA is further degraded and has less chance to produce mutant viruses | Off‐target effect may affect the expression of some host genes, but will not change the plant genome | Engineering resistance to RNA viruses and DNA viruses with RNA intermediates |
| Host factors editing | The gene editing machinery can be removed by backcross, so it is able to engineer virus‐resistant plant which is transgenic‐free | Loss‐of‐function of some host factors may lead to lethality or impaired growth | Need to find a well‐characterized host factor and mutation of the factor will not affect the growth and reproduction of the plants |
RNA viruses cause more serious losses in crops, as compared to DNA viruses, and enormous damage to agricultural production. With the development of more CRISPR/Cas systems from other bacterial strains, several Cas protein variants, such as the Cas9 from Francisella novicida (FnCas9) and the Cas13a from Leptotrichia shahii (LshCas13a) or Leptotrichia wadei (LwaCas13a), have been reported to target RNA in vivo (Abudayyeh et al., 2016, 2017; Sampson et al., 2013), which opens up new possibilities against RNA viruses. In the first case, FnCas9 and its sgRNA were engineered to target CMV and TMV, and reduced virus accumulation and attenuated disease symptoms were observed in tobacco and Arabidopsis expressing the antiviral system (Figure 1, Table 1) (Zhang et al., 2018). Interestingly, RNA binding, but not cleavage capacity of FnCas9, is required for virus inhibition (Figure 2). Then, the LshCas13a system was reprogrammed and employed to antagonize RNA viruses in plants (Figure 2). Two groups successfully used the system to inhibit potyvirus infection in tobacco and potato (Aman et al., 2018; Zhan et al., 2019), while Zhang et al. (2019) established resistance to RNA viruses in both dicot and monocot plants. The LshCas13a system was designed to cleave genomic RNA of TMV in tobacco and to degrade the genomic RNA of Southern rice black‐streaked dwarf virus (SRBSDV) and Rice stripe mosaic virus (RSMV) in rice plants (Figure 1; Zhang et al., 2019). These cases demonstrated that the LshCas13a system can act against different types of RNA virus, including +ssRNA, ‐ssRNA and dsRNA genomes (Table 1). Targeting RNA genomes is superior as it would not lead to heritable off‐target effect in the host genomic DNA, although it may lead to nonspecific RNA cleavage. Furthermore, the cleaved of viral genomic RNA will be further destroyed by RNAi system of plants. Therefore, RNA viruses have less chance to escape the CRISPR/Cas targeting antiviral system by mutating their genomes (Table 2).
In order for the infection to progress, virus needs to recruit many host factors to assist in replication, transcription, translation, etc. This feature provides us with a potential target of genome editing to limit virus infection. For example, viruses lack ribosomes in their virions, and the host translation machinery is essential for viral protein synthesis. In plants, the eukaryotic initiation factor 4E (eIF4E) and its isoform (eIFiso4E) are essential for some viruses to initiate viral protein translation (Sanfaçon, 2015). Previous work showed that the Arabidopsis eIF(iso)4E mutant has enhanced resistance to Turnip mosaic virus (TuMV) (Lellis et al., 2002). By using CRISPR/Cas9, knockout of Arabidopsis eIF(iso)4E resulted in resistance to TuMV without affecting the plant growth (Pyott et al., 2016), although these translation initiation factors are important for growth and reproduction (Table 2). Similar work was performed in cucumber, where the eIF4E gene destructed by CRISPR/Cas9 showed broad‐spectrum resistance to the family Potyviridae, including Cucumber vein yellowing virus (CVYV), PRSV and ZYMV (Chandrasekaran et al., 2016). Cassava encoded five proteins belonging to the eIF4E family, of which only nCBP‐1 (novel cap‐binding protein‐1) and nCBP‐2 interact with VPg (viral genome‐linked protein) of Cassava brown streak virus (CBSV), which is a major constraint on cassava yields in East and Central Africa. The virus showed delayed and attenuated symptoms on the ncbp‐1/ncbp‐2 double mutants generated by CRISPR/Cas9 (Table 1) (Gomez et al., 2019).
