Summary
MYB transcription factors (TFs) have been demonstrated to play diverse roles in plant growth and development through interaction with basic helix‐loop‐helix (bHLH) TFs. MdbHLH33, an apple bHLH TF, has been identified as a positive regulator in cold tolerance and anthocyanin accumulation by activating the expressions of MdCBF2 and MdDFR . In the present study, a MYB TF MdMYB308L was found to also positively regulate cold tolerance and anthocyanin accumulation in apple. We found that MdMYB308L interacted with MdbHLH33 and enhanced its binding to the promoters of MdCBF2 and MdDFR . In addition, an apple RING E3 ubiquitin ligase MYB30‐INTERACTING E3 LIGASE 1 (MdMIEL1) was identified to be an MdMYB308L‐interacting protein and promoted the ubiquitination degradation of MdMYB308L, thus negatively regulated cold tolerance and anthocyanin accumulation in apple. These results suggest that MdMYB308L acts as a positive regulator in cold tolerance and anthocyanin accumulation in apple by interacting with MdbHLH33 and undergoes MdMIEL1‐mediated protein degradation. The dynamic change in MYB‐bHLH protein complex seems to play a key role in the regulation of plant growth and development.
Keywords: apple, MYB transcription factor, cold tolerance, anthocyanin accumulation, E3 ubiquitin ligase
Introduction
Cold stress is an adverse environmental factor that limits plants growth and crop production, which may even lead to the death of plants (Thakur et al., 2010; Thomashow, 1999). At the cellular level, cold stress damages cell membrane structure and affects metabolism of nutrients. At the tissue level, cold stress causes damages to plant roots, leaves, flowers and fruits. To sense and adapt to adverse cold stress conditions, plants have evolved efficient regulatory mechanisms to increase cold tolerance (Xin and Browse, 2000; Zhu, 2001). Extensive investigations have revealed that numerous genes are activated in response to cold stress, including those involved in the biosynthesis of cold‐triggered components, such as soluble sugar, proline, betaine, polyamines, phenylpropanoids and antioxidants (Bajwa et al., 2014; Chinnusamy et al., 2007; Provart et al., 2003; Rivero et al., 2001; Thomashow, 1999). C‐repeat binding factor (CBF) transcription factors (TFs) play essential roles in the cold stress response by binding to the C‐repeat/dehydration‐responsive elements (CRT/DRE) in the promoters of the cold‐regulated (COR) genes and regulating their expressions (Gilmour et al., 2004; Jaglo et al., 2001; Stockinger et al., 1997). So far, three CBFs in Arabidopsis (Medina et al., 1999; Medinab et al., 2011) and five CBFs in apple (An et al., 2018; Wisniewski et al., 2014) have been isolated and characterized. Constitutive expression of CBF genes improves plant cold stress tolerance (Liu et al., 1998; Thomashow, 1999; Wisniewski et al., 2014). CBF genes are regulated at the transcriptional level by the basic helix‐loop‐helix (bHLH) TFs ICE1 and ICE2 in Arabidopsis (Chinnusamy et al., 2003; Fursova et al., 2009). In apple, MdCIbHLH1, the homolog of Arabidopsis ICE1, contributes to the improved cold tolerance by directly activating the CBF genes (Feng et al., 2012). It is recognized that the ICEs‐CBFs‐CORs regulatory pathway plays a central role in the regulation of cold stress response. Besides ICEs, the transcriptions of CBFs are also regulated by MYB TFs (Agarwal et al., 2006; An et al., 2018; Xie et al., 2018), calmodulin‐binding transcription activator 1‐3 (CAMTA1‐3) (Doherty et al., 2009), PSEUDO‐RESPONSE REGULATORs (PRRs) (Nakamichi et al., 2009), CIRCADIAN CLOCKASSOCIATED 1 (CCA1) (Dong et al., 2011), ETHYLENE‐INSENSITIVE 3 (EIN3) (Shi et al., 2012), phytohormone‐interacting factors (PIFs) (Jiang et al., 2017; Lee and Thomashow, 2012) and brassinazole‐resistant 1 (BZR1) (Li et al., 2017a,b). In addition to the CBF cold stress response pathway, many studies have demonstrated that CBF‐independent signalling pathway also plays an important role in the regulation of cold stress response (Li et al., 2017a,b; Zhu et al., 2004).
As the largest TF family, MYB TFs act as important regulators in plant biotic and abiotic stress responses including cold stress response (Chinnusamy et al., 2007; Dubos et al., 2010; Li et al., 2015). MYB TFs involved in the regulation of cold tolerance have been functionally identified in several species. For example, Arabidopsis MYB14 and MYB15 negatively regulate cold tolerance by repressing the expression of CBF genes (Agarwal et al., 2006; Chen et al., 2013). In rice, overexpression of MYB4, MYB3R‐2, MYBS3 and MYB2 improves plant cold tolerance (Dai et al., 2007; Su et al., 2010; Vannini et al., 2004; Yang et al., 2012). Recent investigations in apple show that MdoMYB121, MdSIMYB1, MdMYB4, MdMYB23, MdMYB88 and MdMYB124 are positive regulators of cold tolerance (An et al., 2018; Cao et al., 2013; Wang et al., 2014; Wu et al., 2017; Xie et al., 2018), whereas MdMYB44 and MdMYB15L negatively regulate plant cold tolerance (Wu et al., 2018; Xu et al., 2018a,b).
In addition to functioning in stress response, MYB TFs have been characterized to play key roles in the regulation of plant secondary metabolism including anthocyanin biosynthesis (Allan et al., 2008; Dubos et al., 2010; Liu et al., 2015). In Arabidopsis, MYB75, MYB90, MYB113 and MYB114 contribute to anthocyanin accumulation (Gonzalez et al., 2008; Maier et al., 2013; Teng et al., 2005), whereas MYBL2 represses anthocyanin biosynthesis (Dubos et al., 2008; Matsui et al., 2008). In apple, MdMYB1 and its allelic genes are recognized as the central regulators in regulating anthocyanin biosynthesis (Ban et al., 2007; Espley et al., 2007; Takos et al., 2006). In addition, MdMYB3, MdMYB9, MdMYB11, MdMYB12, MdMYB16, MdMYB22, MdMYB110a and MdMYBPA1 are also identified to regulate flavonoid biosynthesis in apple (An et al., 2014; Umemura et al., 2013; Vimolmangkang et al., 2013; Wang et al., 2017, 2018; Xu et al., 2017). A few of MYB TFs are considered to function in association with bHLH and WD‐repeat proteins (Dubos et al., 2010; Jaakola, 2013; Zimmermann et al., 2004). The MYBs‐bHLHs‐WD‐repeat protein complex plays a decisive role in the regulation of anthocyanin biosynthesis.
Ubiquitination is an important post‐translational modification that extensively regulates plant growth and development including biotic and abiotic stress responses (Lyzenga and Stone, 2011; Sun and Chen, 2004). Ubiquitination consists of tagging the ubiquitin to target proteins, which leads to the 26S proteasome‐mediated degradation of target proteins (Peng et al., 2003; Pickart, 2001). Three enzymatic activities are involved in this action including the E1 ubiquitin activating enzymes, the E2 ubiquitin‐conjugating enzymes and the E3 ubiquitin ligases (Serino and Xie, 2013; Smalle and Vierstra, 2004). Among these three components, E3 ubiquitin ligases play key roles in determining substrate specificity (Ardley and Robinson, 2005; Buetow and Huang, 2016). According to its conserved structural domain, E3 ubiquitin ligases are divided into four types including HECT, RING, SCF and APC ligases (Deshaies and Joazeiro, 2009; Metzger et al., 2012; Morreale and Walden, 2016). Among them, RING domain E3 ubiquitin ligases have been widely studied to modulate abscisic acid signalling (Stone et al., 2006; Zhang et al., 2007), anthocyanin biosynthesis (An et al., 2017a; Li et al., 2012; Maier et al., 2013), cold (Dong et al., 2006), salt (Kim and Kim, 2013; Zhang et al., 2015), drought (Kim and Kim, 2013; Qin et al., 2008; Ryu et al., 2010) and heat tolerances (Liu et al., 2016).
In this study, a cold‐responsive MYB TF named MdMYB308L was characterized in apple. Overexpression of MdMYB308L led to improved cold tolerance and increased anthocyanin accumulation. MdMYB308L physically interacted with MdbHLH33, a bHLH TF that positively regulates cold tolerance and anthocyanin biosynthesis (Xu et al., 2017, 2018a,b). Further studies showed that MdMYB308L enhanced the binding of MdbHLH33 to the MdCBF2 and MdDFR promoters through a direct protein interaction. In addition, an apple RING E3 ubiquitin ligase MdMIEL1 interacted with MdMYB308L and promoted the degradation of MdMYB308L through the 26S‐proteasome pathway, thus negatively regulating cold tolerance and anthocyanin accumulation. Taken together, we have identified and characterized a novel MYB TF that regulates the cold tolerance and anthocyanin accumulation in association with a bHLH TF and undergoes the 26S proteasome‐mediated degradation.
Results
Identification of a cold‐responsive MYB TF MdMYB308L in apple
In our previous study, cold stress transcriptome was performed to identify the cold‐responsive MYB TFs, in which an MYB TF MdMYB23 was characterized as a positive regulator in cold tolerance (An et al., 2018). Similarly, we identified another cold‐responsive MYB TF (GenBank accession number: MDP0000950559), which showed the highest cold‐induced expression only second to MdMYB15 and MdMYB23 (Table S1). Sequence search against the National Center for Biotechnology Information (NCBI) database showed that it was an apple MYB domain 308‐like gene, thus designated as MdMYB308L. Sequence alignment and phylogenetic tree analysis demonstrated that MdMYB308L had the highest protein sequence identity with PbMYB308L from pears (Pyrus x bretschneideri) and contained a conserved MYB DNA‐binding domain and a bHLH binding motif (Figure S1a‐d). In addition, we performed another sequence alignment and phylogenetic tree analysis from MdMYB proteins involved in anthocyanin biosynthesis and cold tolerance (Figure S2a‐b). The result revealed that MdMYB308L had high genetic relationship with MdMYB3 and it showed a large sequence difference compared with other MdMYB proteins, indicating that MdMYB308L might be a novel MYB protein and it might have unique biological functions.
