Abstract
This special communication describes activities, products, and lessons learned from a recent hackathon that was funded by the National Center for Advancing Translational Sciences via the Biomedical Data Translator program (‘Translator’). Specifically, Translator team members self-organized and worked together to conceptualize and execute, over a five-day period, a multi-institutional clinical research study that aimed to examine, using open clinical data sources, relationships between sex, obesity, diabetes, and exposure to airborne fine particulate matter among patients with severe asthma. The goal was to develop a proof of concept that this new model of collaboration and data sharing could effectively produce meaningful scientific results and generate new scientific hypotheses. Three Translator Clinical Knowledge Sources, each of which provides open access (via Application Programming Interfaces) to data derived from the electronic health record systems of major academic institutions, served as the source of study data. Jupyter Python notebooks, shared in GitHub repositories, were used to call the knowledge sources and analyze and integrate the results. The results replicated established or suspected relationships between sex, obesity, diabetes, exposure to airborne fine particulate matter, and severe asthma. In addition, the results demonstrated specific differences across the three Translator Clinical Knowledge Sources, suggesting cohort- and/or environment-specific factors related to the services themselves or the catchment area from which each service derives patient data. Collectively, this special communication demonstrates the power and utility of intense, team-oriented hackathons and offers general technical, organizational, and scientific lessons learned.
Keywords: hackathon, open data, clinical data, team science, application programming interface, multi-institutional collaboration
Graphical Abstract
1. Introduction
The Biomedical Data Translator Consortium currently comprises 11 teams, representing 28 institutions and ~200 team members. Consortium members have been working to overcome challenges in the sharing, reuse, and integration of data as part of the Biomedical Data Translator program (‘Translator’), funded by the National Center for Advancing Translational Sciences (NCATS; The Biomedical Data Translator Consortium 2019a;b). The program aims to “bridge the current symptom-based diagnosis of disease with research-based molecular and cellular characterizations through an informatics platform that enables interrogation of relationships across the full spectrum of data types, from disease names, to clinical signs and symptoms, organ and cell pathology, genomics, and drug effects” (Austin et al. 2019). The Translator program draws on the combined principles and practices of open science and collaboration, open source software development, agile science and software development, team science, and open community engagement (Hammond 2010; Bennet and Gadlin 2012; Fecher and Friesike 2014; National Research Council 2015). Open access to clinical data is critical as part of this effort. However, the many regulations that surround the use of clinical data, while critical to ensure and respect patient privacy, often hinder access to the data for research purposes. Moreover, the ability to share clinical data across institutions is central to open team science and the success of multi-institutional consortia (Burwell et al. 2013). Yet, numerous sociological and technical barriers challenge such efforts, including cultural norms that promote individual achievement over collective advancement, disciplinary nuances that present integration and communication challenges, and disparities and inconsistencies across resources that encumber efforts to share and reuse data (Cutcher-Gershenfeld et al. 2017).
To overcome these challenges and achieve the ambitious goals of the program, NCATS leadership has fostered a multi-institutional culture and community that promotes collaboration and team engagement, including regular in-person hackathons (The Biomedical Data Translator Consortium 2019b). Herein, we describe several open approaches to share clinical data that have been developed as part of the Translator program—Clinical Profiles, the Integrated Clinical and Environmental Exposures Service (ICEES), and Columbia Open Health Data (COHD) (see Ahalt et al. 2019 for overview). We focus on the successful application of these open clinical data sources in a research study that was conceptualized and executed over the course of a five-day Translator hackathon. While the hackathon writ large was focused primarily on software development and testing of the prototype Translator system, we describe the application of Translator Clinical ‘Knowledge Sources’ as one productive activity that took place during the broader hackathon event. We conclude with a discussion of technical, organizational, and scientific lessons learned.
2. Materials and methods
2.1. Hackathon Structure and Methodology
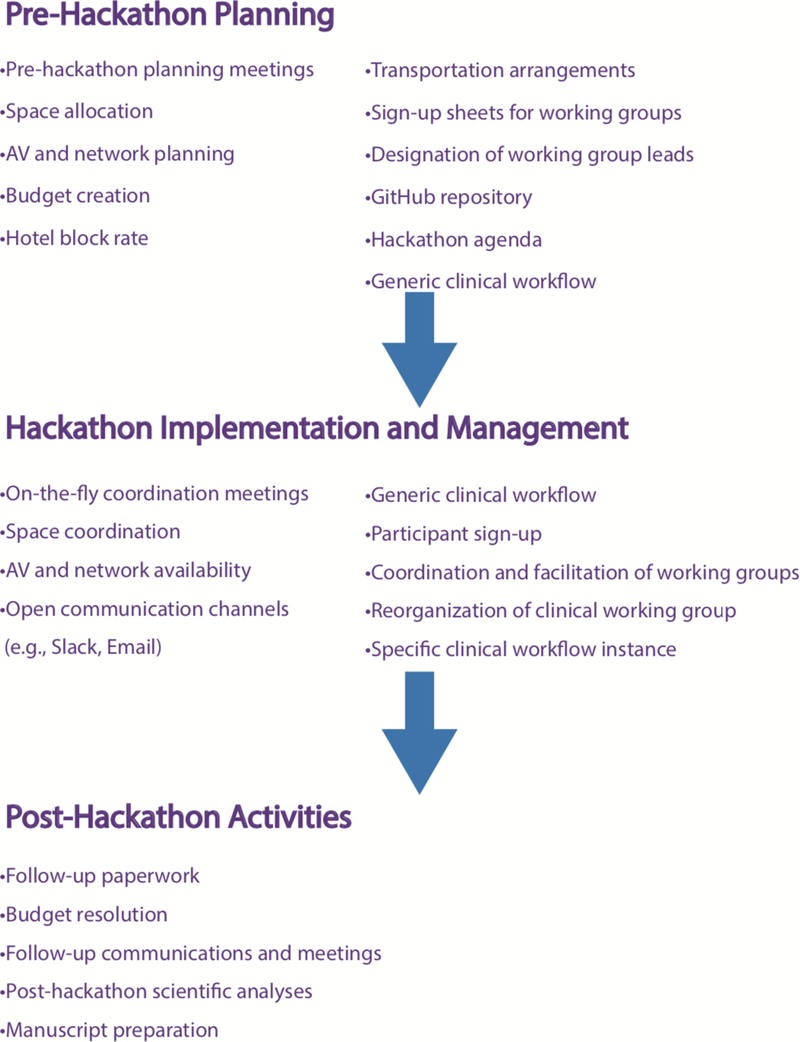
As part of the Translator’s grounding in open science, collaboration, and software development, the Translator program has held regular, in-person cross-team hackathons three times per year, for 2 ½ to 3 days each, from its inception in 2016 through 2018. In 2019, the program’s leadership decided to change the structure of the hackathons and hold two five-day hackathons over the course of the year. This paper describes a subset of the activities and scientific applications that took place during the most recent hackathon, which was held on March 4–8, 2019 at the Renaissance Computing Institute, University of North Carolina, Chapel Hill, North Carolina, USA. (The hackathon agenda and other relevant event information can be found in the supplementary materials.) The overall goal of the hackathon was to continue development and testing of the prototype Translator system in an effort to reveal new connections among existing data sources and facilitate discovery, new research hypotheses, disease classification, and intervention (see graphical abstract). However, the goal of the clinical working group that is the focus of this paper was to evaluate and compare the three open Translator Clinical Knowledge Sources that were developed as part of the Translator program (Clinical Profiles, ICEES, and COHD), and to determine whether those knowledge sources could be used to execute a meaningful research project over the course of the five-day event. The overall structure of the hackathon, as well as the key activities that we believe contributed to the overall success of the hackathon, are provided in Figure 1, and discussed in greater detail in Section 3.
