Summary
Small signalling peptides, generated from larger protein precursors, are important components to orchestrate various plant processes such as development and immune responses. However, small signalling peptides involved in plant immunity remain largely unknown. Here, we developed a pipeline using transcriptomics‐ and proteomics‐based screening to identify putative precursors of small signalling peptides: small secreted proteins (SSPs) in rice, induced by rice blast fungus Magnaporthe oryzae and its elicitor, chitin. We identified 236 SSPs including members of two known small signalling peptide families, namely rapid alkalinization factors and phytosulfokines, as well as many other protein families that are known to be involved in immunity, such as proteinase inhibitors and pathogenesis‐related protein families. We also isolated 52 unannotated SSPs and among them, we found one gene which we named immune response peptide ( IRP ) that appeared to encode the precursor of a small signalling peptide regulating rice immunity. In rice suspension cells, the expression of IRP was induced by bacterial peptidoglycan and fungal chitin. Overexpression of IRP enhanced the expression of a defence gene, PAL1 and induced the activation of the MAPKs in rice suspension cells. Moreover, the IRP protein level increased in suspension cell medium after chitin treatment. Collectively, we established a simple and efficient pipeline to discover SSP candidates that probably play important roles in rice immunity and identified 52 unannotated SSPs that may be useful for further elucidation of rice immunity. Our method can be applied to identify SSPs that are involved not only in immunity but also in other plant functions.
Keywords: immunity, Magnaporthe oryzae, proteomics, transcriptomics, rice, signalling peptide, small secreted protein
Introduction
Plants are sessile organisms inescapably exposed to abiotic and biotic stresses in their living environment. To protect themselves from attack by pathogens, plants have developed effective plasma membrane‐localized pattern recognition receptors (PRRs), which recognize conserved foreign (non‐self) molecules called pathogen‐associated molecular patterns (PAMPs) such as fungal chitin as well as bacterial peptidoglycans (PGNs) and flagellin. Perception of PAMPs by PRRs leads to a response called PAMP‐triggered immunity (PTI), which includes calcium influx, generation of reactive oxygen species (ROS), activation of mitogen‐activated protein kinases (MAPKs) and expression of defence genes (Bigeard et al., 2015; Monaghan and Zipfel, 2012; Nicaise et al., 2009). In addition to non‐self molecules, certain endogenous (self) elements such as cell wall fragments and small peptides, referred to as damage‐associated molecular patterns (DAMPs), are also involved in plant immunity (Choi and Klessig, 2016; Yamaguchi and Huffaker, 2011). Upon injury and pathogen infection, they are released into the extracellular space and locally trigger immune responses. However, compared to well‐studied PAMPs, our knowledge of DAMPs is limited. Several studies have revealed that proteinaceous DAMPs such as plant elicitor peptides (PEPs), systemin, rapid alkalinization factors (RALFs) and PAMP‐induced secreted peptides (PIPs) are induced by wound, PAMPs and/or pathogens (Atkinson et al., 2013; Hou et al., 2014; Huffaker et al., 2006). For example, the fungal elicitor E‐FOL or fungal pathogens Alternaria brassicicola and Sclerotinia sclerotiorum trigger the induction of the precursor of DAMP phytosulfokines (PSKs; Loivamaki et al., 2010). The analysis of flg22‐ and elf18‐induced transcription data led to the identification of PIPs (Hou et al., 2014). Therefore, identifying PAMP‐induced small peptides is an effective strategy to obtain proteinaceous DAMPs.
In general, proteinaceous DAMPs are divided into three major classes: non‐secreted‐type peptides without N‐terminal leader sequences derived from cytosolic precursor proteins, secreted‐type peptides originated from secreted precursors with an N‐terminal signal sequence and degradation peptides from proteins with distinct primary functions (Gust et al., 2017; Yamaguchi and Huffaker, 2011). Non‐secreted‐type peptides include tomato Lycopersicon esculentum (v. Castlemart) systemin, the first small peptide identified in plants and the Arabidopsis thaliana plant elicitor peptide (AtPep) family (Huffaker et al., 2006; Pearce et al., 1991). AtPep1, the mature 23‐amino acid (aa) peptide, is derived from a 92‐aa precursor and is recognized by two leucine‐rich repeat RLK receptors, PEPR1 and PEPR2 (Krol et al., 2010; Yamaguchi et al., 2010). The expression of propeptide of PEP3 is induced after inoculation with bacterial pathogen and PEP3 peptides are released into the medium (Yamada et al., 2016). AtPeps control various immune responses including the expression of defence genes, MAPK activation and promoting the generation of ROS (Krol et al., 2010; Yamada et al., 2016). Secreted‐type peptides are categorized into two subclasses: cysteine‐rich peptides and post‐translationally modified small peptides. Cysteine‐rich peptides contain more than four cysteine residues for formation of intramolecular disulfide bonds, as exemplified by RALFs (Pearce et al., 2001b). Post‐translationally modified small peptides are generated from their precursor proteins by proteolytic processing and possess at least one post‐translational modification such as proline hydroxylation, hydroxyproline arabinosylation or tyrosine sulfation. The PSKs and hydroxyproline‐rich systemin (HypSys) are known examples of post‐translationally modified small peptides (Matsubayashi and Sakagami, 1996; Pearce et al., 2001a). On the other hand, inceptins, originated from chloroplastic ATP synthase γ‐subunits and GmSubPep, derived from a putative subtilisin‐like protein, are degradation peptides (Pearce et al., 2010; Schmelz et al., 2007). In animals, various endogenous small peptides called cytokines, a group that includes interferons and interleukins, act as cell signalling mediators in the modulation of immune responses such as inflammation (Mathew et al., 2016). In plants, knowledge regarding endogenous small peptides analogous to animal cytokines is limited and identification of cytokine‐like peptides in plants is therefore an important and challenging goal.
In the past, three approaches have been used to identify endogenous (self) small peptides in plants: bioassay‐guided purification, forward genetics and bioinformatics. Tomato systemin was isolated through a bioassay‐guided purification process. Since then, many peptide families such as PSKs, RALFs and Peps have also been isolated in the similar way (Huffaker et al., 2006; Matsubayashi and Sakagami, 1996; Pearce et al., 2001b). However, bioassay‐guided purification is often technically difficult because the concentration of small peptides is very low in plants. CLAVATA3 (CLV3): critical in the maintenance and development of shoot apical and floral meristems, was found to be a key gene that encodes a small secreted protein through forward genetics (Fletcher et al., 1999). However, due to functional redundancy, only a limited number of such genes have been isolated using the forward genetic approach. Recently, much attention has been focused on identifying small signalling peptides through bioinformatic analyses. For example, by searching for conserved motifs CLV3/ESR‐related (CLE) family peptides were discovered (Sawa et al., 2008). The limitations of this approach are that consensus motifs are prerequisite and only additional members of known small peptide families can be identified. Recent technological advances have driven the increased use of high‐throughput transcriptome and proteome data in plant research. However, there are limitations when these approaches are used independently of one another. For example, transcriptome analysis cannot capture proteins that are encoded by genes with no obvious change in expression. In addition, using proteome analysis alone, it is difficult to detect low‐abundance proteins. Integrative analysis of these two ‘omics’ data sets may yield complementary information and help to identify novel small peptides, especially those protein families that are not currently known to produce small peptides.
In this study, to identify small signalling peptide candidates involved in rice immunity, we developed an experimental pipeline to obtain their putative precursors, SSPs, by using a combination of transcriptomics‐ and proteomics‐based screenings. We isolated 236 SSP candidates including previously reported immune peptide families, RALFs and PSKs and 52 novel SSP candidates that are functionally uncharacterized. We found one uncharacterized SSP, named immune response peptide (IRP), whose mRNA level was strongly enhanced in suspension cells treated with chitin and PGN. Moreover, the protein level of IRP in medium increased upon exposure to chitin. Overexpression of IRP induced the expression of defence gene phenylalanine ammonia‐lyase 1 (PAL1) and the activation of MAPK, indicating that IRP plays a role in rice immunity. Therefore, our experimental approach is useful for efficient discovery of SSP candidates that participate in immunity and, in principle, may be applied in other biological processes in plants.
Results
Transcriptome analysis to identify SSP candidates induced by M. oryzae and its elicitor chitin
To identify small endogenous peptides functioning in rice immunity, we first tried to isolate genes induced by the virulent rice blast fungus M. oryzae (race 007.0) and fungal chitin in rice plants and rice suspension cells, respectively. Rice plants collected at 1, 2 and 3 days post‐inoculation (dpi) with M. oryzae and suspension cells harvested at 1, 3, 6 and 12 hours post‐treatment (hpt) with chitin, were subjected to RNA sequencing (RNA‐Seq) analysis (Figure 1). Prior to RNA‐Seq, we validated the appropriate induction of three defence genes, Chitinase 1, PAL1 and probenazole‐induced protein1 (PBZ1), in response to M. oryzae or chitin as reported previously (Figure S1; Akamatsu et al., 2013). A total of 1848 (up, 1336; down, 512) genes in plants and 1722 (up, 1420; down, 302) genes in suspension cells were determined to be differentially expressed (>2‐fold change in expression, FDR < 0.05) at one or more time points under rice blast fungus infection and chitin treatment, respectively (Figure 2a). The number of up‐regulated genes was much higher than that of down‐regulated genes in both plant and suspension cell systems. To identify genes that were induced by fungus and/or chitin treatment, we focused only on genes whose expression was enhanced in blast fungus or/and chitin treatment. In plants, similar number of up‐regulated genes was observed at 1 dpi (875) and 2 dpi (852), while the number of up‐regulated genes decreased by about 50% at 3 dpi. In suspension cells treated with chitin, about 80% of up‐regulated genes (1145) were observed at 1 hpt, implying that more synchronized immune responses occur in suspension cells (Figure 2a). Comparing up‐regulated genes between plants and suspension cells, 302 genes were found to be up‐regulated in both samples (Figure 2b). Gene ontology (GO) enrichment analysis suggested that 29 GO terms were enriched in these common genes, including terms belonging to biological processes (e.g. protein phosphorylation, response to biotic stimulus, metabolic process, chitin catabolic process) and molecular functions (e.g. protein tyrosine kinase activity, protein serine/threonine kinase activity, ATP binding; Table S1).
