Summary
Silique number is the most important component of yield in rapeseed (Brassica napus L.). To dissect the mechanism underlying the natural variation of silique number in rapeseed germplasm, a series of studies were performed. A panel of 331 core lines was employed to genome‐wide association study (GWAS), and 27 loci (including 20 novel loci) were identified. The silique number difference between the more‐ and fewer‐silique lines can be attributed to the accumulative differences in flower number and silique setting rate. Each of them accounted for 75.2% and 24.8%, respectively. The silique number was highly associated with the total photosynthesis and biomass. Microscopic analysis showed that the difference between extremely more‐ and fewer‐silique lines normally occurred at the amount of flower bud but not morphology. Transcriptome analysis of shoot apical meristem (SAM) suggested that most of enriched groups were associated with the auxin biosynthesis/metabolism, vegetative growth and nutrition/energy accumulation. By integrating GWAS and RNA‐seq results, six promising candidate genes were identified, and some of them were related to biomass accumulation. In conclusion, the natural variation of silique number is largely affected by the biomass and nutrition accumulation, which essentially reflects the positive regulatory relationship between the source and sink. Our study provides a comprehensive and systematic explanation for natural variation of silique number in rapeseed, which provides a foundation for its improvement.
Keywords: Brassica napus L., silique number, GWAS, RNA‐seq, leaf area, photosynthesis
Background
Among the three components (silique number, seed number per silique and seed weight) of yield in rapeseed, silique number is highly correlated with yield (Shi et al., 2015). To increase the yield of rapeseed, the improvement in silique number is crucial. In rapeseed germplasm resources, silique number shows extensive variation, which is invaluable for its study and improvement. However, the comprehensive and systematic study on this kind of natural variation is poor, which is a constraint to our in‐depth understanding and genetic improvement of this important trait.
From a developmental perspective, silique number is multiplicatively determined by flower number and silique setting rate; however, the relative importance of both on silique number variation has not been analysed in spite of its fundamental interest/significance in the genetics and breeding study. For flower number, flower bud differentiation is the decisive developmental stage (Luo et al., 2018). At the initial stage of reproductive growth, the shoot apical meristem (SAM) transforms to inflorescence meristem (IM), and then, the IM produces floral primordia continuously (Ye et al., 2017). These processes are regulated by many factors including developmental cues, phytohormones, the environment and their interactions (Zhang et al., 2016). The silique setting rate is also a complex trait sensitive to the environment, the flower degeneration and drop result in the decline of silique setting rate and nutrient waste. Although the exact regulatory mechanism underlying the flower degeneration and drop is not well known, some hypotheses in context of nutrient supply, fertilization and phytohormone modulation had been proposed (Sun et al., 2009; Zhang et al., 2016).
As a typical quantitative trait, silique number is easily affected by the environmental conditions with moderate heritability. In rapeseed, the research on silique number mostly focused on QTL positioning. To present, more than 90 loci for silique number have been reported (Lu et al., 2017; Shi et al., 2015; Ye et al., 2017), which were distributed on 18 (excluding C07) of the 19 linkage groups. Given that almost all of these silique number QTLs showed moderate effect, and sensitive to the environment, it is difficult to identify the underlying genes by map‐based cloning. In the model plant Arabidopsis, tens of genes have been identified to affect silique number by regulating flower number, such as WUS, CLV3 and AP (Bartrina et al., 2011; Brand et al., 2000; Chen, 2004; Schoof et al., 2000). Several genes that regulate flower organ number were also identified, such as FON1, FON2, FON4 in rice, SFT in tomato and BIF2 in maize (Bortiri and Hake, 2007; Chu et al., 2006; Krieger et al., 2010; Suzaki et al., 2006). Furthermore, several pathways/mechanisms that regulate silique/flower number have been identified (Brand et al., 2000; Kinoshita et al., 2010; Mu and Bleckmann, 2008; Mu et al., 2006; Sarkar et al., 2007). However, most of these genes were identified through mutant analysis, which could not explain general natural variation.
In the current study, we aimed to identify the key mechanism responsible for silique number variation in rapeseed (Brassica napus L.) germplasm through a systematic study. We investigated the genetic, physiological, microscopic and molecular causes of silique number variation. After the integration of these experimental results, we proposed new insight into the regulatory mechanism for this trait: the natural variation of silique number is largely affected by biomass and nutrition accumulation, which essentially reflected the positive regulatory relationship between the source and sink. The study provides a foundation for genetic improvement of silique number.
Results
GWAS for silique number
Phenotypic variation and heritability of silique number in three environments
Extensive phenotypic variations for silique number of the main inflorescence (SNm), branch inflorescence (SNb) and whole plant (SNw) were observed in the 331 accessions that were grown in three environments (N14, 2014 in Nanchang; W14, 2014 in Wuhan; W16, 2016 in Wuhan). Across the three environments, SNm varied from 20.5 to 149.3, with 4.0‐ to 6.5‐fold variations; SNb varied from 41 to 327, with 5.9‐ to 7.6‐fold variations; SNw varied from 63 to 384, with 5.6‐ to 5.9‐fold variations (Figure 1, Table 1). Analysis of variance showed that genotype, environment and genotype by environment interaction all had significant effects on the three investigated traits (Table S1). The estimated heritability for the three traits was 0.54, 0.58 and 0.58, respectively, which was highly accordant with the relatively low value reported in the previous studies (Shi et al., 2015; Ye et al., 2017). Pearson's correlation coefficient of SNm and SNw, SNb and SNw, SNm and SNb was 0.562**, 0.969** and 0.385**, respectively (Table S2). This indicated that SNw was mainly determined by the SNb, followed by SNm. The low correlation between SNm and SNb reflected the different genetic regulation of the two traits in which SNb was affected by branch number.
Figure 1.
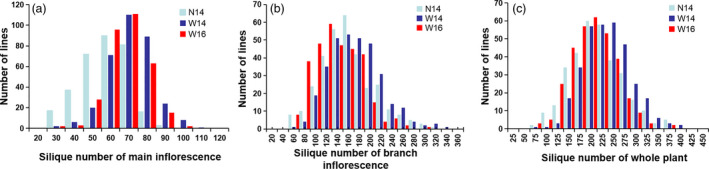
Distribution of silique number in the three environments. The horizontal axis represented the value of silique number for the main inflorescence (a), branch inflorescence (b) and whole plant (c). The vertical axis represented the number of lines within the association population.
Table 1.
Phenotypic variation of SNm, SNb and SNw in the association panel
| Trait | Environment | Minimum | Maximum | Mean | SD | Coefficient of variation (%) | Skewness | Kurtosis | Heritability |
|---|---|---|---|---|---|---|---|---|---|
| SNm | N14 | 20.5 | 82.8 | 51.60 | 12.69 | 24.59 | −0.25 | −0.44 | 0.54 |
| W14 | 24.5 | 105.4 | 65.86 | 12.30 | 18.68 | −0.20 | 0.74 | ||
| W16 | 23.1 | 149.3 | 63.08 | 11.73 | 18.60 | 1.10 | 8.83 | ||
| SNb | N14 | 41.0 | 312.0 | 151.38 | 49.42 | 32.65 | 0.44 | 0.19 | 0.58 |
| W14 | 55.0 | 327.0 | 164.36 | 47.22 | 28.73 | 0.49 | 0.31 | ||
| W16 | 41.0 | 297.0 | 126.89 | 42.80 | 33.73 | 0.53 | 0.33 | ||
| SNw | N14 | 66.0 | 375.0 | 203.51 | 57.47 | 28.24 | 0.29 | 0.12 | 0.58 |
| W14 | 65.0 | 384.0 | 226.41 | 55.21 | 24.38 | 0.64 | 1.9 | ||
| W16 | 63.0 | 372.0 | 187.20 | 49.77 | 26.59 | 0.37 | 0.37 |
SNm, SNb and SNw were the abbreviations of silique number from the main inflorescence, branch inflorescence and whole plant, respectively.
N14, W14 and W16 were the codes of the three environments: 2014 in Nanchang; 2014 in Wuhan; 2016 in Wuhan.
Marker‐trait association
The GWAS analysis was performed using the GLM model. The QQ plot displayed in Figure S1 showed that the model could be used to identify significant SNPs. A total of 191 significantly associated SNPs were identified, of which 75, 88 and 104 were from SNm, SNb and SNw, respectively. Among these associated SNPs, 22, 111 and 16 were identified in N14, W14 and W16, respectively; 62 SNPs were identified using BLUP values in the three environments. (Figure 2 and Table S3).