Next, we tested how the expression of MdMYB308L responded to the cold stress. When apple seedlings were placed at 4°C, the expression of MdMYB308L showed increase starting from only 1 h after the treatment and the increase lasted until 9 h‐12 h after the treatment (Figure 1a). We also set up a marker gene expression system to further examine the cold stress response of MdMYB308L. The promoter sequence of MdMYB308L (Figure S3) was cloned into pCAMBIA1391‐GUS vector to generate the ProMdMYB308L::GUS construct, which was introduced into apple calli. Transgenic apple calli expressing the ProMdMYB308L::GUS construct clearly exhibited higher GUS activity after cold treatment at 4°C for 9 h compared with the control that was placed at 24°C (Figure 1b), indicating that cold stress triggered the expression of MdMYB308L. Furthermore, the MdMYB308L protein was also examined in response to cold treatment (4°C) using an in vitro protein degradation system. Purified MdMYB308L‐GST fusion proteins were incubated with the total proteins extracted from wild‐type apple calli with or without 4°C treatments. It seemed that MdMYB308L‐GST protein was slowly degraded since 2 h after the incubation started, which could be blocked by the presence of MG132 (Figure 1c). However, the cold treatment seemed to have slowed down the degradation process of MdMYB308L‐GST protein and improved its stability (Figure 1c). Taken together, these data suggest that MdMYB308L is responsive to cold stress at both transcriptional and post‐transcriptional levels.
Figure 1.
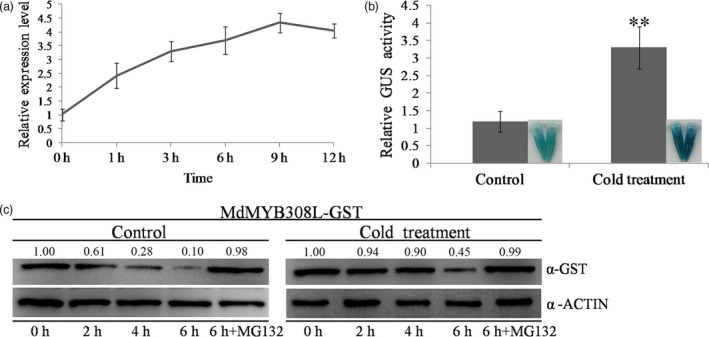
The expression pattern of MdMYB308L in response to cold stress (4°C treatment). (a) MdMYB308L gene expression detected by qRT‐PCR analysis. The value for 0 h was set to 1. (b) GUS staining and relative GUS activity analysis of the MdMYB308L promoter expression construct Pro MdMYB308L ::GUS in transgenic apple calli. Control: GUS staining and activity analysis at 9 h at 24°C. Cold treatment: GUS staining and activity analysis at 9 h under cold stress. (c) Degradation of the MdMYB308L‐GST fusion protein under cold stress. Total proteins extracted from wild‐type apple calli with or without 4°C treatments and the inclusion of 100 μm MG132 were incubated with the purified MdMYB308L‐GST fusion protein. The samples were collected at the indicated time. Control: 24°C; cold treatment: 4°C. ACTIN was used as internal reference. The relative intensity ratio between the GST and the ACTIN was shown. Each experiment was performed in three replicates. Error bars donate standard deviation. Significant differences were detected by t‐test (**P < 0.01).
MdMYB308L plays a positive role in cold stress response
To explore the biological role of MdMYB308L in cold stress, its expression was modified by transforming an overexpressing construct (MdMYB308L‐OX) or an antisense suppressing construct (MdMYB308L‐Anti) into wild‐type apple calli (Figure S4a). There was no growth difference observed among these three types of apple calli under normal growth condition at 24°C (Figure 2a‐b, control). However, when these 8‐day‐old wild‐type and transgenic apple calli were treated with cold conditions (4°C) for 10 days, apple calli with the overexpression construct (MdMYB308L‐OX) showed faster growth (Figure 2a) and higher fresh weight (Figure 2b) compared to the WT control. To the contrary, the apple calli carrying the antisense construct MdMYB308L‐Anti showed the opposite phenotypes with slower growth (Figure 2a) and smaller fresh weight (Figure 2b). These observations indicate that MdMYB308L may play a role in the cold stress response. In this experiment, we also observed that the expressions of some cold‐responsive genes were modified by the alteration of MdMYB308L expression. Among the eight genes tested, MdCBF2, MdKIN1, MdRD29A and MdCOR47 showed the most dramatic increase of expressions in the MdMYB308L‐OX calli and significant decrease of expressions in the MdMYB308L‐Anti calli (Figure 2c; An et al., 2018). Meanwhile, MdCBF1 and MdCBF3 also showed significant increase in expressions in the MdMYB308L‐OX calli, although to a much lesser extent (Figure 2c; An et al., 2018). These results suggest that MdMYB308L may execute its role in cold tolerance by regulating the expression of cold‐responsive genes.
Figure 2.
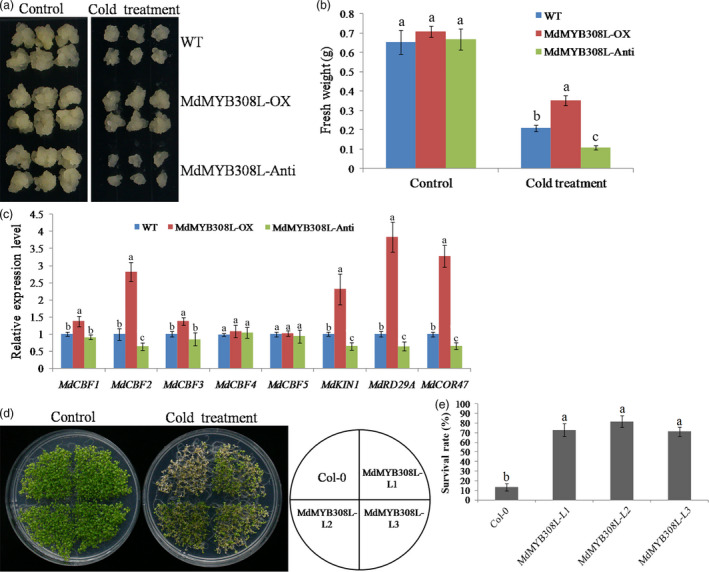
Phenotypes of MdMYB308L transgenic apple calli and Arabidopsis seedlings under cold stress. (a) Appearance of 8‐day‐old apple calli under control (24°C) and cold stress (4°C) conditions for 10 days. WT: wild‐type; MdMYB308L‐OX: MdMYB308L‐overexpression; MdMYB308L‐Anti: MdMYB308L antisense suppression. (b) The fresh weight of apple calli shown in (a). (c) Expressions of MdCBFs and their target genes in apple calli shown in (a). Quantitative real‐time PCR analysis was performed in three biological replicates and three technical replicates. The value for WT was set to 1. (d) Appearance of control (Col‐0) and MdMYB308L overexpression (MdMYB308L‐L1, MdMYB308L‐L2 and MdMYB308L‐L3) Arabidopsis seedlings with or without (control) cold stress treatment. Plants were grown on MS plates at 22°C for 12 days before cold treatment at ‐4°C for 0.5 h. (e) Survival rate of Arabidopsis seedlings shown in (d). Each experiment was performed in three replicates. Error bars denote standard deviation. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
To verify the results in plants, the overexpression construct MdMYB308L‐OX was transformed into wild‐type Arabidopsis Col‐0 seedlings and three independent lines with very high expressions of MdMYB308L (MdMYB308L‐L1, MdMYB308L‐L2 and MdMYB308L‐L3) were selected (Figure S4b). Together with the Col‐0 control, these transgenic Arabidopsis seedlings of 12‐day‐olds were treated at ‐4°C for 0.5 h by gradient cooling. As shown in Figure 2d and 2e, MdMYB308L overexpression seedlings exhibited the higher survival rate compared to the wild type after cold treatments, indicating that MdMYB308L plays a positive role in cold stress response.
Overexpression of MdMYB308L increases anthocyanin accumulation
Anthocyanin functions in cold tolerance by scavenging reactive oxygen species (Hernández et al., 2009; Winkel‐Shirley, 2002), and MYB TFs play key roles in the regulation of anthocyanin biosynthesis (Allan et al., 2008; Dubos et al., 2010). To determine whether the modification of MdMYB308L expression has any impact on the biosynthesis of anthocyanin, anthocyanin contents were examined in 15‐day‐old apple calli treated with high light for 7 days. We found that MdMYB308L‐OX calli accumulated significantly more anthocyanin and MdMYB308L‐Anti calli accumulated less anthocyanin compared to the wild‐type calli (Figure 3a‐b). In addition, the anthocyanin biosynthesis‐related genes MdDFR, MdUF3GT, MdF3H and MdCHS all showed significant increase of expressions in the MdMYB308L‐OX calli, and MdDFR and MdUF3GT also showed decreased expressions in the MdMYB308L‐Anti calli (Figure 3c; Figure S4a). These data suggest that MdMYB308L promotes anthocyanin accumulation by modulating the expression of anthocyanin biosynthesis‐related genes.
Figure 3.

Anthocyanin accumulation in MdMYB308L transgenic apple calli and leaves. (a) Appearance of apple calli of 15‐day‐olds after highlight treatment for 7 days. WT: wild‐type; MdMYB308L‐OX: MdMYB308L‐overexpression; MdMYB308L‐Anti: MdMYB308L antisense suppression. (b) The relative anthocyanin contents of apple calli shown in (a). The value for WT was set to 1. (c) Relative expression levels of anthocyanin biosynthesis‐related genes in apple calli shown in (a). Quantitative real‐time PCR analysis was performed in three biological replicates and three technical replicates. The value for WT was set to 1. (d) Appearance of apple leaves treated with highlight for 5 days. EV: empty vector; MdMYB308L‐OX: MdMYB308L‐overexpression; MdMYB308L‐Anti: MdMYB308L antisense suppression. (e) Relative anthocyanin contents of apple leaves shown in (d). The value for EV was set to 1. Each experiment was performed in three replicates. Error bars denoted standard deviation. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
In a parallel experiment, the MdMYB308L‐OX and MdMYB308L‐Anti constructs were transiently expressed in apple leaves using a vacuum pump (Figure S4c). The apple leaves were treated with high light for 5 days after transformation to induce the anthocyanin deposition. Consistently, overexpression of MdMYB308L promoted, but suppression of MdMYB308L reduced the accumulation of anthocyanin in these apple leaves (Figure 3d–e), confirming that MdMYB308L contributes to anthocyanin accumulation.
