Summary
China is the origin and evolutionary centre of Oriental pears. Pyrus betuleafolia is a wild species native to China and distributed in the northern region, and it is widely used as rootstock. Here, we report the de novo assembly of the genome of P. betuleafolia‐Shanxi Duli using an integrated strategy that combines PacBio sequencing, BioNano mapping and chromosome conformation capture (Hi‐C) sequencing. The genome assembly size was 532.7 Mb, with a contig N50 of 1.57 Mb. A total of 59 552 protein‐coding genes and 247.4 Mb of repetitive sequences were annotated for this genome. The expansion genes in P. betuleafolia were significantly enriched in secondary metabolism, which may account for the organism's considerable environmental adaptability. An alignment analysis of orthologous genes showed that fruit size, sugar metabolism and transport, and photosynthetic efficiency were positively selected in Oriental pear during domestication. A total of 573 nucleotide‐binding site (NBS)‐type resistance gene analogues (RGAs) were identified in the P. betuleafolia genome, 150 of which are TIR‐NBS‐LRR (TNL)‐type genes, which represented the greatest number of TNL‐type genes among the published Rosaceae genomes and explained the strong disease resistance of this wild species. The study of flavour metabolism‐related genes showed that the anthocyanidin reductase (ANR) metabolic pathway affected the astringency of pear fruit and that sorbitol transporter (SOT) transmembrane transport may be the main factor affecting the accumulation of soluble organic matter. This high‐quality P. betuleafolia genome provides a valuable resource for the utilization of wild pear in fundamental pear studies and breeding.
Keywords: Pyrus betuleafolia, De novo assembly, PacBio SMRT , BioNano optical mapping, Hi‐C
Introduction
Pear (Pyrus L.) is a fruit grown worldwide and has a long cultivation history of 2500–3000 years. It is generally believed that the genus Pyrus originated in the west or mountainous regions of southwestern China during the Tertiary period (Rubtsov, 1944) and gradually evolved into two groups: Occidental and Oriental pear (Bailey, 1917). However, no reproductive isolation occurs in pear plants, which results in widespread interspecific hybridization. Therefore, many species, varieties and types that may have developed from 22 recognized primary species (Bell et al., 1996) have been named. China, as the origin and evolutionary centre for Oriental pear, has 13 native species with 5 primary wild species: P. betuleafolia, P. calleryana, P. pashia, P. ussuriensis and P. pyrifolia (Pu and Wang, 1963; Yu, 1979). Chinese white pear (P. bretschneideri), Chinese sand pear (P. pyrifolia), Sinkiang pear (P. sinkiangensis) and Ussurian pear (P. ussuriensis) are commercially cultivated in China.
High‐quality genomes are an important guarantee for the best utilization of genetic resources and the improvement of agronomic traits (Huang et al., 2010; Walker et al., 2014). The emergence and maturity of new sequencing technologies, such as real‐time single‐molecule sequencing (SMRT), 10X Genomics, optical mapping and Hi‐C sequencing, have made the sequencing of high‐quality genomes possible, and their combination for the assembly of genomes has shown good prospects (Bickhart et al., 2017; Jarvis et al., 2017; Zhang et al., 2017). The first Pyrus genome was sequenced by HiSeq Illumina sequencing: P. bretschneideri accession ‘Dangshansu’ (DSHS), the most important commercial Oriental pear cultivar in China (Wu et al., 2013). After that, an Occidental pear cultivar (P. communis ‘Bartlett’) was sequenced, which further enriched the functional genome information of Pyrus plants (Chagné et al., 2014).
Generally, in modern pear production, a pear plant includes two parts: the rootstock and the scion. Domesticated cultivated species are used as scions, and wild species with high stress tolerance are used as rootstocks. Although several pear genomes have been sequenced, and the genome of DSHS provided valuable genetic resources for pear study (Bai et al., 2017; Yin et al., 2015), all of the sequences came from cultivated pears. The lack of a genome from rootstocks limits basic biological research and breeding in pear. Moreover, comparing the genomes of wild species and cultivars provided us with an unparalleled system for functional and evolutionary studies in plants (Chen et al., 2013; Wang et al., 2014).
Pyrus betuleafolia is a wild species native to China, and it is widely distributed in the northern region and bears small fruit with a diameter of ~1 cm and two carpels (Figure 1A). The plants of this species show characteristics of a well‐developed root system, vigorous growth, tolerance to several abiotic and biotic stresses and excellent affinity for Occidental and Oriental pear cultivars (Pu and Wang, 1963). With these peculiarities, P. betuleafolia is widely used as a rootstock in northern China. Pyrus betuleafolia was believed to be an ancestral species linked to Oriental and Occidental pears (Iketani et al., 1998; Kikuchi, 1948). In a phylogenetic study of Pyrus based on chloroplast and nuclear DNA sequences, P. betuleafolia was considered one of four primitive gene pools of Oriental pear, and it was identified as monophyletic (Jiang et al., 2016; Zheng et al., 2014). Our previous study also suggested that P. betuleafolia has the ancient chloroplast haplotype, which is from the centre of pear divergence in Northern China (Chang et al., 2017). Despite its important role in genetic evolution, nurturing seedlings and functional gene mining, P. betuleafolia is still underutilized and its further research is lacking. Thus, a finished, accurate reference genome of P. betuleafolia will provide a platform for elucidating the genomic evolution of Pyrus and mining functional genes for agronomic traits.
Figure 1.

Pyrus betuleafolia‐Shanxi Duli de novo genome assembly. (A) P. betuleafolia‐Shanxi Duli used in this study. (B) Summary of the de novo genome assembly and sequencing analysis of P. betuleafolia‐Shanxi Duli. A, Chromosome number; B, heat map view of genes; C, NBS‐type resistance gene analogues (RGAs); D, repeat density in 200‐kb windows (red, average +1 SD; blue, average −1 SD; yellow, gene and repeat density between red and blue); and E paralogous relationships between P. betuleafolia chromosomes.
Here, we report the sequencing and assembly of P. betuleafolia‐Shanxi Duli (Pbe‐SD) from Qinyuan, Shanxi Province, one of the centres of pear divergence. Furthermore, we conducted gene family evolution analysis and genome‐wide comparative analysis to characterize the functional and structural features of the P. betuleafolia genome. Disease resistance genes were predicted at a genome‐wide scale and compared with those of Rosaceae plants. The genetic differences related to the different fruit flavours between Pbe‐SD and DSHS were compared, and the factors underlying these differences were investigated. This genome will facilitate pear genomic research and the utilization of wild pear resources.
Results
Sequencing and assembly
We sequenced and assembled the genome of Pbe‐SD using a combination of short‐read sequencing from Illumina HiSeq, SMRT from Pacific Biosciences (PacBio, Menlo Park, CA), optical mapping from BioNano Genomics Irys and Hi‐C sequencing (Simão et al., 2015; Figure S1). Based on a 19‐mer analysis, we evaluated the genome size to be 511 Mb, with a heterozygosity of 1.54% (Figure S2). A total of 95.9× coverage of SMRT sequences (52.7 Gb) were used for initial contig assembly (Table S1), and we used the HGAP pipeline to assemble the SMRT sequences, which resulted in 684 Mb of sequences, with a contig N50 size of 572 kb. We also assembled the sequences with Falcon and Canu pipelines, resulting in assembly sizes of 791 and 772 Mb, and contig N50 sizes of 188 and 272 kb, respectively (Table S2). Therefore, the HGAP assembly was eventually used as a reference due to its largest contig N50. We generated 54.6‐fold coverage of Illumina paired‐end (PE) reads (30.0 Gb), with insert sizes of 450 bp (Table S3), which were used for SMRT sequencing correction. Two BioNano genome maps were constructed by using BssSI and BspQI enzymes (Table S4), which were used for hybrid assembly of SMRT sequence genomes. Subsequently, the contigs were scaffolded using data from the two optical maps, and during this step, 376 and 375 contigs containing conflicting connections were identified and broken to resolve conflicts. Gap filling was performed with the Canu‐assembled sequence data, resulting in a contig N50 value of 1.5 Mb and initial scaffold N50 values of 5.2 Mb. Next, when the Hi‐C data for scaffold extension and chromosome mount were used, the final assembly contained 139 scaffolds, with contig N50 values of 1.57 Mb and a scaffold N50 of 28.1 Mb (Table 1 and Figure S3). The total assembly size is 532.7 Mb, and 500 Mb (94%) of the scaffold sequences are anchored onto 17 chromosomes, with maximum and minimum lengths of 45.5 Mb and 18.4 Mb, respectively. Our assembly captured 18 long stretches of telomeric sequences at both ends of five chromosomes and at a single end of eight chromosomes (Table S5). Attesting to the accuracy and completeness of the assembly, full‐length transcripts from four pooled tissues were mapped against our assembly and 95.9% of them were successfully mapped. The continuity and integrity of the assembly for Pbe‐SD is significantly better than those of the published pear genomes (Chagné et al., 2014; Wu et al., 2013). We also used Benchmarking Universal Single‐Copy Orthologs (BUSCO; Daccord et al., 2017) to assess the completeness of gene regions, and the results showed that 94.8% of the plant single‐copy orthologues were complete. Single‐copy and multi‐copy genes in the complete genes accounted for 62.6% and 32.2%, respectively, which is close to the sequenced pear and apple genomes (Table S6). Therefore, these results indicated that our genome assembly is of high quality and has high coverage.
Table 1.