Figure 2.
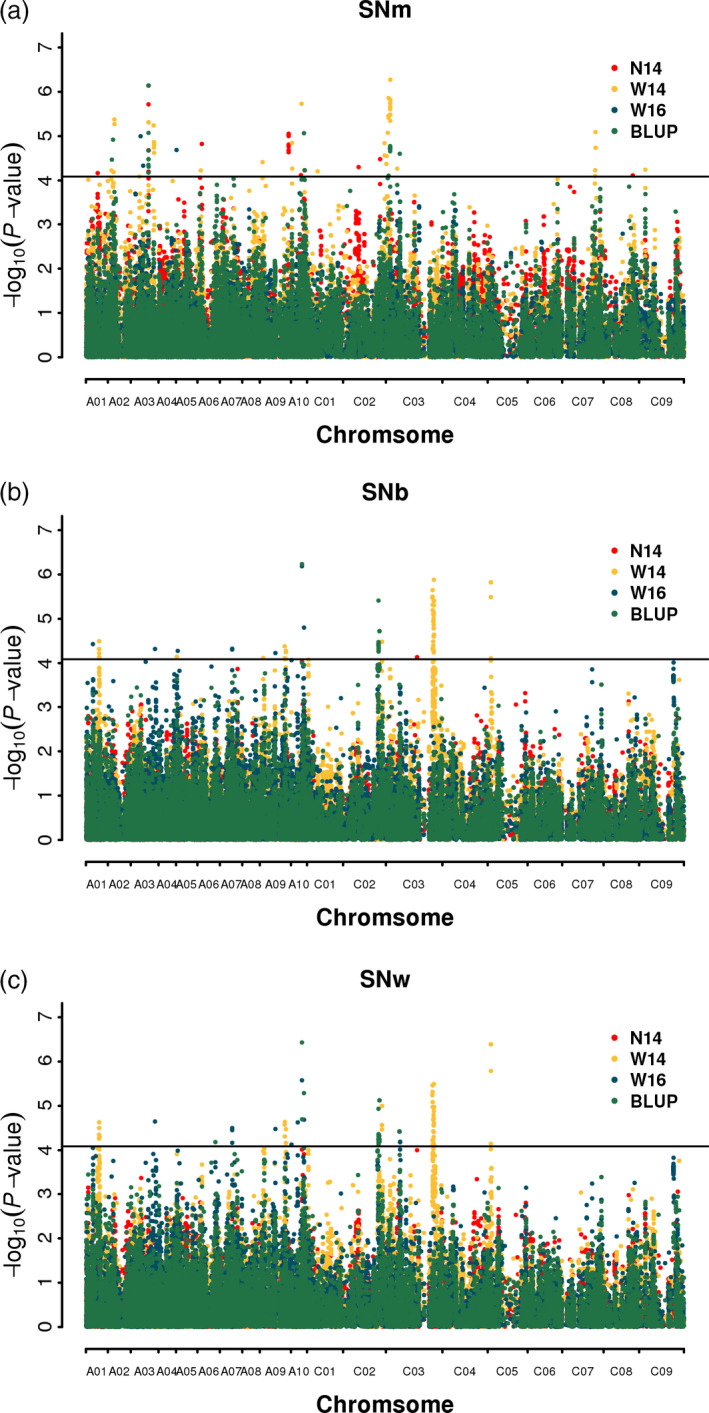
Manhattan plots generated from GWAS results for silique number from the main inflorescence (a), branch inflorescence (b) and whole plant (c).
To integrate significant clustered SNP loci, we employed the previously reported method (Chen et al., 2018). The SNPs were considered the same loci if the lead and following SNPs were within 500 kb or LD statistic r 2 > 0.2. As a result, all the 191 significantly associated SNPs were merged into 47 loci. Of these, 27 loci had one more significant SNPs, which implied high reliability, and were adopted for further analysis. These loci were distributed on 12 of the 19 B. napus chromosomes (excluding A04, A06, A08, A11, C04, C06 and C08), explaining 5.60%–8.71% of the phenotypic variance (Table 2). It should be noted that 12 (44.4%) loci could be repeatedly identified in different environments, with nine loci in two environments and three loci in three environments (including BLUP), indicating their high reliability. In addition, two identified loci were associated with both SNm and SNw simultaneously, eight were identified with SNb and SNw simultaneously, and four were identified with SNm, SNb and SNw simultaneously. All of these loci explained 31.93%, 25.95% and 31.46% of the total phenotypic variance for SNm, SNb and SNw, respectively. These results indicated that the silique number was a genetically complex trait and sensitive to the environment, which was accordant with other studies (Shi et al., 2015; Ye et al., 2017).
Table 2.
Details of 27 loci of silique number detected from GWAS in the different environments
| Loci | Traits | Peak SNP | Chromosome | Position | −Log10(P) | R 2 (%) | Environment | Reports in previous research |
|---|---|---|---|---|---|---|---|---|
| 1 | SNb, SNw | Bn‐A01‐S16094037 | A01 | 14,045,897 | 4.35 | 6.67 | W14 | New |
| 2 | SNm | Bn‐A02‐S7482915 | A02 | 4,511,692 | 4.47 | 6.40 | BLUP, W14 | Ding et al. (2012) |
| 3 | SNm | Bn‐A02‐S8767145 | A02 | 5,739,739 | 4.92 | 6.75 | BLUP,W14 | New |
| 4 | SNm | Bn‐A02‐S10323998 | A02 | 7,170,926 | 5.38 | 7.31 | W14 | New |
| 5 | SNm | Bn‐A03‐S20398340 | A03 | 19,267,907 | 6.14 | 8.31 | BLUP, N14, W14 | Shi et al. (2013) |
| 6 | SNm | Bn‐A03‐S26437883 | A03 | 24,829,819 | 5.24 | 7.14 | W14 | New |
| 7 | SNb,SNw | Bn‐A03‐S29118954 | A03 | 26,262,031 | 4.64 | 6.54 | W16 | New |
| 8 | SNm,SNb | Bn‐A05‐S474257 | A05 | 585,628 | 4.68 | 7.10 | W16,W14 | New |
| 9 | SNb,SNw | Bn‐A02‐S588093 | A07 | 13,217,992 | 4.50 | 6.34 | W16 | Shi et al. (2009) |
| 10 | SNm,SNb | Bn‐A09‐S3097420 | A09 | 3,031,274 | 4.41 | 6.07 | W14 | Ding et al. (2012) |
| 11 | SNb,SNw | Bn‐A09‐S19941634 | A09 | 16,914,495 | 4.48 | 6.31 | W16 | New |
| 12 | SNb,SNw, | Bn‐A09‐S29186255 | A09 | 27,063,736 | 4.31 | 7.32 | W14 | New |
| 13 | SNb,SNw, | Bn‐A09‐S25899875 | A09 | 28,211,039 | 4.47 | 6.16 | W14 | New |
| 14 | SNm | Bn‐A09‐S33976464 | A09 | 31,210,585 | 5.05 | 7.10 | N14 | Shi et al. (2013) |
| 15 | SNm,SNw | Bn‐A10‐S3921433 | A10 | 900,559 | 4.84 | 6.63 | W14,W16 | Lu et al. (2017) |
| 16 | SNm,SNb,SNw | Bn‐A10‐S10298013 | A10 | 11,677,067 | 6.43 | 8.71 | W14, W16,BLUP | New |
| 17 | SNm,SNb,SNw | Bn‐A10‐S13789579 | A10 | 13,825,021 | 5.29 | 7.20 | BLUP, N14,W16 | New |
| 18 | SNm,SNb,SNw | Bn‐scaff_15712_6‐S1336179 | C02 | 38,045,422 | 5.41 | 8.69 | BLUP, N14 | New |
| 19 | SNm,SNb,SNw | Bn‐scaff_17109_1‐S557859 | C02 | 41,808,474 | 5.00 | 7.07 | W14 | Radoev et al. (2008) |
| 20 | SNm | Bn‐scaff_16614_1‐S1480092 | C03 | 660,236 | 4.55 | 6.29 | W14 | New |
| 21 | SNm | Bn‐scaff_18936_1‐S102755 | C03 | 2,745,162 | 5.86 | 8.55 | BLUP,W14 | New |
| 22 | SNm | Bn‐scaff_15877_1‐S926737 | C03 | 4,656,329 | 6.27 | 8.66 | BLUP,W14 | New |
| 23 | SNm,SNw | Bn‐scaff_22466_1‐S754489 | C03 | 14,826,392 | 4.60 | 6.36 | BLUP,W16 | New |
| 24 | SNb, SNw | Bn‐scaff_18602_1‐S270185 | C03 | 51,667,421 | 5.88 | 7.97 | W14 | New |
| 25 | SNb, SNw | Bn‐scaff_20901_1‐S369010 | C05 | 3,670,200 | 6.39 | 8.63 | W14 | New |
| 26 | SNm | Bn‐scaff_15705_3‐S436841 | C07 | 36,219,357 | 5.09 | 6.96 | W14 | New |
| 27 | SNm | Bn‐scaff_17487_1‐S512535 | C09 | 6,772,003 | 4.82 | 7.45 | BLUP, W14 | New |
SNm, SNb and SNw were the abbreviations of silique number from the main inflorescence, branch inflorescence and whole plant, respectively.