MdMYB308L interacts with MdbHLH33
A few of MYB TFs have been known to function in association with the bHLH proteins (Dubos et al., 2010; Jaakola, 2013; Zimmermann et al., 2004). Previous reports have revealed that an apple bHLH TF MdbHLH33 positively regulates both cold stress response and anthocyanin biosynthesis (Xu et al., 2017, 2018a,b). To elucidate whether MdMYB308L interacts with MdbHLH33, yeast two‐hybrid (Y2H) assays were performed (Figure 4a). MdMYB308L‐pGBD and MdbHLH33‐pGAD were transformed into yeast cells. And the empty vectors were used as controls. The result showed that yeast cells expressing both MdMYB308L and MdbHLH33 grew normally in the selective medium (Figure 4a), indicating that MdMYB308L interacts with MdbHLH33 in yeast cells. In addition, pull‐down assays were carried out using the fusion proteins MdMYB308L‐GST and MdbHLH33‐HIS (Figure 4b). The protein mixtures were purified using a glutathione purification kit. As shown in Figure 4b, MdbHLH33‐HIS was pulled down by MdMYB308L‐GST, while HIS alone did not, indicating that MdMYB308L interacts with MdbHLH33 in vitro. To provide more evidence for the interaction between MdMYB308L and MdbHLH33, we conducted bimolecular fluorescence complementation (BiFC) assays, by transforming MdMYB308L‐YFPC and MdbHLH33‐YFPN into onion epidermal cells for YFP fluorescence signals (Figure 4c). The result demonstrated that MdMYB308L interacted with MdbHLH33 in the nucleus. In addition, we found that MdMYB308L did not interact with MdbHLH3, a homologous gene of MdbHLH33 (Figure S3c). Taken together, these data reveal that MdMYB308L indeed interacts with MdbHLH33.
Figure 4.

MdMYB308L interacts with MdbHLH33. (a) Yeast two‐hybrid assays. The open reading frames of MdMYB308L and MdbHLH33 were fused with pGBD and pGAD vectors, respectively. Transformed yeast cells were grown on SD‐Trp/‐Leu (‐T/‐L), SD‐Trp/‐Leu/‐His/‐Ade (‐T/‐L/‐H/‐A) or SD‐Trp/‐Leu/‐His/‐Ade supplementing X‐gal (‐T/‐L/‐H/‐A+X‐gal) media. (b) Pull‐down assays. E. coli‐expressed HIS or MdbHLH33‐HIS proteins were incubated with a cobalt chelate affinity resin containing the immobilized glutathione‐tagged MdMYB308L protein. The protein mixtures were purified using a glutathione purification kit. (c) Bimolecular fluorescence complementation assays. The open reading frames of MdbHLH33 and MdMYB308L were fused to the N‐terminal part of YFP and the C‐terminal part of YFP, respectively. Bars = 10 μm.
MdMYB308L enhances the binding of MdbHLH33 to its target genes
In apple calli expressing either MdMYB308L‐OX or MdMYB308L‐Anti, the expression of MdbHLH33 did not show any change (Figure 3c). Considering the direct interaction between MdMYB308L and MdbHLH33, we determined to investigate the relationship between these two proteins. Previous studies have demonstrated that MdbHLH33 promotes cold tolerance and anthocyanin biosynthesis by directly activating MdCBF2 and MdDFR genes, respectively (Xu et al., 2017, 2018a,b). We verified these results in our study with a gel mobility shift assay using DNA probes carrying the LTR element within the MdCBF2 promoter and the E‐box within the MdDFR promoter (Figure 5a‐b). Moreover, we found that, with the increased addition of MdMYB308L‐GST proteins, the binding intensities of MdbHLH33 to the MdCBF2 and MdDFR promoters increased (Figure 5a‐b; Figures S5‐S7). Alternatively, we constructed effectors and reporters to perform a luciferase assay (Figure 5c), which showed that the MdbHLH33 contributed to the increased LUC/REN activities of pMdCBF2 and pMdDFR, and the LUC/REN activities further increased when MdMYB308L and MdbHLH33 were co‐expressed (Figure 5d‐e). These results suggest that MdMYB308L promotes the binding of MdbHLH33 to its target genes.
Figure 5.

MdMYB308L modifies the binding of MdbHLH33 to its target genes. (a) and (b) Electrophoretic mobility shift assays. The ‘‐’ sign represents the absence of relevant proteins; ‘+’ represents the presence of relevant probes or proteins; ‘2 × ’ and ‘3 × ’ represents increased protein levels. MdCBF2‐Mut is the mutant form of MdCBF2, in which the 5′–CCGAAA–3′ motif was replaced with 5′–CCACAA–3′. MdDFR‐Mut is the mutant form of MdDFR, in which the 5′–CATTTG–3′ motif was replaced with 5′–AATTTC–3′. Biotin‐labelled probes were incubated with MdMYB308L‐GST or MdbHLH33‐GST protein, and the free and bound probes were separated on an acrylamide gel. The GST protein was used to ensure an equal quantity of proteins. (c) Schematic representation of the constructs (effector and reporter vectors) used for the luciferase reporter lines. (d) and (e) LUC/REN activities of constructs pMdCBF2:LUC (d) and pMdDFR:LUC (e) when co‐transformed with different constructs. Empty vector was used as the reference. Empty vector: 62SK+LUC; MdMYB23 + pMdCBF2: MdMYB23‐62SK+pMdCBF2‐LUC; MdMYB308L+pMdCBF2: MdMYB308L‐62SK+pMdCBF2‐LUC; MdbHLH33 + pMdCBF2: MdbHLH33‐62SK+pMdCBF2‐LUC; MdbHLH33 + pMdCBF2‐Mut: MdbHLH33‐62SK+pMdCBF2‐Mut‐LUC; MdMYB308L+MdMYB23 + pMdCBF2: MdMYB308L‐62SK+MdMYB23‐62SK+pMdCBF2‐LUC; MdMYB308L+MdbHLH33 + pMdCBF2: MdMYB308L‐62SK+MdbHLH33‐62SK+pMdCBF2‐LUC; MdMYB1 + pMdDFR: MdMYB1‐62SK+pMdDFR‐LUC; MdMYB308L+pMdDFR: MdMYB308L‐62SK+pMdDFR‐LUC; MdbHLH33 + pMdDFR: MdbHLH33‐62SK+pMdDFR‐LUC; MdbHLH33 + pMdDFR‐Mut: MdbHLH33‐62SK+pMdDFR‐Mut‐LUC; MdMYB308L+MdMYB1 + pMdDFR: MdMYB308L‐62SK+MdMYB1‐62SK+pMdDFR‐LUC; MdMYB308L+MdbHLH33 + pMdDFR: MdMYB308L‐62SK+MdbHLH33‐62SK+pMdDFR‐LUC; each experiment was performed in three replicates. Error bars denoted standard deviation. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
MdbHLH33 is essential for MdMYB308L‐mediated cold tolerance and anthocyanin accumulation
To dissect the genetic interaction between MdMYB308L and MdbHLH33, we generated MdbHLH33 antisense suppressing plasmid (MdbHLH33‐Anti) and transformed it into the MdMYB308L‐overexpressing apple calli and leaves (Figure S4d‐e). The transgenic materials were used for the cold tolerance and anthocyanin accumulation assays. Apple calli with altered expressions of MdMYB308L and/or MdbHLH33 did not show any growth difference under control conditions (Figure 6a‐b). However, when the calli were placed at 4°C for cold treatments, MdbHLH33‐Anti ones showed significantly slower growth (Figure 6a) and smaller fresh weight (Figure 6b), which was overcame by the co‐expression of the MdMYB308L‐OX construct (Figure 6a‐b). Similarly, in both apple calli and apple leaf transient expression systems, the accumulation of anthocyanin was greatly induced by the overexpression of MdMYB308L (Figure 6c‐f). When the two constructs were co‐delivered, the anthocyanin levels were still elevated compared to the controls but significantly lowered than overexpression of MdMYB308L alone (Figure 6c‐f). These results showed that suppression of MdbHLH33 decreased MdMYB308L‐promoted cold tolerance and anthocyanin accumulation, indicating that MdMYB308L regulates cold tolerance and anthocyanin accumulation partially depending on MdbHLH33.
Figure 6.

Phenotypes of apple calli and leaves with modified expressions of MdMYB308L and MdbHLH33. (a) Appearance of 8‐day‐old apple calli grown at 24°C (control) or 4°C (cold treatment) for 10 days. WT: wild‐type; MdMYB308L‐OX: MdMYB308L‐overexpression; MdbHLH33‐Anti: MdbHLH33 antisense suppression; MdMYB308L‐OX/MdbHLH33‐Anti: suppression of MdbHLH33 in the background of MdMYB308L‐overexpression. (b) The fresh weights of apple calli shown in (a). (c) Appearance of apple calli (15‐day‐old) used in (a) when treated with highlight for 5 days. (d) The relative anthocyanin content of apple calli shown in (c). The value for WT was set to 1. (e) Appearance of apple leaves expressing different constructs after highlight treatment for 5 days. EV: empty vector; MdMYB308L‐OX: MdMYB308L‐overexpression; MdbHLH33‐Anti: MdbHLH33 antisense suppression; MdMYB308L‐OX/MdbHLH33‐Anti: suppression of MdbHLH33 in the background of MdMYB308L‐overexpression. (f) The relative anthocyanin content of apple leaves shown in (e). The value for EV was set to 1. Each experiment was performed in three replicates. Error bars denoted standard deviation. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
MdMYB308L interacts with MdMIEL1
Our above results reveal that cold stress influences the stability of the MdMYB308L protein (Figure 1c). To further explore the post‐transcriptional regulatory mechanism of MdMYB308L in response to cold stress, a yeast screening assay was performed using MdMYB308L‐pGBD as the bait (Table S2). As a result, MdMIEL1 (GenBank accession number: MDP0000185659) was identified. Previous studies have shown that MdMIEL1 encodes a RING E3 ubiquitin ligase in apple and acts as a negative regulator in oxidative and salt stresses (An et al., 2017b), as well as anthocyanin accumulation (An et al., 2017a). To confirm the interaction between MdMYB308L and MdMIEL1 in Y2H assays, MdMYB308L‐pGBD and MdMIEL1‐pGAD were transformed into yeast cells. The result showed that yeast cells expressing both MdMYB308L and MdMIEL1 grew normally in the selective medium (Figure 7a), indicating that MdMYB308L interacts with MdMIEL1 in yeast cells. We then performed pull‐down assays using fusion proteins MdMYB308L‐GST and MdMIEL1‐HIS. As shown in Figure 7b, MdMIEL1‐HIS was detected with a HIS antibody in eluted solution, indicating that MdMYB308L interacts with MdMIEL1 in vitro. Furthermore, BiFC assays were carried out to provide another evidence for the interaction between MdMYB308L and MdMIEL1 by transforming MdMYB308L‐YFPC and MdMIEL1‐YFPN constructs into onion epidermal cells together or individually. The YFP fluorescence signals were only detected with the presence of both MdMYB308L‐YFPC and MdMIEL1‐YFPN constructs, indicating that MdMYB308L interacts with MdMIEL1 in the nucleus (Figure 7c).