Comparison of the Pbe‐SD genome with previously published assemblies of the pear genome
| Pbe‐SD | DSHS | Bartlett | |
|---|---|---|---|
| Total assembly size (Mb) | 532.7 | 512.0 | 577.3 |
| Contig number | 595 | 25 312 | 182 196 |
| Contig N50(kb) | 1,571.5 | 35.7 | 6.6 |
| Contig length (Mb) | 497.0 | 501.3 | 507.7 |
| Scaffold number | 139 | 2103 | 142 083 |
| Scaffold N50 (kb) | 28 122.4 | 540.8 | 88.1 |
| % Sequence anchored on chromosome | 94 | 75.5 | 29.7 |
| Gene number | 59 552 | 42 812 | 43 419 |
Genome annotation
We analysed repetitive sequences by combining de novo prediction and homology‐based search at both the DNA and protein levels, and revealed that repetitive sequences occupy 46.43% (247.3 Mb) of Pbe‐SD genome. Long‐terminal repeat (LTR) retrotransposons accounted for 33.1% of the genome as the most common type of transposable elements. The most abundant LTR retrotransposons are the gypsy elements (19.9%), followed by copia elements (11.6%). As another major class of transposable elements, DNA transposons account for 8.3% of the genome, with PIF‐Harbinger, hAT‐Ac, Helitron and MULE‐MuDR being the more abundant types (Table S7). Compared with those of most other species, the LTRs of Pbe‐SD showed relatively recent insertion during the gene expansion period, and most of the LTR retrotransposons were inserted into the genome within the last 0.5 million years (Figure S4). The centromeres of the chromosome consisted of highly repetitive DNA sequences, and the Hi‐C technology allowed us to identify these regions. To evaluate the assembly of these regions, we identified a high proportion of repeat sequences of blocks (repeat sequence ratio between 74.1% and 99.05%) on each chromosome, with a repeating ratio of 5 blocks at greater than 90%. These high repeat regions are assumed to be centromere regions of the chromosome, and the main repeat type is LTR/Gypsy (Table S8). Based on the same method, the transposable elements of the DSHS genome were re‐annotated (262.0 Mb), resulting in a slightly smaller size than the previous evaluation. We compared the genome size and sequence composition of the DSHS and Pbe‐SD pear species, and the results showed that the transposable element (TE) and form distributions of the two pear species as well as the insertion times and intensities were similar (Table S7 and Figure S5). Given that TEs are the main drivers of genome amplification, these results indicate that these genomes evolved with similar evolutionary rates after their split from a common ancestor (Yin et al., 2015).
We performed an ab initio, homology‐based search and RNA‐Seq to predict gene models from the repeat‐masked Pbe‐SD genome sequence (Figure S6). A total of 59 552 protein‐coding genes were predicted (representing 34.12% of the genome assembly), with an average transcript length of 1608 bp, an average coding sequence size of 1346 bp and a mean number of exons per transcript of 5.2 (Table S9). The number of annotated genes is more in this genome than in the two pear genomes that have been sequenced (Table 1). The gene density throughout the genome is approximately 11.2 genes per 100 kb, with 56 553 genes (94.96%) present on chromosomally anchored contigs (Figure 1B). In the GO analysis, 23 678 (39.76%), 19 747(33.16%) and 25 655 (43.08%) annotated genes were assigned to the GO slim terms biological process, cellular component and molecular function, respectively (Figure S7). Then, the annotated genes were subjected to length filtration (filtering out genes <300 bp or larger than 20 kb) and homologous alignment with annotated genes in the DSHS genome and genes in the nonredundant protein database, and a total of 42 520 ‘high‐confidence’ genes were identified.
Evolution and gene family expansion analysis
We performed orthologous clustering on three sequenced pear genomes. In the Pbe‐SD genome, 22 658 gene families were identified, which was far more than that in the DSHS and Bartlett genomes. In addition, 15 055 of those gene families were common to all tree pear genomes, whereas 1536 gene families containing 8033 genes were specific to the Pbe‐SD genome, which is more than the number found in the other two genomes (Figure 2A). This difference is consistent with the large gene number in the Pbe‐SD genome. An analysis of GO terms for these lineage‐specific families revealed that several biological processes, such as DNA metabolic process, DNA integration, DNA recombination and cellulose microfibril organization, are enriched in the Pbe‐SD genome (Table S10). A phylogenetic tree was constructed based on a sequence alignment of the 1310 single‐copy gene families shared by six Rosaceae plants (Chagné et al., 2014; Daccord et al., 2017; Shirasawa et al., 2017; Verde et al., 2013; Wu et al., 2013) and tomato (Figure 2B). It is estimated that Oriental and Occidental pear diverged between 17.4 and 29.7 million years ago (MYA). In addition, the cultivated and wild species of Oriental pear diverged between 8.4 and 26.5 MYA. This result provides direct proof of independent domestication processes for both Oriental and Occidental pears (Wu et al., 2018). We analysed gene family expansion and contraction in the P. betuleafolia lineage. Among all 17 477 gene families of the seven species, 2831 gene families were expanded and 977 gene families were contracted (Figure 2B), after speciation from P. bretschneideri. Compared with those in the other two pear genomes, the number of contracted gene families was the lowest in the P. betuleafolia genome. Functional annotation of the expanded genes demonstrates that they are significantly enriched in functional categories involved in flavonoid metabolic processes including two subcategories for molecular function (‘quercetin 3‐O‐glucosyltransferase activity’ and ‘quercetin 7‐O‐glucosyltransferase activity’) and six subcategories for biological processes (‘uronic acid metabolic process’, ‘glucuronate metabolic process’, ‘cellular glucuronidation’, ‘flavonoid glucuronidation’, ‘flavonoid biosynthetic process’ and ‘flavonoid metabolic process’) (Table S11). Significant gene expansion corresponds to two classes of UDP‐glycosyltransferases (UGT71K2 and UGT87A1), including 22 genes, which show its important influence on the evolution of functional categories in the Pbe‐SD genome. Phylogenetic analysis of UGT71K2 and UGT87A1 from six sequenced Rosaceae genomes revealed that three Pyrus plants tend to cluster together in each subclade (Figure S8). The presence of Pyrus‐specific subclades of UGT71K2 and UGT87A1 indicates lineage‐specific expansion of the UGT family. Gene clustering on the chromosome of Pbe‐SD suggested that these genes evolved mainly through gene duplications. UGTs can glucosylate a diverse array of aglycones, including plant hormones and secondary metabolites, which are involved in stress and defence responses (Cui et al., 2016). UGT79B2 and UGT79B3 contribute to cold, salt and drought stress tolerance by modulating anthocyanin accumulation in Arabidopsis (Li et al., 2017b). UGT83A1 and UGT74E1 have an important regulatory role in pear plants under drought stress (Wang et al., 2018). It is speculated that the expansion of this type of gene affects the accumulation of secondary metabolites and the adaptability of P. betuleafolia.
Figure 2.

Gene family evolution analysis. (A) Venn diagram showing the shared and unique gene families among three pear species. Each number in parentheses represents the number of genes within corresponding families (without parentheses). (B) Expansion and contraction of gene families in six Rosaceae species and tomato. A phylogenetic tree was constructed based on all single‐copy orthologous genes using tomato (Solanum lycopersicum) as the outgroup. Pie diagrams on each branch of the tree represent the proportion of genes undergoing gain (red) or loss (green) events. The numerical value beside each node shows the estimated divergent time.
Variation analysis of the Pyrus betuleafolia genome
Apple and pear belong to the subfamily Maloideae. The nonrepetitive sequences of these two genomes are similar in size, but the apple genome contains a higher proportion of repeat sequences than the pear, which resulted in a larger genome size in apple. A comparison of genomic structure revealed high collinearity between Pbe‐SD and M. domestica (GDDH13), which indicated that the chromosome structure of the two genomes was relatively stable (Figure 3A). This result also proved that there was no large structural variation such as chromosomal rearrangement and fusion, since the differentiation of apple and pear from 22.4 to 39.4 MYA, and the two genomes shared the whole‐genome duplication (WGD) that occurred at ~50 MYA (Figure S9). Previous studies have reported that there is high collinearity between apple and pear genomes (Celton et al., 2009; Pierantoni et al., 2004; Yamamoto et al., 2004), so we further identified the degree and nature for genome organization changes between them. Using a whole‐genome alignment approach, we found 1243 collinear blocks covering 44.77% and 33.92% of the Pbe‐SD and GDDH13 genomes, respectively, and 22 405 and 24 103 genes conserved with gene collinearity, respectively. Therefore, although the collinearity between the two genomes is very high, the conservation of gene organization was destroyed, even in closely related species, due to unequal gene‐duplication events in pedigree‐specific parallel evolution.
Figure 3.

Comparative analysis and evolution events in the Pbe‐ SD genome. (A) Syntenic blocks shared between the Pbe‐ SD and GDDH13 genomes. Grey lines connect matched gene pairs, with one set highlighted in red. (B) Ks distribution for paralogous and orthologous genes in comparisons of the Pbe‐ SD, DSHS and Bartlett genomes and Ka/Ks distribution in comparisons of the Pbe‐ SD and DSHS genomes.
We also performed synteny analyses for the Pbe‐SD genome against DSHS genome, which did not show better collinearity than that of apple (Figure S10). This unexpected result is due to the low quality of the second‐generation sequencing assembly of the DSHS genome. The presence‐absence variation (PAV) analysis identified 7992 presence variations (PVs, only fragments >1000 bp were counted) in Pbe‐SD and 6422 PVs in DSHS, which accounted for 18.77 Mb and 18.88 Mb, respectively. An enrichment analysis showed that DNA integration, nucleic acid binding transcription factor activity and transcription factor activity were found to be significantly enriched in Pbe‐SD‐PV genes (Table S12). We found that certain presence variations are located in the promoter region of genes and may play important biological functions. For instance, a LTR/Gypsy was inserted into the promoter region of WRKY 15 (Chr1.g02239) in the Pbe‐SD genome, and it plays an important role in the process of adversity stress. Another SV was inserted into the promoter region of barely any meristem 1 (BAM 1, Chr12.g42983) in the Pbe‐SD genome, which may affect the morphogenesis and cell differentiation of plant meristems. Then, to further identify the variations between these two pear genomes, we compared the scaffold sequence of DSHS to the Pbe‐SD genome and identified 968 943 variants, of which 850 287 were SNPs and 118 656 were indels (Table S13 and Figure S11). As expected, most of the identified variation is located outside the genes or within introns, with few locations in coding sequences (CDSs). The frequency of variants within the exon, intron and intergenic regions was 3.506%, 8.488% and 25.04%, respectively (Table S14). Among all variants in coding regions, 42.77% were synonymous and 57.23% were nonsynonymous (Table S15). The low frequency of variation in CDSs was attributed to the conservation of protein functions.