N14, W14 and W16 were the codes of the three environments: 2014 in Nanchang; 2014 in Wuhan; 2016 in Wuhan.
In previous studies, more than 90 loci for silique number had been reported, which were distributed on 18 (excluding C07) of the 19 linkage groups (Lu et al., 2017; Shi et al., 2015; Ye et al., 2017). To clarify the positional relationship of silique number loci detected in our and other studies, a comparative analysis was performed based on the physical map of Darmor_V4.1. Of the 27 detected loci, seven loci were overlapped with those reported in previous studies, respectively, whereas the majority of loci detected in our study were novel (Table 2).
The physiological basis of silique number variation in rapeseed
To understand the physiological basis of silique number variation in rapeseed, we chose a batch of extreme lines (10 more‐silique lines and 9 fewer‐silique lines) from the associated population for further research. The SNm of the ten more‐silique and nine fewer‐silique lines ranged from 86.4 to 125.5 and 33.0 to 62.3, respectively, and the mean (102.4) of the former was significantly larger than that (50.9) of the latter (P < 0.01) (Figure 3). As illustrated in Figure S2, the silique number and flower number of the main inflorescence were highly associated with those of per branch. To reduce the workload, the flower and silique number mentioned in the physiological study were both from the main inflorescence.
Figure 3.
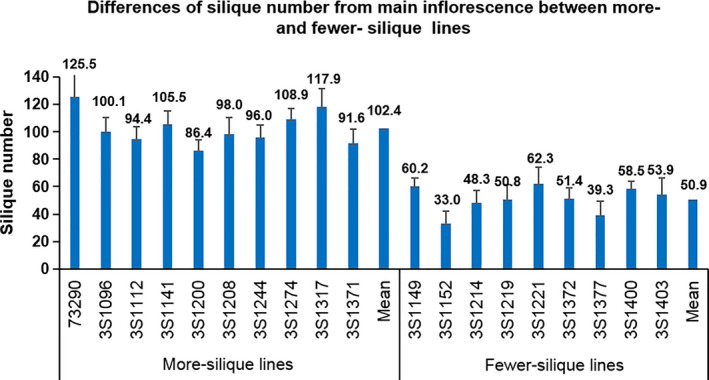
Differences of silique number from main inflorescence between more‐ and fewer‐silique lines.
The silique number variation is caused by both the flower number and silique setting rate
In order to clarify the relative contribution of the two factors to silique number variation in rapeseed, the flower number and silique setting rate (=silique number/flower number) were investigated and compared for the two types of extreme lines. As expected, both flower number and silique setting rate were highly correlated with silique number with the Pearson correlation coefficients of 0.84** and 0.71**, respectively (Figure 4).
Figure 4.
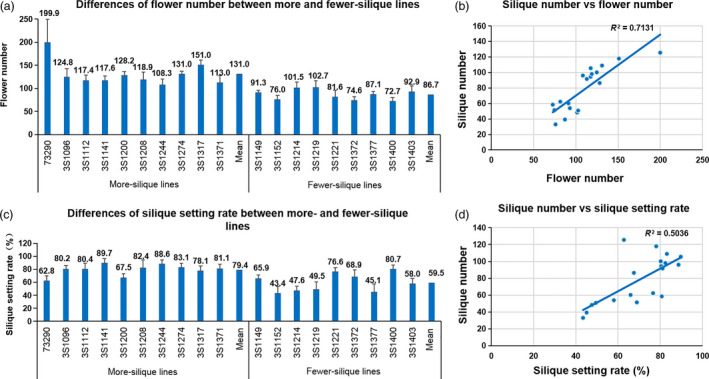
The silique number variation was caused by both flower number and silique setting rate. (a) Differences of flower number between more‐ and fewer‐silique lines. (b) Correlation analysis between silique number and flower number. (c) Differences of silique setting rate between more‐ and fewer‐silique lines. (d) Correlation analysis between silique number and silique setting rate.
The flower number of more‐ and fewer‐silique lines varied from 108.3 to 199.9 and from 74.6 to 102.7, respectively, and the mean (131.0) of the former was significantly higher than that (86.7) of the latter (P < 0.01) (Figure 4). The silique setting rate of more‐silique lines and fewer‐silique lines varied from 62.8% to 89.7% and from 43.4% to 80.7%, respectively, and the mean (79.4%) of the former was also significantly higher than that (59.5%) of the latter (P < 0.01) (Figure 4). The more‐silique lines were 51.1% and 33.4% more than fewer‐silique lines in flower number and silique setting rate, respectively. Furthermore, statistical analysis showed that the silique number difference between the more‐ and fewer‐lines was attributable to the accumulative differences in flower number and silique setting rate, which accounted for 75.2% and 24.8%, respectively.
The silique number is highly associated with total photosynthesis
The source of energy and organic matter required in a series of plant growth and development activities (including flower bud differentiation) was derived from photosynthesis. During the IM differentiation and flowering period, the leaf is the most important photosynthesis organ (Diepenbrock, 2000; Faraji, 2010). To assess the role of leaf photosynthesis in the formation of the silique number difference between the two types of extreme lines, we measured the photosynthetic rate and leaf area at the initial flowering, the time before which the plant grew rapidly, and after which the IM‐differentiated floral primordium could not form effective flower/silique (Mendham et al., 1981; Zhang et al., 2016). The results showed that the leaf photosynthetic rates of the more‐ and fewer‐silique lines ranged from 19.0 to 22.6 and 18.3 to 21.3 μmol/m2/s, respectively, and the mean of two types (20.3 and 19.7 μmol/m2/s) was similar (P < 0.01) (Figure 5). However, the total leaf area of more‐silique lines (1671–4498 cm2) was significantly larger than that (358–1792 cm2) of the fewer‐silique lines, the mean of the former (3001 cm2) was 125.8% larger than that (1329 cm2) of the latter (P < 0.01**), and resulting total leaf photosynthesis of more‐silique lines was 132.7% larger than fewer‐silique lines (P < 0.01**) (Figure 5). These results strongly implied the important role of the total photosynthesis in the determination of silique number.
Figure 5.
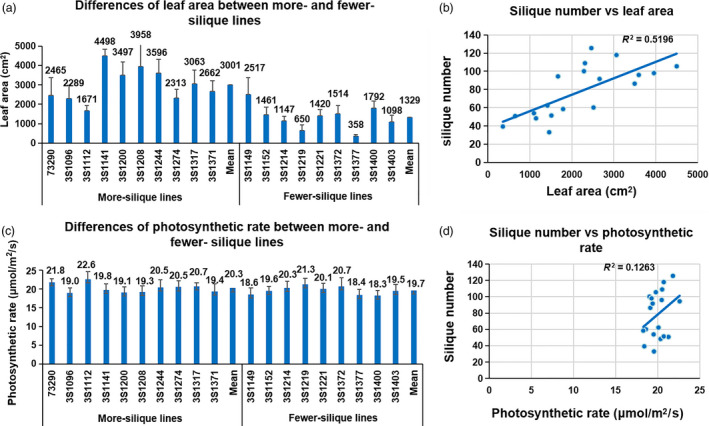
The silique number was highly associated with total photosynthesis. (a) Differences of leaf area between more‐ and fewer‐silique lines. (b) Correlation analysis between silique number and leaf area. (c) Differences of photosynthetic rate between more‐ and fewer‐silique lines. (d) Correlation analysis between silique number and photosynthetic rate.