Figure 1.
Hackathon flowchart, showing the three major operational components of the event and highlighting key example activities associated with each component. Pre-hackathon planning helped to guide group discussions during the first day or two of the hackathon. Hackathon implementation and management was aided by the pre-hackathon planning, although it did involve a nimble approach to respond to unexpected emergent issues. Post-hackathon activities focused largely on scientific outcomes.
2.2. Scientific Structure and Methodology
The structure for the scientific research that is described in this manuscript and that was designed and implemented during the hackathon was minimal. However, a critical factor was that, prior to the hackathon, Translator team members had developed Clinical Profiles; ICEES; and COHD (Table 1). Each of these services is openly accessible via an Application Programming Interface (API) and provides access to data derived from the electronic health record (EHR) systems of different academic institutions. Briefly, Clinical Profiles represent statistical profiles of disease and associated phenotypic presentation, derived from observational data on patients from Johns Hopkins Medicine, adapting the Evidence-Based Medicine draft of the Health Level Seven International Fast Healthcare Interoperability Resources (FHIR) standard. Clinical Profiles are designed to be disease-agnostic and currently offer open access to profiles on patients with asthma, diabetes, or Ehlers-Danlos Syndrome. ICEES provides open access to observational data on patients from UNC Health Care System (Fecho et al. 2019). The clinical data have been integrated with a variety of publicly available data on environmental exposures (e.g., airborne pollutants, socioeconomic factors), using a complex, space- and time-dependent data-extraction and integration pipeline, termed Clinical Asset Mapping Program for FHIR (CAMP FHIR) and FHIR Patient data Integration Tool (FHIR PIT) (Pfaff et al. 2019). Like Clinical Profiles, ICEES is designed as a disease-agnostic service and currently offers access to clinical data on patients with ‘asthma-like’ conditions. COHD provides open access to observational data on patients from Columbia University Irving Medical Center (CUIMC; Ta et al. 2018). Unlike ICEES or Clinical Profiles, COHD offers occurrence and co-occurrence rates of conditions, drugs, and procedures across all patients at CUIMC. Thus, all three open Translator Clinical Knowledge Sources bear similarities and differences, and each draws on patient data from a different catchment area, thereby allowing for comparisons across patient populations and inferences regarding environmental factors that may differentially affect patient subpopulations. (A more detailed description of each knowledge source is available via the hyperlinks provided in column 4 of Table 1.)
Table 1.
Translator Clinical Knowledge Sources
| Translator Clinical Knowledge Source |
EHR System |
Brief Description | Available Cohorts1 | Relevant Hyperlinks |
|---|---|---|---|---|
| Clinical Profiles | Johns Hopkins Medicine | Statistical profiles of disease and associated phenotypic presentation, derived from observational patient data | Asthma (~60,000 patients) Diabetes (~70,000 patients) Ehlers-Danlos syndrome (~1,200 patients) |
Clinical_Profiles_GitHub_repository Clinical_Profiles_Data_Model Clinical_Profiles_Example_Output_Files_crepes LOINC2HPO_tool |
| ICEES | UNC Health Care System | Patient- or visit-level counts of observational patient data integrated at the patient and visit level with a variety of environmental exposures derived from multiple public data sources | Asthma (~160,000 patients with asthma-like conditions; ~23,000 patients in year 2010 study period) |
ICEES_OpenAPI ICEES_GitHub_Documentation ICEES_API_Example_Queries |
| COHD | CUIMC | Counts of observational clinical occurrences and co-occurrences (e.g., co-occurrences of specific diagnoses and prescribed medications), as well as their derived relative frequency and observed-expected frequency ratio | General patient population (5-year dataset of all patients at Columbia University Irving Medical Center) |
COHD_SmartAPI COHD_Initiative |
These are the currently available patient cohorts offered by the existing services. Each service is designed to be disease-agnostic and adaptable for any cohort of interest.
Abbreviations: API = Application Programming Interface; COHD = Columbia Open Health Data; crepes = clinical research engine for profile extraction & summarization; CUIMC = Columbia University Medical Center; EHR = Electronic Health Records; ICEES = Integrated Clinical and Environmental Exposures Service.
While the development of these open Translator Clinical Knowledge Sources was by itself a valuable accomplishment, a detailed plan for how these knowledge sources would be applied during the hackathon in the context of a meaningful scientific question had not been developed prior to the hackathon. However, a generic clinical workflow plan (Unertl et al. 2010) was defined prior to the hackathon (Figure 2).
Figure 2.