To analyse the selective evolution of Pyrus genes, we calculated the substitution rate (Ks) values between orthologous genes for each pair between any two genomes of Pbe‐SD, DSHS and Bartlett. We found two peaks in the Ks distribution: the first one corresponded to a group of genes that might derive from recent genetic exchanges (Ks < 0.02085), and the other one represents genes diverged from the common ancestors of Pyrus (Ks < 0.175) (Figure 3B). We next compared orthologous gene pairs from Pbe‐SD and DSHS to find genes with evidence for selection in Oriental pear. The Ka/Ks ratios for most orthologous genes are close to zero because most nonsynonymous mutations are deleterious and experienced strong purifying selection (Sun et al., 2018). Approximately 286 genes were strictly positively selected, and 116 of them had functional annotations in the evolution of Oriental pears (Table S16). In the positive gene list, we found that those genes are mainly involved in three traits. (i) Fruit size, including one EXP gene (expansin‐a11‐like) and two cyclin‐like genes (cyclin‐u4‐1‐like and cyclin‐b1‐2‐like). Fruit size was selected during the domestication of edible fruit plants, and different genome regions were selected for this trait in Occidental and Oriental pears (Wu et al., 2018). (ii) Sugar metabolism and transport, including one SDH‐like (sorbitol dehydrogenase‐like), and two SWEET genes (bidirectional sugar transporter sweet 10‐like and bidirectional sugar transporter sweet17‐like). Sorbitol dehydrogenase (SDH) can catalyse the irreversible oxidation of sorbitol to fructose with NAD+ as a coenzyme (Park et al., 2002), and the SWEET gene family may be involved in the transport and distribution of soluble sugar in the fruit (Chen, 2014). The positive selection of these two types of genes may affect the accumulation of sugar in the fruit during evolution, which in turn increases the sweet flavour of pear fruit. (iii) Photosynthetic efficiency, including one CAB (chloroplastic chlorophyll ab‐binding) protein, one cytochrome c oxidase (cytochrome c oxidase subunit 6b‐2) and two NADH dehydrogenases (NADH dehydrogenase subunit 5 and NADH dehydrogenase). Photosynthetic phosphorylation and oxidative phosphorylation are two important physiological processes in plants that help release energy for plant growth and development, which was easily selected for during evolution (Zhang et al., 2016). These results suggested that the function of the genes involved in photosynthesis and energy production underwent positive selection during the evolution of P. betuleafolia for adapting to environmental changes.
Identification of disease resistance‐related gene families
With a pipeline for the genome‐wide prediction of RGAs (Li et al., 2016a), we detected 2129 RGAs in the Pbe‐SD genome sequence (Table S17). We identified 573 NBS‐type RGAs, with 459 genes residing on 17 chromosomes and 114 genes residing on unplaced scaffolds (Table S18). Among these, 130 CC‐NBS‐LRR (CNL)‐type and 150 TNL‐type genes were further identified in Pbe‐SD predicted protein sequences. NBS‐type RGAs tended to cluster near the ends of the chromosome, showing a preference for the distal end of the chromosomes (Figure 4A and Figure S12). The genes clustered on chromosomes 2, 5, 7, 10 and 11 accounting for more than half of the identified NBS‐encoding genes are mainly tandem repeats of CNL‐type and TNL‐type genes. For instance, one‐third of the identified CNL‐type genes are on chromosome 11, and over half of TNL‐type genes are on chromosomes 2, 5 and 10.
Figure 4.

Distribution of RGAs in chromosomes. (A) Distribution of RGAs along the Pbe‐ SD chromosomes. The bottom graph shows the absolute number of genes homologous to nucleotide‐binding site–leucine‐rich repeat (NBS‐LRR‐encoding) proteins, receptor‐like protein kinases (RLKs), receptor‐like proteins (RLPs) and transmembrane coiled‐coil (TMCC) proteins along each of the 17 chromosomes. The top four graphs show the ratio of the number of genes in each RGA class to the total number of genes in a sliding window of 10 Mb wide. (B) Collinearity comparison of TNL‐type genes on chromosome 2 of the GDDH13 and Pbe‐ SD genomes.
The RGAs are significantly more similar in the GDDH13 and Pbe‐SD genomes compared with that of the other Rosaceae species (Table S18). In addition, there are markedly more TNL‐type genes in the Pbe‐SD genome than in other Rosaceae genomes. Previous studies have shown that apple and pear resistance genes have high functional synteny (Bouvier et al., 2012). We performed a chromosome‐level collinearity comparison of the RGA distribution between the GDDH13 and Pbe‐SD genomes (Figure S13), and the results showed that 1048 and 1065 genes were conserved in the collinear region of those two genomes, accounting for 56.65% and 52.83% of the total RGAs, respectively. The Rvi15 gene provides full resistance to apple scab; however, the breaking of this resistance has not yet been reported, and this resistance locus is mapped at the top of chromosome 2. Currently, three TNL genes have been cloned from this region, and one of the candidate genes appeared to confer full resistance to apple scab; it is the only candidate gene available (Galli et al., 2010; Schouten et al., 2014). We further analysed the collinearity of the TNL gene in chromosome 2 between the GDDH13 and Pbe‐SD genomes, and RVi15 (Vr2) has a collinear site at the upper position of chromosome 2 in Pbe‐SD genome. Therefore, this gene may be an ancient disease resistance gene in the pome fruit tree. On chromosome 2 of GDDH13 and Pbe‐SD genome, there are 16 and 25 TNL genes, respectively, and 5 pairs of them were collinear (Figure 4B). The TNL gene plays an important role in the disease resistance of Rosaceae plants, and the Pbe‐SD genome has the most TNL genes of Rosaceae genomes, which may be the reason why this wild resource has strong resistance to disease erosion.
A total of 1043 putative pattern‐recognition receptor genes, which encode receptor‐like kinases with an LRR domain (RLK‐LRR), were identified in the Pbe‐SD genome (Table S18). This number is similar to that found in apple (1001), slightly larger than the number in the other two pear species and significantly larger than the number in strawberry, cherry and peach, which have not experienced recent WGD events. Therefore, it is suggested that pattern‐triggered immunity (PTI), a type of ancient innate immunity, is conserved in pome fruit trees and may have an important role in defence against potential pathogens. As with the NBS‐LRR‐encoding genes, LRR‐RLK genes were nonrandomly distributed among the 17 Pbe‐SD chromosomes, with chromosomes 5 (130), 10 (89) and 15 (95) possessing the enriched LRR‐RLK‐encoding gene clusters.
Identification of fruit flavour related gene families
Compared with the fruits of cultivated varieties of Oriental pear, the fruit of P. betuleafolia was sour and astringent. To investigate the bases behind this difference, we analysed genes related to flavour compound biosynthesis. Transcriptome analysis was conducted to compare differentially expressed genes (DEGs) in fruit at maturity between Pbe‐SD and DSHS, and an orthologue search was performed in those two genomes to identify their gene families.
Compared with DSHS fruit, Pbe‐SD fruit has more total soluble solids (35.02% vs. 10.91%) and titratable acid (3.25% vs. 0.051%) and a much lower TSS‐acid ratio (10.78 vs. 213.92), thus generating a very large difference in sweetness/acidity taste between them. The gene copy number of 16 gene families in sugar and acid metabolism processes was compared between the two genomes, and more such genes were present in the DSHS genome (138 vs. 152) (Table S19). Members of these gene families exhibit diverse expression patterns in sugar and acid metabolism, indicating the complexity of regulation in this pathway (Figure 5A). However, we paid particular attention to the SOT gene family among the DEGs, and SOT expression was significantly higher in Pbe‐SD than in DSHS. Sorbitol is the main form of transport for photosynthetic products in Rosaceae, with sorbitol accounting for approximately 70% of the photosynthates produced in the leaves (Wrangham et al., 1998). They are transported from the phloem and taken up into the cytosol of parenchyma cells by a SOT located on the plasma membrane (Park et al., 2002). Then, almost all the sorbitol is converted to fructose and participates in the carbon flux of the fruits. This process may be the main cause of the formation of high amounts of total soluble solids in mature Pbe‐SD fruits. The synthesized fructose is decomposed into malic acid and citric acid through the tricarboxylic acid cycle, which were the main soluble acids in pear fruit (Tanner et al., 2003). The malate dehydrogenase (MDH) gene in this cycle was expressed at a higher level in Pbe‐SD than in DSHS, and the isocitrate dehydrogenase (IDH) gene was expressed at a lower level in Pbe‐SD, and these genes were involved in the synthesis and accumulation of soluble acids. At the same time, the high expression level of neutral invertase (NINV) in Pbe‐SD is beneficial to the accumulation of glucose and fructose for organic acid synthesis, while the lower expressed sucrose synthase (SUSY) and vacuolar acid invertase (VAINV) genes are conducive to the accumulation of sucrose (Figure 5B and Table S20). Based on the results, we speculated that the transmembrane transport capacity of sorbitol carriers may be one of the important factors determining the soluble organic matter content of Pbe‐SD. The regulation of sugar acid metabolism affected the ratio of sugar to acid in the fruit, thus affecting the sweetness/acidity taste of the pear fruits. Because sugar acid metabolism is a process of developmental regulation, future clarification of the temporal and spatial expression of gene family members will help determine the underlying mechanism.