Figure 1.
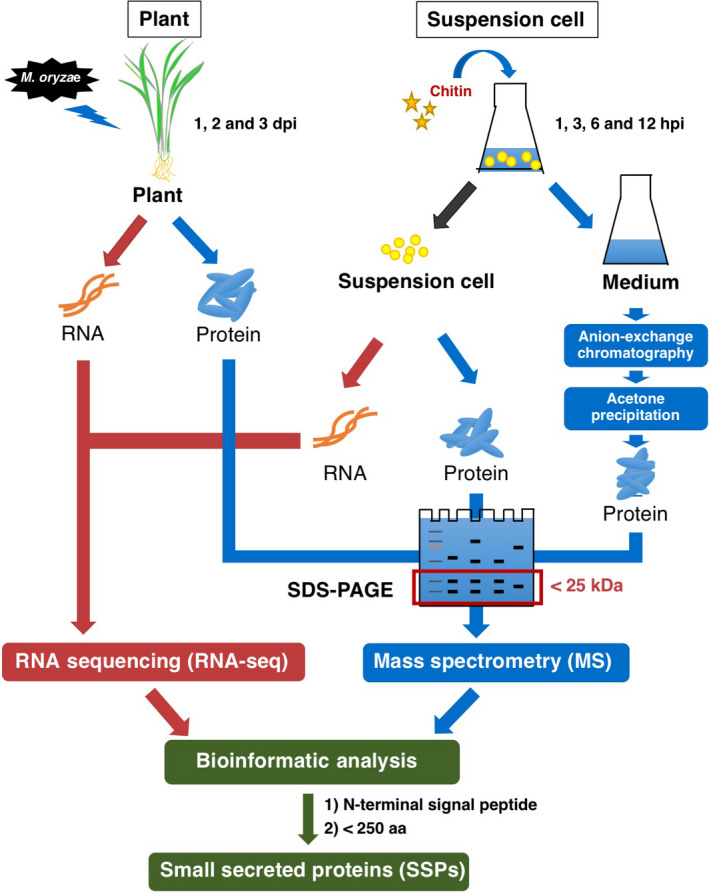
Strategy for identifying SSPs. Rice plants were infected by M. oryzae, samples were collected at 1, 2 and 3 days post‐inoculation and rice suspension cells were treated with chitin for 1, 3, 6 and 12 h. Before sample collection, suspension cells and medium were separated and the medium was passed through an anion‐exchange column. Protein samples were subjected to SDS‐PAGE and proteins smaller than 25 kDa were recovered for PAGE gel. RNA sequencing (RNA‐Seq) and mass spectrometry (MS) results were combined to identify small secreted proteins (SSPs) shorter than 250 aa and containing an N‐terminal signal peptide sequence.
Figure 2.
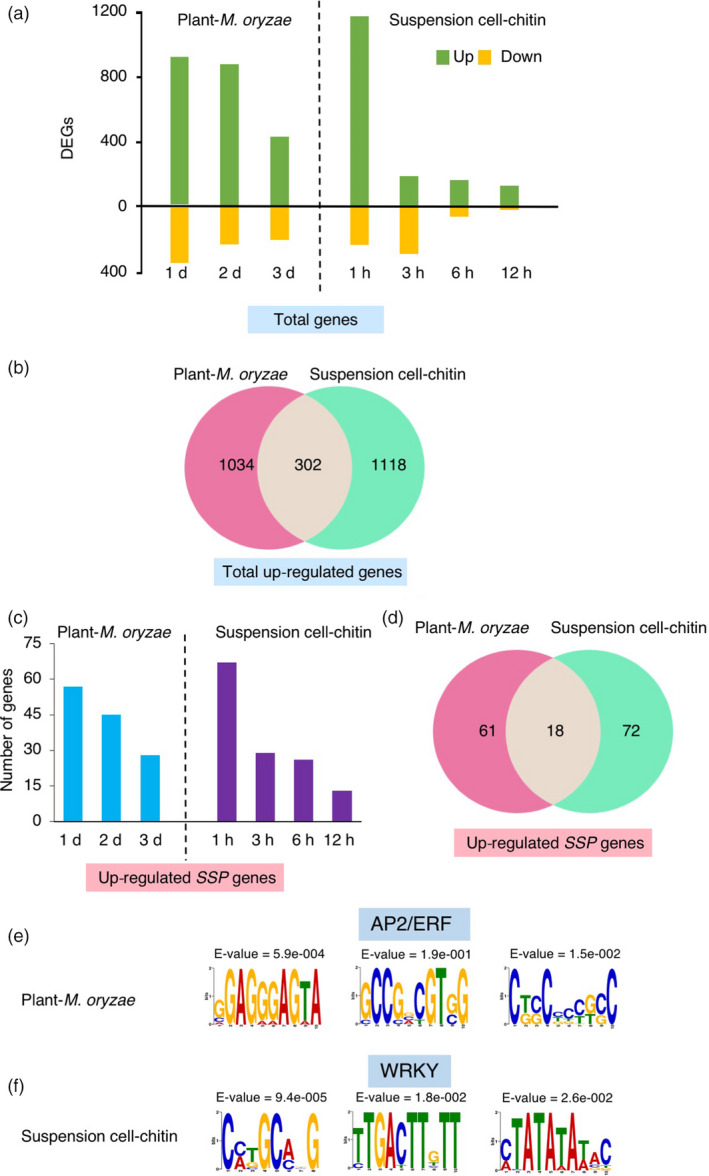
SSPs identified by transcriptome analysis. (a) Number of differentially expressed genes (DEGs) (>2‐fold change, FDR < 0.05) in plants and suspension cells. Yellow and green bars indicate down‐regulated and up‐regulated genes, respectively. (b) Venn diagram showing the overlapping up‐regulated genes between plants and suspension cells. (c) Number of up‐regulated SSP genes (>2‐fold change, FDR < 0.05) in plants and suspension cells, respectively. (d) Venn diagram showing the overlapping up‐regulated SSP genes between plants and suspension cells. (e and f) cis‐regulatory elements identified in the promoter regions of the up‐regulated 79 SSP genes induced in rice plants infected with blast fungus (e) and the 90 up‐regulated SSP genes induced in suspension cells treated with chitin (f).
Precursors of secreted‐type endogenous signalling peptides usually have a length of 50–200 aa and possess an N‐terminal signal sequence (Murphy et al., 2012; Pearce et al., 1991). Based on these findings, we selected up‐regulated SSP genes that encode a protein of less than 250 aa and have an N‐terminal signal sequence predicted by the SignalP 4.0 software (Figure 1). Consequently, we obtained 79 up‐regulated SSP candidates in plants and 90 SSP candidates in suspension cells, respectively (Figure 2c, d, Table S2). In plants, 57, 45 and 28 genes encoding SSPs were induced at 1, 2 and 3 dpi, respectively. In suspension cells, 74% of SSPs were identified at 1 hpt. These results indicate that most of the SSPs were induced at the early stage after blast fungus infection or chitin treatment. Among the SSP candidates identified in the two sources, only 18 were common (Figure 2d).
Transcriptional re‐programming plays a key role in plant immunity and evidence has been accumulating that transcription factors contribute to transcriptional re‐programming (Eulgem, 2005). To identify conserved cis‐regulatory motifs, Multiple EM for Motif Elicitation (MEME) was used to identify conserved motifs within the 1000‐bp upstream regions of the up‐regulated SSP genes in plants and suspension cells, separately. Three cis‐regulatory motifs, GGAGGGAGTA, GCCGCCGTg/cG and Ct/gc/gCCCCg/tc/gC, were over‐represented in the promoters of the 79 SSP genes in plants (Figure 2e). The GCCGCC motif is the core sequence of the GCC box, one of the DNA binding sites for APETELA 2/ethylene response factor (AP2/ERF)‐type transcription factors. GCC boxes are found in the promoter region of ethylene‐inducible pathogenesis‐related genes (PR) in several plant species (Buttner and Singh, 1997; Ohme‐Takagi and Shinshi, 1995). In addition, the rice ERF transcription factor OsERF92 binds specifically to the GCC box sequence and negatively regulates resistance to M. oryzae and salt tolerance (Liu et al., 2012). We identified six ERF transcription factors that were induced by blast fungus treatment in our RNA‐Seq results. In suspension cells, three cis‐regulatory motifs, Cc/aTGCa/cGG, TTGACTTGTT and c/aTATATAt/aAC, were identified in the promoters of up‐regulated SSP genes (Figure 2f). The second cis‐motif overlaps the W box, (T)(T)TGAC(c/t), which is a motif specifically bound by the WRKY transcription factors (Eulgem et al., 2000). WRKY transcription factors are involved in diverse plant processes such as germination, senescence and responses to biotic and abiotic stresses (Rushton et al., 2010). We found that 30 WRKY transcription factors were up‐regulated by fungal or chitin treatment, including WRKY28, WRKY45, WRKY70 and WRKY77, which are known to play key roles in disease resistance (Cheng et al., 2015; Chujo et al., 2013; Lan et al., 2013; Li et al., 2015).