Scientific flowchart, showing key hackathon aspects of clinical workflow development and implementation. (A) Pre-hackathon planning focused on the development of two generic clinical workflows; this planning helped to guide group discussions during the first one and a half days of the hackathon. (B) Initial hackathon activities focused on evaluating the three open Translator Clinical Knowledge Sources that were developed as part of the Translator program, in terms of the clinical workflows and the capabilities and data available from each knowledge source. (C) Subsequent brainstorming in the context of the generic workflows and the capabilities of the clinical knowledge sources led to the development of a specific instance of the generic clinical workflows. (D) The workflow was successfully implemented and executed over days three and four of the hackathon, and a first-pass analysis of the results was conducted by day five.
On day one of the hackathon, the generic workflow plan was reviewed and Translator Clinical Knowledge Sources were evaluated in terms of available data and limitations. This activity led to the development on day two of a specific instance of the generic workflows that aimed to explore the relationship between, obesity, diabetes, exposure to airborne fine particulate matter (particulate matter ≤ 2.5 μm in diameter [PM2.5]), and severe asthma. An analysis plan also was developed and agreed upon during day two of the hackathon.
Specifically, we employed patient use of prednisone as an indicator of severe asthma. A post hoc query of ICEES confirmed that prednisone is a valid surrogate indicator of severe asthma by showing that 16.67% of patients with an asthma-like condition who were prescribed or administered prednisone had ≥ 2 annual emergency department or inpatient visits for respiratory issues versus 5.58% of patients not prescribed or administered prednisone (P < 0.0001; data not shown, same cohort as presented herein).
Jupyter Python notebooks, executed in GitHub repositories, were used to query Clinical Profiles, ICEES, and COHD, using each service’s open API. As a first-step exploratory analysis, the API queries were designed to stratify patients by sex (sex code = male or female), obesity (ICD diagnostic code = yes or no), diabetes (ICD diagnostic code = yes or no), and (in the case of ICEES) exposure to PM2.5, binned using pandas.qcut as [6.77, 47.06] versus (47.06, 114.94] μg/m3 maximum daily exposure). A Chi Square analysis, with the significance level set at α = 0.10, was used to compare associations between the four stratification variables for Clinical Profiles, ICEES, Clinical Profiles + ICEES, and COHD. The Chi Square analyses were conducted in Python. Results for Clinical Profiles and ICEES were examined independently and also jointly because those cohorts were selected specifically to include only patients with severe asthma. Results for COHD were not combined with the results for Clinical Profiles or ICEES because the COHD cohort included all patients at CUIMC and was not restricted to patients with severe asthma due to limitations of the API (see Table 1).
3. RESULTS
3.1. Hackathon Findings
Close to 100 Translator team members attended the hackathon, with most participants joining the hackathon for all five days and self-organizing into six topic-based working groups (see supplementary materials). The largest working group, with over two dozen participants, was the clinical working group.
During the first day and a half of the hackathon, members of the clinical working group brainstormed to: (1) compare and contrast the capabilities and data available through each of the Translator Clinical Knowledge Sources; and (2) develop a specific instance of one or both workflows that could be implemented using all three services and executed by the end of the hackathon. The brainstorming effort was guided by both an expert on asthma and the working group lead, who also was the lead developer of the generic workflows and thus was very familiar with them.
The group recognized that the design of COHD included all patients, without the ability to subset patients with severe asthma, and that only ICEES could provide data on environmental exposures. With those caveats in mind, the specific workflow instance that was developed aimed to examine the relationship between sex, obesity, diabetes, and exposure to fine particulate matter among patients with severe asthma. This decision was made at the end of the morning session on day two of the hackathon and was facilitated by pre-hackathon efforts on the generic workflows. Of note, the decision to focus on asthma largely reflected the fact that a subject matter expert had self-selected to join the group and lead the brainstorming effort. The generic workflows themselves were designed to be disease-agnostic.
Over the next two and a half days of the hackathon, the group implemented and executed the workflow, working largely in Jupyter Python notebooks and GitHub, and then conducted a first-pass analysis of the results. A presentation was then prepared and presented to all hackathon participants on the final day of the event. Of note, the activities and products described here represent just one of many tangible hackathon products of the clinical working group. Smaller sub-groups focused on other activities that were coordinated by the lead of the clinical working group.
3.2. Scientific Findings
Clinical Profiles, ICEES, and COHD were queried for data on sex, obesity, diabetes, and exposure to PM2.5 among patients with asthma or asthma-like conditions (Clinical Profiles, ICEES) or the general patient population (COHD).
When the data were stratified by sex (Table 2), obesity was found to be significantly more common among female patients than male patients across Translator Clinical Knowledge Sources (Clinical Profiles: 30.52% vs 17.50%, P < 0.001; ICEES: 18.01% vs 10.65%, P < 0.0001; Clinical Profiles + ICEES: 27.07% vs 15.29%, P < 0.0001; COHD: 4.80% vs 3.75%, P < 0.001). Moreover, Clinical Profiles and ICEES had higher overall rates of obesity than COHD. Results for diabetes were less consistent across Translator Clinical Knowledge Sources. For instance, diabetes was more common among females than males with Clinical Profiles (18.62% vs 13.13%, P < 0.001), less common among females than males with COHD (4.82% vs 6.35%, P < 0.001), and equally common among females and males with ICEES (22.14% vs 22.52%, N.S.).
Table 2.