Figure 5.

Differential expression of genes involved in sugar/acid metabolism in fruits from DSHS and Pbe‐SD. (A) Sugar/acid synthesis pathway and related structural genes in pear fruit. SUSY: sucrose synthase; SPS: sucrose phosphate synthase; SPP: sucrose‐phosphatase; HK: hexokinase; FK: fructokinase; PK: pyruvate kinase; MDH: malate dehydrogenase; ACO: aconitate hydratase; IDH: isocitrate dehydrogenase; CS: citrate synthesis; NINV: neutral invertase; CWINV: cell wall invertase; vAINV: vacuolar acid invertase; PFK: 6‐phosphofructokinase; SOT: sorbitol transporter; and HT: hexose transporter. (B) Differentially expressed genes in the proanthocyanidin metabolism pathway. Red colour represents higher than log10 (FPKM) data of genes; green colour represents lower than log10 (FPKM) data of genes; and black colour represents log10 (FPKM) = 0. S1, S2 and S3 indicate the three biological replications of Pbe‐SD. D1, D2 and D3 indicate the three biological replications of DSHS.
Proanthocyanidins, also known as condensed tannins, affect the fruit astringency (Xie et al., 2004). The synthesis of anthocyanins and proanthocyanidins occurs through the same metabolic pathway from phenylalanine to leucocyanidin. Then, the synthesis of proanthocyanidins from leucocyanidins follows two specific pathways, whose reactions are catalysed by two enzymes, leucoanthocyanidin reductase (LAR) and ANR (Liao et al., 2015). LAR catalyses the conversion of leucocyanidins into catechin, while ANR catalyses the synthesis of epicatechin from anthocyanins. These two types of flavan‐3‐ols are then condensed into proanthocyanidins (Figure 6A). We searched for genes related to proanthocyanidin synthesis in the Pbe‐SD and DSHS genomes. Seven of the 15 gene families had greater copy numbers in Pbe‐SD than in DSHS, and only two genes showed lower copy numbers in Pbe‐SD (Table S21). In terms of gene expression, all DEGs in the anthocyanin synthesis pathway were preferentially expressed in the Pbe‐SD relative to DSHS, which promotes the synthesis of leucocyanidins and anthocyanins (Figure 6B and Table S22). This result is consistent with the severe astringency in Pbe‐SD. ANR and LAR were believed to be a key enzymes for the synthesis of flavan‐3‐ol monomeric units, which further condense into proanthocyanidins. However, the high expression of the LAR gene in transgenic tobacco does not lead to the production of catechin and proanthocyanidins (Chen et al., 2007; Yamaki and Ino, 1992). Therefore, the genetic evidence for the biosynthesis of proanthocyanidins by plants through the LAR pathway remained to be supplemented. Our results also showed that only the ANR expression level was significantly increased in Pbe‐SD compared to DSHS. It was suggested that the ANR pathway rather than the LAR pathway was mainly responsible for proanthocyanidins in Pyrus.
Figure 6.
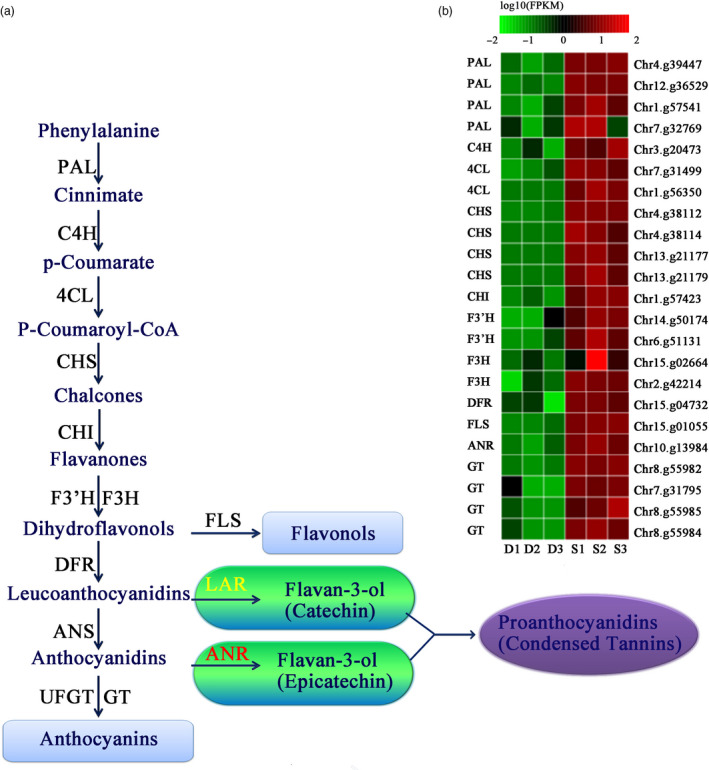
Differential expression of genes involved in proanthocyanidin metabolism in fruits from DSHS and Pbe‐ SD. (A) Proanthocyanidin synthesis pathway and related structural genes in pear fruit. PAL, phenylalanine ammonia lyase; C4H, cinnamate‐4‐hydroxymate; 4CL, 4‐coumarate:coenzyme A ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3‐hydroxylase, F3'H, flavanone 3’‐hydroxylase; DFR, dihydroflavonol 4‐reductase; ANS, anthocyanidin synthase; UFGT, UDP‐glucose flavonoid 3‐O‐glucosyl transferase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; ANR, anthocyanidin reductase; GT, glycosyltransferases; and FLS, flavonol synthase. (B) Differentially expressed genes in the proanthocyanidin metabolism pathway. Red colour represents higher than log10 (FPKM) data of genes; green colour represents lower than log10 (FPKM) data of genes; and black colour represents log10 (FPKM) = 0. S1, S2 and S3 indicate the three biological replications of Pbe‐ SD. D1, D2 and D3 indicate the three biological replications of DSHS.
Discussion
Here, we present a high‐quality genome sequence for Oriental wild species P. betuleafolia. This genome provides important information to understand the evolution of Occidental and Oriental pears and the independent domestication in Oriental pears. This genome sequence also lays the foundation for revealing the genetic basis of quality traits, stress‐resistant traits and disease‐resistant traits of pear.
The UGT gene family is significantly expanded in the Pbe‐SD genome and related to flavonoid metabolic processes that regulate secondary metabolite synthesis. The abundant accumulation of metabolic constituents has apparently played a significant role in supporting environmental adaptations of plants (Xia et al., 2017). Previous research suggests that the UGT gene family plays a regulatory role in plant drought tolerance. Studies on Arabidopsis have shown that the UGT gene modulates stomatal behaviours with ABA to enhance plant adaptation to drought stress (Li et al., 2015). The evidence supports the expansion of UGTs involved in secondary metabolite synthesis and stress responses that may affect the wide environmental adaptability of P. betuleafolia. A phylogenetic tree based on single copies of conserved genes was constructed. It was estimated that P. betuleafolia and P. bretschneideri diverged between 8.4 and 26.5 MYA, later than Asian and European pears diverged, which provided direct evidence for the independent evolution of Oriental pears (Wu et al., 2018). We further detected the selective evolution of genes between P. betuleafolia and P. bretschneideri to provide new insights into genetic events that may have occurred in the domestication of Oriental pear. In the resulting gene list, we found an enrichment of genes involved in fruit size, sugar metabolism and transport, photophosphorylation and oxidative phosphorylation, which is compatible with the traits that evolved in Oriental pear.
We demonstrated the value of the reference genome by identifying genes involved in resistance gene collinearity and flavour compound synthesis. Pear scab is a major disease in commercial orchards worldwide that often causes significant losses to production. Its associated pathogens are Venturia nashicola (Tanaka and Yamamoto, 1964) and Venturia pirina Aderh (Langford, 1942), which specifically infect Oriental pears and Occidental pears, respectively. Both fungal species are classified in the same genus as Venturia inaequalis, the causal agent of apple scab. Numerous studies have proven the existence of potential orthologous scab resistance genes in the highly collinear apple and pear genomes (Bus et al., 2010; Cho et al., 2009; Pierantoni et al., 2007). In apple, chromosome 2 appears to be deeply involved in resistance to scab and also is a highly conserved position of major resistance genes in apple and pear genomes (Bouvier et al., 2012). The resistance gene Rvp1, providing resistance to V. pirina, was mapped close to the distal end of chromosome 2 in the pear genome. In apple, this region contains a major scab resistance gene cluster, which confirms the existence of functional synteny between pear and apple. Rvi15 is a full‐resistance gene located at the proximal end of chromosome 2 (Galli et al., 2010), and we currently identified its homologous gene in the same region of chromosome 2 in the pear genome. Because of the close phylogenetic relationship between apple and pear and between corresponding Venturia species, gene mining based on homologous genes is an effective approach. In addition, we detected the TNL gene that was most abundant in the Pbe‐SD pear genome compared with those in other Rosaceae genomes. In view of the effective functioning of the TNL gene, it may contribute as a wild resource to the high disease resistance of P. betuleafolia.