Challenges and future aspects
Application of modern biotechnology has great potential to overcome the limitations of conventional viral resistance breeding. First, since in the case of both RNAi and genome editing technologies only viral sequence information is required, these approaches are particularly applicable to crops with limited genome sequence information. Second, resistance breeding using RNAi or genome editing does not require genetic crosses and selection of segregating progeny. Therefore, the breeding period can be greatly shortened. For some viral pandemics, rapid emergency response can be provided by exogenous application of dsRNA to induce RNA silencing against the virus. Third, destroying an essential host factor by CRISPR/Cas9 system is an effective way to generate virus‐resistant crops, as in the case of eIF4E (Chandrasekaran et al., 2016; Pyott et al., 2016). Through several generations of backcross and screening, or even using DNA‐free delivery of in vitro transcripts or ribonucleoprotein complexes of CRISPR/Cas9 by particle bombardment (Liang et al., 2018), virus‐resistant crops that are free of transgenic DNA can be generated, thus making it easier to release them for commercial production.
Nevertheless, these new technologies also have certain limitations. Through long‐term evolution, plant viruses have developed a range of counter‐defensive measures against RNA silencing, one of which is the encoded viral suppressors of RNA silencing (VSR), which have become a major problem in RNA silencing approaches (Figure 2, Table 2) (Qu, 2010; Voinnet, 2005). Crop plants are often subjected to mixed viral infection. VSRs from untargeted viruses are able to disrupt the RNAi‐mediated silencing process by targeting key components of RNAi pathways, and sometimes the targeted virus has a strong VSR that can break through the immunity conferred by RNA silencing (Kung et al., 2015). CRISPR/Cas is an immunity system derived naturally from prokaryotes, so eukaryotic viruses have not evolved in the presence of CRISPR/Cas, which implies they are unlikely to have CRISPR/Cas evasion strategies. Hence, using the genome editing strategy, or even combining RNA silencing and CRISPR/Cas system, can potentially and effectively solve the problem.
Zinc finger nuclease and TALEN approaches are not widely used due to their weaknesses related to affordability, simplicity and efficiency. In particular, with the rapid development of CRISPR technology, there are only a few examples of using these genome editing approaches to generate virus‐resistant crops.
The establishment and development of CRISPR/Cas system is a definite milestone in genome editing technology, but it also has some problems in antiviral application. First, when CRISPR/Cas system is used to knockout an essential host factor to obtain viral resistance, such as eIF4E, the loss‐of‐function of these host factors often leads to lethality or impaired growth (Callot and Gallois, 2014; Gauffier et al., 2016). In some crops, the redundancy among eIF4E genes could reduce eIF4E‐based resistance durability by making other members available to viruses (Bastet et al., 2017). The best strategy to develop eIF4E‐based resistance would be to design functional alleles by introducing point mutations in the gene, which does not affect its function in plant growth but prevents interaction with the virus, instead of knocking it out (Bastet et al., 2017). This can be achieved in situ using recently developed CRISPR/Cas base editing technology (Figure 1; Kim, 2018; Rees and Liu, 2018), as well as by de novo design and construction using synthetic biology technologies (Bastet et al., 2018; Liu and Stewart, 2015). Second, as mentioned above, off‐target effect can never be ignored when using genome editing strategies, irrespective of