The silique number is highly associated with biomass
More total photosynthesis led to more assimilates which promoted biomass accumulation and flower/silique formation. To verify this assumption, the biomass was measured at initial flowering. The aboveground, underground and total biomass of more‐silique lines varied from 273.8 g to 534.5 g, from 22.1 g to 60.0 g and from 298.4 g to 581.6 g, respectively (Figure 6). The aboveground, underground and total biomass of fewer‐silique lines varied from 105.6 g to 290.1 g, from 16.0 g to 37.5 g and from 129.4 g to 324.9 g, respectively (Figure 6). Obviously, the average aboveground, underground and total biomass (444.3 g, 40.3 g and 484.6 g) of more‐silique lines were all significantly higher than those (200.0 g, 27.0 g and 227.0 g) of the fewer‐silique lines, with a proportion of 122.2% and 49.3% and 113.5%, respectively (Figure 6). The biomass and silique number were highly correlated, with the Pearson correlation coefficients of 0.88** (aboveground), 0.61** (underground) and 0.88** (total), respectively (Figure 6). The results strongly implied that more accumulation of biomass (from leaf photosynthesis) promoted the formation of silique in rapeseed.
Figure 6.
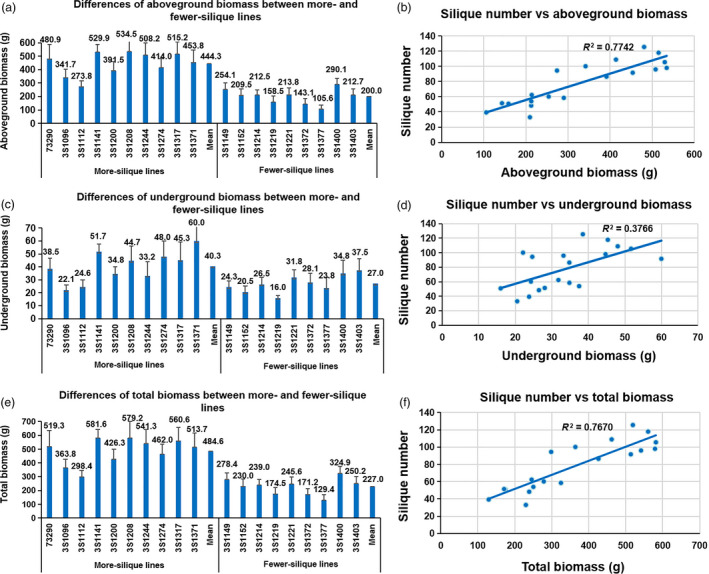
The silique number was highly associated with the biomass. (a) Differences of aboveground biomass between more‐ and fewer‐silique lines. (b) Correlation analysis between silique number and aboveground biomass. (c) Differences of underground biomass between more‐ and fewer‐silique lines. (d) Correlation analysis between silique number and underground biomass. (e) Differences of total biomass between more‐ and fewer‐silique lines. (f) Correlation analysis between silique number and total biomass.
The comparative analysis of inflorescence meristem (IM) differentiation between extreme lines of more‐ and fewer‐siliques
The flower organ is derived from inflorescence meristem (IM) differentiation, and factors that affect IM differentiation should be directly correlated with the silique number (Zhang et al., 2016). To assess the differences in process of IM differentiation between more‐ and fewer‐silique lines, three more‐ and three fewer‐silique lines were chosen for continuously microscopic observation. As illustrated in Figure 7, the process of IM differentiation was divided into five main stages. First, the beginning of IM differentiation, the IM became much larger and rounder, flower primordium formed on the periphery of IM. Second, the seventh day after initial IM differentiation, more new primordia were produced, the outermost flower primordium began to elongate, and the sepal began to differentiate. The third stage, the size of the outermost flower bud reached 0.5mm. The fourth stage, the size of the outermost flower bud reached 1mm. The fifth stage, the size of the outermost flower bud reached 2 mm.
Figure 7.
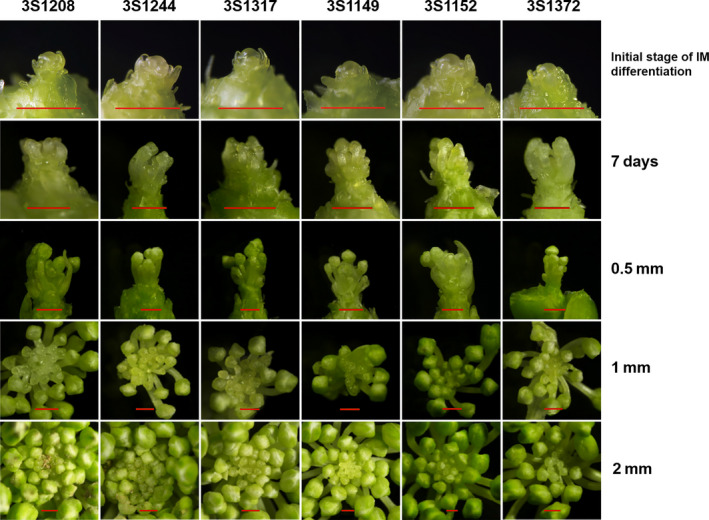
The contrast between more‐ and fewer‐silique lines in process of IM differentiation (Bars equal 1 mm). The more‐silique lines: 3S1208, 3S1244 and 3S1317; the fewer‐silique lines: 3S1149, 3S1152 and 3S1372.
At the beginning of flower bud differentiation, the morphology of IM between extremely more‐ and fewer‐silique lines was similar, with the average diameter of IM 0.31 and 0.33 mm, respectively. At the second stage, obvious differences still could not be found (Figure 7). As the IM continued to differentiate, the accumulative quantity difference of flower bud began to emerge. When the size of outermost flower bud reached 0.5 mm, the number of the more‐silique lines was 11.5% more than that of fewer‐silique lines, but then the difference did not reach the significant level (P > 0.05) (Figure S3). At the fourth and fifth stage, the flower bud number of the more‐silique lines was 26.8% and 57.6% more than that of fewer‐silique lines (P < 0.01**), with average from 30.7 to 52.3 and from 24.2 to 33.2, respectively (Figure S3). In summary, the difference between extremely more‐ and fewer‐silique lines normally occurred at the amount of flower bud but not morphology, and the accumulative quantity difference of flower bud usually emerged at the middle and late period of IM differentiation.
Comparative transcriptome analysis between more‐ and fewer‐silique pools
To dissect the possible molecular mechanism of flower number difference between more‐ and fewer‐silique lines at transcriptome level, transcriptome sequencing was performed using the shoot apical meristem (SAM) of two contrasting pools. Three biological replicates were included, and a high consistency between each other implied the accuracy of transcriptome sequencing (Figure S4). To validate the RNA‐seq results, qRT‐PCR was carried out. The trend of RT‐PCR based expression patterns among these selected genes was generally consistent with those detected by RNA‐seq based method (Figure S5).
A total of 2746 (1422 up‐ and 1324 down‐regulated) differentially expressed genes (DEGs) were identified, and then they were used for the GO enrichment analysis using TBtools software (Chen et al., 2018; Shi et al., 2019).
Among the 23 most enriched classes (corrected P‐value<1.0E‐5), six were auxin‐related biosynthesis/metabolism biological processes, such as indoleacetic acid biosynthetic/metabolic process and auxin biosynthetic/metabolic process (Figure 8), indicating the vital role of auxin in IM differentiation process. It should be noted that, besides the auxin, a considerable proportion of other enriched classes were closely related to the vegetative growth by promoting nutrition synthesis, photoperiodism/circadian rhythm regulation and improving plant comprehensive resistance. They were long‐day photoperiodism‐flowering, long‐day photoperiodism, rhythmic process, circadian rhythm, sulphur compound biosynthetic process, response to stress and response to abiotic stimulus (Figure 8). These results provided further insights into the molecular mechanism responsible for silique number variation in rapeseed: the good vegetative growth could improve the differentiation ability of apical meristem which decided silique number to a large extent.
Figure 8.
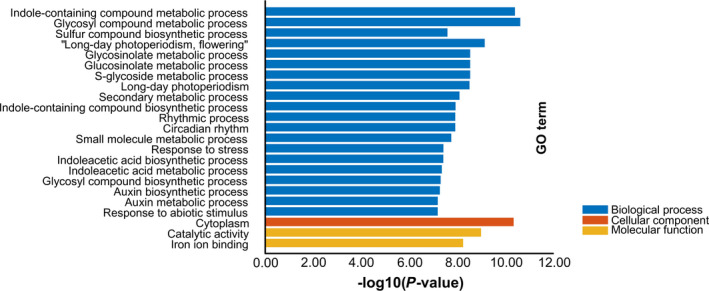
The Gene Ontology (GO) enrichment analysis of differentially expression genes. The top 23 enriched GO terms (P value < 1.0E‐5) were used for diagram.