Associations between obesity, diabetes, and sex as revealed by Translator Clinical Knowledge Sources: stratification by sex1
| Clinical Profiles | ICEES | Clinical Profiles + ICEES | COHD2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Total | X2, P value3 |
Male | Female | Total | X2, P value |
Male | Female | Total | X2, P value |
Male | Female | Total | X2, P value |
|
| Obesity Dx | ||||||||||||||||
| No | 1414 | 2597 | 4011 | 102.50, | 730 | 1170 | 1900 | 21.69, | 2144 | 3767 | 5911 | 132.24, | 721240 | 935389 | 1656629 | 1130.30, |
| 82.50% | 69.48% | 73.57% | P<0.001 | 89.35% | 81.99% | 84.67% | P<0.0001 | 84.71% | 72.93% | 76.81% | P<0.0001 | 96.25% | 95.20% | 95.66% | P<0.001 | |
| Yes | 300 | 1141 | 1441 | 87 | 257 | 344 | 387 | 1398 | 1785 | 28081 | 47148 | 75229 | ||||
| 17.50% | 30.52% | 26.43% | 10.65% | 18.01% | 15.33% | 15.29 | 27.07% | 23.19% | 3.75% | 4.80% | 4.34% | |||||
| Diabetes Dx | ||||||||||||||||
| No | 1489 | 3042 | 4531 | 25.30, | 633 | 1111 | 1744 | 0.04, | 2122 | 4153 | 6275 | 13.30, | 701725 | 935166 | 1636891 | 1921.60, |
| 86.87% | 81.38% | 83.11% | P<0.001 | 77.48% | 77.86% | 77.72% | P=0.9977 | 83.84% | 80.41% | 81.54% | P<0.001 | 93.65% | 95.18% | 94.52% | P<0.001 | |
| Yes | 225 | 696 | 921 | 184 | 316 | 500 | 409 | 1012 | 1421 | 47596 | 47371 | 94967 | ||||
| 13.13% | 18.62% | 16.89% | 22.52% | 22.14% | 22.28% | 16.16% | 19.59% | 18.46% | 6.35% | 4.82% | 5.48% | |||||
| Total | 1714 | 3738 | 5452 | 817 | 1427 | 2244 | 2531 | 5165 | 7696 | 749321 | 982537 | 1731858 | ||||
| 31.44% | 68.56% | 100% | 36.41% | 63.59% | 100.00% | 32.89% | 67.11% | 100.00% | 43.27% | 56.73% | 100% | |||||
Note that column percentages are provided in the table, except for the total row, which row percentages.
Unlike Clinical Profiles and ICEES, COHD results are not restricted to patients with severe asthma, but rather reflect all patients.
Abbreviations: Dx = diagnosis; ICEES = Integrated Clinical and Environmental Exposures Service; COHD = Columbia Open Health Data.
When the data were stratified by obesity (Table 3), the results showed that the proportion of females was significantly higher among obese patients than non-obese patients (Clinical Profiles: 79.18% vs 64.75%, P < 0.001; ICEES: 74.71% vs 61.58%, P < 0.0001; Clinical Profiles + ICEES: 78.32% vs 63.73%, P < 0.0001; COHD: 62.67% vs. 56.46%, P < 0.001). Likewise, diabetes was significantly more prevalent among obese patients than non-obese patients (Clinical Profiles: 29.63% vs 12.32%, P < 0.001; ICEES: 41.57% vs 18.79%, P < 0.0001; Clinical Profiles + ICEES: 31.93% vs 14.40%, P < 0.0001; COHD: 25.06% vs 4.59%, P < 0.001).
Table 3.
Associations between obesity, diabetes, and sex as revealed by Translator Clinical Knowledge Sources: stratification by obesity1
| Clinical Profiles | ICEES | Clinical Profiles + ICEES | COHD2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obesity Dx = No |
Obesity Dx = Yes |
Total | X2, P value |
Obesity Dx = No |
Obesity Dx = Yes |
Total | X2, P value |
Obesity Dx = No |
Obesity Dx = Yes |
Total | X2, P value |
Obesity Dx = No |
Obesity Dx = Yes |
Total | X2, P value |
|
| Sex | ||||||||||||||||
| Male | 1414 | 300 | 1714 | 102.50, | 730 | 87 | 817 | 21.69, | 2144 | 387 | 2531 | 132.24, | 721240 | 28081 | 749321% | 1130.30, |
| 35.25% | 20.82% | 31.44% | P<0.001 | 38.42% | 25.29% | 36.41% | P<0.0001 | 36.27% | 21.68% | 32.89% | P<0.0001 | 43.54% | 37.33% | 43.27% | P<0.001 | |
| Female | 2597 | 1141 | 3738 | 1170 | 257 | 1427 | 3767 | 1398 | 4024 | 935389 | 47148 | 982537 | ||||
| 64.75% | 79.18% | 68.56% | 61.58% | 74.71% | 63.59% | 63.73% | 78.32% | 52.29% | 56.46% | 62.67% | 56.73% | |||||
| Diabetes Dx | ||||||||||||||||
| No | 3517 | 1014 | 4531 | 226.40, | 1543 | 201 | 1744 | 87.28, | 5060 | 1215 | 6275 | 280.03, | 1580517 | 56374 | 1636891 | 58173.80, |
| 87.68% | 70.37% | 83.11% | P<0.001 | 81.21% | 58.43% | 77.72% | P<0.0001 | 85.60% | 68.07% | 81.54% | P<0.0001 | 95.41% | 74.94% | 94.52% | P<0.001 | |
| Yes | 494 | 427 | 921 | 357 | 143 | 500 | 851 | 570 | 1421 | 76112 | 18855 | 94967 | ||||
| 12.32% | 29.63% | 16.89% | 18.79% | 41.57% | 22.28% | 14.40% | 31.93% | 18.46% | 4.59% | 25.06% | 5.48% | |||||
| Total | 4011 | 1441 | 5452 | 1900 | 344 | 2244 | 5911 | 1785 | 7696 | 1656629 | 75229 | 1731858 | ||||
| 73.57% | 26.43% | 100% | 84.67% | 15.33% | 100.00% | 76.81% | 23.19% | 100.00% | 95.66% | 4.34% | 100% | |||||
Note that column percentages are provided in the table, except for the total row, which row percentages.
Unlike Clinical Profiles and ICEES, COHD results are not restricted to patients with severe asthma, but rather reflect all patients.
Abbreviations: Dx = diagnosis; ICEES = Integrated Clinical and Environmental Exposures Service; COHD = Columbia Open Health Data.
