To the Editor:
Clustered regularly interspaced short palindromic repeat/CRISPR‐associated Cas9 (CRISPR/Cas9) systems of bacteria and archaea have engineered for genome editing in eukaryotic genomes. In such CRISPR/Cas9 system, CRISPR RNA (crRNA) and trans‐activating CRISPR RNA (tracrRNA) were engineered into a simplified single guide RNA (sgRNA). Cas9 and sgRNA form a complex that scans through genome for the protospacer adjacent motif (PAM) sequence (predominantly 5′‐NGG‐3′) and for the sequence (ca. 18–20 nucleotides) complementary to the sgRNA, leading to double‐stranded DNA breaks (DSBs) that are exploited for site‐specific DNA alterations (Jinek et al., 2012).
Sorghum is the fifth most important cereal crop across the globe. Genome‐editing platform in sorghum was lagging behind other cereal crop plants due to its low stable transformation efficiency. Cas9/sgRNA activity was demonstrated for mutagenesis in sorghum callus cells derived from immature embryos using reporter gene encoding for red fluorescence protein (Jiang et al., 2013). One recent study has reported the application of CRISPR/Cas9 that targeted the centromere‐specific histone 3 (SbCENH3) gene, while the inheritance of the CRISPR‐induced mutations has yet to be demonstrated (Che et al., 2018). More recently, targeted mutagenesis of the k1C gene family encoding for α‐kafirins with a single guide RNA to target the conserved region of multiple k1C genes led to increased protein digestibility and lysine content in sorghum grain (Li et al., 2018a). Here, we report the development of a CRISPR/Cas9 system tailored for Agrobacterium‐mediated transfer and validation of its efficiency for targeted mutagenesis in two endogenous genes SbFT (Sb10G045100) and SbGA2ox5 (Sb09G230800). The CRISPR‐induced mutations were able to pass on to the T1 generation. The continuing activity of Cas9/sgRNA can lead to more site‐specific mutations in next generation as long as the Cas9/sgRNA transgene is still present in the sorghum plants. The SbFT mutant plants exhibit significant difference in flowering time.
We first adapted a schematic workflow and cloning strategy for sorghum CRISPR/Cas9 system (Figure 1a). To assemble the CRISPR/Cas9 system for sorghum mutagenesis through Agrobacterium‐mediated gene transfer, three vectors were designed and constructed. An intermediate vector (pgRNA1) was used for constructing guide RNA genes through sequentially inserting two spacer sequences each fused with the guide RNA scaffold. Another pENTR4‐derived intermediate vector (pCas9:GFP) contains a Cas9 gene directed by the maize ubiquitin 1 gene promoter and a cloning site (BssHII) for accepting the sgRNA cassette. The destination vector was adapted from a binary vector (pTF101.1), suitable for sorghum transformation (Paz et al., 2004), to accept the Cas9/sgRNA cassette through a Gateway recombination reaction, resulting in a single Agrobacterium‐borne Cas9/sgRNA construct. The pTF101.1‐derived sorghum CRISPR/Cas9 construct also has the bar gene that is driven by 2× CaMV 35S promoter for selection of glufosinate or bialaphos‐resistant callus cells and plants in transformation process. The genotype P898012 of sorghum was transformed by following a protocol as described previously (Do et al., 2016).
Figure 1.