To study the transcriptional profiles of our SSPs, a hierarchical clustering analysis was conducted on the log2‐transformed fold change values (logFCs) of 79 SSPs identified in plants and 90 SSPs identified in suspension cells using RNA‐Seq (Figure S2). The 79 SSPs in plants were classified into seven clusters. Cluster 2 consisted of 24 SSPs induced at 1 and 2 dpi. GO analysis revealed that several SSPs in cluster 2 were in general located in the extracellular region, including three cupin domain‐containing proteins, three protease inhibitor/seed storage/LTP family proteins and one thaumatin. A total of 21 SSPs belonged to cluster 4 and the expression of its members was elevated at 1 dpi. GO analysis indicated that many SSPs in cluster 4 were associated with response to stress, such as two thaumatin proteins (Figure S2a). This feature is consistent with the characteristics of PR proteins (van Loon et al., 2006). In contrast, cluster 7 consists of seven SSPs induced at 2 and 3 dpi, including three early light‐induced proteins (ELIPs) (LOC_Os01g14410, LOC_Os07g08150 and LOC_Os07g08160). They are homologues of Arabidopsis ELIPs, AtELIP1 and AtELIP2, the major light‐responsive genes that contribute to tolerance to photooxidative stress and photoinhibition (Rossini et al., 2006). However, unlike AtELIP1 and AtELIP2, our identified OsELIP proteins have N‐terminal signal sequences and appear to be secreted proteins. In suspension cells, six clusters of SSPs were obtained (FigureS2b). The GO analysis of cluster 4 revealed that several SSPs are in the extracellular region, including six cupin proteins and many other SSPs, such as dirigent, that are involved in stress responses. In particular, the induction of four plastocyanin‐like domain‐containing proteins was observed. Phytocyanins are blue copper proteins that bind to a single copper atom, act as electron transporters and are responsive to abiotic stresses (Ma et al., 2011).
Proteome analysis to identify SSPs challenged with M. oryzae and chitin
We also performed proteome analysis to identify candidate SSPs. It is technically difficult to isolate SSPs from apoplastic space of rice plants. Thus, we also prepared the liquid medium sample from suspension cells in addition to rice plant and suspension cell samples, expecting that SSPs may be secreted into the medium. In our study, we employed the following strategies to increase the efficiency of identifying SSPs in the medium: (i) suspension cells were washed with fresh medium before chitin treatment to remove pre‐existing secreted proteins in the medium; (ii) considering protein concentration in the liquid medium is low, we concentrated the liquid medium using an anion‐exchange column followed by acetone precipitation (Figure 1, Figure S6); (iii) to eliminate large‐size non‐target proteins and increase the efficiency of identifying small proteins, we performed SDS‐PAGE and collected the portion of the gel containing proteins smaller than 25 kDa, since precursors of small peptides are usually less than 200 aa (Murphy et al., 2012; Pearce et al., 1991); and (iv) two in‐gel digestions, single trypsin digestion and double enzyme digestion combining trypsin with Asp‐N, were used for each sample to increase the coverage of proteins (Figure 3e). Finally, we counted proteins that were detected specifically in samples treated with rice blast fungus or chitin but not in mock samples.
Figure 3.
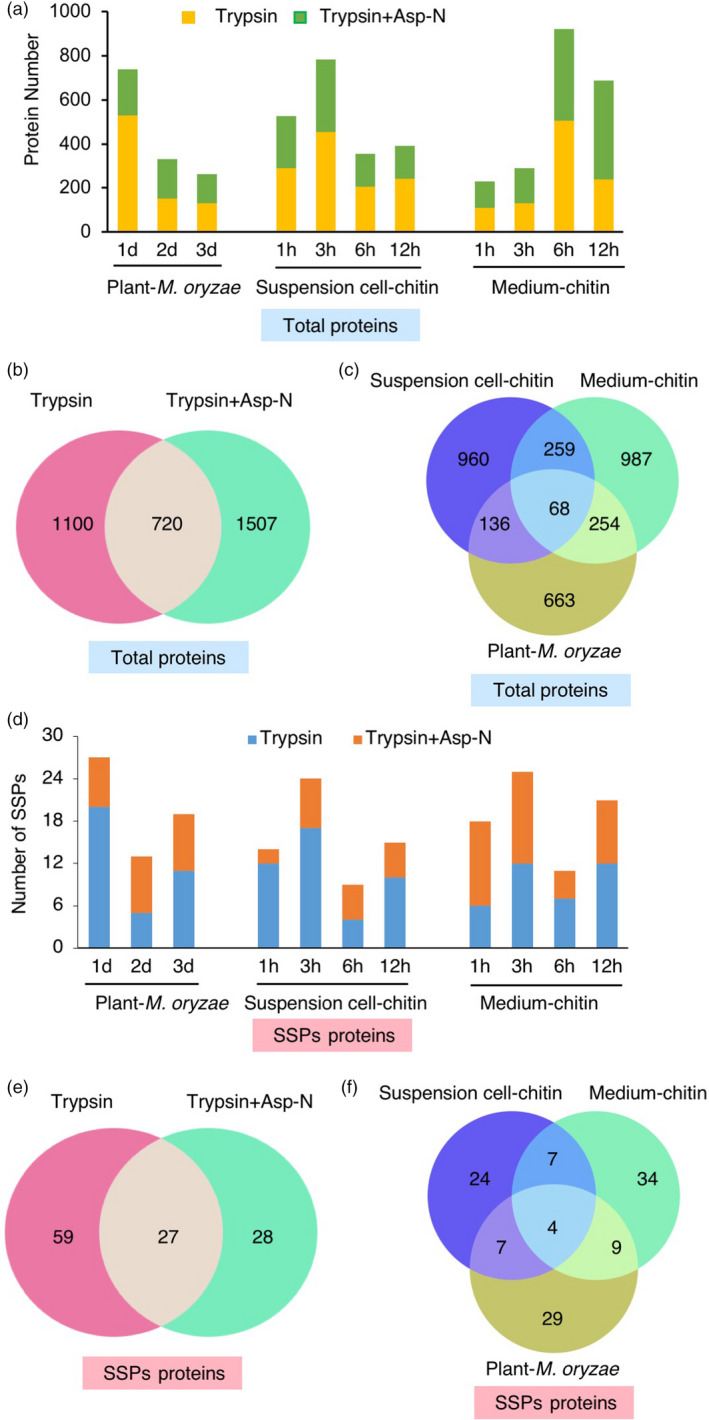
SSPs identified by proteome analysis. (a) Number of proteins identified by MS analysis in plants infected by M. oryzae and suspension cells and medium treated by chitin, respectively. Yellow and green bars indicate the results from trypsin digestion and double enzyme digestion combining trypsin and Asp‐N, respectively. (b) Venn diagram showing the number of overlapping proteins identified by trypsin digestion and double enzyme digestion. (c) Venn diagram showing the number of overlapping proteins identified from plants, suspension cells and medium. (d) Number of SSPs identified in plants, suspension cells and medium, respectively. Blue and orange bars indicate the results of trypsin digestion and double enzyme digestion combining trypsin and Asp‐N, respectively. (e) Venn diagram showing the number of overlapping SSPs identified by trypsin digestion and double enzyme digestion. (f) Venn diagram showing the number of overlapping SSPs identified in plants, suspension cells and medium.
As a result, we identified 1121, 1423 and 1568 proteins in plants, suspension cells and medium, respectively. In total, 3327 proteins remained after eliminating redundancy (Figure 3a). In addition to the 1820 proteins identified by single trypsin digestion, 1507 proteins were found by double enzyme digestion particularly, indicating that the combination of two kinds of proteases for MS analysis dramatically increases the number of identified proteins (Figure 3b). A Venn diagram analysis of proteins identified in plants, suspension cells and medium showed that only 18%–29% of proteins were shared between two of three samples and 68 proteins were common to all samples (Figure 3c), including several related to defence response, such as PAL1 and a PR Bet VI family protein.
Next, we screened SSPs from proteome samples using the same criteria we employed in transcriptome analysis (Figure 1). From the proteome data, we obtained 49, 42 and 54 SSPs in plants, suspension cells and medium, respectively. In plants, a total of 27, 13 and 19 SSPs were identified at 1, 2 and 3 dpi, respectively (Figure 3d and Table S2). In suspension cells and medium, the number of identified SSPs was highest at 3 hpt. Venn diagram analysis revealed that 24% of SSPs were uniquely identified in the double‐enzyme‐digested samples (Figure 3e). Only four SSPs were shared among plants, suspension cells and medium (Figure 3f). Most of the previously identified signal peptides are post‐translationally modified and the modifications are important for their physiological activity (Matsubayashi, 2014). Therefore, in our analysis we considered three post‐translational modifications, sulfation on tyrosine, serine and threonine; oxidation on proline and methionine; and hydroxyproline arabinosylation, in MS analysis. Among 114 SSPs identified by MS, we detected 54 SSPs with post‐translational modifications (Table S3). Interestingly, most of the peptides possess multiple modifications, implying that these SSP candidates are favourite targets of post‐translational modifications.
Protein families identified by transcriptome and proteome analyses
Combining transcriptome with proteome analyses, we obtained 236 SSPs: 117, 130 and 54 in plants, suspension cells and medium, respectively (Figure 4a). There are only seven overlapping SSPs in all three sources. On the other hand, in total 151 and 114 SSPs were harvested by transcriptomics and proteomics analysis using three different materials, respectively. Only 29 common SSPs were identified in both transcriptomics and proteomics analyses (Figure 4b). Interestingly, we were able to detect members of two known SSP families: RALFs and PSKs (Table 1). RALF was originally identified in tobacco as a 5‐kDa cysteine‐rich secreted peptide from a 115‐aa precursor that causes rapid alkalinization of culture medium (Pearce et al., 2001b). Several studies indicate that RALFs are important in a range of plant processes such as root development and abiotic and biotic stress responses. To date, 39 and 43 RALF genes have been identified in Arabidopsis and rice, respectively (Sharma et al., 2016). We detected five RALFs in our experimental conditions. OsRALFL6 (LOC_Os01g25540) was identified in suspension cells by MS and OsRALFL26 (LOC_Os10g41980) and OsRALFL31 (LOC_Os12g38360) were found in plants by RNA‐Seq. OsRALFL7 (LOC_Os01g25560) and OsRALFL8 (LOC_Os02g44940) were detected in medium by MS, implying that they were secreted from suspension cells into medium upon chitin treatment. Interestingly, the mRNA levels of OsRALFL7 and OsRALFL8 were induced not only by fungal chitin but also by bacterial peptidoglycan (PGN) treatment (Figure 5a, b), suggesting that OsRALFs are involved in the rice response to both fungal and bacterial infections. The alignment analysis revealed that OsRALF6, OsRALFL7 and OsRALFL8 have the signal peptide sequence at their N‐terminus, the four conserved cysteines and tyrosine–isoleucine–serine–tyrosine (YISY) motif, that is important for attachment of the RALF protein(s) to their putative receptors (Figure S3a). Interestingly, our identified OsRALFLs contain a conserved arginine–arginine (RR) motif which is the recognition motif of subtilase family proteases. Previous studies indicated that cleaved RALF members inhibit immunity while non‐cleaved members promote immunity in Arabidopsis (Stegmann et al., 2017).