When the data were stratified by diabetes (Table 4), obesity was found to be more prevalent among patients with diabetes than among patients without diabetes (Clinical Profiles: 46.36% vs 22.38%, P < 0.001; ICEES: 28.60% vs 11.53%, P < 0.0001; Clinical Profiles + ICEES: 40.11% vs. 19.36%, P < 0.0001; COHD: 19.85% vs. 3.44%, P < 0.001). Clinical Profiles and ICEES had higher overall rates of obesity among patients with diabetes than COHD. As with the results shown in Table 2, the relationship between diabetes and sex was not consistent across Translator Clinical Knowledge Sources.
Table 4.
Associations between obesity, diabetes, and sex as revealed by Translator Clinical Knowledge Sources: stratification by diabetes1
| Clinical Profiles | ICEES | Clinical Profiles + ICEES | COHD2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diabetes Dx = No |
Diabetes Dx = Yes |
Total | X2, P value |
Diabetes Dx = No |
Diabetes Dx = Yes |
Total | X2, P value |
Diabetes Dx = No |
Diabetes Dx = Yes |
Total | X2, P value |
Diabetes Dx = No |
Diabetes Dx = Yes |
Total | X2, P value |
|
| Sex | ||||||||||||||||
| Male | 1489 | 225 | 1714 | 25.30, | 633 | 184 | 817 | 0.04, | 2122 | 409 | 2531 | 13.30, | 701725 | 47596 | 749321 | 1921.60, |
| 32.86% | 24.42% | 31.44% | P<0.001 | 36.30% | 36.80% | 36.41% | P=0.9977 | 33.82% | 28.78% | 32.89% | P<0.0003 | 42.87% | 50.12% | 43.27% | P<0.001 | |
| Female | 3042 | 696 | 3738 | 1111 | 316 | 1427 | 4153 | 1012 | 5165 | 935166 | 47371 | 982537 | ||||
| 67.14% | 75.57% | 68.56% | 63.70% | 63.20% | 63.59% | 66.18% | 71.22% | 67.11% | 57.13% | 49.88% | 56.73% | |||||
| Obesity Dx | ||||||||||||||||
| No | 3517 | 494 | 4011 | 226.40, | 1543 | 357 | 1900 | 87.28, | 5060 | 851 | 5911 | 280.03, | 1580517 | 76112 | 1656629 | 58173.8, |
| 77.62% | 53.64% | 73.57% | P<0.001 | 88.47% | 71.40% | 84.67% | P<0.0001 | 80.64% | 59.89% | 76.81% | P<0.0001 | 96.56% | 80.15% | 95.66% | P<0.001 | |
| Yes | 1014 | 427 | 1441 | 201 | 143 | 344 | 1215 | 570 | 1785 | 56374 | 18855 | 75229 | ||||
| 22.38% | 46.36% | 26.43% | 11.53% | 28.60% | 15.33% | 19.36% | 40.11% | 23.19% | 3.44% | 19.85% | 4.34% | |||||
| Total | 4531 | 921 | 5452 | 1744 | 500 | 2244 | 6275 | 1421 | 7696 | 1636891 | 94967 | 1731858 | ||||
| 83.11% | 16.89% | 100% | 77.72% | 22.28% | 100.00% | 81.54% | 18.46% | 100.00% | 94.52% | 5.48% | 100% | |||||
Note that column percentages are provided in the table, except for the total row, which row percentages.
Unlike Clinical Profiles and ICEES, COHD results are not restricted to patients with severe asthma, but rather reflect all patients.
Abbreviations: Dx = diagnosis; ICEES = Integrated Clinical and Environmental Exposures Service; COHD = Columbia Open Health Data.
ICEES was used to examine the relationship between exposure to PM2.5 and sex, obesity, and diabetes among patients with severe asthma (Table 5). The results showed that obesity and diabetes were more common among patients exposed to relatively high levels of PM2.5 than among those exposed to relative low levels of PM2.5 (obesity: 17.83% vs 13.62%, P = 0.0593; diabetes: 26.21% vs 19.59%, P < 0.01). There was no relationship between exposure to PM2.5 and sex.
Table 5.
Associations between sex, obesity, diabetes, and airborne fine particulate matter among patients with severe asthma, as revealed by ICEES: stratification by exposure to high levels of fine particulate matter1
| ICEES | ||||
|---|---|---|---|---|
| Average Maximum Daily PM2.5 Exposure Bin: [6.77, 47.06 μg/m3] |
Average Maximum Daily PM2.5 Exposure Bin: (47.06, 114.94 μg/m3] |
Total | X2, P value |
|
| Sex | ||||
| Male | 461 | 352 | 817 | 1.52, |
| 35.27% | 37.81% | 36.41% | P=0.6781 | |
| Female | 846 | 579 | 1427 | |
| 64.73% | 62.19% | 63.59% | ||
| Obesity | ||||
| No | 1129 | 765 | 1900 | 7.43, |
| 86.38% | 82.17% | 84.67% | P=0.0593 | |
| Yes | 178 | 166 | 344 | |
| 13.62% | 17.83% | 15.33% | ||
| Diabetes | ||||
| No | 1051 | 687 | 1744 | 13.77, |
| 80.41% | 73.79% | 77.72% | P<0.01 | |
| Yes | 256 | 244 | 500 | |
| 19.59% | 26.21% | 22.28% | ||
| Total | 1307 | 931 | 2244 | |
| 58.24% | 41.49% | 100.00% | ||
Note that column percentages are provided in the table, except for the “total” row, which lists row percentages.
4. DISCUSSION
4.1. Hackathon Lessons Learned
We assert that intensive hackathons provide an opportunity for large, multi-institutional consortia to engage in team science and collaboratively address relevant scientific questions, producing results that are informative and scientifically meaningful (Figure 1). The clinical working group that convened as part of the Translator hackathon was highly productive; we believe that several key aspects of the hackathon contributed to this success.
First, participants embraced the tenets of open, agile, and team-based science and software development (Hammond 2010; Bennet and Gadlin 2012; Fecher and Friesike 2014; National Research Council 2015), as promoted by the Translator program. In that spirit, the hackathon was largely self-organized and self-led, and an open GitHub repository served as the focal point for software development and related activities. The choice to participate in the hackathon was partially driven by geographical location and the availability of travel funds, but it was largely driven by enthusiasm and desire to participate, as well as positive experiences during prior hackathons.