Identification of candidates for silique number by integrating genome‐wide association and transcriptome analysis
A total of 142 DEGs were located in the regions of the 27 loci, of which, 101 DEGs (71.1%) showed significant homology with Arabidopsis genes and had the functional annotation. Referring to the Arabidopsis homologous annotation, six DEGs underlying significant loci were identified as candidate genes, they were BnaA02g08800D, BnaA02g08900D, BnaA05g01050D, BnaA09g05170D, BnaA10g00780D and BnaA10g14470D, distributing on A02, A05, A09 and A10 linkage. The six DEGs were known to regulate flower number by affecting IM meristem activities, floral organ development, leaf enlarge or biomass accumulation. Our study also indicated that integrating analysis of GWAS and RNA‐seq could be an efficient method to identify candidate genes within the silique number loci.
Discussion
Novel loci identified for silique number
The silique number is crucial for seed yield which is an important breeding goal (Ali et al., 2003; Tokatlidis, 2017). Obtaining the loci of related traits, combined with molecular marker‐assisted breeding, is an effective way to increase the silique of rapeseed variety (Kamaluddin et al., 2017). Numerous loci of silique number in Brassica napus L. have been identified by QTL mapping and GWAS (Lu et al., 2017; Shi et al., 2015; Ye et al., 2017). Ye et al. (2017) identified eight QTLs for silique number using BnaZNRIL population, explaining 5.8%–11.9% of phenotypic variance. Lu et al. (2017) identified 11 loci for silique number by GWAS using data of 520 diverse Brassica napus L. accessions from two different environments. Most of loci for silique number showed a moderate effect and could not be detected repeatedly. In the present study, 20 novel loci for silique number were identified. In addition, based on the physical map of rapeseed, all of the currently and previously reported silique number QTLs in rapeseed were integrated, and more than 120 loci were successfully mapped in the physical map which represented the most comprehensive genetic architecture of silique number in rapeseed until now (Table S4). This study established the basis for gene cloning of silique number in Brassica napus L. and marker‐assisted selection of this trait in breeding programme.
The physiological basis underlying difference of silique number between extremely more‐ and fewer‐silique lines
As we know, the silique number is determined by flower number and silique setting rate together, but the relative importance of the two factors has not been analysed in past. In this study, we proved the silique number difference between the more‐ and fewer‐silique lines was attributable to the accumulative difference in its components, of which, the flower number and the silique setting rate accounted for 75.2% and 24.8%, respectively.
During flower bud differentiation, a considerable level of nutrients would be absorbed from other tissues (Zhang et al., 2016). The source of energy and organic matter required in flower bud differentiation was derived from photosynthesis of leaf (Diepenbrock, 2000; Faraji, 2010). The decrease in the production of carbon assimilates by shading or leaf removal led to fewer flowers and siliques (Clarke, 1978). Nitrogen generally stimulates plant growth by means of an enlarged leaf canopy and a greater rate of leaf expansion, which may result in more siliques (Diepenbrock, 2000). It was repeatedly reported that the silique number of each plant was negatively correlated with planting density (Chay and Thurling, 1989; Leach et al., 1999). The explanation to this phenomenon is that, with planting density increasing, the leaf area of each plant is reduced, and the leaves are overlapped, which results in the decrease of photosynthetic area and photosynthetic product. In the present study, we found that though the photosynthetic rates were similar between more‐ and fewer‐silique lines, the mean leaf area of the former was 125.8% larger than that of the latter (P < 0.01**), and the calcualted total leaf photosynthesis of more‐silique lines was 132.7% larger than that of fewer‐silique lines (P < 0.01**). It explained the physiological cause of silique number variation in natural germplasm to a large extent. For breeding purposes, physiologically based selection criteria about silique number should include light absorption, the leaf area index which increase the total photosynthesis.
The silique setting rate of more‐ and fewer‐silique lines varied from 62.8% to 89.7% and from 43.4% to 80.7%. Those flowers could not develop to silique, which represents a considerable waste of resources. Therefore, improving the silique setting rate is very meaningful with respect to nutrient utilization and silique formation. The silique setting rate is influenced by a series of biological processes such as flower fertilization and silique development. Some hypotheses had been proposed for flower degeneration and drop, such as nutrient supply, fertilization and phytohormone modulation (Boldingh et al., 2016; Monerri et al., 2011; Zhang et al., 2016). Regarding nutrient supply hypothesis, flower degeneration and drop occurred because of the nutrition competition among the flowers of different position. In the present study, the flower number and silique setting rate of 73 290 were the largest (199.9) and lowest (62.8%) in more‐silique lines, respectively, which was consistent with nutrient supply hypothesis well. Fertilization and phytohormone modulation also play an important role in flower degeneration and drop. Gamer and Lovatt reported that the majority of flowers drop was due to a lack of pollen germination and subsequent fertilization, and ABA accumulation was related to ovule/seed abortion and abscising fruit in avocado (Gamer and Lovatt, 2016). Poze et al. reported that the proximal abscission zones of abscising fruits had higher content of ABA and ABA‐like substances, and the cultivars with stronger early fruit abscission had higher content of jasmonic acid‐like compounds in floral organs and developing fruits, which suggested that ABA‐like and jasmonic acid‐like substances could play a shared role in fruit set and early fruit abscission (Pozo, 2001). Furthermore, bad weather condition after flowering, such as continuously rainy weather, the high temperature rapidly rising, all of these speed up the degeneration of developing flowers (Selak et al., 2014).
Microscopic analysis
Microscopic analysis showed that the difference among extremely lines normally occurred at the amount of flower bud but not morphology, and the accumulative quantity difference emerged at the middle and late period of flower bud differentiation. These results showed that the general cause of flower number difference in natural rapeseed germplasm was not specific morphology of the SAM (larger or smaller), as some previous reported mutants, such as wus, clv3 mutant in Arabidopsis (Brand et al., 2000; Schoof et al., 2000) or fon1, fon4 mutant in rice (Chu et al., 2006; Moon et al., 2006; Suzaki et al., 2006), but the ability of the apical meristem continuous differentiation. Furthermore, the efficient supply of energy and nutrient to SAM may effect on this ability.
Characteristics of SAM DEGs
As a critical plant hormone, auxin influences many aspects of plant growth and development including IM differentiation (Woodward and Bartel, 2005). Shen et al. (2017) found that the down‐regulation of the genes involved in the IAA biosynthesis pathway led to lower concentration of IAA in SAM and more flowers/siliques in Brassica napus L. This predominant role of auxin in IM differentiation activities relies on its critical involvement in the regulation and coordination of cell division and differentiation (Tromas et al., 2013). In our study, of the 23 most enriched classes, six were auxin‐related biosynthesis/metabolism biological processes. The result indicated that the auxin‐related biological processes could affect flower/silique number to a certain extent by regulating inflorescence meristem activities.
Besides auxin, a considerable proportion of enriched classes could affect flower/silique number by regulating vegetative growth. Among above enriched classes, four classes were related to circadian clock and photoperiodism. Circadian clock produced an internal estimate of time that synchronized biological events with external day–night cycles (Dodd et al., 2005). The correctly matched circadian clock conferred a competitive advantage and allowed plants to increase photosynthesis and biomass (Dodd et al., 2005). Ni et al. (2009) also found that epigenetic modification of the circadian oscillators was associated with growth vigour in Arabidopsis thaliana hybrids. Sulphur compound biosynthesis enriched class was also closely related to nutrition accumulation. Sulphur provides an essential nutrient for the synthesis of many metabolites (Herrmann et al., 2014; Kopriva et al., 2012; Krishnan, 2005; Ravilious and Jez, 2012; Takahashi et al., 2011), which provide the basis of energy and nutrition for IM differentiation. In this enriched class, some of DEGs were homologous to APK in Arabidopsis thaliana, the mutant of which had the smaller leaves, the lower rate of vegetative development compared with wild type (Mugford et al., 2009). The response to stress and abiotic stimulus enriched classes was closely related to the plant resistance. Improvement of plant comprehensive resistance helps plants adapt to various environments and maintain the good growth.
Comparative transcriptomic analysis proved the differences of silique number between more‐ and fewer‐silique lines were mainly due to biomass/nutrition accumulation and auxin‐related regulation at the molecular level.