Figure 4.
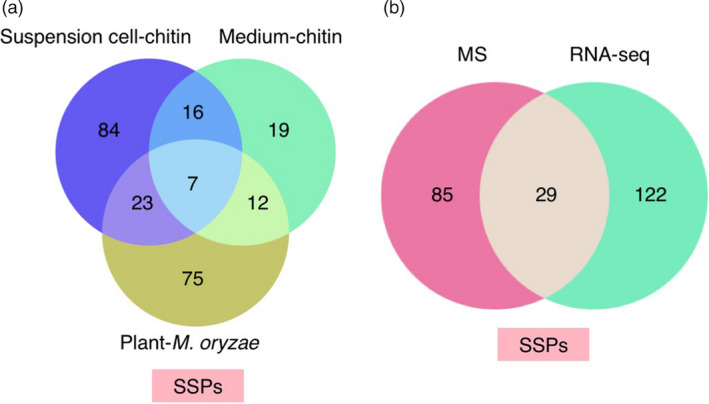
Identified SSPs number. (a) Venn diagram showing the number of overlapping SSPs identified in plants, suspension cells and medium after combining RNA‐Seq with MS results. (b) Venn diagram showing the number of overlapping SSPs identified by RNA‐Seq and MS.
Table 1.
Protein families of SSPs
| Protein family | Total | Medium |
|---|---|---|
| Cupin/germin (including PR‐15 and PR‐16) | 22 | 9 |
| Protease inhibitor/seed storage/LTP family protein (including PR‐14) | 18 | 4 |
| Plastocyanin‐like domain‐containing protein | 11 | 5 |
| Glycine‐rich cell wall protein | 8 | 0 |
| Thaumatin family domain‐containing protein (including PR‐5) | 7 | 3 |
| Dirigent family protein | 6 | 2 |
| Rapid alkalinization factor (RALF) family protein | 5 | 2 |
| SCP‐like extracellular protein (including PR‐1) | 5 | 0 |
| Cysteine proteinase inhibitor | 4 | 1 |
| Barwin domain‐containing protein (including PR‐4) | 4 | 0 |
| Bowman‐Birk‐type bran trypsin inhibitor | 4 | 0 |
| Early light‐induced protein | 3 | 0 |
| Thionin family protein (including PR‐13) | 3 | 0 |
| Pollen Ole e I allergen and extensin family protein | 3 | 1 |
| Gibberellin‐regulated GASA/GAST/Snakin family | 2 | 1 |
| Glycine‐rich protein family protein | 2 | 0 |
| Peptidyl‐prolyl cis–trans isomerase | 2 | 1 |
| Ribonuclease T2 family domain‐containing protein | 2 | 1 |
| Phytosulfokine (PSK) | 1 | 0 |
Figure 5.
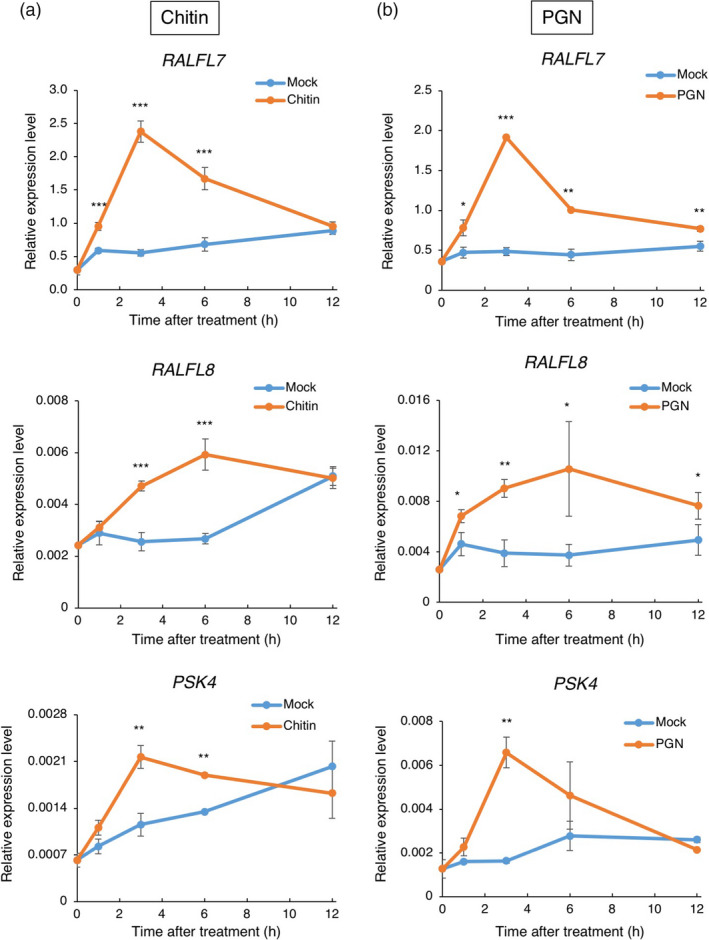
Expression of OsRALFL s and OsPSK4 induced by PAMPs. OsRALFL7, OsRALFL8 and OsPKS4 were induced in suspension cells treated with chitin (a) or PGN (b). Expression levels of OsRALFL7, OsRALFL8 and OsPKS4 were quantified by qRT‐PCR with six biological replicates. Error bars indicate standard error (S.E). Statistically significant differences between treated suspension cells and mock control are depicted with asterisks (*, P < 0.05; **, P < 0.01 and ***, P < 0.001) according to the two‐tailed t‐test.
Another small secreted peptide family we identified is PSK, which has a highly conserved pentapeptide sequence YIYTQ at the C‐terminus (Figure S4b) (Matsubayashi and Sakagami, 2006; Srivastava et al., 2008). PSKs have multiple functions such as promoting cell growth, contributing to pollen tube guidance and plant immunity (Sauter, 2015). We detected OsPSK4 (LOC_Os07g03200) in the plant sample by RNA‐Seq and further confirmed by qRT‐PCR assay (Figure S4a). OsPSK4 also possesses signal peptide sequence at their N‐terminus, the RR motif, a recognition site for a subtilisin‐like serine protease and conserved mature peptide sequence (Figure S4b). AtPSK4 is cleaved by the serine protease AtSBT1.1 and the cleaved form of AtPSK4 is secreted from callus into the callus induction medium (Srivastava et al., 2008). OsPSK4 mRNA expression was induced in suspension cells by both chitin and PGN treatment (Figure 5a, b). Taken together, these data suggest that both RALFs and PSKs are involved in rice immunity.
The 236 identified SSP candidates encompass seven PR protein families (Table 1). PR proteins are inducible defence‐related proteins in infected plants that are classified into 17 families and play important roles in immunity (van Loon et al., 2006). We identified 22 proteins in the cupin/germin family and nine of them were found in the medium. Cupin/germin proteins comprise two PR protein families, PR‐15 and PR‐16. Rice chromosome (chr) 8 contains one of the major quantitative trait loci for disease resistance to rice blast fungus, encompassing a cluster of 12 germin‐like genes (OsGLPs) (Manosalva et al., 2009). In our experimental conditions, we detected the induction of ten OsGLP genes that are located in the same region of chr 8. The suppression of the 12 germin‐like genes in chr 8 by multiplex RNAi makes the rice plants more susceptible to two distinct fungal diseases, rice blast and sheath blight (Manosalva et al., 2009), indicating that our OsGLPs in chr 8 are involved in disease resistance. We also identified 18 non‐specific lipid transfer proteins (nsLTPs) that transfer phospholipids and fatty acids between membranes in vitro (Kader, 1996). Several studies have demonstrated the extracellular localization of nsLTPs in various plant species (Liu et al., 2015). Plant nsLTPs contain the PR‐14 family and participate in various plant processes including sexual reproduction, seed development and germination, as well as resistance to biotic and abiotic stresses (Liu et al., 2015). In addition, seven thaumatin‐like proteins were identified, including three in the medium (Table 1). The plant thaumatin family is classified as PR‐5 and it appears to have antifungal activity. The overexpression of PR‐5 family member proteins inhibits the growth of phytopathogenic fungi, Scleretonia sclerotiorum and Botrytis cinerea in Arabidopsis and improves downy mildew resistance in Vitis vinifera grapevine (He et al., 2017; Misra et al., 2016). We also found several proteinase inhibitor (PI) families such as protease inhibitor/seed storage/LTP family protein, Bowman‐Birk‐type bran trypsin inhibitors (BBI) and cysteine proteinase inhibitors. BBIs participate in plant resistance to insects and salt stress (Dantzger et al., 2015; Shan et al., 2008). In addition, several functional studies have reported that cysteine PIs have insecticidal and antifungal effect on plants (Popovic et al., 2013).
IRP regulates rice immunity
To identify novel SSP families participating in rice immunity, we analysed the 52 unannotated SSPs and selected 20 SSPs that have homologues in other plant species. Then, we checked whether the 20 SSPs were induced by chitin treatment using a qRT‐PCR assay. A strong induction of one gene, LOC_Os04g28390, was detected by both RNA‐Seq and qRT‐PCR analyses done on chitin‐treated (1 h) samples (Figure 6b). Similarly, the expression of this gene was also found to be enhanced 1 h after PGN treatment. We named the gene immune response peptide (IRP). It is a single‐copy gene in the rice genome and encodes a 66‐aa protein. IRP homologues are found only in Poaceae (Figure 6a, Figure S5). All IRPs have an N‐terminal signal sequence and conserved C‐terminus. We tested whether IRP contributes to defence responses and generated IRP‐overexpressing suspension cell lines using the maize ubiquitin promoter. Interestingly, IRP overexpression enhanced the expression of the defence gene PAL1 without chitin treatment. After chitin treatment, transcription of PAL1 and IRP itself was further induced (Figure 6c). Overexpression of IRP induced the activation of two MAP kinases MPK3 and MPK6, suggesting that IRP regulates rice immunity through the regulation of MAPK activity (Figure 6d).