Astringency is one of the most important components of fruit taste quality. Astringency mainly comes from condensed tannins (proanthocyanidins) and causes the drying, roughening and puckering of the mouth epithelia attributed to an interaction between tannins and salivary proteins. The gradual disappearance of astringency from wild species to cultivars is also a part of domestication. We interpret the genetic background of this domestication process in pear. Previous research has reported that both the LAR and ANR pathways are assigned to proanthocyanidin biosynthesis in plants (Tanner et al., 2003). Our finding suggests that the ANR pathway is the main contributor, which is also in accordance with reports on Arabidopsis thaliana and apple (Liao et al., 2015; Xie et al., 2004). In addition to the genes in the metabolic pathway, GST, MATE and ATPase transporters are involved in the intracellular transport of proanthocyanidin monomers, and transcription factors such as WIP‐ZF, MYB, bHLH, WRKY and MADS are involved in the regulation of proanthocyanidin synthesis and accumulation (Gonzalez et al., 2008; Pourcel et al., 2005; Sharma and Dixon, 2005; Yamazaki et al., 2003). Therefore, the interaction mechanism between them still requires in‐depth research and exploration.
Pyrus betuleafolia is widely used as a rootstock and has strong resistance to stress. It is a good material for anti‐reverse regulation mechanism research and functional gene characterization (Duan et al., 2016; Li et al., 2016b,2016c, 2017a). Variants such as structural variations (SVs) are widely present in different species, populations or individuals, and they have a great impact on species divergence and trait determination (Lv et al., 2018; Studer et al., 2011). The publication of this genome will provide important assistance for breakthroughs in Pyrus plant research.
Methods
Plant materials
Pyrus betuleafolia‐Shanxi Duli, which was originally collected in Qinyuan, Shanxi Province, was preserved in the Chinese National Pear Germplasm Repository (Xingcheng, Liaoning). The materials used for genome sequencing assembly were healthy and young leaves, and they were used for DNA extraction immediately after collection.
Short‐read Illumina sequencing and genome size evaluation
The 450‐bp PE libraries were constructed using the NEBNext Ultra DNA Library Prep Kit and sequenced on the Illumina HiSeq 2500 platform. All raw Illumina sequencing reads were cut and filtered using the Trimmomatic program v0.33 to remove adaptors, reads with >3% N and low‐quality reads with more than 50% of bases with a quality score Q < 3 (Vurture et al., 2017). Genome size estimation was conducted via a 19 bp k‐mer frequency analysis with JELLYFISH v2.1.4 (Marçais and Kingsford, 2011).
PacBio sequencing and assembly
High‐quality DNA was extracted from fresh leaf tissue via the CTAB method (Porebski et al., 1997). PacBio SMRTbell libraries (20 kb inserts) were prepared using the Template Prep Kit. Then, 11 SMRT cells were run on the PacBio Sequel system with P6‐C4 chemistry (Chin et al., 2013). De novo assembly was conducted using Falcon v0.3.0 (https://github.com/PacificBiosciences/FALCON-integrate), with the parameters ‘length_cutoff=5000, length_cutoff_pr=12000’; Canu v1.6 (https://github.com/marbl/canu/releases), with the parameters ‘genomeSize=550m, minReadLength=2000’; and HGAP3 (https://github.com/PacificBiosciences/smrtmake), with the parameters ‘genomeSize=550m, minReadLength=2000’ and other parameters ‘default’. The computing platform was a SGE cluster with multiple computer nodes, with each computer node consisting of 120 CPUs and 500 Gb memory. The assembly by the HGAP pipeline resulted in the optimal assembly and was used as a reference. Finally, the Illumina data were aligned to the assembly contigs with bwa mem v0.7.12 (https://sourceforge.net/projects/bio-bwa/files/), and single‐base errors and small indels were corrected using Pilon v1.16 (Walker et al., 2014).
BioNano optical map construction and hybrid assembly
High‐molecular‐weight DNA was prepared from fresh leaf tissue of Pbe‐SD using the IrysPrep Plant Tissue DNA Isolation Kit. The DNA was labelled with the single‐stranded nicking endonucleases Nb.BssSI and Nt.BspQI according to the IrysPrep Reagent Kit protocol. The labelled DNA was then loaded into an IrysChip and linearized DNA molecules were imaged automatically using the Irys system (BioNano Genomics, San Diego, CA). The IrysView (BioNano Genomics) software package was used to produce single‐molecule maps, and de novo assemble them into consensus physical maps. The IrysSlove software was used to hybrid scaffolds with the two BioNano maps and PacBio assembly contigs, resulting in super scaffolds. Some contigs within a super scaffold had high‐quality overlap sequences, and the two contigs were merged to improve the performance of the hybrid scaffolding assembly results when the overlap length ≥ 1 KB and identity ≥ 95%. We performed gap filling using FGAP v1.7 (https://sourceforge.net/projects/fgap/) using the Canu assembly sequences and the filtering parameters ‘score ≥ 500, identity ≥ 85%’.
Hi‐C assembly
To anchor hybrid scaffolds onto the chromosome, we constructed the Hi‐C library and obtained sequencing data via the HiSeq X Ten platform (Illumina, San Diego, CA) (Lieberman‐Aiden et al., 2009; Louwers et al., 2009). The sequencing data were aligned to the assembled scaffold using Bowtie2 in HiC‐Pro_2.9.0 (https://github.com/nservant/HiC-Pro), and then, the scaffold was clustered onto chromosomes with LACHESIS (https://github.com/shendurelab/LACHESIS), with parameters CLUSTER_MIN_RE_SITES=94, CLUSTER_MAX_LINK_DENSITY=4. Finally, we performed artificial correction of the LACHESIS‐assembled results and gap filling or sequence de‐duplication to increase the accuracy and completeness of the assembled genome (Servant et al., 2015).
Full‐length transcriptome sequences
Tissues of flowers, young leaves, stems and mature fruits from Pbe‐SD were collected, and total RNA was extracted from each sample according to the TRIzol (Invitrogen) manufacturer's protocol. cDNA was synthesized using a SMARTer PCR cDNA Synthesis Kit, optimized for preparing full‐length cDNA. Size fractionation and selection (<1 kb, 1–2 kb, 2–3 kb, 3–10 kb) were performed using the BluePippin Size Selection System (Sage Science, Beverly, MA). The SMRT bell libraries were constructed with the Template Prep Kit. Each library underwent SMRT sequencing using one SMRT cell line. SMRT sequencing was then performed on the PacBio Sequel platform.
Genome evaluation
The BUSCO v3.0.0 (http://busco.ezlab.org/) evaluation used a single‐copy orthologous gene library combined with tblastn (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/), AUGUSTUS (http://augustus.gobics.de) and hmmer (http://hmmer.org/download.html) to evaluate the completeness and accuracy of the assembled genome (Daccord et al., 2017). To validate the quality of the assembled genome, full‐length transcripts from four different tissues were mapped to the assembled genome with GMAP 2018‐07‐04 (http://research-pub.gene.com/gmap/).
Repeat annotations
An ab initio repeat library was predicted with LTR Finder v1.05 (http://tlife.fudan.edu.cn/ltr_finder/), RepeatScout v1.0.5 (http://www.repeatmasker.org) and PILER v1.0 (http://www.drive5.com/piler). The predicted repeats were aligned to the SwissProt Database (ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase/uniprot_sprot.fasta.gz) and the Rfam 11.0 database (http://lab.ylog.org/2014/04/09/rfam_scan/) to remove non‐TE protein sequences and ncRNA sequences, respectively. Then, the predicted repeats were aligned to RepBase (http://www.girinst.org/repbase) and the TE protein database with the WU‐BLAST search engine and classified by the RepeatClassifier (http://www.repeatmasker.org/RepeatModeler/). We found and classified the predicted repeats and known repeats (RepBase) with the RepeatMasker Database (http://www.girinst.org/server/RepBase/index.php) and known TE proteins with RepeatProteinMask (http://www.repeatmasker.org/RMDownload.html) in scaffolds. Then, the assembled genome was subjected to repeated sequence screening using the repeat sequences obtained above for the ab initio gene search.
Telomere sequences were identified by searching the 10 kb sequence at both ends of the pseudochromosomes for high copy number repeats with the repeat unit 5‐TTTAGGG‐3. A sliding window (1 Mb windows, 250 kb step) was used to search for high repeat regions in each chromosome, which are assumed to be heterochromatin regions.
Gene annotations
A comprehensive strategy combining ab initio gene prediction, homology‐based gene prediction and Iso‐Seq reads was used for annotation of protein‐coding genes. cDNA protein sequences of P. bretschneideri (ftp://ftp.bioinfo.wsu.edu/species/Pyrus_x_bretschneideri/Pbretschneideri-genome.v1.1), P. communis (ftp://ftp.bioinfo.wsu.edu/species/Pyrus_communis/Pcommunis-draft_genome.v1.0), M. domestica (https://iris.angers.inra.fr/gddh13/downloads/) and Prunus persica (ftp://ftp.bioinfo.wsu.edu/species/Prunus_persica/Prunus_persica-genome.v2.0.a1) were used to predict homologous genes by performing GeMoMa‐1.4.2 (http://www.jstacs.de/index.php/GeMoMa). The reads of Iso‐Seq were aligned to scaffolds using GMAP for the Iso‐Seq‐based gene prediction. The transcripts were used to predict ORFs by PASA v2.0.1 (https://sourceforge.net/projects/pasa/files/stats/timeline), and full‐length cDNA was screened as a training set. AUGUSTUS v3.0.3 (http://augustus.gobics.de/binaries/), SNAP v2013‐02‐16 (http://snap.stanford.edu/snap/download.html), GeneMark‐ET v4.212 (http://topaz.gatech.edu/GeneMark/license_download.cgi) and GlimmerHMM v3.0.4 (http://ccb.jhu.edu/software/glimmerhmm/) were used in ab initio gene prediction. All the gene structures predicted by the above methods were combined into consensus gene models using EVM (https://sourceforge.net/projects/evidencemodeler/).
After removing genes shorter than 300 bp or longer than 20 kb, gene sequences were aligned with annotated gene in the DSHS genome using the blastn program of Blast v2.2.28 and with the parameters ‘‐max_target_seqs 1 ‐evalue 1E–5’. Then, the remaining gene sequences were submitted to the blastx program of Blast2GO v3.0 with an e‐value of 1e3 to blast with the DB of nonredundant protein database. Finally, ‘high‐confidence’ genes were identified.