Identification of candidate genes
Given that it is difficult to identify silique number genes by map‐based cloning approach as the trait is sensitive to the environment and the identified loci are almost all with moderate effetcs. Therefore, integration of GWAS and RNA‐seq was used for identification of candidate genes. A total of six DEGs were identified as the candidate genes of silique number.
During flower bud differentiation, phytohormones play an important role (Ye et al., 2017). Of the above‐mentioned six candidates, BnaA10g14470D is the orthologue of EIL (Arabidopsis), which was shown to correlate with ethylene insensitivity in flowers, and affect flower organ development. EIN3‐ or EIL1‐overexpressing Arabidopsis plants showed greatly reduced fertility, and the gynoecium protruded from a flower much smaller than wild type (Chao et al., 1997). Ethylene can delay flowering by regulating EIN3/EIL1 to inhibit the accumulation of gibberellin (Achard et al., 2007). Liu et al. (2008) found the sex‐determining gene was co‐separated from cucumber CsEIL1 and inhibited stamen development, which suggested that EIN3/EILs may play a role in ethylene‐promoting female flower formation in Cucurbitaceae (Byers et al., 1972). However, the role of EIN3/EILs in the flower organ forming and development of B. napus may need to be further investigated.
Transcription factors also play a key role in the shoot apical and floral meristem activities (Ye et al., 2017). BnaA10g00780D is homologous to LHY (AT1G01060), which is a transcription factor involved in circadian rhythm, playing an essential role in flower bud differentiation initiation, flowering time and vegetative growth. LHY, TOC1 and CCA1 formed the negative feedback loop of the circadian clock (McClung et al., 2006). The circadian clock proteins LHY repressed the floral transition under short‐day and long‐day conditions (Fujiwara et al., 2008). Overexpression of LHY led to hypocotyl growth and late flowering. Adams et al. (2018) found LHY not only promoted the expression of ABA‐responsive genes responsible for increased tolerance to drought and osmotic stress but alleviated the inhibitory effect of ABA on plant growth (Adams et al., 2018). In our study, the expression level of BnaA10g00780D in more‐lines was higher than in fewer‐lines, which suggested that BnaA10g00780D may positively regulate the flower number.
Some genes may regulate flower number by affecting biomass accumulation, floral organ development and the transition from vegetative growth to flowering. For example, BnaA02g08900D is homologous to BAM4 (AT5G55700), which may play a regulatory role in the starch breakdown (Li et al., 2009). The previous study showed that BAM family genes apparently interacted genetically in leaf starch metabolism (Francisco et al., 2010), which was closed with biomass accumulation. BnaA09g05170D is homologous to RUP2 (AT5G23730). Under noninductive short‐day conditions, RUP2 inhibits the UVR8‐mediated induction of flowering and thus provides a key mechanism of photoperiodic flowering control. (Arongaus et al., 2018). BnaA02g08800D is homologous to HAP8 (AT5G56250), which regulates the male gametophyte, the female gametophyte and short pollen tube growth, during flower organ development (Johnson et al., 2004). BnaA05g01050D is homologous to CCA1 (AT2G46830) which encodes a transcriptional repressor performing overlapping functions with LHY in a regulatory feedback loop that is closely associated with the circadian oscillator, flower bud differentiation and flowering in Arabidopsis (Fujiwara et al., 2008; McClung et al., 2006).
To follow up on the genes identified in this study, we plan to clone the six candidate genes in order to confirm their roles in silique number difference generating in Brassica napus L.
Conclusion
Our study provided a comprehensive and multilevel explanation for the natural variation of silique number in rapeseed. In this context, we conducted systematic research (encompassing GWAS and gene expression analysis, microscopic analysis, physiological experiments) to demonstrate the natural variation of silique number is largely affected by the biomass and nutrition accumulation in rapeseed. Therefore, improving the ability of biomass and nutrient accumulation is beneficial to increase the silique number in rapeseed. Our study provides new ideas for the genetic improvement of silique number in rapeseed.
Material and methods
Research materials and field experiments
The association mapping population used in this study consisted of 331 diverse rapeseed accessions. The association mapping population was grown over the course of 2 years (2014, 2016) in Wuhan (Hubei province, China) and 1 year (2014) in Nanchang (Jiangxi province, China) followed a randomized complete block design with two replications. Each plot contained three rows of 2‐m length and 33‐cm spacing, with 15 plants in each row. The field management followed standard agriculture practice. At maturity, ten representative plants from each plot were harvested. The effective silique number was investigated according to Shi et al. (2015). Three traits of silique number were measured: SNm, SNb and SNw. Statistical analysis of phenotypic data was performed in SPSS v22 (IBM SPSS, Armonk, NY, USA). The best linear unbiased prediction (BLUP) for the traits was calculated by the R package lme4 (Merk et al., 2012). Moreover, the extremely more‐ and fewer‐silique lines selected from the association population were planted twenty rows with two random replications in Wuhan (Oct 2016 to May 2017) and Xining (May to Sep 2017) for further physiological research.
Genome‐wide association study (GWAS)
The Brassica 60K Illumina R Infinium SNP array was used to evaluate the genotypes of the 331 rapeseed accessions. The association analysis was conducted in TASSEL v.5.2.41 (Bradbury et al., 2007) using the general linear model (GLM). The population structure and relative kinship study of this association population had been previously published (Zheng et al., 2017). Especially, the linkage disequilibrium (LD) was measured by calculating the squared correlation coefficient (r 2) between pairs of SNPs on each chromosome. The threshold for significantly associated SNP markers was set to P < 4.08 × 10−5 (P = 1/24508, −log10 P = 4.08), in accordance with the published literature (Wei et al., 2016).
Measurement of flower number, biomass, leaf area and photosynthetic rate
Ten plants in each plot were sampled to measure the flower number and silique number. At the beginning of flowering, the plants of each plot with uniform growth were sampled by the excavation method and the roots of sampled plants were rinsed with water to remove the soil. Then, the plants were brought to the laboratory for measuring fresh aboveground biomass, fresh underground biomass and leaf area. The method for biomass and leaf area measurement was described in Hu et al. (2017). The photosynthetic parameter was measured in the field using a portable photosynthesis system (LI‐6800XT, LI‐COR). The measurement was always performed between 9:30–11:00 AM and 14:30–16:00 PM. The measurement condition was consistent with the previous report (Li et al., 2019).
Microscopic analysis
To study the differences between extremely lines at the microscopic level, three more‐silique and three fewer‐silique lines were chosen for microscopic analysis during the period of flower bud differentiation. From the early stage of flower bud differentiation, ten plants from each line were continuously sampled for observation of flower bud differentiation. All leaves of the sampled plants were removed until they were difficult to distinguish with the naked eye (Zhang et al., 2016). The young leaves wrapped around the IM were carefully peeled off with an anatomic needle under the perspective of a stereomicroscope (NikonSMZ25). The morphology of IM and the process of flower bud differentiation were photographed using camera that accompanied the stereomicroscope.
RNA sequencing and transcriptome analysis
Two pools (each consisting of 10 and 9 accessions, respectively, with extremely more siliques or fewer siliques) were analysed at the transcriptomic level using HiSeq/Illumina sequencing. To gain credible data, two pools’ RNA was isolated from SAM at beginning of reproductive bud swell (code 511) according to Biologische Bundesanstalt, Bundessortenamt and Chemical Industrie (BBCH) scale and then equally mixed for more‐ and fewer‐silique pools, respectively. Methods for RNA extraction, purification and quantification were as described in Ye et al. (2017). After the final cDNA library was synthesized, the libraries were sequenced and the raw reads were generated. The clean reads which passed quality control were used for subsequent analysis. All reads of each library were mapped to the reference genome (Chalhoub et al., 2014), using software Hisat2 (https://ccb.jhu.edu/software/hisat2/index.shtml), and the uniquely mapped reads were selected for transcript quantification. The gene expression level was measured by FPKM value using software RSEM (http://www.biomedsearch.com/nih/RSEM-accurate-transcript-quantification-from/21816040.html). In this study, FDR ≤ 0.05 and the absolute value of log2FC ≥ 1 were used to characterize DEGs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interesting.
Author contributions
HW and JS designed the experiments. GL and XW provided the research materials. R K Varshney revised the manuscript. SL sampled the SAM for RNA‐seq. JS, YZ, JZ and XZ collected the phenotypic data. SL and JS analysed the data. SL, JS and HW wrote the manuscript.