using ZFN, TALEN or CRISPR/Cas (Tsai and Joung, 2016). Using a virus‐inducible genome editing system could effectively reduce off‐target effect in antiviral breeding (Ji et al., 2018). In addition, CRISPR/Cas base editing technology, which chemically modifies the nucleotide instead of producing DSB (Kim, 2018; Rees and Liu, 2018), could be widely applied in crop antiviral breeding. Moreover, the RNA binding and cleaving of CRISPR/Cas13 system could target RNA viruses and the RNA intermediates of DNA viruses, and is ideal to avoid off‐target genomic modifications in the host genome. Last but not least, when using CRISPR/Cas9 system in developing crops resistant to DNA viruses, the targeted viruses evolve mutations that escape from CRISPR/Cas9 cleavage (Ali et al., 2016; Mehta et al., 2019). Bacteria and archaea use the CRISPR system to defend against phages and plasmids, but most bacteria lack NHEJ as a DNA repair mechanism, and the cleaved invading DNA is usually degraded rather than repaired (Wigley, 2012). However, DNA repair mechanisms differ substantially in eukaryotes, and NHEJ enables efficient genome editing by effectively repairing cleaved DNA (Lieber, 2010). Thus, this efficient repair mechanism in eukaryotes makes CRISPR/Cas9‐mediated DNA virus resistance more prone to evolving mutant viruses (Ali et al., 2016; Mehta et al., 2019). This mechanism will not only make the antiviral crops lose their effective virus defense ability, but it may even promote the evolution of viruses to produce super‐viruses, which have greater pathogenicity and produce more severe symptoms (Table 2). Optimal selection of sgRNA target sites in the viral genome may help reduce the viral mutation rate, for example, targeting certain viral genomic regions, such as the noncoding intergenic sequences, within some plant virus genomes led to the formation of mutations that were deleterious to virus replication. Furthermore, multiplex targeting of different regions of the viral genome in one CRISPR/Cas system, which will cause deletion of large fragments that is difficult to repair, can minimize the generation of escapee mutants. Finally, as outlined above, CRISPR/Cas base editors can introduce a mis‐sense mutation in a critical codon sequence outside of the protospacer seed sequence, thereby maintaining the ability of the sgRNA to recognize and bind to the targeted sequence, as well as preventing the formation of NHEJ‐mediated escapee viruses.
Although various biotechnologies have their advantages and disadvantages (Table 2), the full potential of RNAi and CRISPR/Cas systems for engineering resistance against eukaryotic viruses has not yet been exploited. More research is needed to improve these systems for virus interference, including improving their accuracy, durability, convenience and safety of delivery. To overcome the shortcomings of each strategy, the combination of RNA silencing and CRISPR/Cas strategies has great potential in antiviral breeding.
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
Y.Z. and T.Z. jointly developed the conceptual structure of manuscript. Y.Z. and X.Y. were involved in the compilation of relevant literature and drafting the manuscript. T.Z. and G.Z. provided a critical feedback and edited the final manuscript.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31871928, 31671993), the Pearl River S&T Nova Program of Guangzhou (201906010093), Research and Development Project in Major Fields of Guangdong (2019B020238001) and Guangdong Provincial Innovation Team for General Key Technologies in Modern Agricultural Industry (2019KJ133). We apologize to all investigators whose researches are not cited in the review owing to space limitations.
Contributor Information
Guohui Zhou, Email: ghzhou@scau.edu.cn.
Tong Zhang, Email: zhangtong@scau.edu.cn.