Figure 6.
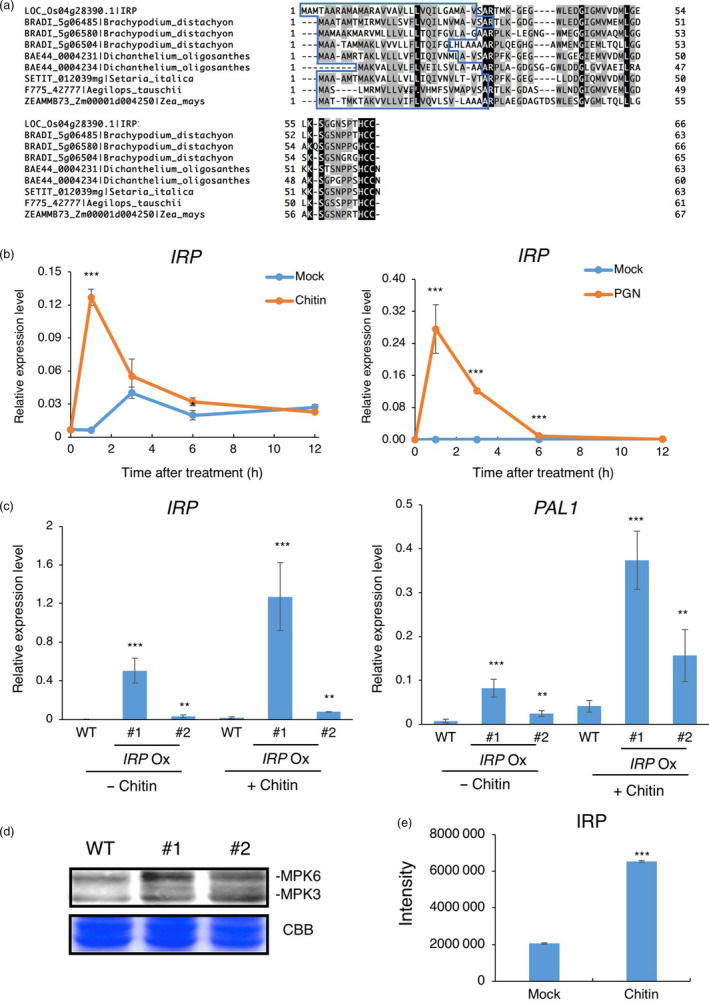
IRP is involved in rice immunity. (a) Protein alignment of IRP and its homologues. Predicted N‐terminal signal sequences are in a blue box. (b) IRP expression was induced in suspension cells treated with chitin or PGN. The expression level of IRP was quantified by qRT‐PCR with six biological replicates. Error bars indicate S.E. Statistically significant differences between the mock and chitin‐treated cells are depicted with asterisks (*, P < 0.05; **, P < 0.01 and ***, P < 0.001) according to the two‐tailed t‐test. (c) Effect of overexpression of IRP on the defence gene PAL1. Rice suspension cells overexpressing IRP were treated with chitin for 1 h. #1 and #2 are two independent lines of transgenic rice suspension cells overexpressing IRP . The expression levels of IRP and PAL1 were quantified by qRT‐PCR with four independent replicates. Error bars indicate S.E. Statistically significant differences between wild‐type (WT) and overexpression lines are depicted with asterisks (*, P < 0.05; **, P < 0.01; and ***, P < 0.001) according to the two‐tailed t‐test. (d) Overexpression of IRP induces MAPK activation. MAPK activation was detected in WT and IRP overexpression lines #1 and #2 suspension cells using anti‐phospho‐p44/42 MAPK antibody. CBB, Coomassie Brilliant Blue. Similar results were obtained in four independent experiments. (e) The protein abundance of IRP in the medium increased following chitin exposure. The 20‐aa peptide GEGWLEDGIGMVVDMLGELK of IRP was used as a spectral to match the MS/MS spectrum of the peptide acquired using the parallel reaction monitoring (PRM) method. The rice suspension cells were treated with chitin for 2 h in two independent experiments. The total intensity of the peptide in the medium was determined in the mock and chitin‐treated sample, respectively. Error bars indicate S.E. Statistically significant differences between the mock and chitin treatments are depicted with asterisks (***, P < 0.001) according to the two‐tailed t‐test.
In order to confirm the secretion characteristic of IRP, we performed parallel reaction monitoring (PRM) assays, as PRM analysis generates full MS/MS data with high resolution and high mass accuracy and is widely used for the quantification of targeted proteins/peptides (Majovsky et al., 2014). Using a suspension cell system, we detected the 20‐aa peptide GEGWLEDGIGMVVDMLGELK region of IRP in the medium and found that a much larger amount of IRP protein was detected in the medium sample after 2 h of chitin treatment, confirming that IRP protein is released to the extracellular spaces upon chitin treatment (Figure 6e).
Discussion
In this study, we established a simple and efficient pipeline to identify endogenous small signalling peptides that are involved in rice immunity. Small signalling peptides are produced from larger 50–200‐aa polypeptide precursors by proteolytic processing (Matsubayashi and Sakagami, 2006). Considering the difficulties in purifying mature small signalling peptides from plants, we tried to isolate small signalling peptides by identifying their putative precursor SSPs, which encode proteins of <250 aa with an N‐terminal signal peptide sequence (Figure 1). To increase the number of identified SSPs, we used three different sources: plants, suspension cells and medium, and combined transcriptome and proteome analyses. Notably, the combination of three different sources drastically enhanced the coverage of SSPs (Figure 4a) and only 12.3% of total SSPs are identified in both transcriptome and proteome analyses (Figure 4b). Taken together, our results demonstrate that our integrative analysis using the different sources and the methods is an effective way for identifying SSP candidates.
Recently, the suspension cell system has become a useful resource for studying the secretome of various plants, such as Arabidopsis (Oh et al., 2005; Tran and Plaxton, 2008) and rice (Chen et al., 2016; Kim et al., 2009). Kim et al. identified 21 secreted proteins induced by rice blast fungus M. grisea and its elicitor by a combination of 2‐DE and MS analyses. A total of 34 secreted proteins, including 10 up‐regulated and 24 down‐regulated proteins, were identified to respond to a rice bacterial blight disease pathogen Xanthomonas oryzae pv. oryzae by a proteomic study combining 2‐DE with MS assay. However, the number of identified secreted proteins in those studies is small, which prompted us to test new procedures for SSP identification. We isolated 54 SSPs in media, including cupin/germin (Manosalva et al., 2009), thaumatin (Misra et al., 2016), dirigent (Shi et al., 2012), SCP‐like extracellular protein (Alexander et al., 1993), wound‐induced protein/Barwin domain (Kim and Hwang, 2015), Bowman‐Birk (Qu, 2003), thionin (Ji et al., 2015), GASA/GAST/snakin family (Oliveira‐Lima et al., 2017) and peptidyl‐prolyl cis–trans isomerase family proteins (Zhu et al., 2011). These protein families have been shown to act as positive regulators in plant immunity, demonstrating that our pipeline was effective in identifying SSP candidates that are important for plant immunity. Besides, we built the database considering three post‐translational modifications, hydroxyproline arabinosylation on proline; oxidation on methionine and proline; and sulfation on serine, threonine and tyrosine, as post‐translationally modified small signalling peptides possess at least one post‐translational modification (Matsubayashi, 2014). Among 114 SSPs identified by proteomics approach, we found that 54 SSPs had at least one of the three post‐translational modification (Table S3), indicating that our database increases the coverage of identification of SSPs. To the best of our knowledge, this is the first report in plants to identify post‐translationally modified SSPs considering hydroxyproline arabinosylation on proline; oxidation on methionine and proline; and sulphation on serine, threonine and tyrosine using global proteomics analysis.
The expression levels of the members of two known small signalling peptide families, RALFs and PSKs, were increased in plants challenged with rice blast fungus and/or suspension cells treated with chitin. In our study, we identified five RALF members. Interestingly, OsRALFL6‐8 are clustered in a rice‐specific subclade in the phylogenic tree of RALF proteins in rice and Arabidopsis, suggesting that they may have originated in rice after the separation of two species (Figure S3b; Cao and Shi, 2012). The expression of OsRALFL7 and OsRALFL8 was strongly induced by chitin and PGN (Figure 5a, b), indicating that OsRALFL7 and OsRALFL8 play a role in the rice response to fungal and bacterial infections. A previous study showed that AtRALFL8, AtRALFL23, AtRALFL33 and AtRALFL34 are up‐regulated upon simultaneous water deficit and nematode stress in root tissue (Atkinson et al., 2013). In addition, AtRALFL8 overexpression confers susceptibility to drought stress and nematode infection. Furthermore, AtRALF23 inhibits PTI through its receptor, the malectin‐like receptor kinase FERONIA (Stegmann et al., 2017). Although the rice genome encodes seven OsPSKs, we identified only OsPSK4 in the plant sample by RNA‐Seq (Figure S4c). PSKs are post‐translationally modified small peptides that mature by tyrosine sulfation and proteolytic processing (Matsubayashi and Sakagami, 1996). PSK appears to have an antagonistic manner in regulating the immunity of Arabidopsis to biotrophic and necrotrophic pathogens (Mosher et al., 2013). Further studies are needed to confirm whether our OsRALFs and OsPSK4 play key roles in rice immunity.