Gene family and phylogenetic analyses
We used the OrthoMCL package v2.0.9 (http://orthomcl.org/orthomcl/) to identify gene families/clusters. The longest proteins for each gene were aligned to one another. The species‐specific gene families were determined according to the presence or absence of genes for a given species. The shared family expansion and contraction analysis were conducted with CAFÉ v3.1 (https://sourceforge.net/projects/cafehahnlab/). Phylogenetic relationships were resolved using PhyML v3.1 (http://www.atgc-montpellier.fr/phyml/versions.php) based on 1310 high‐quality 1:1 single‐copy orthologous genes. The CodeML utility in the PAML software package was used to analyse divergence times (http://abacus.gene.ucl.ac.uk/software/paml.html). Fossil‐derived timescales and evolutionary history were obtained from the TimeTree database (http://www.timetree.org).
Genomic evolution analysis
To assess the degree of collinearity, a BLASTP search (with an E‐value cutoff of 1 × 10−5) was performed to identify paralogous genes between Pbe‐SD and GDDH13. Syntenic blocks (with at least five genes per block) were identified by MCscan (http://chibba.pgml.uga.edu/mcscan2/). The syntenic blocks were confirmed to represent orthologous blocks between P. betuleafolia and M. domestica. Genes were then classified as collinear or noncollinear according to whether they have a homologous gene in the orthologous regions.
Presence‐absence variation sequences in the Pbe‐SD and DSHS genomes were identified via scanPAV v1.0 (https://github.com/wtsi-hpag/scanPAV). For variants in the Pbe‐SD genome, first, the Pbe‐SD genome was shred into 1 kb fragments after the removal of N‐bases, and the obtained fragments were mapped against the DSHS genome using BWA, and then, the alignments were processed to filter out small repeats and identify the mapping coordinates. Then, the 1 kb fragments from the Pbe‐SD genome missing in the DSHS genome were extracted and the adjacent ones were merged into a single sequence. The absence variations were identified through the same method after DSHS genome mapping against the Pbe‐SD genome. We further aligned the Pbe‐SD sequencing reads to the DSHS genome and DSHS Illumina reads to the Pbe‐SD genome with BWA mem v0.7.12 (https://sourceforge.net/projects/bio-bwa/files/) to exclude potential false positives.
The scaffold of P. bretschneideri was rearranged, based on the P. betuleafolia chromosome. The scaffolds were aligned using the MUMmer (http://mummer.sourceforge.net/) Toolkits nucmer, delta‐filter and show‐coords for the assembled genomes and were called for SNPs and indels, using the Genome Analysis MUMmer Toolkits show‐snp and show‐diff. Classification and annotation of DNA variations were performed using SnpEff (Cingolani et al., 2012).
To detect genes of Pyrus that might be under evolutionary pressure, we first calculated the synonymous nucleotide change rate (Ks) between pairs of orthologous genes between any two genomes of the P. bretschneideri, P. communis and P. betuleafolia genomes. We then calculated the Ka/Ks ratio for orthologous genes between P. bretschneideri and P. betuleafolia to find genes under positive selection (Ka/Ks ratio > 1.0).
Identification of NBS‐LRR and LRR‐RLK resistance genes
The entire gene set was screened for the presence of RGAs using the RGAugury pipeline (https://bitbucket.org/yaanlpc/rgaugury). The default P‐value cut‐off for initial RGA filtering was set to le‐5 for BLASTP. Four classes of RGAs were analysed: NBS‐encoding proteins, RLKs, RLPs and transmembrane coiled‐coil proteins. The symmetry of functional resistance genes between apple and pear was identified using MCScanx (http://chibba.pgml.uga.edu/mcscan2/).
RNA‐Seq data analysis
Using the apple gene as a bait, we searched for genes related to sugar acid and proanthocyanidin anabolism through blastx (Henry‐Kirk et al., 2012; Li et al., 2012). Target genes with sequence coverage ≥ 50% in length, alignment E‐value <10−5 and identity ≥ 50% were selected and classified into the corresponding gene families according to the best hit query sequence. The transcriptome of pulp from mature Pbe‐SD and DSHS was sequenced using the Illumina NovaSeq 6000 platform. RNA clean reads were aligned to the reference genome using HISAT2 (http://ccb.jhu.edu/software/hisat2/faq.shtml). The expression level of each gene in terms of FPKM was computed by RSEM v1.2.15 (http://deweylab.github.io/RSEM/). A gene was considered to be expressed if FPKM > 0. Differential gene expression analysis was conducted using edgeR with the following parameters: FDR < 0.05 and log2|FoldChange|>1. A gene that was considered to be differentially expressed must have at least twofold expression change.
Authors’ contributions
X.D., Z.W., Z.T. and Y.C designed the project; L.T. and H.H collected the experimental materials; W.S.A., C.L. and S.Z. prepared and purified the DNA samples; Y.Z., D.Q. and H.H. identified the phenotypic and metabolic data; Z.L., R. L. and M. S. performed the genome assembly and genome annotation; Z.L., Z.W. and X.D. performed the genomic evolution and variation analysis; X.D. and R. L. performed the transcriptome analysis; and X.D., Z.W., Z.T. and Y.C wrote the manuscript.
Conflict of interest
No conflicts of interest are declared.
Supporting information
Figure S1 Schematic workflow for the genome assembly of P. betuleafolia‐Shanxi Duli (Pbe‐SD) from China.
Figure S2 K‐mer frequency distribution curve (k‐mer = 19) of Illumina short reads of the Pbe‐SD genome.
Figure S3 Mutogram between all chromosomes in the Pbe‐SD genome.
Figure S4 Estimated insertion times of the LTR and two main elements (Copia and Gypsy) with the complete structure of Pbe‐SD.
Figure S5 Comparison between the Pbe‐SD and DSHS genomes for repetitive elements.
Figure S6 Schematic workflow for the gene annotation of the Pbe‐SD genome.
Figure S7 Gene ontology categories of the annotated genes.
Figure S8 Phylogenetic analysis of the UGT71K2 and UGT87A1 genes in the Pbe‐SD genome and 5 additional Rosaceae plant genomes.
Figure S9 Gene collinearity between the Pbe‐SD and GDDH13 genomes.
Figure S10 Gene collinearity between the Pbe‐SD and DSHS genomes.
Figure S11 Nucleotide alignments between Pbe‐SD chromosomes and DSHS scaffolds.
Figure S12 Distribution of RGA loci along the 17 Pbe‐SD chromosomes.
Figure S13 Collinearity comparison of RGAs on the GDDH13 and Pbe‐SD genomes.
Table S1 Statistics of 11 cell single‐molecule real‐time (SMRT) sequencing from PacBio.
Table S2 Initial assembly of SMRT sequencing data using three software programs.
Table S3 Statistics of short‐read sequencing from Illumina HiSeq.
Table S4 BioNano map constructed from two restriction enzymes.
Table S5 Statistical results for each chromosome on the genome.
Table S6 Comparison of the BUSCO single‐copy genes between the Pbe‐SD genome and other published Rosaceae genomes.
Table S7 Detailed TE analysis of P. betuleafolia and P. bretschneideri.
Table S8 Possible heterochromatin regions on the chromosome of P. betuleafolia.
Table S9 Gene prediction results statistics.
Table S10 GO enrichment of specific gene families in P. betuleafolia compared to P. bretschneideri and P. communis.
Table S11 Significantly expanded gene families in the P. betuleafolia genome.
Table S12 GO enrichment of the Pbe‐SD‐presence variation genes.
Table S13 Summary of the identified SNPs and INS or INDEL in the P. betuleafolia genome compared with P. bretschneideri.
Table S14 Number of variants by region.
Table S15 Number of effects by functional class.
Table S16 Positive selection genes involved in three traits.
Table S17 Chromosome distribution of disease resistance genes.
Table S18 Resistance genes in Pyrus betuleafolia and comparison with the other six sequenced genomes.
Table S19 Comparison of the number of gene families related to sugar acid metabolism in different species.
Table S20 Differentially expressed genes involved in sugar/acid metabolism in fruits from DSHS and Pbe‐SD.
Table S21 Comparison of the number of proanthocyanidin synthesis‐related gene families.
Table S22 Differentially expressed genes involved in proanthocyanidin metabolism in fruits from DSHS and Pbe‐SD.
Acknowledgements
This work was supported by the Earmarked Fund for China Agriculture Research System (CARS‐29‐01), the Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS‐ASTIP) and the Fundamental Research Funds for Central Non‐profit Scientific Institution.
Contributor Information
Zhixi Tian, Email: zxtian@genetics.ac.cn.
Yufen Cao, Email: yfcaas@263.net.
Data availability
Data generated during the study were deposited in the NCBI under BioProject number PRJNA529328. For genomic sequencing data, the BioSample accession number is SAMN11264821. For transcriptome data on pulp, the BioSample accession numbers are SAMN11521738 and SAMN11521725. The genome assembly and gene annotations have also been deposited in the Genome Warehouse of the BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences under accession number GWHAAYT00000000.