References
- Abel, P. , Nelson, R. , De, B. , Hoffmann, N. , Rogers, S. , Fraley, R. and Beachy, R. (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science, 232, 738–743. [DOI] [PubMed] [Google Scholar]
- Abudayyeh, O.O. , Gootenberg, J.S. , Konermann, S. , Joung, J. , Slaymaker, I.M. , Cox, D.B.T. , Shmakov, S. et al. (2016) C2c2 is a single‐component programmable RNA‐guided RNA‐targeting CRISPR effector. Science, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh, O.O. , Gootenberg, J.S. , Essletzbichler, P. , Han, S. , Joung, J. , Belanto, J.J. , Verdine, V. et al. (2017) RNA targeting with CRISPR–Cas13. Nature, 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Abulfaraj, A. , Idris, A. , Ali, S. , Tashkandi, M. and Mahfouz, M.M. (2015) CRISPR/Cas9‐mediated viral interference in plants. Genome Biol. 16, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Ali, S. , Tashkandi, M. , Zaidi, S.S.‐E.‐A. and Mahfouz, M.M. (2016) CRISPR/Cas9‐mediated immunity to geminiviruses: differential interference and evasion. Sci. Rep. 6, 26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman, R. , Ali, Z. , Butt, H. , Mahas, A. , Aljedaani, F. , Khan, M.Z. , Ding, S. et al. (2018) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignus, Y. , Vunsh, R. , Lachman, O. , Pearlsman, M. , Maslenin, L. , Hananya, U. and Rosner, A. (2004) Truncated Rep gene originated from Tomato yellow leaf curl virus‐Israel [Mild] confers strain‐specific resistance in transgenic tomato. Ann. Appl. Biol. 144, 39–44. [Google Scholar]
- Baltes, N.J. , Hummel, A.W. , Konecna, E. , Cegan, R. , Bruns, A.N. , Bisaro, D.M. and Voytas, D.F. (2015) Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants, 1, 15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet, A. , Robaglia, C. and Gallois, J.‐L. (2017) eIF4E Resistance: natural Variation Should Guide Gene Editing. Trends Plant Sci. 22, 411–419. [DOI] [PubMed] [Google Scholar]
- Bastet, A. , Lederer, B. , Giovinazzo, N. , Arnoux, X. , German‐Retana, S. , Reinbold, C. , Brault, V. et al. (2018) Trans‐species synthetic gene design allows resistance pyramiding and broad‐spectrum engineering of virus resistance in plants. Plant Biotechnol. J. 16, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau, H.‐J. , Cheng, Y.‐H. , Yu, T.‐A. , Yang, J.‐S. and Yeh, S.‐D. (2003) Broad‐spectrum resistance to different geographic strains of papaya ringspot virus in coat protein gene transgenic papaya. Phytopathology, 93, 112–120. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (1994) Novel strategies for engineering virus resistance in plants. Curr. Opin. Biotechnol. 5, 117–124. [Google Scholar]
- Baulcombe, D.C. (1996) Mechanisms of pathogen‐derived resistance to viruses in transgenic plants. Plant Cell, 8, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Beachy, R.N. (1993) Transgenic resistance to plant viruses. In: Seminars in Virology (United Kingdom). [Google Scholar]
- Bhaya, D. , Davison, M. and Barrangou, R. (2011) CRISPR‐cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. [DOI] [PubMed] [Google Scholar]
- Bloom, K. , Mussolino, C. and Arbuthnot, P. (2015) Transcription activator‐like effector (TALE) nucleases and repressor TALEs for antiviral gene therapy. Curr. Stem Cell Rep. 1, 1–8. [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. et al. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Callot, C. and Gallois, J.‐L. (2014) Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal Behav. 9, e27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran, J. , Brumin, M. , Wolf, D. , Leibman, D. , Klap, C. , Pearlsman, M. , Sherman, A. et al. (2016) Development of broad virus resistance in non‐transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, J. (2016) 7 ‐ Food security in the face of salinity, drought, climate change, and population growth. In Halophytes for Food Security in Dry Lands ( Khan, M.A. , Ozturk, M. , Gul, B. and Ahmed, M.Z. , eds), pp. 111–123. San Diego: Academic Press. [Google Scholar]
