Abstract
Introduction:
The currently available anti-HIV-1 drugs can control the infection but do not eradicate the virus. Their long-term use can lead to side effects and resistance to therapy. Therefore, eradication of the virus has been a major goal of research. Biological therapeutics including broadly neutralizing monoclonal antibodies (bnAbs) are promising tools to reach this goal. They could also help design novel vaccine immunogens potentially capable of eliciting bnAbs targeting the HIV-1 envelope glycoproteins (Envs).
Areas covered:
We review HIV-1 bnAbs and their potential as candidate prophylactics and therapeutics used individually, in combination, or as bispecific fusion proteins. We also discuss their potential use in the ‘activation-elimination’ approach for HIV-1 eradication in infected patients receiving antiretroviral treatment as well as current vaccine design efforts based on understanding of interactions of candidate vaccine immunogens with matured bnAbs and their putative germline predecessors, and related antibody maturation pathways.
Expert opinion:
Exploration of HIV-1 bnAbs has provided and will continue to provide useful knowledge that helps develop novel types of biotherapeutics and vaccines. It is possible that bnAb-based candidate therapeutics could help eradicate HIV-1. Development of vaccine immunogens capable of eliciting potent bnAbs in humans remains a fundamental challenge.
Keywords: broadly neutralizing monoclonal antibodies, eradication, HIV-1, prophylactics, therapeutics, vaccines
1. Introduction
Human immunodeficiency virus type 1 (HIV-1) is a member of the family retroviridae and can infect cells of the human immune system such as CD4+ T cells, macrophages, and dendritic cells. If resting CD4+ T cells are infected by HIV-1 or activated CD4+ T cells are infected and transition to memory cells before being killed, HIV-1 latency is established [1–4]. The latently infected cells thus become a long-lived reservoir which cannot be eliminated by highly active antiretroviral therapy (HAART). Although HAART is highly effective in suppressing viral replication, it can result in resistance to therapy and toxicity with unpredictable effects on health; if the therapy is interrupted, the virus rebounds [5, 6]. Monoclonal antibodies (mAbs) have been successful, providing favorable clinical benefits for patients with various diseases including some cancers, immune disorders, and infectious diseases [7, 8]. They are generally safer than small molecule drugs and long-term use may not lead to toxicities. However, administration of HIV-1 broadly neutralizing mAbs (bnAbs) to infected patients results in only modest and transient efficacy [9, 10]. Viral rebound occurs in all patients receiving the antibodies, suggesting that resistant viruses emerge and the antibodies have limited effects on the control of infected cells where HIV-1 replicates and spreads.
Traditional prophylactic vaccines such as those against smallpox, measles, and hepatitis A use either inactivated or attenuated viruses to elicit protective humoral and/or cellular immune responses similar to those observed in natural infections [11]. Rather than introducing whole viruses to the immune system, subunit vaccines composed of only a part of virus particle (e.g., surface proteins) can also induce sterilizing immunity [12, 13]. However, both inactivated HIV-1 and recombinant viral envelope glycoproteins (Envs) have not yet been successful to elicit potent broadly neutralizing responses [14]. The only HIV-1 vaccine trial that showed some indications of efficacy is the Phase III RV114 Thai trial where a recombinant canarypox vector vaccine combined with recombinant Env gp120 for boosting was used but its efficacy remains to be confirmed [15].
On the other battlefield against HIV – the fight for a cure, there is significant progress. An HIV-1-positive patient (Berlin, Germany) received a bone marrow transplant for treatment of leukemia from a donor who has a 32 base pair deletion in the HIV-1 coreceptor CCR5 gene [16]. The ‘Berlin patient’ showed no evidence of residual HIV-1 after the transplantation even though the patient stopped taking anti-HIV-1 drugs and still remains virus-free 5 years thereafter (http://articles.nydailynews.com/2012-07-25/news/32853069_1_berlin-patient-berlin-patient-hiv-infection). It appears that two other patients receiving similar treatments have also cleared the virus (http://www.aids2012.org/WebContent/File/AIDS2012_Media_Release_HIV_Cure_26_July_2012_EN.pdf). Importantly, in these two cases, the donors of bone marrow do not have the CCR5 mutation, suggesting that the new immune cells in the recipients could have gained control of the infection although they are fully susceptible to the virus. These discoveries raise hope that a cure for HIV is possible and urge pursuit of scalable cost-effective treatment strategies.