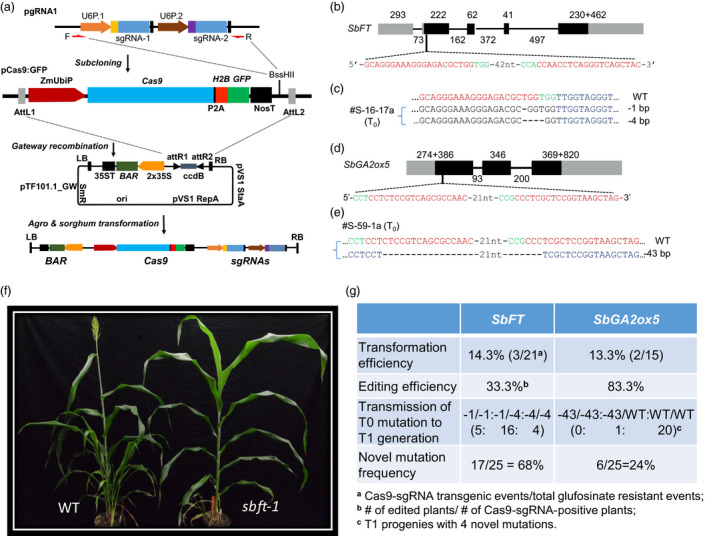
(a) Cloning strategy to construct sorghum CRISPR/Cas9. pgRNA1, a guide RNA cloning vector that contains Btg ZI and BsaI sites for sequential insertions of two double‐stranded oligos for spacer sequences of two sgRNAs. Two different rice U6 promoters are used to drive each of the sgRNAs. The sgRNA cassette was PCR‐amplified with primers F and R; the amplicon is cloned into pCas9:GFP vector at Bss HII site through Gibson assembly. Through Gateway recombination, the Cas9 and sgRNA expression cassettes are mobilized into plant expression vector pTF101.1_GW for Agrobacterium‐mediated sorghum transformation. (b) Gene structure of SbFT on chromosome 10 with sgRNA target sites (in red) and PAM sequences (in green). Forty‐two nucleotides (42nt) indicate sequence not shown between two target sites. Bars in black colour denote coding regions, and bars in grey colour represent untranslated regions. Numbers indicate the nucleotides in exons (above bars) and introns (below lines). (c) Biallelic mutants (1‐bp and 4‐bp deletion) at the first gRNA target site detected in a T0 plant #S‐16‐17a. The nucleotide changes were indicated (dashed lines for deletion and WT for wild type). Dots represent nucleotides not shown. (d) Gene structure of SbGA2ox5 on chromosome 9 with sgRNA target sequences (in red colour) and PAM sequences (in green). Number 21 indicates nucleotides between two target sites not shown. Bars (in solid black) denote coding regions and bars (in grey colour) represent the 5′ and 3′ UTR, respectively. Numbers of nucleotides are denoted as exons (above bars) and introns (below lines). (e) The mono‐allelic mutation (43‐bp deletion) occurring across both gRNA target sites detected in a T0 plant #S‐59‐1a. The nucleotide changes were indicated (dashed lines for deletion and WT for wild type). Dots represent nucleotides not shown. (f) Flowering time was affected by CRISPR‐induced mutation. Morphological characterization of wild type and mutants in the T1 generation. The frameshift mutation created by CRISPR/Cas9 shows the mutant plant (sbft‐1, n = 16, right side) flowered 10 days later on average compared with wild‐type plant (n = 34, left side). (g) Summary of transformation efficiency, CRISPR/Cas9 induced gene‐editing efficiency, transmission of T0 mutation to T1 generation and novel mutations happen in T1 progeny of SbFT and SbGA2ox5.
In this study, two candidates, based on literatures and bioinformatic analyses, as the genes underlying quantitative trait loci (QTLs) for flowering time and plant height, were selected for targeted mutagenesis. The target region of SbFT is located at the first exon, 13‐bp downstream of the translation start site (Figure 1b). Similarly, target region of SbGA2ox5 is also located in exon one, 329‐bp downstream of the translation start site (Figure 1d). For pCas9‐gSbFT, three out of twenty‐one transformation events (multiple plants/event) were transgene positive at a frequency of 14.3% (Figure 1g), while two out of fifteen events were transgene positive at a rate of 13.3% for pCas9‐gSbGA2ox5 (Figure 1g). The high number of non‐transgenic escapes might be due to the lower concentration of glufosinate applied during the in vitro selection processes of callus induction and plant regeneration.