Funding
This research was supported by the National Key Research and Development Program (2016YFD0100305), the National Key Basic Research Development Program of China (2015CB150203), the Natural Science Foundation of Hubei Province (2018CFA075), the Natural Science Foundation (31101181), the Rapeseed Industry Technology System (CARS‐13), the Agricultural Science and Technology Innovation Project (CAAS‐ASTIP‐2013‐OCRI), the Core Research Budget of the Non‐profit Governmental Research Institution (1610172017001).
Supporting information
Figure S1 The QQ plots resulting from the GWAS for silique number from the main inflorescence (a), branch inflorescence (b), and whole plant (c).
Figure S2 Correlation analysis between main inflorescence and individual branch. (a) Silique number. (b) Flower number.
Figure S3 The comparative analysis of the flora organ number in process of IM differentiation between more ‐ and fewer‐silique lines.
Figure S4 The consistency between different sample of transcriptome sequencing.
Figure S5 Comparison of the relative expression abundance measured by qRT‐PCR and RNA‐seq.
Table S1 ANOVA analysis of silique number in different environments.
Table S2 Phenotypic correlations between the silique number in the three environments.
Table S3 The significant SNPs identified from GWAS.
Table S4 List of loci identified in our and previous studies for silique number in rapeseed.
Contributor Information
Jiaqin Shi, Email: shijiaqin@caas.cn.
Hanzhong Wang, Email: wanghz@oilcrops.cn.
References
- Achard, P. , Baghour, M. , Chapple, A. , Hedden, P. , Straeten, D. , Genschik, P. , Moritz, T. et al. (2007) The plant stress hormone ethylene controls floral transition via DELLA‐dependent regulation of floral meristem‐identity genes. Proc. Natl. Acad. Sci. USA, 104, 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, S. , Grundy, J. , Veflingstad, S.R. , Dyer, N.P. , Hannah, M.A. , Ott, S. and Carre, I.A. (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome‐wide analysis of LHY binding targets. New Phytol. 220, 893–907. [DOI] [PubMed] [Google Scholar]
- Ali, N. , Javidfar, F. , Elmira, J.Y. and Mirza, M.Y. (2003) Relationship among yield components and selection criteria for yield improvement in winter rapeseed (Brassica napus L.). Pak. J. Bot. 35, 167–174. [Google Scholar]
- Arongaus, A.B. , Chen, S. , Pireyre, M. , Glöckner, N. , Galvão, V.C. , Albert, A. , Winkler, B. et al. (2018) Arabidopsis RUP2 represses UVR8 mediated flowering in noninductive photoperiods. Gene Dev. 32, 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina, I. , Otto, E. , Strnad, M. , Werner, T. and Schmülling, T. (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana . Plant Cell, 23, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldingh, H.L. , Alcaraz, M.L. , Thorp, T.G. , Minchin, P.E.H. , Gould, N. and Hormaza, J.I. (2016) Carbohydrate and boron content of styles of ‘Hass’ avocado (Persea americana Mill.) flowers at anthesis can affect final fruit set. Sci. Hortic. 198, 125–131. [Google Scholar]
- Bortiri, E. and Hake, S. (2007) Flowering and determinacy in maize. J. Exp. Bot. 58, 909–916. [DOI] [PubMed] [Google Scholar]
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) Tassel: software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Brand, U. , Fletcher, J.C. , Hobe, M. , Meyerowitz, E.M. and Simon, R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science, 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Byers, R.E. , Baker, L.R. , Sell, H.M. , Herner, R.C. and Dilley, D.R. (1972) Ethylene: a natural regulator of sex expression of Cucumis melo L. Proc. Natl Acad. Sci. USA, 69, 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S.Y. , Parkin, I.A. , Tang, H.B. , Wang, X.Y. , Chiquet, J. et al. (2014) Early allopolyploid evolution in the post Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chao, Q. , Rothenberg, M. , Solano, R. , Roman, G. , Terzaghi, W. and Ecker, J.R. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ethylene insensitive3 and related proteins. Cell, 59, 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chay, P. and Thurling, N. (1989) Variation in pod length in spring rape (Brassica napus) and its effect on seed yield and yield components. J. Agric. Sci. Camb. 113, 139–147. [Google Scholar]
- Chen, X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Wan, H. , Qian, J. , Guo, J. , Sun, C. , Wen, J. , Yi, B. et al. (2018) Genome wide association study of cadmium accumulation at the seedling stage in rapeseed (Brassica napus L.). Front. Plant Sci. 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Qian, Q. , Liang, W. , Yin, C. , Tan, H. , Yao, X. , Yan, Z. et al. (2006) The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.K. (1978) The effects of leaf removal on yield and yield components of Brassica napus . Can. J. Plant Sci. 58, 1103–1105. [Google Scholar]
- Diepenbrock, W. (2000) Yield analysis of winter oilseed rape (Brassica napus L.): a review. Field. Crop. Res. 67, 35–49. [Google Scholar]
- Ding, G. , Zhao, Z. , Liao, Y. , Hu, Y. , Shi, L. , Long, Y. and Xu, F. (2012) Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus . Ann. Bot. 109, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N. , Salathia, N. , Hall, A. , Kevei, E. , Toth, R. , Nagy, F. , Hibberd, J.M. et al. (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science, 309, 630–633. [DOI] [PubMed] [Google Scholar]
- Faraji, A. (2010) Flower formation and pod/flower ratio in canola (Brassica napus L.) affected by assimilates supply around flowering. Int. J. Plant Prod. 4, 271–280. [Google Scholar]
- Francisco, P. , Li, J. and Smith, S.M. (2010) The gene encoding the catalytically inactive β amylase BAM4 involved in starch breakdown in Arabidopsis leaves is expressed preferentially in vascular tissues in source and sink organs. J. Plant Physiol. 167, 890–895. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S. , Oda, A. , Yoshida, R. , Niinuma, K. , Miyata, K. , Tomozoe, Y. , Tajima, T. et al. (2008) Circadian clock proteins LHY and CCA1 regulate SVP Protein accumulation to control flowering in Arabidopsis . Plant Cell, 20, 2960–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamer, L.C. and Lovatt, C.J. (2016) Physiological factors affecting flower and fruit abscission of ‘Hass’ avocado. Sci. Hortic. 199, 32–40. [Google Scholar]
- Herrmann, J. , Ravilious, G.E. , McKinney, S.E. , Westfall, C.S. , Lee, S.G. , Baraniecka, P. , Giovannetti, M. et al. (2014) Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation. J. Biol. Chem. 289, 10919–10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M. , Li, X. , Wang, Z. , You, Q. and Lu, J. (2017) Effects of sowing date on biomass and nutrient accumulation of oilseed rape as green manure. Hubei Agr. Sci. 55, 657–660. [Google Scholar]
- Johnson, M.A. , Besser, K.V. , Zhou, Q. , Smith, E. , Aux, G. , Patton, D. , Levin, J.Z. et al. (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics, 168, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaluddin, M.A. , Kiran, U. , Ali, A. , Abdin, M.Z. , Zargar, M.Y. , Ahmad, S. , Sofi, P.A. et al. (2017) Molecular Markers and Marker Assisted Selection in Crop Plants. In: Plant Biotechnology: Principles and Applications, pp. 295–328. Singapore: Springer. [Google Scholar]
- Kinoshita, A. , Betsuyaku, S. , Osakabe, Y. , Mizuno, S. , Nagawa, S. , Stahl, Y. , Simon, R. et al. (2010) RPK2 is an essential receptor like kinase that transmits the CLV3 signal in Arabidopsis . Development, 137, 3911–3920. [DOI] [PubMed] [Google Scholar]
- Kopriva, S. , Mugford, S.G. , Baraniecka, P. , Lee, B.R. , Matthewman, C.A. and Koprivova, A. (2012) Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis . Front. Plant Sci. 3, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, U. , Lippman, Z.B. and Zamir, D. (2010) The flowering gene single flower truss drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. [DOI] [PubMed] [Google Scholar]
- Krishnan, H.B. (2005) Engineering soybean for enhanced sulfur amino acid content. Crop Sci. 45, 454–461. [Google Scholar]
- Leach, J.E. , Stevenson, H.J. , Rainbow, A.J. and Mullen, L.A. (1999) Effects of high plant populations on the growth and yield of winter oilseed rape (Brassica napus). J. Agric. Sci. Camb. 132, 173–180. [Google Scholar]