Figure 7.
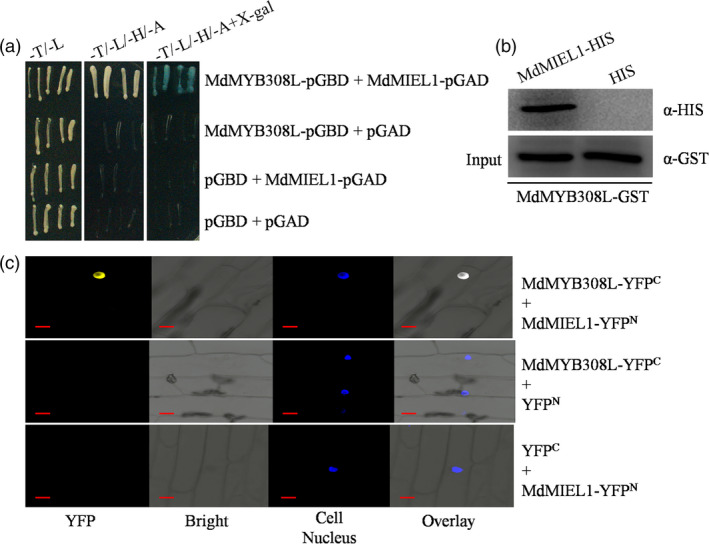
MdMYB308L interacts with MdMIEL1. (a) Yeast two‐hybrid assays. The open reading frames of MdMYB308L and MdMIEL1 were fused with pGBD and pGAD vectors, respectively. Transformed yeast cells were grown on SD‐Trp/‐Leu (‐T/‐L), SD‐Trp/‐Leu/‐His/‐Ade (‐T/‐L/‐H/‐A) or SD‐Trp/‐Leu/‐His/‐Ade supplementing X‐gal (‐T/‐L/‐H/‐A+X‐gal) media. (b) Pull‐down assays. E. coli‐expressed HIS or MdMIEL1‐HIS proteins were incubated with a cobalt chelate affinity resin containing the immobilized glutathione‐tagged MdMYB308L protein. The protein mixtures were purified using a glutathione purification kit. (c) Bimolecular fluorescence complementation assays. The open reading frames of MdMIEL1 and MdMYB308L were fused to the N‐terminal part of YFP and the C‐terminal part of YFP, respectively. Bars = 10 μm.
MdMIEL1 is a repressor of cold tolerance and anthocyanin accumulation
To study the molecular function of MdMIEL1 in cold stress, we first examined the expression pattern of MdMIEL1 in response to cold stress (4°C). MdMIEL1 was repressed when apple calli were exposed to cold stress (Figure 8a). In addition, the promoter sequence of MdMIEL1 was inserted into pCAMBIA1391‐GUS vector to generate the ProMdMIEL1::GUS reporter construct (Figure S8). The GUS activity of ProMdMIEL1::GUS transgenic apple calli was reduced significantly after cold treatment (4°C) (Figure 8b), indicating that cold stress repressed the expression of MdMIEL1. Furthermore, the MdMIEL1 protein level was examined in response to cold treatment (4°C) using an in vitro protein degradation system. The results showed that cold treatment accelerated the degradation of the MdMIEL1 protein (Figure 8c).
Figure 8.

The expression pattern of MdMIEL1 in response to cold stress (4°C treatment). (a) MdMIEL1 gene expression detected by qRT‐PCR analysis. Experiments were performed in three biological replicates and three technical replicates. The value for 0 h was set to 1. (b) GUS staining and relative GUS activity analysis of the MdMIEL1 promoter expression construct Pro MdMIEL1 ::GUS in transgenic apple calli. Control: GUS staining and activity analysis at 9 h at 24°C. Cold treatment: GUS staining and activity analysis at 9 h under cold stress. (c) Degradation of the MdMIEL1‐HIS fusion protein under cold stress. Total proteins extracted from wild‐type apple calli with or without 4°C treatments and the inclusion of 100 μm MG132 were incubated with the purified MdMIEL1‐HIS fusion protein. The samples were collected at the indicated time. Control: 24°C; cold treatment: 4°C. ACTIN was used as internal reference. The relative intensity ratio between the HIS and the ACTIN was shown. Each experiment was performed in three replicates. Error bars denoted standard deviation. Significant differences were detected by t‐test (**P < 0.01).
To explore the physiological role of MdMIEL1 in cold stress and anthocyanin accumulation, transgenic apple calli (MdMIEL1‐OX: overexpression of MdMIEL1; MdMIEL1‐Anti: suppression of MdMIEL1), Arabidopsis plants (MdMIEL1‐L1, MdMIEL1‐L2 and MdMIEL1‐L3) and apple leaves (MdMIEL1‐OX: overexpression of MdMIEL1; MdMIEL1‐Anti: suppression of MdMIEL1) were generated (Figure S4f‐h). In all testing systems, the overexpression of MdMIEL1 significantly delayed plant material growth (Figure 9a‐d) and the anthocyanin accumulation (Figure 9e‐h) under cold stress conditions. The overexpression of MdMIEL1 also decreased the anthocyanin biosynthesis in both apple calli and leaves, while the suppression of MdMIEL1 expression showed the opposite effect (Figure 9e‐h), which is consistent with our previous report (An et al., 2017a). These observations suggest that MdMIEL1 is a repressor of both cold tolerance and anthocyanin accumulation.
Figure 9.

Phenotypes of MdMIEL1 transgenic apple calli, Arabidopsis seedlings and apple leaves. (a) Appearance of 8‐day‐old apple calli under control (24°C) and cold stress (4°C) conditions for 10 days. WT: wild‐type; MdMIEL1‐OX: MdMIEL1‐overexpression; MdMIEL1‐Anti: MdMIEL1 antisense suppression. (b) The fresh weight of apple calli shown in (a). (c) The appearance of control (Col‐0) and MdMIEL1 overexpression (MdMIEL1‐L1, MdMIEL1‐L2 and MdMIEL1‐L3) Arabidopsis seedlings with or without (control) cold stress treatment. Plants were grown on MS plates at 22°C for 12 days before cold treatment at −4°C for 10 min. (d) Survival rate of Arabidopsis seedlings shown in (c). (e) Appearance of apple calli (15‐day‐old) used in (a) when treated with highlight for 7 days. WT: wild‐type; MdMIEL1‐OX: MdMIEL1‐overexpression; MdMIEL1‐Anti: MdMIEL1 antisense suppression. (f) The relative anthocyanin content of apple calli shown in (e). The value for WT was set to 1. (g) Appearance of apple leaves expressing different constructs after highlight treatment for 5 days. EV: empty vector; MdMIEL1‐OX: MdMIEL1‐overexpression; MdMIEL1‐Anti: MdMIEL1 antisense suppression. (h) The relative anthocyanin content of apple leaves shown in (g). The value for EV was set to 1. Each experiment was performed in three replicates. Error bars denoted standard deviation. Significant differences were detected by t‐test (**P < 0.01). Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
MdMIEL1 ubiquitinates the MdMYB308L protein and accelerates its degradation
MIEL1 encodes an E3 ubiquitin ligase, and MYB30, MYB98 and MYB1 have been verified as ubiquitination substrates of MIEL1 (An et al., 2017a; Lee and Seo, 2016; Marino et al., 2013). Since MdMYB308L and MdMIEL1 proteins interacted with each other directly (Figure 7), and the overexpression of them showed the opposite phenotypes for cold stress response and anthocyanin accumulation (Figures 6 and 9), we examined whether MdMIEL1 regulates the protein stability of MdMYB308L by ubiquitination modification. To test the hypothesis, we performed an ubiquitination detection assay in vitro. The ubiquitination pattern of MdMYB308L‐GST was detected by anti‐GST probing in the presence or absence of ATP, ubiquitin, E1, E2 and MdMIEL1‐HIS, to test the E3 ubiquitin ligase activity of MdMIEL1‐HIS (Figure 10a). The result demonstrated that MdMYB308L‐GST could be ubiquitinated by MdMIEL1‐HIS in the present of all necessary components (Figure 10a). In addition, an in vivo ubiquitination detection assay was performed using MdMYB308L‐MYC and MdMYB308L‐MYC/MdMIEL1‐OX apple calli. Protein gel blot analysis showed that MdMIEL1 enhanced the ubiquitination modification of the MdMYB308L‐MYC protein (Figure 10b). These findings suggest that MdMIEL1 ubiquitinates the MdMYB308L protein in vitro and in vivo.
Figure 10.