Genotyping three pCas9‐gSbFT‐positive plants, each from a transformation event, identified one plant (S‐16‐17a) had a genotype of biallelic mutants, 1‐bp and 4‐bp deletions (‐1bp/‐4 bp), at the first target site (Figure 1c), whereas plants S‐66‐8b and S‐66‐9a both had the wild‐type allele of SbFT. Among the two transformation events positive for pCas9‐gSbGA2ox5, all five plants (S‐59‐1a, ‐1b, ‐1c, ‐1d and ‐1e) of S‐59‐1 possessed the identical mono‐allelic mutation of 43‐bp deletion (Figure 1e); no mutation was detected in S‐59‐6a. Our results showed that the site‐specific mutations at the SbFT and SbGA2ox5 loci in sorghum genotype P898012 were induced by the two CRISPR/Cas9 constructs.
Twenty‐five progeny plants from S‐16‐17a were used to analyse for the transmission of mutations. We observed a Mendelian segregation ratio of 1:2:1 for ‐1/‐1, ‐1/‐4 and ‐4/‐4 genotypes along the 25 T1 progeny plants (P = 0.36, df = 2, χ2 = 2.04, chi‐square test; Figure 1g). For SbGA2ox5, similar analysis indicated that the mono‐allelic 43‐bp deletion in S‐59‐1a was passed on to only one T1 progeny plant (Figure 1g). Additionally, new heterozygous mutations occurred in some progeny plants in S‐59‐1a (Figure 1g). It is worthwhile to notice that recent studies indicate that SbGA2ox5 is not the underlying gene for the corresponding plant height QTL, but unexpectedly, we failed to detect any homozygous 43‐bp deletion in the T1 progenies, suggesting a potential lethality of homozygous knockout of SbGA2ox5 (Hilley et al., 2016; Yamaguchi et al., 2016). Coincidently, no SbGA2ox5 homozygous knockout mutants were detected in the 25 T1 progeny plants from event S‐59‐6a described below. All the novel discovered mutations are mono‐allelic mutations including + 1/WT, ‐50/WT, ‐1/WT and ‐15/WT. Further verification of the relevance of the knockout genotype to the biological significance is needed.
To determine whether CRISPR/Cas9 expression continuously induced new mutations in T1 generation, twenty‐five plants derived from self‐pollinated S‐66‐9a (containing sgRNA for SbFT) and twenty‐five plants from event S‐59‐6a (containing sgRNA for SbGA2ox5) were genotyped. These two lines were chosen because they carried the CRISPR transgenes and also possessed the unedited target genes in T0 generation. The progeny plants were characterized for newly induced mutations at the CRISPR target sites. The presence of Cas9 and sgRNA in the gametes of sorghum plants during meiosis or in zygotes might result in additional mutations in the unedited alleles. Sanger sequencing analyses from the PCR amplicons of relevant regions showed that novel mutations were induced in both SbFT and SbGA2ox5 genes. Seventeen out of twenty‐five plants (68%) with novel mutations were detected in SbFT, and six out of twenty‐five (24%) with newly induced mutations were identified in SbGA2ox5 genomic target site (Figure 1g).
Expression profile of SbFT indicates that it is the most plausible florigen‐coding gene in sorghum and the gene underlies the flowering time QTL (Li et al., 2018b; Wolabu et al., 2016). The T1 plants from S‐16‐17a and S‐66‐9a events were grown in the greenhouse, and we planted them on two different dates. For the first planting date, plants containing knockout mutations flowered 8 days later than wild‐type plants on average (P = 0.020, df = 13, two‐sample t test). The flowering time difference for the second planting date between edited plants and wild types was 10 days (P < 0.001, df = 33, two‐sample t test) (Figure 1f). These frameshift mutations induced by CRISPR/Cas9 verified that Sobic.010G045100 is the gene underlying the detected flowering time QTL. The results also support our previous conclusion that the PIF/Harbinger insertion at 4 kb upstream in P898012 appeared to be the functional polymorphism disrupting the expression of SbFT (Li et al., 2018b).