- Chen, W. , Qian, Y. , Wu, X. , Sun, Y. , Wu, X. and Cheng, X. (2014) Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral‐conserved nucleotide motif. Virus Genes, 48, 494–501. [DOI] [PubMed] [Google Scholar]
- Cheng, X. , Li, F. , Cai, J. , Chen, W. , Zhao, N. , Sun, Y. , Guo, Y. et al. (2015) Artificial TALE as a convenient protein platform for engineering broad‐spectrum resistance to begomoviruses. Viruses, 7, 4772–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christou, P. (2013) Plant genetic engineering and agricultural biotechnology 1983–2013. Trends Biotechnol. 31, 125–127. [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al. (2013) Multiplex genome engineering using CRISPR/Cas Systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.‐W. (2010) RNA‐based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Ding, S.‐W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C.‐G. , Wang, C.‐H. and Guo, H.‐S. (2012) Application of RNA silencing to plant disease resistance. Silence, 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elayabalan, S. , Kalaiponmani, K. , Subramaniam, S. , Selvarajan, R. , Panchanathan, R. , Muthuvelayoutham, R. , Kumar, K.K. et al. (2013) Development of Agrobacterium‐mediated transformation of highly valued hill banana cultivar Virupakshi (AAB) for resistance to BBTV disease. World J. Microbiol. Biotechnol. 29, 589–596. [DOI] [PubMed] [Google Scholar]
- Fire, A. , Xu, S. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Fitch, M.M.M. , Manshardt, R.M. , Gonsalves, D. , Slightom, J.L. and Sanford, J.C. (1992) Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Bio/Technology, 10, 1466–1472. [Google Scholar]
- Fuentes, A. , Ramos, P.L. , Fiallo, E. , Callard, D. , Sánchez, Y. , Peral, R. , Rodríguez, R. et al. (2006) Intron–hairpin RNA derived from replication associated protein C1 gene confers immunity to Tomato yellow leaf curl virus infection in transgenic tomato plants. Transgenic Res. 15, 291–304. [DOI] [PubMed] [Google Scholar]
- Gan, D. , Zhang, J. , Jiang, H. , Jiang, T. , Zhu, S. and Cheng, B. (2010) Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 29, 1261–1268. [DOI] [PubMed] [Google Scholar]
- Gao, L. , Ding, X. , Li, K. , Liao, W. , Zhong, Y. , Ren, R. , Liu, Z. et al. (2015) Characterization of Soybean mosaic virus resistance derived from inverted repeat‐SMV‐HC‐Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 128, 1489–1505. [DOI] [PubMed] [Google Scholar]
- Gauffier, C. , Lebaron, C. , Moretti, A. , Constant, C. , Moquet, F. , Bonnet, G. , Caranta, C. et al. (2016) A TILLING approach to generate broad‐spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 85, 717–729. [DOI] [PubMed] [Google Scholar]
- Gomez, M.A. , Lin, Z.D. , Moll, T. , Chauhan, R.D. , Hayden, L. , Renninger, K. , Beyene, G. et al. (2019) Simultaneous CRISPR/Cas9‐mediated editing of cassava eIF4E isoforms nCBP‐1 and nCBP‐2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves, D. (2006) Transgenic papaya: development, release, impact and challenges. Adv. Virus Res. 67, 317–354. [DOI] [PubMed] [Google Scholar]
- Guo, S. and Kemphues, K.J. (1995) Par‐1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell, 81, 611–620. [DOI] [PubMed] [Google Scholar]
- Guo, H.S. , Cervera, M.A.T. and García, J.A. (1998) Plum pox potyvirus resistance associated to transgene silencing that can be stabilized after different number of plant generations. Gene, 206, 263–272. [DOI] [PubMed] [Google Scholar]