MdMYB308L is an ubiquitination substrate of MdMIEL1. (a) MdMIEL1 ubiquitinates MdMYB308L in vitro. MdMIEL1‐HIS was tested for E3 ubiquitin ligase activity in the presence and absence of ATP, ubiquitin, E1, E2, MdMIEL1‐HIS and MdMYB308L‐GST. The protein gel blot was analysed using a GST antibody. (b) MdMIEL1 ubiquitinates MdMYB308L in vivo. MdMYB308L‐MYC was immunoprecipitated using MYC antibody from the two transgenic apple calli (MdMYB308L‐MYC and MdMYB308L‐MYC/MdMIEL1‐OX). Immunoblotting using an ubiquitin antibody was shown on the top and using a MYC antibody at below. (c) MdMIEL1 promotes the degradation of MdMYB308L‐GST protein in vitro. Total proteins extracted from wild‐type and transgenic apple calli with or without 100 μm MG132 treatment were incubated with the purified MdMYB308L‐GST fusion protein. The samples were collected at the indicated time. ACTIN was used as internal reference. The relative intensity ratio between the GST and the ACTIN was shown.
We then carried out in vitro protein degradation assays to assess whether MdMIEL1 affects the stability of the MdMYB308L protein. Total proteins extracted from wild‐type and MdMIEL1 transgenic apple calli were incubated with the purified MdMYB308L‐GST fusion protein. As expected, overexpression of MdMIEL1 accelerated the degradation of the MdMYB308L‐GST protein, while suppression of the MdMIEL1 expression showed the opposite effect (Figure 10c). Collectively, these results illustrated that MdMIEL1 accelerates MdMYB308L protein degradation by modulating the ubiquitination modification of MdMYB308L.
MdMIEL1 negatively regulates MdMYB308L‐promoted cold tolerance and anthocyanin accumulation
To provide genetic evidence for the above conclusion, the MdMIEL1 overexpressing plasmid (MdMIEL1‐OX) and antisense suppressing plasmid (MdMIEL1‐Anti) were transformed into MdMYB308L‐overexpressing apple calli, Arabidopsis seedlings and apple leaves (Figure S4f‐h). We then examined the cold tolerance and anthocyanin accumulation of co‐transformed plant materials. In apple calli, the cold‐induced suppression of growth was smaller in the MdMYB308L overexpression materials compared to the WT and the calli overexpressing both MdMYB308L and MdMIEL1 (Figure 11a‐b). However, the calli with both the MdMYB308L overexpression construct and the MdMIEL1 suppression construct showed the best growth under cold condition among the four different plant materials (Figure 11a‐b). In Arabidopsis young seedlings, a similar pattern was observed. While overexpression of MdMYB308L helped the plant growth under cold condition, seedlings overexpressing both MdMYB308L and MdMIEL1 behaved similarly to the Col‐0 control with cold stress (Figure 11c‐d).
Figure 11.

MdMIEL1 decreases MdMYB308L‐promoted cold tolerance and anthocyanin accumulation. (a) Appearance of apple calli of 8‐day‐olds expressing different constructs under control condition (24°C) or cold treatment condition (4°C) for 10 days. WT: wild‐type; MdMYB308L‐OX: MdMYB308L‐overexpression; MdMYB308L‐OX/MdMIEL1‐OX: overexpression of MdMIEL1 in the background of MdMYB308L‐overexpression; MdMYB308L‐OX/MdMIEL1‐Anti: suppression of MdMIEL1 in the background of MdMYB308L‐overexpression. (b) The fresh weight of apple calli shown in (a). (c) Appearance of wild‐type (Col‐0) and transgenic Arabidopsis seedlings under control condition or cold treatment condition. Arabidopsis seedlings were grown on MS plates at 22°C for 12 days before being treated at −4°C for 0.5 h. MdMYB308L‐L1 and MdMYB308L‐L2: MdMYB308L‐overexpression Arabidopsis seedlings. MdMYB308L‐L1/MdMIEL1‐L1 and MdMYB308L‐L2/MdMIEL1‐L1: MdMIEL1 and MdMYB308L co‐transformed Arabidopsis seedlings. Control: no treatment; cold treatment: grow at 4°C for 2 days, followed by cold treatment at −4°C for 0.5 h. (d) Survival rate of Arabidopsis seedlings after cold stress treatment. (e) Appearance of 15‐day‐old apple calli treated with highlight for 5 days. (f) The relative anthocyanin content of apple calli shown in (e). The value for WT was set to 1. (g) Appearance of apple leaves treated with highlight for 5 days. EV: empty vector; MdMYB308L‐OX: MdMYB308L‐overexpression; MdMYB308L‐OX/MdMIEL1‐OX: overexpression of MdMIEL1 in the background of MdMYB308L‐overexpression; MdMYB308L‐OX/MdMIEL1‐Anti: suppression of MdMIEL1 in the background of MdMYB308L‐overexpression. (h) The relative anthocyanin content of apple leaves shown in (g). The value for EV was set to 1. Each experiment was performed in three replicates. Error bars denoted standard deviation. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
In a similar manner, apple calli overexpressing MdMYB308L showed significant increase in anthocyanin accumulation, but co‐expression of both MdMYB308L and MdMIEL1 at the same showed less increase in anthocyanin accumulation (Figure 11e‐f). However, suppressing the expression of MdMIEL1 in the MdMYB308L overexpression background increased the anthocyanin deposition even more dramatically (Figure 11e‐f). The same trend of anthocyanin accumulation was observed in apple leaves (Figure 11g‐h). These findings demonstrate that MdMIEL1 negatively regulates MdMYB308L‐promoted cold tolerance and anthocyanin accumulation.
Discussion
Cold stress affects plant growth and development. In agricultural production, cold stress can cause crop freezing damage and large area of yield reduction and even lead to crop failure under severe circumstances (Thakur et al., 2010). As an important economic crop, apple production is also affected by cold stress. Therefore, it is of great significance to study the cold stress response mechanism of apple for improving apple yield and farmers’ income. We previously performed transcriptome analyses to identify the cold‐responsive MYB TFs, in which an apple MYB TF MdMYB23 was characterized to positively regulate cold tolerance (An et al., 2018). Here, we investigated the cold response mechanism of another MYB TF MdMYB308L (GenBank accession number: MDP0000950559), whose induction by cold was second only to MdMYB23 (Table S1; An et al., 2018). Our present data demonstrate that MdMYB308L is a positive regulator of cold tolerance and anthocyanin accumulation and functions in association with MdbHLH33 in apple. In addition, MdMYB308L is confirmed as a target protein of MdMIEL1 and undergoes MdMIEL1‐mediated ubiquitination degradation.
MdMYB308L interacts with MdbHLH33 to regulate cold tolerance and anthocyanin accumulation
Cold stress induces the reactive oxygen species production and anthocyanin accumulation (Jaakola, 2013; Suzuki and Mittler, 2006). In return, anthocyanin contributes to reactive oxygen species scavenging and improved cold tolerance (Hernández et al., 2009; Winkel‐Shirley, 2002). Multiple types of TFs play key roles in the regulation of both cold stress response and anthocyanin accumulation including MYB TFs (Allan et al., 2008; Chinnusamy et al., 2007; Dubos et al., 2010; Li et al., 2015; Liu et al., 2015). For example, in apple, MdMYB23, MdMYB88 and MdMYB124 positively regulate both cold tolerance and flavonoids accumulation (An et al., 2018; Xie et al., 2018), whereas MdMYB15L is a negative regulator of both cold tolerance and anthocyanin accumulation (Xu et al., 2018a,b). In the present study, a cold‐induced MYB TF MdMYB308L was isolated and overexpression of MdMYB308L improved cold tolerance and anthocyanin biosynthesis (Figures 1, 2, 3). It is acknowledged that MYBs‐bHLHs‐WD‐repeat protein complex play a central role in the regulation of anthocyanin biosynthesis (Jaakola, 2013; Xu et al., 2015). Previous reports have revealed that an apple bHLH TF MdbHLH33 regulates both cold stress response and anthocyanin biosynthesis (Xu et al., 2017, 2018a,b). A series of physiological and biochemical data indicated that MdMYB308L physically interacted with MdbHLH33 (Figure 4), and MdMYB308L improved cold tolerance and anthocyanin accumulation by enhancing the binding of MdbHLH33 to its downstream target genes (Figures 5, 6). These findings indicate that MdMYB308L is a novel cold‐responsive gene, which functions through a ‘MYB‐bHLH’ module.
MdMIEL1 negatively regulates cold tolerance and anthocyanin accumulation by degrading MdMYB308L
Since the MdMYB308L protein stability was regulated by cold treatments (Figure 1c), the post‐transcriptional regulatory mechanism of MdMYB308L was explored here. An apple RING ubiquitin ligase MdMIEL1 was identified as an MdMYB308L‐interacting protein (Figure 7), whose function was characterized previously (An et al., 2017a,b). Arabidopsis MIEL1 negatively regulates plant defence and ABA signalling by degrading MYB30 and MYB96 proteins, respectively (Lee and Seo, 2016; Marino et al., 2013). Arabidopsis MIEL1 also inhibits the biosynthesis of stem cuticular wax (Gil et al., 2017). In rice, OsSRFP1, the homologous gene of MIEL1, is involved in multiple abiotic stress tolerance responses to salt, cold and oxidative stresses (Fang et al., 2015). Our previous studies in apple have shown that MdMIEL1 negatively regulates oxidative and salt stresses (An et al., 2017b) and inhibits the anthocyanin accumulation by degrading MdMYB1 (An et al., 2017a). Here, we found that cold stress inhibited the expression of MdMIEL1 (Figure 8), and MdMIEL1 acted as a repressor of cold tolerance and anthocyanin accumulation by degrading the MdMYB308L protein (Figures 9, 10, 11). Taken together, our data reveal that MYB308L is a specific ubiquitination target of MIEL1, and MIEL1 may mediate plant growth and development by regulating different MYB proteins.
In addition, MdMIEL1 seems to undergo an ubiquitination modification in response to cold stress (Figure 8c), similar as MdMYB308L (Figure 1c), which indicates that E3 ligases may also undergo the ubiquitination modification (Serino and Xie, 2013). And this observation may be indicative of the connector role of the E3 ligases in response to external stresses and regulating their downstream substrates. However, which E3 ubiquitin ligase regulates the ubiquitination of the MdMIEL1 protein remains unknown and it will be an interesting research topic in the future.