Second, while NCATS developed a loose agenda for the hackathon (see supplementary materials), the event was mostly unstructured and comprised of pure ‘hacking’ sessions, although key logistics were carefully prepared prior to the event, including space allocation, hotel reservations, transportation, sign-up sheets for working groups, etc. NCATS set the goals for the hackathon and assigned a lead to each working group in advance of the event, but other team members were free to assign themselves to a working group of their choice.
Third, in addition to the working group lead, the specific scientific question that is the focus of this manuscript was guided by an expert on the use case, i.e., asthma. This helped to keep the interdisciplinary team focused on clinically relevant questions. Note, however, that the choice of subject matter simply reflected the fact that an expert on asthma self-selected to join the group and lead the brainstorming effort. The generic workflows and specific workflow instance that was implemented during the hackathon could very well have focused on another subject.
Fourth, the self-organized clinical working group was quite large at the start of the hackathon, and it quickly became clear that the entire group would not be able to efficiently coordinate their efforts on any single activity and thereby produce a viable hackathon product. Thus, with some direction from the group leader, and with the willingness of the group, as well as support from NCATS to reorganize on an ad hoc basis and function as more of an ‘unconference’ (Budd et al. 2015), the larger group divided into smaller sub-groups focused on different scientific questions and hackathon activities. This nimble ‘divide-and-conquer’ approach proved to be quite successful, and other outcomes of the clinical working group will be reported elsewhere. Moreover, the discussions that took place during the hackathon, and the experience itself, led to the conceptualization and initiation of several new projects. For example, several members of the clinical working group are now working to develop a shared Translator Clinical Knowledge Sources, one that adheres to the Translator program’s open API standards and allows users to select functionalities and/or services and directly compare results across services, without requiring separate API calls.
We would be amiss to ignore the organizational and technical challenges presented by the hackathon. First, the logistics of organizing a hackathon, particularly one involving a relatively large number of participants, need to be sorted out well in advance of the event itself.
Second, the physical location of the event needs to be conducive to participation by as many team members as possible. This is especially important for multi-institutional consortia such as The Biomedical Data Translator Consortium. In this regard, NCATS has rotated the physical location of hackathons between the East and West Coast of the continental United States. While NCATS has opted for in-person hackathons, other options such as online hackathons or community DREAM challenges are possible. These alternatives have the benefit of avoiding travel costs, but they have the potential disadvantages of remote, asynchronous participation. Careful consideration of cost versus efficiency and anticipated outcomes should be considered prior to organizing and hosting any in-person hackathon.
Third, the length of the hackathon needs to be carefully considered. For the Translator project, moving from a 2 ½- or 3-day hackathon to a 5-day hackathon presented a risk, in terms of lack of participation and hackathon burnout (Siva 2018; Swanner 2018). Most participants attended all five days of the event, although the majority of participants were from the East Coast, i.e., close to the physical location and therefore requiring less travel time to attend the event. In terms of burnout, informal discussions suggest that participants experienced a certain amount of mid-week burnout, but the energy of the event led to renewed engagement toward the end of the week.
Fourth, prior to the hackathon, a high-level clinical workflow had been developed by the leader of the clinical working group and other team members. In addition, the group leader spent time considering specific instantiations of the workflow and other possible hackathon activities and preparing for them. A challenge was that the composition of the group was not finalized until day one of the hackathon. Nonetheless, the pre-hackathon preparation allowed the group leader to suggest possible workflow instantiations and other hackathon activities at the beginning of the event, something which proved critical when the larger group divided into smaller subgroups.
In terms of evaluating the success of the hackathon, we received informal feedback, but NCATS did not conduct a formal post-hackathon survey. This decision was made largely because prior post-hackathon surveys received a low response rate and were not very informative. We did, however, receive quite a bit of positive verbal and email feedback. For example, participants provided email comments such as: “this was the best Hackathon yet”, “really liked the meeting spaces and the food”, “the meeting felt ‘chill’ despite the intensity and the high bar set by NCATS regarding expected hackathon deliverables”, “the whole event was awesome”, “this was the best…most engaging and collaborative”. While this feedback was qualitative and perhaps biased, the indication is that participants viewed the hackathon favorably and derived value from it.
4.2. Scientific Lessons Learned
With Clinical Profiles and ICEES, we were able to target queries on sex, obesity, and diabetes to patients with severe asthma and compare results to those of the general patient population, using COHD. We used these services to replicate established or suspected interactions between sex, obesity, diabetes, exposure to particulate matter, and asthma. For instance, Assad et al. (2013) found that body mass index predicts incident asthma (unadjusted hazard ratio of 1.17 per five index units; P<0.001). When that group stratified patients by sex, a significant effect was identified for women (unadjusted hazard ratio of 1.19 per five index units; P <0.001), but not men (unadjusted hazard ratio of 0.98; P = 0.60). Greenblatt et al. (2019) used EHR data to examine the incidence of asthma exacerbations (defined by ICD code for asthma and an order for oral corticosteroid) and identify contributing factors. This group found that females were overrepresented among patients with asthma exacerbations (74.7% versus 59.5% in general population). After controlling for sex and several other demographic factors, significant predictors of asthma exacerbations included chronic bronchitis, sinusitis, emphysema, fluid and electrolyte disorders, class 3 obesity, and diabetes (odds ratios of 2.70, 1.50, 1.39, 1.35, 1.32, and 1.28, respectively). A study by Requia et al. (2017) examined energy generation and fuel sales in 117 regions in Canada between 2007 and 2014 and used an over-dispersed spatiotemporal Poisson regression model to estimate risk of diabetes, asthma, and high blood pressure. These authors found a significant association between a two-year increase of 10 μg/m3 PM2.5 and increased risk in the incidence of diabetes, asthma, and high blood pressure (increased risk [95% confidence intervals] of 5.34% [2.28, 12.53], 2.24% [0.93, 5.38], and 8.29% [3.44, 19.98], respectively).