To address the issue of potential off‐targets with gRNAs targeting SbFT and SbGA2ox5, we searched the potential sequence homology between on‐target sequences (both spacer and PAM) of gRNAs and the sorghum Tx623 reference genome. For the SbFT gRNAs, the closest match to the 21‐nt targeting site is only 15‐nt with PAM sequence missing, suggesting a less likely off‐target site in the transgenic sorghum plants. The similar results were found in the second target site. For SbGA2ox5, the first 19‐nt on‐target sequence matches one potential off‐target sites of 16 nt which misses the PAM sequence, an essential component for Cas9 recognition. The second 20‐nt target site of SbGA2ox5 is unique since there is only one 15‐nt hit also with PAM sequence missing. Therefore, the potential off‐target effects in the sorghum CRISPR/Cas9 system can be greatly reduced by carefully choosing and designing gRNAs for the target genes.
Authors’ contribution
SNC, XL, JY and BY designed the experiment; SNC, JW and QM performed the experiments; ZJZ supervised sorghum transformation; SNC, JW, QM, XL, ZJZ, JY and BY analysed the data and wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank the National Institute of Food and Agriculture of the US Department of Agriculture (2014‐67013‐21720 to B.Y.), the Iowa State University Presidential Initiative for Interdisciplinary Research (B.Y.) and the Iowa State University Plant Sciences Institute (J.Y.) for providing funding for this research. We thank Dr. Kan Wang for providing pTF101.1 binary vector in establishing sorghum CRISPR/Cas9 system. The authors thank the Iowa State University Crop Bioengineering Center for publication subvention. We also thank Dr. Hyeyoung Lee (Plant Transformation Core Facility at the University of Missouri) for generating transgenic sorghum events and Gregory Schoenbaum for taking care of the sorghum plants in the ISU Agronomy greenhouse.
References
- Che, P. , Anand, A. , Wu, E. , Sander, J.D. , Simon, M.K. , Zhu, W. , Sigmund, A.L. et al. (2018) Developing a flexible, high‐efficiency Agrobacterium‐mediated sorghum transformation system with broad application. Plant Biotechnol. J. 16, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, P.T. , Lee, H. , Mookkan, M. , Folk, W.R. and Zhang, Z.J. (2016) Rapid and efficient Agrobacterium‐mediated transformation of sorghum (Sorghum bicolor) employing standard binary vectors and bar gene as a selectable marker. Plant Cell Rep. 35, 2065–2076. [DOI] [PubMed] [Google Scholar]
- Hilley, J. , Truong, S. , Olson, S. , Morishige, D. and Mullet, J. (2016) Identification of Dw1, a regulator of sorghum stem internode length. PLoS ONE, 11, e0151271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Zhou, H. , Bi, H. , Fromm, M. , Yang, B. and Weeks, D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA‐mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. , Jia, S. , Yobi, A. , Ge, Z. , Sato, S.J. , Zhang, C. , Angelovici, R. et al. (2018a) Editing of an alpha‐kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Guo, T. , Mu, Q. and Yu, J. (2018b) Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proc. Natl Acad. Sci. USA, 115, 6679–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz, M. , Shou, H. , Guo, Z. , Zhang, Z. , Banerjee, A. and Wang, K. (2004) Assessment of conditions affecting Agrobacterium ‐mediated soybean transformation using the cotyledonary node explant. Euphytica, 136, 167–179. [Google Scholar]
- Wolabu, T.W. , Zhang, F. , Niu, L. , Kalve, S. , Bhatnagar‐Mathur, P. , Muszynski, M.G. and Tadege, M. (2016) Three FLOWERING LOCUS T‐like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol. 210, 946–959. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M. , Fujimoto, H. , Hirano, K. , Araki‐Nakamura, S. , Ohmae‐Shinohara, K. , Fujii, A. , Tsunashima, M. et al. (2016) Sorghum Dw1, an agronomically important gene for lodging resistance, encodes a novel protein involved in cell proliferation. Sci. Rep. 6, 28366. [DOI] [PMC free article] [PubMed] [Google Scholar]