References
- Bai, S.L. , Sun, Y.W. , Qian, M.J. , Yang, F.X. , Ni, J.B. , Tao, R.Y. , Li, L. et al. (2017) Transcriptome analysis of bagging‐treated red Chinese sand pear peels reveals light‐responsive pathway functions in anthocyanin accumulation. Sci. Rep. 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, L.H. (1917) Pyrus. Standard cyclopedia of horticulture, vol. V, pp. 2865–2878. New York: Macmillan. [Google Scholar]
- Bell, R. , Quamme, H. , Layne, R. and Skirvin, R. (1996). Pears. In Fruit breeding, vol 1: tree and tropical fruits ( Janick, J. and Moore, J.N. , eds), pp. 441–514. New York: Wiley. [Google Scholar]
- Bickhart, D.M. , Rosen, B.D. , Koren, S. , Sayre, B.L. , Hastie, A.R. , Chan, S. , Lee, J. et al. (2017) Single‐molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 49, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, L. , Bourcy, M. , Boulay, M. , Tellier, M. , Guérif, P. , Denancé, C. , Durel, C.E. et al. (2012) The new pear scab resistance gene Rvp1 from the European pear cultivar ‘Navara’ maps in a genomic region syntenic to an apple scab resistance gene cluster on linkage group 2. Tree Genet. Genomes, 8, 53–60. [Google Scholar]
- Bus, V. , Bassett, H. , Bowatte, D. , Chagné, D. , Ranatunga, C. , Ulluwishewa, D. , Wiedow, C. et al. (2010) Genome mapping of an apple scab, a powdery mildew and a woolly apple aphid resistance gene from open‐pollinated mildew immune selection. Tree Genet. Genomes, 6, 477–487. [Google Scholar]
- Celton, J.M. , Chagné, D. , Tustin, S.D. , Terakami, S. , Nishitani, C. , Yamamoto, T. and Gardiner, S.E. (2009) Update on comparative genome mapping between Malus and Pyrus . BMC Res. Notes, 2, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné, D. , Crowhurst, R.N. , Pindo, M. , Thrimawithana, A. , Deng, C. , Ireland, H. , Fiers, M. et al. (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS ONE, 9, e92644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.J. , Cao, Y.F. , Zhang, J.M. , Tian, L.M. , Dong, X.G. , Zhang, Y. , Qi, D. et al. (2017) Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genet. Genomes, 13, 44. [Google Scholar]
- Chen, L.Q. (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 201, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Chen, J.L. , Wang, Z.F. , Wu, J.H. , Wang, Q. and Hu, X.S. (2007) Chemical compositional characterization of eight pear cultivars grown in China. Food Chem. 104, 268–275. [Google Scholar]
- Chen, J.F. , Huang, Q.F. , Gao, D.Y. , Wang, J.Y. , Lang, Y.S. , Liu, T.Y. , Li, B. et al. (2013) Whole‐genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat. Commun. 4, 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, C.S. , Alexander, D.H. , Marks, P. , Klammer, A.A. , Drake, J. , Heiner, C. , Clum, A. et al. (2013) Nonhybrid, finished microbial genome assemblies from long‐read SMRT sequencing data. Nat. Methods, 10, 563. [DOI] [PubMed] [Google Scholar]
- Cho, K.H. , Shin, I.S. , Kim, K.T. , Suh, E.J. , Hong, S.S. and Lee, H.J. (2009) Development of AFLP and CAPS markers linked to the scab resistance gene, Rvn2, in an inter‐specific hybrid pear (Pyrus spp.). J. Hortic. Sci. Biotech. 84, 619–624. [Google Scholar]
- Cingolani, P. , Platts, A. , Wang, L.L. , Coon, M. , Nguyen, T. , Wang, L. , Land, S.J. et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, L.L. , Yao, S.B. , Dai, X.L. , Yin, Q.G. , Liu, Y.J. , Jiang, X.L. , Wu, Y.H. et al. (2016) Identification of UDP‐glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 67, 2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord, N. , Celton, J.M. , Linsmith, G. , Becker, C. , Choisne, N. , Schijlen, E. , van de Geest, H. et al. (2017) High‐quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099. [DOI] [PubMed] [Google Scholar]
- Duan, X.W. , Zhang, W.N. , Huang, J. , Hao, L. , Wang, S.N. , Wang, A.D. , Meng, D. et al. (2016) PbWoxT1 mRNA from pear (Pyrus betuleafolia) undergoes long‐distance transport assisted by a polypyrimidine tract binding protein. New Phytol. 210, 511–524. [DOI] [PubMed] [Google Scholar]
- Galli, P. , Patocchi, A. , Broggini, G.A.L. and Gessler, C. (2010) The Rvi15 (Vr2) apple scab resistance locus contains three TIR‐NBS‐LRR genes. Mol. Plant Microbe Interact. 23, 608–617. [DOI] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J.M. and Lloyd, A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Henry‐Kirk, R.A. , McGhie, T.K. , Andre, C.M. , Hellens, R.P. and Allan, A.C. (2012) Transcriptional analysis of apple fruit proanthocyanidin biosynthesis. J. Exp. Bot. 63, 5437–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.H. , Wei, X.H. , Sang, T. , Zhao, Q. , Feng, Q. , Zhao, Y. , Li, C.Y. et al. (2010) Genome‐wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961. [DOI] [PubMed] [Google Scholar]
- Iketani, H. , Manabe, T. , Matsuta, N. , Akihama, T. and Hayashi, T. (1998) Incongruence between RFLPs of chloroplast DNA and morphological classification in east Asian pear (Pyrus spp.). Genet. Resour. Crop Evol. 45, 533–539. [Google Scholar]
- Jarvis, D.E. , Ho, Y.S. , Lightfoot, D.J. , Schmöckel, S.M. , Li, B. , Borm, T.J. , Ohyanagi, H. et al. (2017) The genome of Chenopodium quinoa . Nature, 542, 307–312. [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Zheng, X.Y. , Yu, P.Y. , Yue, X.Y. , Ahmed, M. , Cai, D.Y. and Teng, Y.W. (2016) Primitive genepools of Asian pears and their complex hybrid origins inferred from fluorescent sequence‐specific amplification polymorphism (SSAP) markers based on LTR retrotransposons. PLoS ONE, 11, e0149192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, A. (1948) Horticulture of Fruit Trees. Tokyo, Japan: Yokendo Press. [Google Scholar]
- Langford, M.H. (1942) Heterothallism and variability in Venturia pirina . Phytopathology, 32, 357–369. [Google Scholar]
- Li, M.J. , Feng, F.J. and Cheng, L.L. (2012) Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE, 7, e33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.J. , Wang, B. , Dong, R.R. and Hou, B.K. (2015) AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Sci. 236, 157–167. [DOI] [PubMed] [Google Scholar]
- Li, P. , Quan, X. , Jia, G. , Xiao, J. , Cloutier, S. and You, F.M. (2016a) RGAugury: a pipeline for genome‐wide prediction of resistance gene analogs (RGAs) in plants. BMC Genom. 17, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Han, J.L. , Chang, Y.H. , Lin, J. and Yang, Q.S. (2016b) Gene characterization and transcription analysis of two new ammonium transporters in pear rootstock (Pyrus betuleafolia). J. Plant. Res. 129, 737–748. [DOI] [PubMed] [Google Scholar]
- Li, K.Q. , Xu, X.Y. and Huang, X.S. (2016c) Identification of differentially expressed genes related to dehydration resistance in a highly drought‐tolerant pear, Pyrus betuleafolia, as through RNA‐Seq. PLoS ONE, 11, e0149352. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li, K.Q. , Xing, C.H. , Yao, Z.H. and Huang, X.S. (2017a) Pbr MYB 21, a novel MYB protein of Pyrus betuleafolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 15, 1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Li, Y.J. , Zhang, F.J. , Zhang, G.Z. , Jiang, X.Y. , Yu, H.M. and Hou, B.K. (2017b) The Arabidopsis UDP‐glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103. [DOI] [PubMed] [Google Scholar]
- Liao, L. , Vimolmangkang, S. , Wei, G. , Zhou, H. , Korban, S.S. and Han, Y. (2015) Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 6, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden, E. , Van Berkum, N.L. , Williams, L. , Imakaev, M. , Ragoczy, T. , Telling, A. , Amit, I. et al. (2009) Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers, M. , Bader, R. , Haring, M. , van Driel, R. , de Laat, W. and Stam, M. (2009) Tissue‐and expression level–specific chromatin looping at Maize b1 epialleles. Plant Cell, 21, 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, S.W. , Wu, W.G. , Wang, M.H. , Meyer, R.S. , Ndjiondjop, M.N. , Tan, L.B. , Zhou, H.Y. et al. (2018) Genetic control of seed shattering during African rice domestication. Nat. Plants, 4, 331. [DOI] [PubMed] [Google Scholar]
- Marçais, G. and Kingsford, C. (2011) A fast, lock‐free approach for efficient parallel counting of occurrences of k‐mers. Bioinformatics, 27, 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.W. , Song, K.J. , Kim, M.Y. , Hwang, J.H. , Shin, Y.U. , Kim, W.C. and Chung, W.I. (2002) Molecular cloning and characterization of four cDNAs encoding the isoforms of NAD‐dependent sorbitol dehydrogenase from the Fuji apple. Plant Sci. 162, 513–519. [Google Scholar]
- Pierantoni, L. , Cho, K.H. , Shin, I.S. , Chiodini, R. , Tartarini, S. , Dondini, L. , Kang, S.J. et al. (2004) Characterisation and transferability of apple SSRs to two European pear F 1 populations. Theor. Appl. Genet. 109, 1519–1524. [DOI] [PubMed] [Google Scholar]
- Pierantoni, L. , Dondini, L. , Cho, K.H. , Shin, I.S. , Gennari, F. , Chiodini, R. , Tartarini, S. et al. (2007) Pear scab resistance QTLs via a European pear (Pyrus communis) linkage map. Tree Genet. Genomes, 3, 311. [Google Scholar]
- Porebski, S. , Bailey, L.G. and Baum, B.R. (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. [Google Scholar]