- Hanley‐Bowdoin, L. , Bejarano, E.R. , Robertson, D. and Mansoor, S. (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hily, J.‐M. , Ravelonandro, M. , Damsteegt, V. , Bassett, C. , Petri, C. , Liu, Z. and Scorza, R. (2007) Plum pox virus coat protein gene Intron‐hairpin‐RNA (ihpRNA) constructs provide resistance to plum pox virus in Nicotiana benthamiana and Prunus domestica . J. Am. Soc. Hortic. Sci. 132, 850–858. [Google Scholar]
- Ipsaro, J.J. and Joshua‐Tor, L. (2015) From guide to target: molecular insights into eukaryotic RNA‐interference machinery. Nat. Struct. Mol. Biol. 22, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Zhang, H. , Zhang, Y. , Wang, Y. and Gao, C. (2015) Establishing a CRISPR–Cas‐like immune system conferring DNA virus resistance in plants. Nat. Plants, 1, 15144. [DOI] [PubMed] [Google Scholar]
- Ji, X. , Si, X. , Zhang, Y. , Zhang, H. , Zhang, F. and Gao, C. (2018) Conferring DNA virus resistance with high specificity in plants using virus‐inducible genome‐editing system. Genome Biol. 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis, A. , Berbati, M. , Melita, O. , Reppa, C. , Holeva, M. , Otten, P. and Voloudakis, A. (2018) Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol. Plant Pathol. 19, 883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐S. (2018) Precision genome engineering through adenine and cytosine base editing. Nat. Plants, 4, 148–151. [DOI] [PubMed] [Google Scholar]
- Kim, Y.G. , Cha, J. and Chandrasegaran, S. (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. 93, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis, A. , Hamar, É. , Tholt, G. , Bán, R. and Havelda, Z. (2019) Creating highly efficient resistance against Wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 17, 1004–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, Y.‐J. , You, B.‐J. , Raja, J.A.J. , Chen, K.‐C. , Huang, C.‐H. , Bau, H.‐J. , Yang, C.‐F. et al. (2015) Nucleotide sequence‐homology‐independent breakdown of transgenic resistance by more virulent virus strains and a potential solution. Sci. Rep. 5, 9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S.E. , Mazumdar, P. , Hee, T.W. , Song, A.L.A. , Othman, R.Y. and Harikrishna, J.A. (2014) Crude extracts of bacterially‐expressed dsRNA protect orchid plants against Cymbidium mosaic virus during transplantation from in vitro culture. J. Hortic. Sci. Biotechnol. 89, 569–576. [Google Scholar]
- Legg, J.P. , Shirima, R. , Tajebe, L.S. , Guastella, D. , Boniface, S. , Jeremiah, S. , Nsami, E. et al. (2014) Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag. Sci. 70, 1446–1453. [DOI] [PubMed] [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Liang, Z. , Chen, K. , Zhang, Y. , Liu, J. , Yin, K. , Qiu, J.‐L. and Gao, C. (2018) Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413. [DOI] [PubMed] [Google Scholar]
- Lieber, M.R. (2010) The mechanism of double‐strand DNA break repair by the Nonhomologous DNA end‐joining pathway. Annu. Rev. Biochem. 79, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. and Stewart, C.N. (2015) Plant synthetic biology. Trends Plant Sci. 20, 309–317. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Soyars, C.L. , Li, J. , Fei, Q. , He, G. , Peterson, B.A. , Meyers, B.C. et al. (2018) CRISPR/Cas9‐mediated resistance to cauliflower mosaic virus. Plant Direct, 2, e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff, G.P. (1995) Pathogen‐derived resistance to plant viruses. Annu. Rev. Phytopathol. 33, 323–343. [DOI] [PubMed] [Google Scholar]
- Mahas, A. and Mahfouz, M. (2018) Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virol. 32, 1–8. [DOI] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , Norville, J.E. et al. (2013) RNA‐guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, D. , Stürchler, A. , Anjanappa, R.B. , Zaidi, S.S.‐E.‐A. , Hirsch‐Hoffmann, M. , Gruissem, W. and Vanderschuren, H. (2019) Linking CRISPR‐Cas9 interference in cassava to the evolution of editing‐resistant geminiviruses. Genome Biol. 20, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter, N. , Worrall, E.A. , Robinson, K.E. , Li, P. , Jain, R.G. , Taochy, C. , Fletcher, S.J. et al. (2017a) Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants, 3, 16207. [DOI] [PubMed] [Google Scholar]