Among the 52 unannotated SSPs that have not been reported before, we focused on IRP. The expression of IRP was enhanced not only by chitin but also by PGN, implying that IRP is involved in disease resistance to fungi as well as bacteria. In addition, IRP overexpression in rice sufficiently induced defence gene expression as well as MAPK activation in the absence of chitin treatment (Figure 6), suggesting that IRP functions as a positive regulator in plant immunity. We also generated IRP overexpression plants. After chitin and PGN treatment, the expression of endogenous IRP was up‐regulated to 23‐ and 217‐fold (Figure 6b), but unfortunately it is very difficult to obtain plants with the high expression of IRP. The plants having only threefold (Figure S7, IRP Ox line 14) and fivefold expression levels (Figure S7, IRP Ox line 13) exhibited severe dwarf phenotype and therefore, they were not suitable for an infection assay. It is very likely that IRP overexpression can confer pleiotropic effects in addition to defence activation. Until now, some small peptides, HypSys, PSK, PIPs and RALFs, have been shown to contribute to plant immunity. Interestingly, like HypSys, which was identified only in some plants in the Solanaceae and in sweet potato in the Convolvulaceae family (Chen et al., 2008), homologues of our IRP are identified only in Poaceae. It is of interest to understand why plants generate peptide families with such a narrow phylogenetic distribution. On the other hand, the mechanism underlying IRP function in rice immunity still needs to be explored. It will be important to identify IRP receptor(s) and investigate the interactions between IRP and its receptor(s), as well as other relevant elements and how these interactions activate the rice immune response.
In this study, we established a pipeline using transcriptomics‐ and proteomics‐based screening and identified 236 SSP candidates including known 19 protein families involved in plant immunity (Table S2 and Table 1). Further investigation of the functions of these 236 SSP candidates is needed, in order to understand the detailed mechanisms of their function in plant immunity. We identified two known peptide families, RALFs and PSKs, as well as a novel peptide candidate IRP. Some studies revealed that pretreated peptides reduced pathogen growth (Gully et al., 2019; Stegmann et al., 2017; Yamaguchi et al., 2010). It may be useful to apply such peptides into crops for pathogen resistance. Notably, some small peptides are involved in multiple processes and functions in plants. For example, the overexpression of AtPep1 precursor, PROPEP1, enhances disease resistance to pathogens and defence responses while promoting root and aerial growth (Huffaker et al., 2006; Yamaguchi et al., 2010). AtPep3 contributes to not only plant immunity but also salinity stress tolerance (Nakaminami et al., 2018). Moreover, CLE25 controls stomatal closure through abscisic acid in long‐distance signalling. These examples demonstrate that in addition to immunity, endogenous small peptides have pleiotropic actions on various events such as development as well as abiotic and biotic tolerance (Takahashi et al., 2018). It is thus interesting to study the functions of IRP peptide in plant immunity, as well as in other processes such as development and biotic and abiotic stress responses.
Experimental procedures
Plant materials and suspension cells
Rice plants (Oryza sativa L. ssp. japonica cv. Nipponbare) were grown in a greenhouse under long‐day conditions (16 : 8 h, light:dark, 28 : 22 °C). To generate transgenic plants, Agrobacterium‐mediated transformation of rice calli was performed according to an established method (Hiei et al., 1994). To produce suspension cells expressing IRP, the IRP coding region was introduced into the p2K1 vector to control expression with the maize ubiquitin promoter. Rice suspension cells derived from Nipponbare calli were grown in R2 medium (Hayakawa et al., 1992).
Treatment of plants with M. oryzae and suspension cells with PAMPs
For the Magnaporthe oryzae (race 007.0) infection assay, 4‐week‐old rice plants were sprayed with a conidial suspension of 105 conidia/mL in 0.02% Tween‐20. Infected plants were covered with a glass box to maintain high humidity and the leaves were harvested at 1, 2 and 3 days post‐inoculation (dpi). Three biological replicates were prepared for each time point and each sample consisted of five plants. For PAMP treatment, rice suspension cells were cultured in R2 medium and treated with 2 μg/mL chitin (hexa‐o‐acetylchitoheptaose, Carbosynth, 38854‐46‐5) or 100 μg/mL PGN (Sigma, 69554). Three biological replicates were prepared for each PAMP treatment.
RNA and protein isolation
Liquid‐cultured suspension cells were separated into cells and medium by centrifugation at 100 g for 10 min. Leaves and suspension cells were ground in liquid nitrogen to a fine powder for RNA isolation and protein extraction. Total RNA was extracted by TRIzol reagent (Life Technologies). For protein samples of leaves and suspension cells, 100 mg of powder was homogenized in 200 μL SDS sample buffer (160 mm Tris‐Cl, pH 6.8, 130 mm SDS, 2 m glycerol, 10 mm DTT) with protease inhibitors (cOmplete EDTA‐free, Roche). Four independent 25‐mL flasks of suspension cells for each time point were combined into one sample, which was passed through a 20‐μm filter to remove the cells. To concentrate proteins in the medium, the pH was adjusted to 8.5 with NaOH and the samples were loaded onto an anion‐exchange chromatography column (HiTrap Q HP, GE Healthcare). Bound proteins were recovered with elution buffer (50 mm Tris‐HCl, pH 8.5, 1 m NaCl, 1 mm DTT) and the eluted fraction was concentrated by acetone precipitation and dissolved in 100 μL of SDS sample buffer.
Quantitative real‐time PCR
Total RNA was isolated and treated with DNase I (Life Technologies) and 2 μg RNA was reverse‐transcribed by a cDNA Synthesis Kit (1708891, Bio‐Rad). For qRT‐PCR, gene‐specific primers were designed (see Table S4). Rice Ubiquitin (LOC_Os03g13170.1) was used as an internal control to normalize gene expression across different samples. Reactions were performed on a 6500 Real‐Time PCR System (Applied Biosystems) using SYBR (Life Technologies). Relative expression level values were calculated using the 2−△CT method (Livak and Schmittgen, 2001).
RNA‐Seq and clustering analysis
RNA was quantified using the Qubit RNA HS Assay Kit (Life Technologies) and its integrity was examined using the Bioanalyzer 2100 (Agilent). Only RNA samples with RIN > 7 were used for library preparation, which was performed using a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) following the standard protocol. Pooled libraries were sequenced in paired‐end 125‐bp mode on a HiSeq 2500 (Illumina) at the Genomics Core Facility of the Shanghai Center for Plant Stress Biology. High‐throughput sequencing data were filtered by SolexaQA (Q > 17 and length ≥ 25 bp; Cox et al., 2010) and cutadapt (Criscuolo and Brisse, 2014) to remove low‐quality regions and adapter sequences. Clean reads were mapped to the rice genome (MSU7) using TopHat (Trapnell et al., 2009) with default parameters and the following adjustments: ‐p 28 –library‐type fr‐firststrand. Gene expression levels were summarized by HTSeq (Anders et al., 2015). Weakly expressed genes were removed and only genes with an expression level of at least 1 read per million (RPM) in at least three samples were retained.
The R package edgeR (Robinson et al., 2010) was used to identify genes that were differentially expressed in treated plant samples or suspension cell samples compared to the corresponding mock samples. False discovery rate (FDR) < 0.05 and at least twofold change were used as the significance cut‐off for differential expression. The k‐means clustering method was used to cluster up‐regulated SSPs in plants (k = 7) or suspension cells (k = 6), using the Pearson correlation coefficient as the distance metric. And the desired number of clusters, k, was specified by the elbow method in advance. GO enrichment analysis was performed with topGO (Aibar et al., 2015) with P‐value < 0.01 as the cut‐off.
Gel electrophoresis and liquid chromatography–tandem mass spectrometry (LC‐MS/MS) analysis
To eliminate large non‐target proteins, SDS‐PAGE using 16.5% gel (Mini‐PROTEAN Tris‐Tricine Gel, Bio‐Rad) was performed on the isolated proteins. After Coomassie Brilliant Blue staining, the area of the gel containing proteins < 25 kDa was collected. Following destaining with 100 mm NH4HCO3 and dehydration with acetonitrile (ACN), the gel pieces were reduced with 10 mm dithiothreitol (DTT) at 56 °C and alkylated with 55 mm iodoacetamide in the dark at room temperature for 1 h. Proteins in the gel were digested either with trypsin (Promega) alone or with both trypsin and Asp‐N (Promega). The resulting peptides were extracted and dried using a refrigerated CentriVap concentrator (Labconco) and then dissolved in 0.1% formic acid (FA). The samples were analysed by online nano‐ACQUITY Ultra Performance LC (Waters) coupled with an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific). The peptides were trapped by a 2G‐V/MT Symmetry C18 Trap Column at a flow rate of 5 μL/min for 3 min and separated on a BEH130 C18 analytical column at 350 nL/min. Peptides were eluted on a reversed‐phase column using mobile phases consisting of solvent A (0.1% FA) and solvent B (ACN/0.1% FA) through a linear gradient from 4% to 30% and then 85% solvent B with a duration of 65 min. Data‐dependent MS/MS acquisition was performed following a full MS survey scan by Orbitrap at a resolution of 60 000 over the m/z range of 350–1800 and MS/MS measurements of the top 20 most intense precursor ions. The target values of automatic gain controls were set to 2e5 for Orbitrap MS and 1e4 for ion‐trap MS/MS detection. The selected multiply charged peptide ions were fragmented by high‐energy collision‐induced dissociation using nitrogen gas at a normalized collision energy of 35% and dynamic exclusion was enabled for 60 s.
The spectra obtained were compared with a protein database (MSU7) using the MASCOT server (version 2.4). The mascot search parameters were as follows: set off the threshold at 0.05 in the ion‐score cut‐off, peptide tolerance at 15 p.m., MS/MS tolerance at ±0.5 Da, peptide charge of 2+ or 3+, trypsin with/without endopeptidase Asp‐N as the enzyme allowing up to one missed cleavage, carbamidomethylation on cysteine as a fixed modification and hydroxyproline arabinosylation on proline, oxidation on methionine and proline and sulphation on serine, threonine and tyrosine as a variable modification.
PRM
The suspension cell medium was condensed using an anion‐exchange chromatography column and precipitated by acetone. The pellets were lysed in GdmCl lysis buffer (6M guanidine hydrochloride [GdmHCl], 100 mm Tris‐HCl, pH 8.5) and dissolved completely. The samples were quantified by bicinchoninic acid assay method. A total of 100 μg of each sample was digested with 2 μg Trypsin overnight at 37 °C. The pH was adjusted below 3 and samples were heated at 95 °C for 5 min. Afterwards, the detergents were removed by centrifuging at 14 000 g at 4 °C and the supernatant was collected desalted and dried.