We identified similar results that are shared among the three Translator Clinical Knowledge Sources. We also identified specific differences in findings across the services, suggesting cohort- or environment-specific factors related to the catchment area from which each service derives patient data. For instance, we identified higher rates of obesity and diabetes for Clinical Profiles and ICEES when compared to COHD, suggesting that obesity and diabetes are more common among patients with severe asthma than among the general population, similar to the findings of Greenblatt et al. (2019). COHD has low rates of recall for most diagnoses, however, so any conclusions should be tempered. Nonetheless, a post hoc analysis of the raw COHD data behind the API confirmed higher rates of obesity and diabetes among patients with asthma or severe asthma (data not shown). In terms of sex differences, we found that obesity was more common among females than among males for all three Translator Clinical Knowledge Sources, although the association was more pronounced for patients with severe asthma than among the general patient population, similar to the findings of Assad et al. (2013). We also found that diabetes was more common among males than females when using COHD, a finding that is established in the literature (e.g., Kautzky-Willer et al. 2016); however, when we focused on patients with severe asthma, we found the reverse relationship with Clinical Profiles (i.e., higher rates of diabetes among females than males) and no relationship between diabetes and sex with ICEES, suggesting that the presence of asthma influences the relationship between sex and diabetes. Finally, with ICEES, we were able to confirm the results of Mirabelli et al. (2016) and Requia et al. (2017) on exposure to PM2.5 and risk of diabetes and asthma, and we extended the results to demonstrate an association between PM2.5 exposure and obesity.
4.3. Conclusion
We demonstrated as proof of concept that intense, five-day, in-person hackathons can be used to conduct meaningful science, in addition to their traditional application for software development. While hackathons are not always viewed favorably (Siva 2018; Swanner 2018), if executed properly, and with the appropriate level of leadership and team commitment, these events can be quite successful. Specifically, the use case described herein demonstrates the ability to leverage the nimble structure of the hackathon and the engagement of participants to conceptualize and execute a research study that used three open Translator Clinical Knowledge Sources derived from the EHR systems of three major academic institutions to replicate known or suspected associations between sex, obesity, diabetes, and exposure to airborne fine particulate matter among patients with severe asthma.
Supplementary Material
Highlights.
The Biomedical Data Translator Program was launched in October 2016
The Biomedical Data Translator Consortium comprises 11 teams and ~200 team members
Regular in-person hackathons have proven effective in promoting team science
We describe a hackathon activity focused on open Translator clinical data sources
Our ‘lessons learned’ have broad applicability across scientific domains
Acknowledgements
The authors wish to thank the staff at the Renaissance Computing Institute for hosting the hackathon. The authors also acknowledge and appreciate the leadership and support provided by the National Center for Advancing Translational Sciences, including Christine Colvis, Noel Southall, Tyler Beck, Grayson Donley, Tyler Peryea, Sarah Stemann, and Mark Williams. The authors note that Christine Colvis, Tyler Beck, and Sarah Stemann, in particular, were instrumental in the planning, implementation, management, and overall success of the hackathon. Finally, the authors also wish to acknowledge the intellectual input and hackathon camaraderie provided by Debbi Adelakun, Vinicius Alves, Stephen Appold, Alejandro Valencia, Joyce Borba, Maureen Hoatlin, Eugene Muratov, Charles Schmitt, Eric Sid, Lisa Stillwell, Nicholas Tatonetti, and Alexander Tropsha. While these persons did not contribute to the work described herein, they were members of the clinical working group and engaged in other productive activities during the hackathon.
Funding Statement
This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, grant numbers OT3TR002026, OT3TR002020, OT3TR002025, OT3TR002019, OT3TR002027, OT2TR002517, OT2TR002514, OT2TR002515, OT2TR002584, OT2TR002520, and UL1TR002489.
Abbreviations:
- API
Application Programming Interface
- COHD
Columbia Open Health Data
- CUIMC
Columbia University Irving Medical Center
- EHR
electronic health record
- FHIR
Health Level Seven International Fast Healthcare Interoperability Resources
- ICEES
Integrated Clinical and Environmental Exposures Service
- NCATS
National Center for Advancing Translational Sciences
- PM2.5
Particulate matter of size ≤ 2.5-microns in diameter
Footnotes
Competing Interests Statement
The authors have no competing interests to declare.
REFERENCES
- Ahalt SC, Chute CG, Fecho K, Glusman G, Hadlock J, Solbrig H, Overby-Taylor C, Pfaff E, Ta C, Tatonetti N, Weng C,* and The NCATS Biomedical Data Translator Consortium. Clinical data: sources and types, regulatory constraints, applications. Clin Transl Sci, 2019. [E-pub ahead of print] doi: 10.1111/cts.12638 *Authors are listed alphabetically https://ascpt.onlinelibrary.wiley.com/doi/full/10.1111/cts.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assad N, Qualls C, Smith LJ, Arynchyn A, Thyagarajan B, Schuyler M, Jacobs DR Jr, Sood A. Body mass index is a stronger predictor than the metabolic syndrome for future asthma in women. The longitudinal CARDIA study. Am J Respir Crit Care Med 2013;188(3):319–326. https://www.ncbi.nlm.nih.gov/pubmed/23905525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, Colvis CM, Southall NT. Deconstructing the translational tower of babel. Clin Transl Sci 2019;12(2):85. doi 10.1111/cts.12595 https://ascpt.onlinelibrary.wiley.com/doi/10.1111/cts.12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LM, Gadlin H. Collaboration and team science: from theory to practice. J Investig Med 2012;60(5):768–775. https://www.ncbi.nlm.nih.gov/pubmed/22525233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd A, Dinkel H, Corpas M, Fuller JC, Rubinat L, Devos DP, Khoueiry PH, Förstner KU, Georgatos F, Rowland F, Sharan M, Binder JX, Grace T, Traphagen K, Gristwood A, Wood NT. Ten simple rules for organizing an unconference. PloS Comput. Biol. 11, e1003905 (2015). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4310607/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell SM, VanRoekel S, Park T, Mancini DJ, Office of Management and Budget, Executive Office of the President. Memorandum M-13–13, Open Data Policy-Managing Information as an Asset, May 9, 2013. https://www.whitehouse.gov/sites/whitehouse.gov/files/omb/memoranda/2013/m-13-13.pdf