2. HIV-1 bnAbs
We and others had previously reviewed the identification and characterization of several rare HIV-1 bnAbs including b12, 2G12, 2F5, and 4E10, and their potential as candidate therapeutics and/or prophylactics [9, 10, 17, 18]. Here, we discuss in more details a panel of newly identified exceptionally potent bnAbs against HIV-1 (Table 1).
Table 1.
Known bnAbs targeting the HIV-1 Envs.
| Epitope | Antibody | Year reported | Comments | In vitro efficacy* | In vivo efficacy |
|---|---|---|---|---|---|
| CD4bs | b12 | 1991 | First reported HIV-1 bnAb targeting the CD4bs | Neutralizes 41% of 190 HIV-1 isolates tested [24] | Protects macaques against SHIV vaginal challenge |
| VRC01–03 | 2010 | VRC01–02closely mimic CD4 and enhance 17b binding to gp120 | VRC01–02 and VRC03 neutralize about 91% and 57% of 190 HIV-1 isolates tested, respectively [24] | VRC01 protects macaques against SHIV mucosal challenge | |
| NIH45–46 | 2011 | A clonal variant of VRC01 | Neutralizes 96% of 118 HIV-1 isolates tested [25] | NA | |
| 3BNC117 | 2011 | Neutralizes 96% of 118 HIV-1 isolates tested [25] | NA | ||
| VRC-PG04 | 2011 | Neutralizes 76% of 178 HIV-1 isolates tested [27] | NA | ||
| A12 | 2008 | First reported llama VHH targeting the CD4bs | Neutralizes 42% of 65 HIV-1 isolates tested [34] | NA | |
| J3 | 2012 | Llama VHH | Neutralizes 96% of 100 HIV-1 Isolates tested [35] | NA | |
| CoRbs | m36 | 2008 | First reported human VH against HIV-1 | Neutralizes 10 of 11 HIV-1 isolates tested with IC50s < 10 μg/ml [33] | m36.4, an affinity-matured version of m36, protects four of six humanized mice against high-titer HIV-1 intrasplenic challenge |
| Carbohydrate | PGT125–128, 130–131 | 2011 | Neutralize 40 – 72% of 162 HIV-1 isolates tested [28] | NA | |
| 2G12 | 1996 | First reported HIV-1 bnAb targeting carbohydrate | Neutralizes 32% of 162 HIV-1 isolates tested [28] | In combination with 2F5, reduces viral loads and increases CD4+ T cell counts in HIV-1-infected individuals | |
| Quaternary structure | PG9, PG16 | 2009 | Neutralize 79% and 73% of 190 HIV-1 isolates tested [23] | NA | |
| PGT141 −145 | 2011 | Neutralize 38 – 78% of 162 HIV-1 isolates tested [28] | NA | ||
| MPER | 2F5 | 1992 | First reported HIV-1 bnAb targeting gp41 | Neutralize 57% of 177 HIV-1 isolates tested [29] | See 2G12 in vivo efficacy |
| 4E10 | 2001 | Neutralizes 98% of 181 HIV-1 isolates tested [29] | In combination with 2F5 and 4E10, delays viral rebound in HIV-1-infected individuals who stopped HAART before antibody infusion | ||
| 10E8 | 2012 | Non-autoreactive | Neutralizes 98% of 180 HIV-1 isolates tested [29] | NA | |
| m66.6 | 2011 | Less mutated than other bnAbs targeting MPER | Neutralizes 24% of 164 HIV-1 isolates tested [30] | NA |
Unless specified, neutralized viruses are those with IC50s ≤ 50 μg/ml in either pseudovirus/cell line-based assays or PBMC-based assays. Percentages of neutralization are derived from the referenced publications.
NA: Not available
2.1. B12, 2G12, 2F5, and 4E10
In nearly three decades since the discovery of HIV-1, b12, 2G12, 2F5, and 4E10 were the only HIV-1 bnAbs targeting the viral Envs. B12 was identified through phage display of an antibody library constructed from a clade-B HIV-1-infected individual [19]. It is one of the best characterized bnAbs that targets the Env gp120 at the CD4 binding site (CD4bs). 2G12 was selected from hybridomas producing human mAbs against HIV-1 which were established by EBV transformation and cell fusion [20]. It recognizes a unique cluster of high-mannose oligosaccharides on gp120. 2F5 and 4E10 are directed against relatively short amino acid sequences in the membrane-proximal external region (MPER) of the Env gp41 [21, 22].