Tryptic peptides were loaded on C18 stage tips for desalting prior to reversed‐phase chromatography on an Easy nLC‐1200 System (Thermo Fisher Scientific) with 60‐min liquid chromatography gradients with acetonitrile ranging from 5% to 35% in 45 min. PRM analysis was performed on an Orbitrap Fusion Mass Spectrometer (Thermo Scientific). The mass spectrometer was operated in positive ion mode, with the following parameters: a full MS1 scan was acquired with a resolution of 70 000 (200 m/z), automatic gain control (AGC) target values of 3.0 × 106 and a 250 ms maximum ion injection time. Full MS scans were followed by 20 PRM scans at a 35 000 resolution (200 m/z), with AGC 3.0 × 106 and a maximum ion injection time of 200 ms. The targeted peptides were isolated with a 2‐Th window and fragmented at a normalized collision energy of 27 in a higher energy dissociation (HCD) collision cell. The raw data were analysed using Skyline (MacCoss Lab, University of Washington; Sherrod et al., 2012), where signal intensities for individual peptide sequences for each of the significantly altered proteins were quantified relative to each sample and normalized to a reference standard.
Detection of MAPK phosphorylation
Mitogen‐activated protein kinases phosphorylation detection was performed as per a previous assay with little changes (Yamaguchi and Kawasaki, 2017). Suspension cell samples were grinded in liquid nitrogen and the ground powder was added to extraction buffer (50 mm HEPES (pH 7.4), 5 mm EDTA, 5 mm EGTA, 50 mm β‐glycerophosphate, 10 mm Na3VO4, 10 mm NaF, 2 mm DTT, cOmplete EDTA‐free protease inhibitor cocktail (Roche), 1% SDS or Triton X‐100). The Pierce 660 nm Protein Assay was used to determine total protein concentration (Thermo Scientific). Immunoblot of phosphorylated MAPKs was performed using anti‐phospho‐p44/42 MAPK antibody.
Competing financial interests
The authors declare that they have no competing financial interests.
Author contribution statement
P. W. and Y. K. designed the study; P. W., T.G., J.L, S. Y., K. K., Y. F., Y. Z., H. Z., Y. S., P. W., R. L. and Y. K. performed experiments and analysed data; P. W., R. L. and Y. K. wrote the manuscript; W.X., Y. F., H. Z., T.K.K., Y. S., K. H., R. L. and Y. K. gave technical support and conceptual advice.
Supporting information
Figure S1 Defence genes expression during fungus infection or chitin treatment.
Figure S2 Clustering analysis of the expression profiles of up‐regulated SSP genes.
Figure S3 Phylogenetic analysis of RALFs in rice and Arabidopsis.
Figure S4 Expression analysis of OsPSK4 and phylogenetic analysis of PSKs in rice and Arabidopsis.
Figure S5 Phylogenetic tree of IRP.
Figure S6 SDS‐PAGE analysis of protein in medium sample after anion‐exchange chromatography and acetone precipitation.
Figure S7 Phenotype of plants overexpressing IRP.
Table S1 GO terms of up‐regulated genes overlapping in plants and suspension cells.
Table S2 SSPs identified in our study.
Table S3 SSPs with post translational modifications.
Table S4 Primer list.
Acknowledgements
We are grateful to Dr. Rahul Mohan Singh for editing our manuscript, Mr. Junfei Pan and Mr. Aojun Chen for performing the RNA‐Seq experiments and Mr. Jun Ma for performing LC‐MS/MS experiments. We thank the members of the Core Facility of Transformation and the Laboratory of Signal Transduction and Immunity at PSC for valuable supports. This work was supported by Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences, Shanghai Center for Plant Stress Biology, CAS Center of Excellence for Molecular Plant Sciences, Strategic Priority Research Program of the Chinese Academy of Sciences (B) (XDB27040202), Chinese Academy of Sciences Hundred Talents Program (173176001000162114), the National Natural Science Foundation in China (31572073 and 31772246), JSPS KAKENHI and the Takeda Science Foundation. K. K. was supported by CAS President's International Fellowship Initiative (2019PB0056).
Contributor Information
Renyi Liu, Email: ryliu@fafu.edu.cn.
Yoji Kawano, Email: yoji.kawano@icloud.com.
References
- Aibar, S. , Fontanillo, C. , Droste, C. and De Las Rivas, J. (2015) Functional Gene Networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics, 31, 1686–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu, A. , Wong, H.L. , Fujiwara, M. , Okuda, J. , Nishide, K. , Uno, K. , Imai, K. et al. (2013) An OsCEBiP/OsCERK1‐OsRacGEF1‐OsRac1 module is an essential early component of chitin‐induced rice immunity. Cell Host Microbe, 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Alexander, D. , Goodman, R.M. , Gut‐Rella, M. , Glascock, C. , Weymann, K. , Friedrich, L. , Maddox, D. et al. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis‐related protein 1a. Proc. Natl Acad. Sci. USA, 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, N.J. , Lilley, C.J. and Urwin, P.E. (2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162, 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Molecular Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Buttner, M. and Singh, K.B. (1997) Arabidopsis thaliana ethylene‐responsive element binding protein (AtEBP), an ethylene‐inducible, GCC box DNA‐binding protein interacts with an ocs element binding protein. Proc. Natl Acad. Sci. USA, 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. and Shi, F. (2012) Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol. Bioinf. Online, 8, 271–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.C. , Siems, W.F. , Pearce, G. and Ryan, C.A. (2008) Six peptide wound signals derived from a single precursor protein in Ipomoea batatas leaves activate the expression of the defense gene sporamin. J. Biol. Chem. 283, 11469–11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Dong, Y. , Yu, C. , Fang, X. , Deng, Z. , Yan, C. and Chen, J. (2016) Analysis of the proteins secreted from the Oryza meyeriana suspension‐cultured cells induced by Xanthomonas oryzae pv. oryzae . PLoS ONE, 11, e0154793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. , Liu, H. , Deng, Y. , Xiao, J. , Li, X. and Wang, S. (2015) The WRKY45‐2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.W. and Klessig, D.F. (2016) DAMPs, MAMPs and NAMPs in plant innate immunity. BMC Plant Biol. 16, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo, T. , Miyamoto, K. , Shimogawa, T. , Shimizu, T. , Otake, Y. , Yokotani, N. , Nishizawa, Y. et al. (2013) OsWRKY28, a PAMP‐responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. [DOI] [PubMed] [Google Scholar]
- Cox, M.P. , Peterson, D.A. and Biggs, P.J. (2010) SolexaQA: at‐a‐glance quality assessment of Illumina second‐generation sequencing data. BMC Bioinform. 11, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo, A. and Brisse, S. (2014) AlienTrimmer removes adapter oligonucleotides with high sensitivity in short‐insert paired‐end reads. Commentary on Turner Assessment of insert sizes and adapter content in FASTQ data from NexteraXT libraries. Front. Genet. 5, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzger, M. , Vasconcelos, I.M. , Scorsato, V. , Aparicio, R. , Marangoni, S. and Macedo, M.L. (2015) Bowman‐Birk proteinase inhibitor from Clitoria fairchildiana seeds: isolation, biochemical properties and insecticidal potential. Phytochemistry, 118, 224–235. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C. , Brand, U. , Running, M.P. , Simon, R. and Meyerowitz, E.M. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science, 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Gully, K. , Pelletier, S. , Guillou, M.C. , Ferrand, M. , Aligon, S. , Pokotylo, I. , Perrin, A. et al. (2019) The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana . J. Exp. Bot. 70, 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust, A.A. , Pruitt, R. and Nurnberger, T. (2017) Sensing danger: key to activating plant immunity. Trends Plant Sci. 9, 779–791. [DOI] [PubMed] [Google Scholar]
- Hayakawa, T. , Yamaya, T. , Kamachi, K. and Ojima, K. (1992) Purification, characterization and immunological properties of NADH‐dependent glutamate synthase from rice cell cultures. Plant Physiol. 98, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, R. , Wu, J. , Zhang, Y. , Aguero, C.B. , Li, X. , Liu, S. , Wang, C. et al. (2017) Overexpression of a thaumatin‐like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine . Protoplasma, 254, 1579–1589. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hou, S. , Wang, X. , Chen, D. , Yang, X. , Wang, M. , Turra, D. , Di Pietro, A. et al. (2014) The secreted peptide PIP1 amplifies immunity through receptor‐like kinase 7. PLoS Pathog. 10, e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Pearce, G. and Ryan, C.A. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl Acad. Sci. USA, 103, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, H. , Gheysen, G. , Ullah, C. , Verbeek, R. , Shang, C. , De Vleesschauwer, D. , Hofte, M. et al. (2015) The role of thionins in rice defence against root pathogens. Molecular Plant Pathol. 16, 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader, J.C. (1996) Lipid‐transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 627–654. [DOI] [PubMed] [Google Scholar]
- Kim, N.H. and Hwang, B.K. (2015) Pepper pathogenesis‐related protein 4c is a plasma membrane‐localized cysteine protease inhibitor that is required for plant cell death and defense signaling. Plant J. 81, 81–94. [DOI] [PubMed] [Google Scholar]
- Kim, S.T. , Kang, Y.H. , Wang, Y. , Wu, J. , Park, Z.Y. , Rakwal, R. , Agrawal, G.K. et al. (2009) Secretome analysis of differentially induced proteins in rice suspension‐cultured cells triggered by rice blast fungus and elicitor. Proteomics, 9, 1302–1313. [DOI] [PubMed] [Google Scholar]
- Krol, E. , Mentzel, T. , Chinchilla, D. , Boller, T. , Felix, G. , Kemmerling, B. , Postel, S. et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, A. , Huang, J. , Zhao, W. , Peng, Y. , Chen, Z. and Kang, D. (2013) A salicylic acid‐induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana . Front. Plant Sci. 15, 452–461. [DOI] [PubMed] [Google Scholar]