- Mitter, N. , Worrall, E.A. , Robinson, K.E. , Xu, Z.P. and Carroll, B.J. (2017b) Induction of virus resistance by exogenous application of double‐stranded RNA. Curr. Opin. Virol. 26, 49–55. [DOI] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Namgial, T. , Kaldis, A. , Chakraborty, S. and Voloudakis, A. (2019) Topical application of double‐stranded RNA molecules containing sequences of Tomato leaf curl virus and Cucumber mosaic virus confers protection against the cognate viruses. Physiol. Mol. Plant Pathol. 108, 101432. [Google Scholar]
- Napoli, C. , Lemieux, C. and Jorgensen, R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co‐suppression of homologous genes in trans. Plant Cell, 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke, E.C. and Dehne, H.W. (2004) Safeguarding production—losses in major crops and the role of crop protection. Crop Prot. 23, 275–285. [Google Scholar]
- Ordiz, M.I. , Magnenat, L. , Barbas, C.F. and Beachy, R.N. (2010) Negative regulation of the RTBV promoter by designed zinc finger proteins. Plant Mol. Biol. 72, 621–630. [DOI] [PubMed] [Google Scholar]
- Price, A.A. , Grakoui, A. and Weiss, D.S. (2016) Harnessing the prokaryotic adaptive immune system as a eukaryotic antiviral defense. Trends Microbiol. 24, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyott, D.E. , Sheehan, E. and Molnar, A. (2016) Engineering of CRISPR/Cas9‐mediated potyvirus resistance in transgene‐free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F. (2010) Plant viruses versus RNAi: simple pathogens reveal complex insights on plant antimicrobial defense. Wiley Interdiscip. Rev. RNA, 1, 22–33. [DOI] [PubMed] [Google Scholar]
- Ravelonandro, M. , Scorza, R. , Michel, H.J. and Briard, P. (2014) The efficiency of RNA interference for conferring stable resistance to Plum pox virus. Plant Cell, Tissue Organ Cult. 118, 347–356. [Google Scholar]
- Rees, H.A. and Liu, D.R. (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, K.E. , Worrall, E.A. and Mitter, N. (2014) Double stranded RNA expression and its topical application for non‐transgenic resistance to plant viruses. J. Plant Biochem. Biotechnol. 23, 231–237. [Google Scholar]
- Romano, N. and Macino, G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. [DOI] [PubMed] [Google Scholar]
- Sampson, T.R. , Saroj, S.D. , Llewellyn, A.C. , Tzeng, Y.‐L. and Weiss, D.S. (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature, 497, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon, H. (2015) Plant translation factors and virus resistance. Viruses, 7, 3392–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.C. and Johnston, S.A. (1985) The concept of parasite‐derived resistance—Deriving resistance genes from the parasite's own genome. J. Theor. Biol. 113, 395–405. [Google Scholar]
- Scorza, R. , Callahan, A. , Levy, L. , Damsteegt, V. , Webb, K. and Ravelonandro, M. (2001) Post‐transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum pox potyvirus coat protein gene. Transgenic Res. 10, 201–209. [DOI] [PubMed] [Google Scholar]
- Sera, T. (2005) Inhibition of virus DNA replication by Artificial Zinc Finger Proteins. J. Virol. 79, 2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhawat, U.K. , Ganapathi, T.R. and Hadapad, A.B. (2012) Transgenic banana plants expressing small interfering RNAs targeted against viral replication initiation gene display high‐level resistance to banana bunchy top virus infection. J. Gen. Virol. 93, 1804–1813. [DOI] [PubMed] [Google Scholar]