- Pourcel, L. , Routaboul, J.M. , Kerhoas, L. , Caboche, M. , Lepiniec, L. and Debeaujon, I. (2005) TRANSPARENT TESTA10 encodes a laccase‐like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell, 17, 2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu, F. and Wang, Y. (1963). Pomology of China. Pears, vol 3. Shanghai: Shanghai Science and Technology Press. [Google Scholar]
- Rubtsov, G.A. (1944) Geographical distribution of the genus Pyrus and trends and factors in its evolution. Am. Nat. 78, 358–366. [Google Scholar]
- Schouten, H.J. , Brinkhuis, J. , van der Burgh, A. , Schaart, J.G. , Groenwold, R. , Broggini, G.A. and Gessler, C. (2014) Cloning and functional characterization of the Rvi15 (Vr2) gene for apple scab resistance. Tree Genet. Genomes, 10, 251–260. [Google Scholar]
- Servant, N. , Varoquaux, N. , Lajoie, B.R. , Viara, E. , Chen, C.J. , Vert, J.P. , Heard, E. et al. (2015) HiC‐Pro: an optimized and flexible pipeline for Hi‐C data processing. Genome Biol. 16, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S.B. and Dixon, R.A. (2005) Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana . Plant J. 44, 62–75. [DOI] [PubMed] [Google Scholar]
- Shirasawa, K. , Isuzugawa, K. , Ikenaga, M. , Saito, Y. , Yamamoto, T. , Hirakawa, H. and Isobe, S. (2017) The genome sequence of sweet cherry (Prunus avium) for use in genomics‐assisted breeding. DNA Res. 24, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão, F.A. , Waterhouse, R.M. , Ioannidis, P. , Kriventseva, E.V. and Zdobnov, E.M. (2015) BUSCO: assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics, 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- Studer, A. , Zhao, Q. , Ross‐Ibarra, J. and Doebley, J. (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S.L. , Zhou, Y.S. , Chen, J. , Shi, J.P. , Zhao, H.M. , Zhao, H.M. , Zhao, H.N. et al. (2018) Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50, 1289. [DOI] [PubMed] [Google Scholar]
- Tanaka, S. and Yamamoto, S. (1964) Studies on pear scab. J. Biol. Chem. 29, 128–136. [Google Scholar]
- Tanner, G.J. , Francki, K.T. , Abrahams, S. , Watson, J.M. , Larkin, P.J. and Ashton, A.R. (2003) Proanthocyanidin biosynthesis in plants purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 278, 31647–31656. [DOI] [PubMed] [Google Scholar]
- Verde, I. , Abbott, A.G. , Scalabrin, S. , Jung, S. , Shu, S. , Marroni, F. , Zhebentyayeva, T. et al. (2013) The high‐quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. [DOI] [PubMed] [Google Scholar]
- Vurture, G.W. , Sedlazeck, F.J. , Nattestad, M. , Underwood, C.J. , Fang, H. , Gurtowski, J. and Schatz, M.C. (2017) GenomeScope: fast reference‐free genome profiling from short reads. Bioinformatics, 33, 2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, B.J. , Abeel, T. , Shea, T. , Priest, M. , Abouelliel, A. , Sakthikumar, S. , Cuomo, C.A. et al. (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W.Q. , Feng, B.X. , Xiao, J.F. , Xia, Z.Q. , Zhou, X.C. , Li, P.H. , Zhang, W.X. et al. (2014) Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 10, 5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Wang, Z.Y. , Zhang, M. , Jia, B. , Heng, W. , Ye, Z.F. , Zhu, L.W. et al. (2018) Transcriptome sequencing analysis of two different genotypes of Asian pear reveals potential drought stress genes. Tree Genet. Genomes, 14, 40. [Google Scholar]
- Wrangham, R.W. , Conklin‐Brittain, N.L. and Hunt, K.D. (1998) Dietary response of chimpanzees and cercopithecines to seasonal variation in fruit abundance. I. Antifeedants. Int. J. Primatol. 19, 949–970. [Google Scholar]
- Wu, J. , Wang, Z.W. , Shi, Z.B. , Zhang, S. , Ming, R. , Zhu, S.L. , Khan, M.A. et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Wang, Y.T. , Xu, J.B. , Korban, S.S. , Fei, Z.J. , Tao, S.T. , Ming, R. et al. (2018) Diversification and independent domestication of Asian and European pears. Genome Biol. 19, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, E.H. , Zhang, H.B. , Sheng, J. , Li, K. , Zhang, Q.J. , Kim, C. , Zhang, Y. et al. (2017) The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 10, 866–877. [DOI] [PubMed] [Google Scholar]
- Xie, D.Y. , Sharma, S.B. and Dixon, R.A. (2004) Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana . Arch. Biochem. Biophys. 422, 91–102. [DOI] [PubMed] [Google Scholar]
- Yamaki, S. and Ino, M. (1992) Alteration of cellular compartmentation and membrane permeability to sugars in immature and mature apple fruit. J. Am. Soc. Hortic. Sci. 117, 951–954. [Google Scholar]
- Yamamoto, T. , Kimura, T. , Saito, T. , Kotobuki, K. , Matsuta, N. , Liebhard, R. , Gessler, C. et al. (2004) Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hort. 663, 51–56. [Google Scholar]
- Yamazaki, M. , Makita, Y. , Springob, K. and Saito, K. (2003) Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa . Biochem. Eng. J. 14, 191–197. [Google Scholar]
- Yin, H. , Du, J.C. , Wu, J. , Wei, S.W. , Xu, Y.X. , Tao, S.T. , Wu, J.Y. et al. (2015) Genome‐wide annotation and comparative analysis of long terminal repeat retrotransposons between pear species of P. bretschneideri and P. communis . Sci. Rep. 5, 17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. (1979) Taxonomy of the fruit tree in China. Beijing: Agriculture Press. [Google Scholar]
- Zhang, F.T. , Xu, T. , Mao, L.Y. , Yan, S.Y. , Chen, X.W. , Wu, Z.F. , Chen, R. et al. (2016) Genome‐wide analysis of Dongxiang wild rice (Oryza rufipogon Griff.) to investigate lost/acquired genes during rice domestication. BMC Plant Biol. 16, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.j. , Li, X.X. , Ma, B. , Gao, Q. , Du, H.L. , Han, Y.H. , Li, Y. et al. (2017) The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol. Plant., 10, 1224–1237. [DOI] [PubMed] [Google Scholar]
- Zheng, X.Y. , Cai, D.Y. , Potter, D. , Postman, J. , Liu, J. and Teng, Y.W. (2014) Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol. Phylogenet. Evol. 80, 54–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Schematic workflow for the genome assembly of P. betuleafolia‐Shanxi Duli (Pbe‐SD) from China.
Figure S2 K‐mer frequency distribution curve (k‐mer = 19) of Illumina short reads of the Pbe‐SD genome.
Figure S3 Mutogram between all chromosomes in the Pbe‐SD genome.
Figure S4 Estimated insertion times of the LTR and two main elements (Copia and Gypsy) with the complete structure of Pbe‐SD.
Figure S5 Comparison between the Pbe‐SD and DSHS genomes for repetitive elements.
Figure S6 Schematic workflow for the gene annotation of the Pbe‐SD genome.
Figure S7 Gene ontology categories of the annotated genes.
Figure S8 Phylogenetic analysis of the UGT71K2 and UGT87A1 genes in the Pbe‐SD genome and 5 additional Rosaceae plant genomes.
Figure S9 Gene collinearity between the Pbe‐SD and GDDH13 genomes.
Figure S10 Gene collinearity between the Pbe‐SD and DSHS genomes.
Figure S11 Nucleotide alignments between Pbe‐SD chromosomes and DSHS scaffolds.
Figure S12 Distribution of RGA loci along the 17 Pbe‐SD chromosomes.
Figure S13 Collinearity comparison of RGAs on the GDDH13 and Pbe‐SD genomes.
Table S1 Statistics of 11 cell single‐molecule real‐time (SMRT) sequencing from PacBio.
Table S2 Initial assembly of SMRT sequencing data using three software programs.
Table S3 Statistics of short‐read sequencing from Illumina HiSeq.
Table S4 BioNano map constructed from two restriction enzymes.
Table S5 Statistical results for each chromosome on the genome.
Table S6 Comparison of the BUSCO single‐copy genes between the Pbe‐SD genome and other published Rosaceae genomes.
Table S7 Detailed TE analysis of P. betuleafolia and P. bretschneideri.
Table S8 Possible heterochromatin regions on the chromosome of P. betuleafolia.
Table S9 Gene prediction results statistics.
Table S10 GO enrichment of specific gene families in P. betuleafolia compared to P. bretschneideri and P. communis.
Table S11 Significantly expanded gene families in the P. betuleafolia genome.
Table S12 GO enrichment of the Pbe‐SD‐presence variation genes.
Table S13 Summary of the identified SNPs and INS or INDEL in the P. betuleafolia genome compared with P. bretschneideri.
Table S14 Number of variants by region.
Table S15 Number of effects by functional class.
Table S16 Positive selection genes involved in three traits.
Table S17 Chromosome distribution of disease resistance genes.
Table S18 Resistance genes in Pyrus betuleafolia and comparison with the other six sequenced genomes.
Table S19 Comparison of the number of gene families related to sugar acid metabolism in different species.
Table S20 Differentially expressed genes involved in sugar/acid metabolism in fruits from DSHS and Pbe‐SD.
Table S21 Comparison of the number of proanthocyanidin synthesis‐related gene families.
Table S22 Differentially expressed genes involved in proanthocyanidin metabolism in fruits from DSHS and Pbe‐SD.
Data Availability Statement
Data generated during the study were deposited in the NCBI under BioProject number PRJNA529328. For genomic sequencing data, the BioSample accession number is SAMN11264821. For transcriptome data on pulp, the BioSample accession numbers are SAMN11521738 and SAMN11521725. The genome assembly and gene annotations have also been deposited in the Genome Warehouse of the BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences under accession number GWHAAYT00000000.