- Li, R. , Zhang, J. , Li, J. , Zhou, G. , Wang, Q. , Bian, W. , Erb, M. et al. (2015) Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non‐target herbivores. Elife, 4, e04805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Chen, X. , Liu, J. , Ye, J. and Guo, Z. (2012) The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Zhang, X. , Lu, C. , Zeng, X. , Li, Y. , Fu, D. and Wu, G. (2015) Non‐specific lipid transfer proteins in plants: presenting new advances and an integrated functional analysis. J. Exp. Bot. 66, 5663–5681. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loivamaki, M. , Stuhrwohldt, N. , Deeken, R. , Steffens, B. , Roitsch, T. , Hedrich, R. and Sauter, M. (2010) A role for PSK signaling in wounding and microbial interactions in Arabidopsis. Plant Physiol. 139, 348–357. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Ma, H. , Zhao, H. , Liu, Z. and Zhao, J. (2011) The phytocyanin gene family in rice (Oryza sativa L.): genome‐wide identification, classification and transcriptional analysis. PLoS ONE, 6, e25184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majovsky, P. , Naumann, C. , Lee, C.W. , Lassowskat, I. , Trujillo, M. , Dissmeyer, N. and Hoehenwarter, W. (2014) Targeted proteomics analysis of protein degradation in plant signaling on an LTQ‐Orbitrap mass spectrometer. J. Proteome Res. 13, 4246–4258. [DOI] [PubMed] [Google Scholar]
- Manosalva, P.M. , Davidson, R.M. , Liu, B. , Zhu, X. , Hulbert, S.H. , Leung, H. and Leach, J.E. (2009) A germin‐like protein gene family functions as a complex quantitative trait locus conferring broad‐spectrum disease resistance in rice. Plant Physiol. 149, 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, D.J. , Lucy, M.C. and D Geisert, R. (2016) Interleukins, interferons and establishment of pregnancy in pigs. Reproduction, 151, R111–R122. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. (2014) Posttranslationally modified small‐peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. and Sakagami, Y. (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl Acad. Sci. USA, 93, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y. and Sakagami, Y. (2006) Peptide hormones in plants. Annu. Rev. Plant Biol. 57, 649–674. [DOI] [PubMed] [Google Scholar]
- Misra, R.C. , Sandeep, Kamthan, M. , Kumar, S. and Ghosh, S. (2016) A thaumatin‐like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis . Sci. Rep. 6, 25340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan, J. and Zipfel, C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Mosher, S. , Seybold, H. , Rodriguez, P. , Stahl, M. , Davies, K.A. , Dayaratne, S. , Morillo, S.A. et al. (2013) The tyrosine‐sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73, 469–482. [DOI] [PubMed] [Google Scholar]
- Murphy, E. , Smith, S. and De Smet, I. (2012) Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell, 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami, K. , Okamoto, M. , Higuchi‐Takeuchi, M. , Yoshizumi, T. , Yamaguchi, Y. , Fukao, Y. , Shimizu, M. et al. (2018) AtPep3 is a hormone‐like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl Acad. Sci. USA, 115, 5810–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. , Roux, M. and Zipfel, C. (2009) Recent advances in PAMP‐triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150, 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, I.S. , Park, A.R. , Bae, M.S. , Kwon, S.J. , Kim, Y.S. , Lee, J.E. , Kang, N.Y. et al. (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola . Plant Cell, 17, 2832–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme‐Takagi, M. and Shinshi, H. (1995) Ethylene‐inducible DNA binding proteins that interact with an ethylene‐responsive element. Plant Cell, 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira‐Lima, M. , Benko‐Iseppon, A.M. , Neto, J.R. , Rodriguez‐Decuadro, S. , Kido, E.A. , Crovella, S. and Pandolfi, V. (2017) Snakin: structure, roles and applications of a plant antimicrobial peptide. Curr. Protein Pept. Sci. 18, 368–374. [DOI] [PubMed] [Google Scholar]
- Pearce, G. , Strydom, D. , Johnson, S. and Ryan, C.A. (1991) A polypeptide from tomato leaves induces wound‐inducible proteinase inhibitor proteins. Science, 253, 895–897. [DOI] [PubMed] [Google Scholar]
- Pearce, G. , Moura, D.S. , Stratmann, J. and Ryan, C.A. (2001a) Production of multiple plant hormones from a single polyprotein precursor. Nature, 411, 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce, G. , Moura, D.S. , Stratmann, J. and Ryan, C.A. (2001b) RALF, a 5‐kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl Acad. Sci. USA, 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G. , Yamaguchi, Y. , Barona, G. and Ryan, C.A. (2010) A subtilisin‐like protein from soybean contains an embedded, cryptic signal that activates defense‐related genes. Proc. Natl Acad. Sci. USA, 107, 14921–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic, M. andjelkovic, U. , Burazer, L. , Lindner, B. , Petersen, A. and Gavrovic‐Jankulovic, M. (2013) Biochemical and immunological characterization of a recombinantly‐produced antifungal cysteine proteinase inhibitor from green kiwifruit (Actinidia deliciosa). Phytochemistry, 94, 53–59. [DOI] [PubMed] [Google Scholar]
- Qu, L.J. (2003) Molecular cloning and functional analysis of a novel type of Bowman‐Birk inhibitor gene family in rice. Plant Physiol. 133, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D. , McCarthy, D.J. and Smyth, G.K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini, S. , Casazza, A.P. , Engelmann, E.C. , Havaux, M. , Jennings, R.C. and Soave, C. (2006) Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 141, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Sauter, M. (2015) Phytosulfokine peptide signalling. J. Exp. Bot. 66, 5161–5169. [DOI] [PubMed] [Google Scholar]
- Sawa, S. , Kinoshita, A. , Betsuyaku, S. and Fukuda, H. (2008) A large family of genes that share homology with CLE domain in Arabidopsis and rice. Plant Signaling Behavior, 3, 337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A. , LeClere, S. , Carroll, M.J. , Alborn, H.T. and Teal, P.E. (2007) Cowpea chloroplastic ATP synthase is the source of multiple plant defense elicitors during insect herbivory. Plant Physiol. 144, 793–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L. , Li, C. , Chen, F. , Zhao, S. and Xia, G. (2008) A Bowman‐Birk type protease inhibitor is involved in the tolerance to salt stress in wheat. Plant Cell Environ. 31, 1128–1137. [DOI] [PubMed] [Google Scholar]
- Sharma, A. , Hussain, A. , Mun, B.G. , Imran, Q.M. , Falak, N. , Lee, S.U. , Kim, J.Y. et al. (2016) Comprehensive analysis of plant rapid alkalization factor (RALF) genes. Plant Physiol. Biochem. 106, 82–90. [DOI] [PubMed] [Google Scholar]
- Sherrod, S.D. , Myers, M.V. , Li, M. , Myers, J.S. , Carpenter, K.L. , Maclean, B. , Maccoss, M.J. et al. (2012) Label‐free quantitation of protein modifications by pseudo selected reaction monitoring with internal reference peptides. J. Proteome Res. 11, 3467–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Liu, Z. , Zhu, L. , Zhang, C. , Chen, Y. , Zhou, Y. , Li, F. et al. (2012) Overexpression of cotton (Gossypium hirsutum) dirigent 1 gene enhances lignification that blocks the spread of Verticillium dahliae . Acta Biochim. Biophys. Sin. 44, 555–564. [DOI] [PubMed] [Google Scholar]
- Srivastava, R. , Liu, J.X. and Howell, S.H. (2008) Proteolytic processing of a precursor protein for a growth‐promoting peptide by a subtilisin serine protease in Arabidopsis . Plant J. 56, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann, M. , Monaghan, J. , Smakowska‐Luzan, E. , Rovenich, H. , Lehner, A. , Holton, N. , Belkhadir, Y. et al. (2017) The receptor kinase FER is a RALF‐regulated scaffold controlling plant immune signaling. Science, 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Suzuki, T. , Osakabe, Y. , Betsuyaku, S. , Kondo, Y. , Dohmae, N. , Fukuda, H. et al. (2018) A small peptide modulates stomatal control via abscisic acid in long‐distance signalling. Nature, 556, 235–238. [DOI] [PubMed] [Google Scholar]
- Tran, H.T. and Plaxton, W.C. (2008) Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics, 8, 4317–4326. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. and Salzberg, S.L. (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. , Yamashita‐Yamada, M. , Hirase, T. , Fujiwara, T. , Tsuda, K. , Hiruma, K. and Saijo, Y. (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen‐induced depletion of BAK1. EMBO J. 35, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. and Huffaker, A. (2011) Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351–357. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K. and Kawasaki, T. (2017) Chitin‐triggered MAPK activation and ROS generation in rice suspension‐cultured cells. Methods Mol. Biol. 1578, 309–316. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Huffaker, A. , Bryan, A.C. , Tax, F.E. and Ryan, C.A. (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis . Plant Cell, 22, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C. , Wang, Y. , Li, Y. , Bhatti, K.H. , Tian, Y. and Wu, J. (2011) Overexpression of a cotton cyclophilin gene (GhCyp1) in transgenic tobacco plants confers dual tolerance to salt stress and Pseudomonas syringae pv. tabaci infection. Plant Physiol. Biochem. 49, 1264–1271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Defence genes expression during fungus infection or chitin treatment.
Figure S2 Clustering analysis of the expression profiles of up‐regulated SSP genes.
Figure S3 Phylogenetic analysis of RALFs in rice and Arabidopsis.
Figure S4 Expression analysis of OsPSK4 and phylogenetic analysis of PSKs in rice and Arabidopsis.
Figure S5 Phylogenetic tree of IRP.
Figure S6 SDS‐PAGE analysis of protein in medium sample after anion‐exchange chromatography and acetone precipitation.
Figure S7 Phenotype of plants overexpressing IRP.
Table S1 GO terms of up‐regulated genes overlapping in plants and suspension cells.
Table S2 SSPs identified in our study.
Table S3 SSPs with post translational modifications.
Table S4 Primer list.