- Shepherd, D.N. , Mangwende, T. , Martin, D.P. , Bezuidenhout, M. , Thomson, J.A. and Rybicki, E.P. (2007) Inhibition of maize streak virus (MSV) replication by transient and transgenic expression of MSV replication‐associated protein mutants. J. Gen. Virol. 88, 325–336. [DOI] [PubMed] [Google Scholar]
- Soler, N. , Plomer, M. , Fagoaga, C. , Moreno, P. , Navarro, L. , Flores, R. and Peña, L. (2012) Transformation of Mexican lime with an intron‐hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of Citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 10, 597–608. [DOI] [PubMed] [Google Scholar]
- Suweis, S. , Carr, J.A. , Maritan, A. , Rinaldo, A. and D'Odorico, P. (2015) Resilience and reactivity of global food security. Proc. Natl Acad. Sci. 112, 6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado, F. , Martínez‐García, B. , Vargas, M. and Díaz‐Ruíz, J.R. (2003) Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. and Befort, B.L. (2011) Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S.Q. and Joung, J.K. (2016) Defining and improving the genome‐wide specificities of CRISPR–Cas9 nucleases. Nat. Rev. Genet. 17, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov, F.D. , Rebar, E.J. , Holmes, M.C. , Zhang, H.S. and Gregory, P.D. (2010) Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Eggenberger, A.L. , Nutter, F.W. and Hill, J.H. (2001) Pathogen‐derived transgenic resistance to soybean mosaic virus in soybean. Mol. Breeding, 8, 119–127. [Google Scholar]
- Wigley, D.B. (2012) Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9. [DOI] [PubMed] [Google Scholar]
- Wilson, T.M. (1993) Strategies to protect crop plants against viruses: pathogen‐derived resistance blossoms. Proc. Natl Acad. Sci. 90, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittner, A. , Palkovics, L. and Balázs, E. (1998) Nicotiana benthamiana plants transformed with the plum pox virus helicase gene are resistant to virus infection. Virus Res. 53, 97–103. [DOI] [PubMed] [Google Scholar]
- Worrall, E.A. , Bravo‐Cazar, A. , Nilon, A.T. , Fletcher, S.J. , Robinson, K.E. , Carr, J.P. and Mitter, N. (2019) Exogenous application of RNAi‐inducing double‐stranded RNA inhibits aphid‐mediated transmission of a plant virus. Front. Plant Sci. 10, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, C. and Kanaar, R. (2006) DNA double‐strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383. [DOI] [PubMed] [Google Scholar]
- Ye, C. and Li, H. (2010) 20 Years of transgenic research in China for resistance to Papaya ringspot virus. Transgenic Plant J. 4, 58–63. [Google Scholar]
- Yin, K. and Qiu, J.‐L. (2019) Genome editing for plant disease resistance: applications and perspectives. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, S.S.‐E.‐A. , Tashkandi, M. , Mansoor, S. and Mahfouz, M.M. (2016) Engineering plant immunity: using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 7, 1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, X. , Zhang, F. , Zhong, Z. , Chen, R. , Wang, Y. , Chang, L. , Bock, R. et al. (2019) Generation of virus‐resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17, 1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z.‐Y. , Fu, F.‐L. , Gou, L. , Wang, H.‐G. and Li, W.‐C. (2010) RNA interference‐based transgenic maize resistant to maize dwarf mosaic virus. J. Plant Biol. 53, 297–305. [Google Scholar]
- Zhang, Z.‐Y. , Wang, Y.‐G. , Shen, X.‐J. , Li, L. , Zhou, S.‐F. , Li, W.‐C. and Fu, F.‐L. (2013) RNA interference‐mediated resistance to maize dwarf mosaic virus. Plant Cell Tissue Organ. Cult. 113, 571–578. [Google Scholar]
- Zhang, T. , Zheng, Q. , Yi, X. , An, H. , Zhao, Y. , Ma, S. and Zhou, G. (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y. , Ye, J. , Cao, X. , Xu, C. , Chen, B. , An, H. et al. (2019) Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 17, 1185–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]