- Li, J. , Francisco, P. , Zhou, W. , Edner, C. , Steup, M. , Ritte, G. , Bond, C.S. et al. (2009) Catalytically inactive β amylase BAM4 required for starch breakdown in Arabidopsis leaves is a starch binding protein. Arch. Biochem. Biophys. 489, 92–98. [DOI] [PubMed] [Google Scholar]
- Li, N. , Song, D. , Peng, W. , Zhan, J. , Shi, J. , Wang, X. , Liu, G. et al. (2019) Maternal control of seed weight in rapeseed (Brassica napus L.): the causal link between the size of pod (mother, source) and seed (offspring, sink). Plant Biotechnol. J. 17, 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Xu, L. , Jia, Z. , Xu, Y. , Yang, Q. , Fei, Z. , Lu, X. et al. (2008) Genetic association of ETHYLENE INSENSITIVE3‐like sequence with the sex determining M locus in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 117, 927–933. [DOI] [PubMed] [Google Scholar]
- Lu, K. , Peng, L. , Zhang, C. , Lu, J. , Yang, B. , Xiao, Z. , Liang, Y. et al. (2017) Genome wide association and transcriptome analyses reveal candidate genes underlying yield determining traits in Brassica napus . Front. Plant Sci. 8, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, T. , Zhang, J. , Khan, M.N. , Liu, J. , Xu, Z. and Hu, L. (2018) Temperature variation caused by sowing dates significantly affects floral initiation and floral bud differentiation processes in rapeseed (Brassica napus L.). Plant Sci. 271, 40–51. [DOI] [PubMed] [Google Scholar]
- McClung, C.R. (2006) Plant circadian rhythms. Plant Cell, 18, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendham, N.J. , Shipway, P.A. and Scottt, R.K. (1981) The effects of delayed sowing and weather on growth, development and yield of winter oil seed rape (Brassica napus). J. Agr. Sci. 96, 389–416. [Google Scholar]
- Merk, H.L. , Yarnes, S.C. , Van Deynze, A. , Tong, N.K. , Menda, N. , Mueller, L.A. , Mutschler, M.A. et al. (2012) Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm. J. Am. Soc. Hortic. Sci. 137, 427–437. [Google Scholar]
- Monerri, C. , Fortunato Almeida, A. , Molina, R.V. , Nebauer, S.G. , Garcia Luis, A. and Guardiola, J.L. (2011) Relation of carbohydrate and reserves with the forthcoming crop, flower formation and photosynthetic rate, in the alternate bearing ‘Salustiana’ sweet orange (Citrus sinensis L.). Sci. Hortic. 129, 71–78. [Google Scholar]
- Moon, S. , Jung, K. , Lee, D. , Lee, D. , Lee, J. , An, K. , Kang, H. et al. (2006) The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size. Mol. Cells, 21, 147–152. [PubMed] [Google Scholar]
- Mu, R. and Bleckmann, A. (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell‐limiting signal CLAVATA3 independently of CLAVATA1. Plant cell, 20, 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, R. , Borghi, L. , Kwiatkowska, D. and Laufs, P. (2006) Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 Signaling. Plant cell, 18, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford, S.G. , Yoshimoto, N. , Reichelt, M. , Wirtz, M. , Hill, L. , Mugford, S.T. , Nakazato, Y. et al. (2009) Disruption of adenosine 5'phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell, 21, 910–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z. , Kim, E.D. , Ha, M. , Lackey, E. , Liu, J. , Zhang, Y., Sun, Q. et al. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature, 457, 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo, L.V. (2001) Endogenous hormonal status in citrus flowers and fruitlets: relationship with postbloom fruit drop. Sci. Hortic. 91, 251–260. [Google Scholar]
- Radoev, M. , Becker, H.C. and Ecke, W. (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravilious, G.E. and Jez, J.M. (2012) Structural biology of plant sulfur metabolism: from assimilation to biosynthesis. Nat. Prod. Rep. 29,1138–1152. [DOI] [PubMed] [Google Scholar]
- Sarkar, A.K. , Luijten, M. , Miyashima, S. , Lenhard, M. , Hashimoto, T. , Nakajima, K. , Scheres, B. et al. (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature, 446, 811–814. [DOI] [PubMed] [Google Scholar]
- Schoof, H. , Lenhard, M. , Haecker, A. , Mayer, K.F.X. , Jürgens, G. and Laux, T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell, 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Selak, G.V. , Cuevas, J. , Ban, S.G. , Pinillos, V. , Dumicic, G. and Perica, S. (2014) The effect of temperature on the duration of the effective pollination period in ‘Oblica’ olive (Olea europaea) cultivar. Ann. Appl. Biol. 164, 85–94. [Google Scholar]
- Shen, Y. , Sun, S. , Hua, S. , Shen, E. , Ye, C.Y. , Cai, D. , Timko, M.P. et al. (2017) Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid brassica napus uncovers key roles for small RNAs. Plant J. 91, 874–893. [DOI] [PubMed] [Google Scholar]
- Shi, T. , Li, R. , Zhao, Z. , Ding, G. , Long, Y. , Meng, J. , Xu, F. et al. (2013) QTL for yield traits and their association with functional genes in response to phosphorus deficiency in Brassica napus . Plos One 8, e54559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Li, R. , Qiu, D. , Jiang, C. , Long, Y. , Morgan, C. , Bancroft, I. et al. (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus . Genetics 182, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Zhan, J. , Yang, Y. , Ye, J. , Huang, S. , Li, R. , Liu, G. et al. (2015) Linkage and regional association analysis reveal two new tightly linked major QTLs for pod number and seed number per pod in rapeseed (Brassica napus L.). Sci. Rep. 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L. , Song, J. , Guo, C. , Wang, B. , Guan, Z. , Yang, P. , Chen, X. et al. (2019) A CACTA‐like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Liu, F. , Wang, H. and Müller, R. (2009) Effects of ethylene and 1MCP (1‐methylcyclopropene) on bud and flower drop in mini phalaenopsis cultivars. Plant Growth Regul. 59, 83–91. [Google Scholar]
- Suzaki, T. , Toriba, T. , Fujimoto, M. , Tsutsumi, N. , Kitano, H. and Hirano, H.Y. (2006) Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47, 1591–1602. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Kopriva, S. , Giordano, M. , Saito, K. and Hell, R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Tokatlidis, I.S. (2017) Crop adaptation to density to optimise grain yield: breeding implications. Euphytica, 213, 1–25. [Google Scholar]
- Tromas, A. , Paque, S. , Stierle, V. , Quettier, A. , Muller, P. , Lechner, E. , Genschik, P. et al. (2013) Auxin‐binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway. Nat. Commun. 4, 2496. [DOI] [PubMed] [Google Scholar]
- Wei, L. , Jian, H. , Lu, K. , Filardo, F. , Yin, N. , Liu, L. , Qu, C. et al. (2016) Genome wide association analysis and differential expression analysis of resistance to sclerotinia stem rot in Brassica napus . Plant Biotechnol. J. 14, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A.W. and Bartel, B. (2005) Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Yang, Y. , Chen, B. , Shi, J. , Luo, M. , Zhan, J. , Wang, X. et al. (2017) An integrated analysis of QTL mapping and RNA sequencing provides further insights and promising candidates for pod number variation in rapeseed (Brassica napus L.). BMC Genom. 18. 10.1186/s12864-016-3402-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, D. , Yu, H. , Lin, B. , Fu, Y. and Hua, S. (2016) Floral initiation in response to planting date reveals the key role of floral meristem differentiation prior to budding in canola (Brassica napus L.). Front. Plant Sci. 7, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. , Peng, C. , Liu, H. , Tang, M. , Yang, H. , Li, X. , Liu, J. et al. (2017) Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Front. Plant Sci. 8, 1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The QQ plots resulting from the GWAS for silique number from the main inflorescence (a), branch inflorescence (b), and whole plant (c).
Figure S2 Correlation analysis between main inflorescence and individual branch. (a) Silique number. (b) Flower number.
Figure S3 The comparative analysis of the flora organ number in process of IM differentiation between more ‐ and fewer‐silique lines.
Figure S4 The consistency between different sample of transcriptome sequencing.
Figure S5 Comparison of the relative expression abundance measured by qRT‐PCR and RNA‐seq.
Table S1 ANOVA analysis of silique number in different environments.
Table S2 Phenotypic correlations between the silique number in the three environments.
Table S3 The significant SNPs identified from GWAS.
Table S4 List of loci identified in our and previous studies for silique number in rapeseed.