‘MIEL1‐MYB308L‐bHLH33’ module is a novel dynamic cold stress response mechanism
Post‐translational modifications, such as ubiquitination, sumoylation and phosphorylation, affect the plant cold stress response. The E3 ubiquitin ligase HOS1 negatively regulates plant cold response by degrading the ICE1 protein in Arabidopsis (Dong et al., 2006). An apple BTB protein MdBT2 mediates the ubiquitination and degradation of MdMYB23 to suppress cold tolerance (An et al., 2018). SIZ1 modulates sumoylation of ICE1 to modify the Arabidopsis freezing tolerance (Miura et al., 2007). In phosphorylation modification, Arabidopsis OPEN STOMATA 1 (OST1) and MAP kinases regulate the stability of ICE1 to mediate plant cold tolerance (Ding et al., 2015; Li et al., 2017a,b; Liu and Zhou, 2018; Zhao et al., 2017). Our current results show that apple MdMIEL1 promotes the ubiquitination and degradation of the MdMYB308L protein, which leads to decreases in MdMYB308L‐activated expressions of MdCBF2 and MdDFR, thus negatively regulates cold tolerance and anthocyanin accumulation. When exposed to cold stress, the transcription of MdMYB308L is induced, but the expression of MdMIEL1 is repressed at both transcriptional and post‐transcriptional levels. This in turn releases the inhibition effect of MdMIEL1 on MdMYB308L. As a result, the accumulation of MdMYB308L promotes the expressions of MdCBF2 and MdDFR through its interaction with MdbHLH33, which leads to increased cold tolerance and anthocyanin accumulation (Figure 12). The dynamic regulatory module of ‘MIEL1‐MYB308L‐bHLH33’ shows the flexibility of cold stress response mechanism. We propose that ‘MIEL1‐MYB308L‐bHLH33’ signalling regulatory mechanism may play an important role in coordinating the cold stress response with other response mechanisms.
Figure 12.

A working model illustrating that MdMYB308L functions in cold stress response and anthocyanin accumulation. MdMYB308L interacts with MdbHLH33 to increase its transcriptional activity and enhance its binding to the MdCBF2 and MdDFR promoters, thus promoting cold tolerance and anthocyanin accumulation. In the absence of cold stress, MdMIEL1 interacts with MdMYB308L to ubiquitinate and degrade it, thus negatively regulating MdMYB308L‐promoted cold tolerance and anthocyanin accumulation. On one hand, cold stress up‐regulates the transcription of MdMYB308L, which promotes the cold tolerance and anthocyanin accumulation. On the other hand, cold stress inhibited MdMIEL1 expression, thus to release the MdMIEL1‐enhanced protein degradation of MdMYB308L, which also contributes to cold tolerance and anthocyanin accumulation. 26S proteasome represents that MdMYB308L undergoes 26S proteasome‐mediated degradation by MdMIEL1. The green line represents transcriptional regulation. The blue line represents post‐translational regulation. The red line represents transcriptional and post‐translational regulations.
Experimental procedures
Plant materials and growth conditions
Apple calli (Malus domestica, ‘Orin’), leaves of apple tissue culture seedlings (Malus domestica, ‘GL3’) and Arabidopsis seedlings (Arabidopsis thaliana, ‘Col‐0’) were used for the present studies. Apple calli were grown at 24°C under dark conditions, and subcultured at a 20‐day interval. Apple tissue culture seedlings were grown at 24°C for a 16‐h light/8‐h dark cycle and subcultured at a 30‐day interval. Arabidopsis seedlings were grown at 22°C for a 16‐h light/8‐h dark cycle.
Phylogenetic tree and sequence alignment
The MYB308L protein sequences of 16 different species and MYB proteins from different species were obtained from the NCBI database. Phylogenetic tree was constructed with MEGA 5.0. Sequence alignment was performed using DNAMAN.
Plasmid construction and genetic transformation
The promoter sequences of MdMYB308L and MdMIEL1 genes were inserted into pCAMBIA1391‐GUS vector to generate ProMdMYB308L::GUS and ProMdMIEL1::GUS. The promoter sequences of MdCBF2 and MdDFR genes were inserted into pGreen0800‐LUC to generate pMdCBF2‐LUC and pMdDFR‐LUC. The open reading frame (ORF) of MdMYB308L, MdbHLH33 or MdMIEL1 was cloned into pGAD or pGBD vector to generate MdMYB308L‐pGBD, MdbHLH33‐pGAD and MdMIEL1‐pGAD. The ORF of MdMYB308L, MdbHLH33 or MdMIEL1 was cloned into pGEX4T‐1 or pET32a vector to generate MdMYB308L‐pGEX4T‐1, MdbHLH33‐pET32a, MdbHLH33‐pGEX4T‐1 and MdMIEL1‐pET32a. The ORF of MdMYB308L, MdbHLH33 or MdMIEL1 was cloned into YFPC or YFPN vector to generate MdMYB308L‐YFPC, MdbHLH33‐YFPN and MdMIEL1‐YFPN. The ORF of MdMYB308L, MdbHLH33, MdMYB23 or MdMYB1 was cloned into pCXSN‐MYC or pGreen 62‐SK to generate MdMYB308L‐MYC, MdMYB308L‐62SK, MdbHLH33‐62SK, MdMYB23‐62SK and MdMYB1‐62SK. The ORF of MdMIEL1 was cloned into pRI101 to generate MdMIEL1‐OX. The fragments of MdMYB308L, MdbHLH33 and MdMIEL1 were cloned into pCXSN to generate MdMYB308L‐Anti, MdbHLH33‐Anti and MdMIEL1‐Anti. Primers used in this study are listed in Table S3.
The transgenic apple calli were obtained as described previously (An et al., 2018). To generate transient transgenic apple leaves, leaves were incubated with Agrobacterium carrying overexpression or antisense suppression plasmids of MdMYB308L, MdbHLH33 and MdMIEL1, and vacuumed using a vacuum pump for about 15 minutes. qRT‐PCR was performed to examine the expression levels of transient transgenic apple leaves (An et al., 2019). The transgenic Arabidopsis seedlings were generated as described previously (Clough and Bent, 1998).
Quantitative real‐time‐PCR (qRT‐PCR) analysis
Apple calli, apple leaves and Arabidopsis seedlings were collected, and full ground with liquid nitrogen and total RNAs were extracted using a RNA extraction kit (TIANGEN, Beijing, China). Single‐stranded cDNA was obtained using a reverse transcription kit (TaKaRa, Shiga, Japan). All qRT‐PCR analyses were performed with three biological repeats and three technical repeats. All primers used in this study are listed in Table S3.
GUS staining and activity analysis
ProMdMYB308L::GUS and ProMdMIEL1::GUS transgenic apple calli were used for cold stress treatment and GUS staining. GUS staining and activity analysis were performed as described previously (An et al., 2018).
Cold tolerance assays
Cold tolerance assays were performed as described previously (An et al., 2018). For apple calli, 8‐day‐old apple calli were treated at 4°C for 10 days. Apple calli growing at 24°C for 10 days were used as controls. The fresh weights of apple calli under normal and cold stress conditions were recorded. For the cold stress treatment of Arabidopsis seedlings, 12‐day‐old Arabidopsis seedlings grown at 22°C for 12 days were treated at 4°C for 2 days for cold acclimation. And then, seedlings were transferred to −4°C for indicated time (10 min or 0.5 h) by gradient cooling and finally grown at 22°C for 2 days. Gradient cooling was carried out in the incubator. In the incubator, the temperature can reduce slowly from 22°C to −4°C and can also increase slowly to 22°C from −4°C. And the processing time can be adjusted according to the actual situation. Survival rate of Arabidopsis seedlings after cold stress treatment was recorded.
Anthocyanin accumulation assays
Apple calli and leaves of 15‐day‐olds were transferred in a highlight phytotron (photon flux density: 100 μmol/m2/s) for indicated time (5 days or 7 days). Anthocyanin was extracted using anthocyanin extraction buffer. Anthocyanin content was determined as described previously (An et al., 2018).
Screening the potential interacting proteins of MdMYB308L
Yeast two‐hybrid screening assays were performed to identify the interacting proteins of MdMYB308L using MdMYB308L‐pGBD as the bait. MdMYB308L‐pGBD and an apple library (Shanghai OE Biotech. Co., Ltd.) were mixed and transformed into yeast cells ‘Y2H Gold’. The transformed yeast strains were grown in the SD‐Trp/‐Leu/‐His/‐Ade (‐T/‐L/‐H/‐A) medium for 4 days. The cDNA fragments of positive yeast strains were identified by sequencing. As a result, 19 potential interacting proteins were obtained (Table S2).
Due to the false‐positive phenomenon of yeast two‐hybrid screening, we further confirmed the interactions through a one‐to‐one yeast two‐hybrid assays. The results showed that only MDP0000185659 (MdMIEL1) and DQ266451 (MdbHLH33) were the direct interacting proteins of MdMYB308L.
Y2H, pull‐down and BiFC assays
Y2H, pull‐down and BiFC assays were performed as described previously (An et al., 2018). In brief, for Y2H assays, MdMYB308L‐pGBD, MdbHLH33‐pGAD, MdbHLH3‐pGAD and MdMIEL1‐pGAD were transformed into yeast ‘Y2H Gold’. The transformed yeast strains were grown in SD medium. For pull‐down assays, the fusion proteins of MdMYB308L‐GST, MdbHLH33‐HIS and MdMIEL1‐HIS were prepared. The protein mixtures were purified using a glutathione purification kit (Thermo Fisher Scientific, Waltham, MA, USA). The eluted solution was detected using HIS or GST antibodies (Abmart, Shanghai, China). For BiFC assay, MdMYB308L‐YFPC, MdbHLH33‐YFPN and MdMIEL1‐YFPN were transformed into onion epidermal cells by Agrobacterium‐mediated genetic transformation.
Electromobility shift assays (EMSA)
EMSAs were performed as described previously (An et al., 2018). MdMYB308L‐GST, MdbHLH33‐GST and biotin‐labelled probes (MdCBF2‐probe and MdDFR‐probe) were prepared. Biotin‐labelled probes were incubated with MdMYB308L‐GST or MdbHLH33‐GST protein in the binding buffer for 25 minutes, and the free and bound probes were separated on an acrylamide gel. Unlabelled probes were used as competitors. The GST protein was used to ensure an equal quantity of proteins.