- Cutcher-Gershenfeld J, Baker KS, Berente N, Flint C, Gershenfeld G, Grant B, Haberman M, King JL, Kirkpatrick C, Lawrence B, Lewis S, Lenhardt WC, Mayernik M, McElroy C, Mittleman B, Shin N, Stall S, Winter S, Zaslavsky. Five ways consortia can catalyse open science. Nature 2017;543(7647):615–617. https://www.nature.com/polopoly_fs/1.21706!/menu/main/topColumns/topLeftColumn/pdf/543615a.pdf [DOI] [PubMed] [Google Scholar]
- Dixon AE, Holquin F. Diet and metabolism in the evolution of asthma and obesity. Clin Chest Med 2019;40(1):97–106. https://www.ncbi.nlm.nih.gov/pubmed/30691720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze IC, Hemkens LG, Bucher HC, Hoffmann B, Schindler C, Künzil N, Schikowski T, Probst-Hensch NM. Association between ambient air pollution and diabetes mellitus in Europe and North America: systematic review and meta-analysis. Environ Health Perspect 2015;123(5): 381–389. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4421762/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze IC, Schaffner E, Foraster M, Imboden M, von Eckardstein A, Gerbase MW, Rothe T, Rochat T, Künzil N, Schindler C, Probst-Hensch N. Long-term exposure to ambient air pollution and metabolic syndrome in adults. PloS ONE 2015;10(6): e0130337. doi: 10.1371/journal.pone.0130337 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0130337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecher B, Friesike S (2014). Open science: one term, five schools of thought. In Opening Science, pp. 17–47. doi: 10.1007/978-3-319-00026-8_2 http://book.openingscience.org.s3-website-eu-west-1.amazonaws.com/basics_background/open_science_one_term_five_schools_of_thought.html. [DOI] [Google Scholar]
- Fecho K, Pfaff E, Xu H, Champion J, Cox S, Stillwell L, Bizon C, Peden D, Krishnamurthy A, Tropsha A, Ahalt SC. A novel approach for exposing and sharing clinical data: the Translator Integrated Clinical and Environmental Exposures Service. J Am Med Inform Assoc., 2019. [E-pub ahead of print]. doi: 10.1093/jamia/ocs042 https://academic.oup.com/jamia/advance-article-abstract/doi/10.1093/jamia/ocz042/5480568?redirectedFrom=fulltext. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt RE, Zhao EJ, Henrickson SE, Apter AJ, Hubbard RA, Himes BE. Factors associated with exacerbations among adults with asthma according to electronic health record data. Asthma Res Pract 2019;5:1. doi: 10.1186/s40733-019-0048-7 eCollection 2019 https://www.ncbi.nlm.nih.gov/pubmed/30680222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JS (2009). Best practices: improve development effectiveness through strategic adoption of open source. Cambridge, MA: Forrester Research, Inc. [Google Scholar]
- Kautzky-Willer A, Harreiter J, Pacini G. Sex and gender differences in risk, pathophysiology and complications of type 2 diabetes mellitus. Endocr Rev 2016;37(3):278–316. doi: 10.1210/er.2015-1137 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4890267/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynyk JA, Mastronarde JG, McCallister JW. Asthma, the sex difference. Curr Opin Pulm Med 2011;17:6–11. [DOI] [PubMed] [Google Scholar]
- Mirabelli MC, Vaidyanathan A, Flanders WD, Qin X, Garbe P. Outdoor PM2.5, ambient air temperature, and asthma symptoms in the past 14 days among adults with active asthma. Environ Health Perspect 2016;124(12):1882–1890. doi: 10.1289/EHP92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, Committee on the Science of Team Science, Board on Behavioral, Cognitive, and Sensory Sciences, Division of Behavioral and Social Sciences and Education. Enhancing the effectiveness of team science. Cooke NJ and Hilton ML, Editors. Washington, DC: The National Academies Press; (2015). https://www.nap.edu/catalog/19007/enhancing-the-effectiveness-of-team-science. [PubMed] [Google Scholar]
- Pfaff ER, Champion J, Cox S, Xu H, Fecho K, Krishnamurthy A, Chute CG, Overby Taylor C, Ahalt S. All roads lead to FHIR: an extensible clinical data conversion pipeline. Accepted as a conference paper and podium presentation for the AMIA 2019 Informatics Summit, March 25–28, 2019, San Francisco, CA, USA Pfaff et al. AMIA Summit 2019 Abstract [Last accessed March 19, 2019]. [Google Scholar]
- Requia WJ, Adams MD, Koutrakis P. Association of PM2.5 with diabetes, asthma, and high blood pressure incidence in Canada: a spatiotemporal analysis of the impacts of the energy generation and fuel sales. Sci Total Environ 2017;584–585:1077–1083. [DOI] [PubMed] [Google Scholar]
- Siva V. Are hackathons good, bad, or overrated? hackerearth blog, June 6, 2018. https://www.hackerearth.com/blog/innovation-management/hackathons/good-bad-overrated/. [Google Scholar]
- Swanner N. Is it time to rethink the hackathon? Dice, February 21, 2018. https://insights.dice.com/2018/02/21/time-rethink-hackathon/ [Google Scholar]
- Ta C, Dumontier M, Hripcsak G, Tatonetti N, Weng C. Columbia Open Health Data, clinical concept prevalence and co-occurrence from electronic health records. Sci Data 2018; 5:180273. doi: 10.1038/sdata.2018.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Biomedical Data Translator Consortium. The Biomedical Data Translator program: conception, culture, and community. Clin Transl Sci 2019;12(2):91–94. doi: 10.1111/cts.12592 https://ascpt.onlinelibrary.wiley.com/doi/10.1111/cts.12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Biomedical Data Translator Consortium. Toward a universal biomedical data translator. Clin Transl Sci 2019;12(2):86–90. doi 10.1111/cts.12591 https://ascpt.onlinelibrary.wiley.com/doi/10.1111/cts.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unertl KM, Novak LL, Johnson KB, Lorenzi NM. Traversing the many paths of workflow research: developing a conceptual framework of workflow terminology through a systematic literature review. JAMIA 2010;17(3):265–273. doi: 10.1135/jamia.2010.004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.