2.2. Recently discovered HIV-1 bnAbs
Since 2009, new HIV-1 bnAbs have been identified by using novel selection approaches including high-throughput B-cell sorting and functional screening. These antibodies are on average more potent and broadly neutralizing than b12, 2G12, 2F5, and 4E10. PG9 and PG16 were the first reported representatives of the new breed bnAbs; they were isolated from memory B cells of a clade-A HIV-1-infected African donor [23]. They target conserved regions of the variable loops of the gp120 subunit preferentially expressed on trimeric Envs. VRC01, VRC02, and VRC03 are three CD4bs antibodies isolated from a chronically HIV-1-infected individual [24]. VRC01 and VRC02 are somatic variants of the same IgG1 clone and are capable of neutralizing over 90% of HIV-1 strains, while VRC03 blocks 57% of the circulating strains tested in vitro. NIH45–46 is a more potent clonal variant of VRC01. They differ mostly in that NIH45–46 has a four-residue insertion in the heavy chain complementarity determining region 3 (HCDR3), which contributes to gp120 binding [25]. NIH45–46G54W, an improved version of NIH45–46, was created via structure-based design [26]. It has a single substitution in the HCDR2 that increases interaction of the antibody with the gp120 bridging sheet and improves significantly the neutralizing breadth and potency. 3BNC117 [25] and VRC-PG04 [27] are also CD4bs bnAbs that exhibit breadth and potency comparable with those of VRC01. In addition, a series of PGT antibodies were isolated from the repertoires of four ‘elite’ HIV-1-infected donors [28]. PGT 141–145 target glycan-dependent quaternary epitopes similarly to PG9 and PG16. PGT 125–128 and PGT 130–131 interact specifically with the Man8/9 glycans on gp120. Some of the PGT antibodies are reported to be almost 10-fold more potent than VRC01, PG9, and PG16, and 100-fold more potent than the older bnAbs although at the expense of somewhat narrower breadth of neutralization. While most of the above-mentioned new bnAbs are directed against gp120, an MPER-specific antibody, 10E8, was recently reported to neutralize 98% of the tested viruses [29]. 10E8 does not bind to phospholipids and is not autoreactive. Another 2F5-like antibody, m66.6, exhibits relatively low potency, narrow neutralization breadth, and polyspecific reactivity, but is significantly less divergent from its putative germline predecessors than 2F5 and 10E8 [30]. Note however, that most of these antibodies were tested in only one in vitro assay based on cell-free pseudotyped viruses. Recently, it has been shown that bnAbs targeting the CD4bs including b12 and VRC01 exhibit significantly decreased neutralizing activity when tested for inhibitory activity against cell-to-cell transmission [31] which is likely to be dominant in many tissue cultures and in vivo [32].
2.3. Other antibody-based potent HIV-1 inhibitors
Engineered single antibody domains (eAds) (size, ~ 15 kDa) are emerging as promising drug candidates for treatment of HIV-1 infection because of their small molecular size and other excellent properties [17]. m36 is the first reported human antibody heavy chain variable domain (VH)-based eAd that targets the coreceptor-binding site (CoRbs) on gp120 and potently neutralizes genetically diverse HIV-1 isolates in vitro [33]. A12 [34] and J3 [35], llama heavy chain-only antibody (HCAb) variable domains (VHHs) achieve broad and potent neutralization of HIV-1 via interaction with the CD4bs.
Another novel class of HIV-1 inhibitors is bispecific fusion proteins containing two different binding moieties including bnAbs, peptide inhibitors, or soluble forms of human CD4 (sCD4) [9]. PG9-iMab, VRC01-iMab, and iMab-m36 are representatives of such fusion proteins that consist of a humanized CD4-specific antibody ibalizumab joined via a polypeptide linker to PG9, VRC01, and m36, respectively. They exhibit enhanced potency and breadth compared to the individual antibodies alone or in combination, and are active against ibalizumab-resistant viruses (http://www.vaccineenterprise.org/conference/2011/sites/default/files/OA07.01,%20Pace.pdf) [36].