LUC/REN activity analysis
LUC/REN activity was determined as described previously (An et al., 2018). MdbHLH33‐62SK, MdMYB308L‐62SK, MdMYB23‐62SK, MdMYB1‐62SK pMdCBF2‐LUC and pMdDFR‐LUC were prepared. Agrobacterium carrying indicated plasmids was injected into the back of tobacco leaves. MdMYB23‐62SK and MdMYB1‐62SK were used as positive controls.
Ubiquitination detection and protein degradation assays
Ubiquitination detection was performed as described previously (An et al., 2017a). For in vitro ubiquitination detection, MdMIEL1‐HIS was tested for E3 ubiquitin ligase activity in the presence or absence of ATP, ubiquitin, E1, E2, MdMIEL1‐HIS or MdMYB308L‐GST. The protein gel blot was analysed using a GST antibody. For in vivo ubiquitination detection, MdMYB308L‐MYC was immunoprecipitated using MYC antibody from the MdMYB308L‐MYC and MdMYB308L‐MYC/MdMIEL1‐OX apple calli. The protein gel blot was analysed using MYC and Ubi antibodies. Total proteins extracted from apple calli were incubated with the purified MdMYB308L‐GST or MdMIEL1‐HIS fusion protein. The samples were collected at the indicated periods. The protein gel blot was analysed using GST and HIS antibodies.
Statistical analysis
Each experiment was performed in three replicates. Experimental results were analysed using GraphPad Prism 6.02 or DPS software. Error bars denote standard deviations. Significant differences were detected by t‐test: **P < 0.01. Different letters above the bars indicated significant difference (P < 0.05) as obtained by one‐way ANOVA and LSD test.
Author contributions
Y.J.H. and J.P.A. conceived and designed the experiments. J.P.A. performed the research. J.P.A., X.X.W., X.W.Z., H.F.X., S.Q.B. and C.X.Y. analysed the data. J.P.A. and Y.J.H. wrote the paper.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Sequence analysis of MYB308L proteins.
Figure S2 MdMYB308L is a novel MYB protein in apple.
Figure S3 The promoter sequence of MdMYB308L.
Figure S4 Gene expression analysis of transgenic plant materials using qRT‐PCR.
Figure S5 The promoter sequence of MdCBF2.
Figure S6 The promoter sequence of MdDFR.
Figure S7 MdMYB308L does not affect MYB‐related genes expression and not bind to the MdCBF2 and MdDFR promoters.
Figure S8 The promoter sequence of MdMIEL1.
Table S1 Cold‐inducible MYB transcription factors identified from a cold stress transcriptome study using cDNA samples extracted from apple seedlings treated with or without cold stress (4°C) for 9 h.
Table S2 Screening the potential interacting proteins of MdMYB308L.
Table S3 Primers used for gene expression analysis and vector construction.
Acknowledgements
This work was financially supported by grants from the Ministry of Science and Technology of China (2018YFD1000200), Natural Science Foundation of China (31430074), Shandong Province Government (SDAIT‐06‐03), Natural Science Foundation of Shandong Province (ZR2019PC004) and Ministry of Agriculture of China (CARS‐28).
Contributor Information
Chun‐Xiang You, Email: haoyujin@sdau.edu.cn.
Yu‐Jin Hao, Email: youchunxiang@sdau.edu.cn.
References
- Agarwal, M. , Hao, Y. , Kapoor, A. , Dong, C.H. , Fujii, H. , Zheng, X. and Zhu, J.K. (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. [DOI] [PubMed] [Google Scholar]
- Allan, A.C. , Hellens, R.P. and Laing, W.A. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. [DOI] [PubMed] [Google Scholar]
- An, X.H. , Tian, Y. , Chen, K.Q. , Liu, X.J. , Liu, D.D. , Xie, X.B. , Cheng, C.G. et al. (2014) MdMYB9 and MdMYB11 are involved in the regulation of the JA‐induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Liu, X. , Li, H.H. , You, C.X. , Wang, X.F. and Hao, Y.J. (2017a) Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol. 58, 1953–1962. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Liu, X. , Song, L.Q. , You, C.X. , Wang, X.F. and Hao, Y.J. (2017b) Apple RING finger E3 ubiquitin ligase MdMIEL1 negatively regulates salt and oxidative stresses tolerance. J Plant Biol. 60, 137–145. [Google Scholar]
- An, J.P. , Li, R. , Qu, F.J. , You, C.X. , Wang, X.F. and Hao, Y.J. (2018) R2R3‐MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Zhang, X.W. , Bi, S.Q. , You, C.X. , Wang, X.F. and Hao, Y.J. (2019) MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 222, 735–751. [DOI] [PubMed] [Google Scholar]
- Ardley, H.C. and Robinson, P.A. (2005) E3 ubiquitin ligases. Essays Biochem. 41, 15–30. [DOI] [PubMed] [Google Scholar]
- Bajwa, V.S. , Shukla, M.R. , Sherif, S.M. , Murch, S.J. and Saxena, P.K. (2014) Role of melatonin in alleviating cold stress in Arabidopsis thaliana . J. Pineal Res. 56, 238–245. [DOI] [PubMed] [Google Scholar]
- Ban, Y. , Honda, C. , Hatsuyama, Y. , Igarashi, M. , Bessho, H. and Moriguchi, T. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. [DOI] [PubMed] [Google Scholar]
- Buetow, L. and Huang, D.T. (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 17, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Z.H. , Zhang, S.Z. , Wang, R.K. , Zhang, R.F. and Hao, Y.J. (2013) Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE, 8, e69955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Chen, Z. , Kang, J. , Kang, D. , Gu, H. and Qin, G. (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 31, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy, V. , Ohta, M. , Kanrar, S. , Lee, B.H. , Hong, X. , Agarwal, M. and Zhu, J.K. (2003) ICE1: a regulator of cold‐induced transcriptome and freezing tolerance in Arabidopsis . Genes Dev. 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy, V. , Zhu, J. and Zhu, J.K. (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dai, X. , Xu, Y. , Ma, Q. , Xu, W. , Wang, T. , Xue, Y. and Chong, K. (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R‐2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis . Plant Physiol. 143, 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. and Joazeiro, C.A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Li, H. , Zhang, X. , Xie, Q. , Gong, Z. and Yang, S. (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis . Dev. Cell 32, 278–289. [DOI] [PubMed] [Google Scholar]
- Doherty, C.J. , Van Buskirk, H.A. , Myers, S.J. and Thomashow, M.F. (2009) Roles for Arabidopsis CAMTA transcription factors in cold‐regulated gene expression and freezing tolerance. Plant Cell. 21, 972–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.H. , Agarwal, M. , Zhang, Y. , Xie, Q. and Zhu, J.K. (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl Acad. Sci. USA 103, 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, M.A. , Farre, E.M. and Thomashow, M.F. (2011) CIRCADIAN CLOCK‐ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate expression of the C‐REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis . Proc. Natl Acad. Sci. USA 108, 7241–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C. , Le Gourrierec, J. , Baudry, A. , Huep, G. , Lanet, E. , Debeaujon, I. , Routaboul, J.M. et al. (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana . Plant J. 55, 940–953. [DOI] [PubMed] [Google Scholar]
- Dubos, C. , Stracke, R. , Grotewold, E. , Weisshaar, B. , Martin, C. and Lepiniec, L. (2010) MYB transcription factors in Arabidopsis . Trends Plant Sci. 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Espley, R.V. , Hellens, R.P. , Putterill, J. , Stevenson, D.E. , Kutty‐Amma, S. and Allan, A.C. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, H. , Meng, Q. , Xu, J. , Tang, H. , Tang, S. , Zhang, H. and Huang, J. (2015) Knock‐down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol. Biol. 87, 441–458. [DOI] [PubMed] [Google Scholar]
- Feng, X.M. , Zhao, Q. , Zhao, L.L. , Qiao, Y. , Xie, X.B. , Li, H.F. , Yao, Y.X. et al. (2012) The cold‐induced basic helix‐loop‐helix transcription factor gene MdCIbHLH1 encodes an ICE‐like protein in apple. BMC Plant Biol. 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova, O.V. , Pogorelko, G.V. and Tarasov, V.A. (2009) Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana . Gene, 429, 98–103. [DOI] [PubMed] [Google Scholar]
- Gil, H.L. , Kim, J. , Chung, M.S. and Joon, P.S. (2017) The MIEL1 E3 ubiquitin ligase negatively regulates cuticular wax biosynthesis in Arabidopsis stems. Plant Cell Physiol. 58, 1249–1259. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J. , Fowler, S.G. and Thomashow, M.F. (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J.M. and Lloyd, A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Hernández, I. , Alegre, L. , Van Breusegem, F. and Munné‐Bosch, S. (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14, 125–132. [DOI] [PubMed] [Google Scholar]
- Jaakola, L. (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. [DOI] [PubMed] [Google Scholar]
- Jaglo, K.R. , Kleff, S. , Amundsen, K.L. , Zhang, X. , Haake, V. , Zhang, J.Z. , Deits, T. et al. (2001) Components of the Arabidopsis C‐repeat/dehydration‐responsive element binding factor cold‐response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127, 910–917. [PMC free article] [PubMed] [Google Scholar]
- Jiang, B. , Shi, Y. , Zhang, X. , Xin, X. , Qi, L. , Guo, H. and Yang, S. (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA 114, E6695–E6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H. and Kim, W.T. (2013) The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high salt and drought stress responses. Plant Physiol. 162, 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.G. and Seo, P.J. (2016) The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat. Commun. 7, 12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.M. and Thomashow, M.F. (2012) Photoperiodic regulation of the C‐repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana . Proc. Natl Acad. Sci. USA 109, 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.Y. , Mao, K. , Zhao, C. , Zhao, X.Y. , Zhang, H.L. , Shu, H.R. and Hao, Y.J. (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light‐induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Ng, C.K.Y. and Fan, L.M. (2015) MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. [Google Scholar]
- Li, H. , Ding, Y. , Shi, Y. , Zhang, X. , Zhang, S. , Gong, Z. and Yang, S. (2017a) MPK3‐and MPK6‐mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis . Dev. Cell 43, 630–642. [DOI] [PubMed] [Google Scholar]
- Li, H. , Ye, K. , Shi, Y. , Cheng, J. , Zhang, X. and Yang, S. (2017b) BZR1 positively regulates freezing tolerance via CBF‐dependent and CBF‐independent pathways in Arabidopsis . Mol Plant. 10, 545–559. [DOI] [PubMed] [Google Scholar]
- Liu, Y. and Zhou, J. (2018) MAPping kinase regulation of ICE1 in freezing tolerance. Trends Plant Sci. 23, 91–93. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Kasuga, M. , Sakuma, Y. , Abe, H. , Miura, S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought‐and low‐temperature‐responsive gene expression, respectively, in Arabidopsis . Plant Cell. 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Osbourn, A. and Ma, P. (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 8, 689–708. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Zhang, C. , Wei, C. , Liu, X. , Wang, M. , Yu, F. , Xie, Q. et al. (2016) The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2‐induced stomatal closure in rice. Plant Physiol. 170, 429–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga, W.J. and Stone, S.L. (2011) Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616. [DOI] [PubMed] [Google Scholar]
- Maier, A. , Schrader, A. , Kokkelink, L. , Falke, C. , Welter, B. , Iniesto, E. , Rubio, V. et al. (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis . Plant J. 74, 638–651. [DOI] [PubMed] [Google Scholar]
- Marino, D. , Froidure, S. , Canonne, J. , Khaled, S.B. , Khafif, M. , Pouzet, C. , Jauneau, A. et al. (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nature Commun. 4, 1476. [DOI] [PubMed] [Google Scholar]
- Matsui, K. , Umemura, Y. and Ohme‐Takagi, M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis . Plant J. 55, 954–967. [DOI] [PubMed] [Google Scholar]
- Medina, J. , Bargues, M. , Terol, J. , Perez‐Alonso, M. and Salinas, J. (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain‐containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medinab, J. , Cataláa, R. and Salinasa, J. (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci. 180, 3–11. [DOI] [PubMed] [Google Scholar]
- Metzger, M.B. , Hristova, V.A. and Weissman, A.M. (2012) HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Jin, J.B. , Lee, J. , Yoo, C.Y. , Stirm, V. , Miura, T. , Ashworth, E.N. et al. (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . Plant Cell. 19, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreale, F.E. and Walden, H. (2016) Types of ubiquitin ligases. Cell 165, 248–248. [DOI] [PubMed] [Google Scholar]
- Nakamichi, N. , Kusano, M. , Fukushima, A. , Kita, M. , Ito, S. , Yamashino, T. , Saito, K. et al. (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. [DOI] [PubMed] [Google Scholar]
- Peng, J. , Schwartz, D. , Elias, J.E. , Thoreen, C.C. , Cheng, D. , Marsischky, G. , Roelofs, J. et al. (2003) A proteomics approach to understanding protein ubiquitination. Nature Biotech. 21, 921. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001) Mechanisms underlying ubiquitination. Annual Rev Biochem. 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Provart, N.J. , Gil, P. , Chen, W. , Han, B. , Chang, H.S. , Wang, X. and Zhu, T. (2003) Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 132, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, F. , Sakuma, Y. , Tran, L.S.P. , Maruyama, K. , Kidokoro, S. , Fujita, Y. , Fujita, M. et al. (2008) Arabidopsis DREB2A‐interacting proteins function as RING E3 ligases and negatively regulate plant drought stress‐responsive gene expression. Plant Cell. 20, 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero, R.M. , Ruiz, J.M. , Garcıa, P.C. , Lopez‐Lefebre, L.R. , Sánchez, E. and Romero, L. (2001) Resistance to cold and heat stress: accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 160, 315–321. [DOI] [PubMed] [Google Scholar]
- Ryu, M.Y. , Cho, S.K. and Kim, W.T. (2010) The Arabidopsis C3H2C3‐type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an ABA‐dependent response to drought stress. Plant Physiol. 154, 1983–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G. and Xie, Q. (2013) The ever expanding role of ubiquitin and SUMO in plant biology. J. Integr. Plant Biol. 55, 5–6. [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Tian, S. , Hou, L. , Huang, X. , Zhang, X. , Guo, H. and Yang, S. (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type‐A ARR genes in Arabidopsis . Plant Cell. 24, 2578–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J. and Vierstra, R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J. , Gilmour, S.J. and Thomashow, M.F. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain‐containing transcriptional activator that binds to the C‐repeat/DRE, a cis‐acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S. , Williams, L.A. , Farmer, L.M. , Vierstra, R.D. and Callis, J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, C.F. , Wang, Y.C. , Hsieh, T.H. , Lu, C.A. , Tseng, T.H. and Yu, S.M. (2010) A novel MYBS3‐dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. and Chen, Z.J. (2004) The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16, 119–126. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. and Mittler, R. (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plantarum. 126, 45–51. [Google Scholar]
- Takos, A.M. , Jaffé, F.W. , Jacob, S.R. , Bogs, J. , Robinson, S.P. and Walker, A.R. (2006) Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. , Keurentjes, J. , Bentsink, L. , Koornneef, M. and Smeekens, S. (2005) Sucrose‐specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, P. , Kumar, S. , Malik, J.A. , Berger, J.D. and Nayyar, H. (2010) Cold stress effects on reproductive development in grain crops: an overview. Environ. Exp. Bot. 67, 429–443. [Google Scholar]
- Thomashow, M.F. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Umemura, H. , Otagaki, S. , Wada, M. , Kondo, S. and Matsumoto, S. (2013) Expression and functional analysis of a novel MYB gene, MdMYB110a_JP, responsible for red flesh, not skin color in apple fruit. Planta, 238, 65–76. [DOI] [PubMed] [Google Scholar]
- Vannini, C. , Locatelli, F. , Bracale, M. , Magnani, E. , Marsoni, M. , Osnato, M. , Mattana, M. et al. (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 37, 115–127. [DOI] [PubMed] [Google Scholar]
- Vimolmangkang, S. , Han, Y. , Wei, G. and Korban, S.S. (2013) An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.K. , Cao, Z.H. and Hao, Y.J. (2014) Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol Plantarum. 150, 76–87. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Xu, H. , Jiang, S. , Zhang, Z. , Lu, N. , Qiu, H. and Chen, X. (2017) MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red‐fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Qu, C. , Jiang, S. , Chen, Z. , Xu, H. , Fang, H. , Su, M. et al. (2018) The proanthocyanidin‐specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low‐temperature conditions in red‐fleshed apples. Plant J. 96, 39–55. [DOI] [PubMed] [Google Scholar]
- Winkel‐Shirley, B. (2002) Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Wisniewski, M. , Nassuth, A. , Teuli eres, C. , Marque, C. , Rowland, J. , Cao, P.B. and Brown, A. (2014) Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33, 92–124. [Google Scholar]
- Wu, R. , Wang, Y. , Wu, T. , Xu, X. and Han, Z. (2017) MdMYB4, an R2R3‐Type MYB transcription factor, plays a crucial role in cold and salt stress in apple calli. J. Am. Soc. Hortic. Sci. 142, 209–216. [Google Scholar]
- Wu, R. , Wang, Y. , Wu, T. , Xu, X. and Han, Z. (2018) Functional characterisation of MdMYB44 as a negative regulator in the response to cold and salt stress in apple calli. J Hortic Sci Biotech. 93, 347–355. [Google Scholar]
- Xie, Y. , Chen, P. , Yan, Y. , Bao, C. , Li, X. , Wang, L. , Shen, X. et al. (2018) An atypical R2R3 MYB transcription factor increases cold hardiness by CBF‐dependent and CBF‐independent pathways in apple. New Phytol. 218, 201–218. [DOI] [PubMed] [Google Scholar]
- Xin, Z. and Browse, J. (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell Environ. 23, 893–902. [Google Scholar]
- Xu, W. , Dubos, C. and Lepiniec, L. (2015) Transcriptional control of flavonoid biosynthesis by MYB‐bHLH‐WDR complexes. Trends Plant Sci. 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Wang, N. , Liu, J. , Qu, C. , Wang, Y. , Jiang, S. and Chen, X. (2017) The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Wang, N. , Wang, Y. , Jiang, S. , Fang, H. , Zhang, J. , Su, M. et al. (2018a) Overexpression of the transcription factor MdbHLH33 increases cold tolerance of transgenic apple callus. Plant Cell Tiss Org. 134, 131–140. [Google Scholar]
- Xu, H. , Yang, G. , Zhang, J. , Wang, Y. , Zhang, T. , Wang, N. , Jiang, S. et al. (2018b) Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red‐fleshed callus. Biochem. Biophys. Res. Commun. 500, 405–410. [DOI] [PubMed] [Google Scholar]
- Yang, A. , Dai, X. and Zhang, W.H. (2012) A R2R3‐type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Yang, C. , Li, Y. , Zheng, N. , Chen, H. , Zhao, Q. , Gao, T. et al. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress‐responsive abscisic acid signaling in Arabidopsis . Plant Cell. 19, 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Cui, F. , Wu, Y. , Lou, L. , Liu, L. , Tian, M. , Ning, Y. et al. (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1‐INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis . Plant Cell. 27, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Wang, P. , Si, T. , Hsu, C.C. , Wang, L. , Zayed, O. , Yu, Z. et al. (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2001) Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Shi, H. , Lee, B.H. , Damsz, B. , Cheng, S. , Stirm, V. and Bressan, R.A. (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF‐independent pathway. Proc. Natl Acad. Sci. USA 101, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, I.M. , Heim, M.A. , Weisshaar, B. and Uhrig, J.F. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B‐like BHLH proteins. Plant J. 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequence analysis of MYB308L proteins.
Figure S2 MdMYB308L is a novel MYB protein in apple.
Figure S3 The promoter sequence of MdMYB308L.
Figure S4 Gene expression analysis of transgenic plant materials using qRT‐PCR.
Figure S5 The promoter sequence of MdCBF2.
Figure S6 The promoter sequence of MdDFR.
Figure S7 MdMYB308L does not affect MYB‐related genes expression and not bind to the MdCBF2 and MdDFR promoters.
Figure S8 The promoter sequence of MdMIEL1.
Table S1 Cold‐inducible MYB transcription factors identified from a cold stress transcriptome study using cDNA samples extracted from apple seedlings treated with or without cold stress (4°C) for 9 h.
Table S2 Screening the potential interacting proteins of MdMYB308L.
Table S3 Primers used for gene expression analysis and vector construction.