2.4. Prophylactic and therapeutic potential of HIV-1 bnAbs and their derivatives
Currently, there are no mAbs approved for clinical use to treat HIV-1 infection. B12, 2G12, 2F5, and 4E10 have been tested in both animal models and human clinical trials [9]. Passive administration of a single antibody or a dual, triple or quadruple combination of the antibodies confers protection against simian-human immunodeficiency virus (SHIV) challenge in non-human primates but the antibodies do not exhibit favorable antiviral efficacy in humans with established HIV-1 infection. HIV-1 uses a number of strategies to escape antibodies generated by the human immune system. These include the high genetic diversity and mutability of the Envs [9]. In addition, some structural features of the Envs, e.g., steric occlusion of conserved neutralizing epitopes, mimicry of human self-antigens, and encoding of immunogenic non-neutralizing epitopes on the Envs, may further diminish the neutralizing activity of therapeutic antibodies or lead to elicitation of autoreactive antibodies or antibodies that could antagonize bnAbs in vivo.
Some of the newly identified bnAbs have largely improved properties in terms of specificity, neutralizing potency, and breadth, and therefore, could be more promising than the older known bnAbs for HIV-1 therapy and prevention. To date, however, there are not much data from in vivo experiments that could prove this possibility. Recently, a preclinical testing of VRC01 has been conducted in non-human primates [37]. The animals were first administered with VRC01 at a dose of 20 mg/kg and then mucosally challenged with SHIV. All four animals receiving VRC01 were protected while three of four animals in the control groups were infected. m36.4, an affinity-matured version of m36, provided sterilizing protection of four of six humanized NOD/SCID/γcnull mice against intrasplenical challenge with high-titer HIV-1 (> 1000 TCID50s) while extensive infection was detected in all four control animals [38]. In addition, a combination of five newly discovered bnAbs was shown to effectively control HIV-1 infection and suppress viral load to levels below detection in humanized mice [39]. To develop an alternative immunization approach, adeno-associated virus (AAV) was used as a vector for delivery of bnAb-encoding genes to achieve long-lasting production of the antibodies in vivo. A recent study showed that a single intramuscular injection of such vectors induced stable high-level expression of VRC01, b12, 2G12, 2F5, and 4E10 in humanized mice during the 64-week experiment [40]. The animals receiving VRC01- or b12-expressing AAVs were fully protected from high-dose replication-competent HIV-1 challenge.
3. HIV-1 bnAbs as templates for vaccine immunogen design
Knowledge gained from structures of antibodies in complex with antigens appears to be useful for vaccine immunogen design. By identifying fine epitopes on antigens recognized by bnAbs, tailoring antigens for more efficient presentation of epitopes to the immune system or engineering unrelated molecules to carry antigen-specific epitopes becomes feasible. This methodology, however, has met with limited success in the case of HIV-1. In this section, we discuss the attempts to design HIV-1 vaccine immunogens using bnAbs as templates, challenges, which could hamper the eventual success of this strategy, and possible opportunities to overcome those challenges.
3.1. Gp41-based vaccine design
2F5 and 4E10 recognize linear epitopes on the MPER of gp41, which makes them particularly attractive for vaccine design. The finding that MPER-specific antibodies account for the broad neutralization activity of plasma from some chronically infected individuals further validates the potential of the MPER as vaccine immunogens [41, 42]. Gp41 is unstable in the absence of gp120. A fusion protein of the gp4189.6 ectodomain with antibody Fc (gp41Fc) appeared to be stable and reacted not only with 2F5 and 4E10 but also with cross-reactive neutralizing antibodies (e.g., m44, m46, and m48) against conformational epitopes, suggesting that it could retain the conformational state on functional viral spikes [43]. Immunization of rabbits with gp41Fc elicited antibodies that neutralized some HIV-1 and HIV-2 isolates. However, they predominantly bound to peptides derived from the immunodominant loop regions but not to the MPER.
Attempts have been made to improve the antigenicity and immunogenicity of the MPER epitopes. The most straightforward strategy is to use synthetic MPER peptides or to fuse the peptides with unrelated carrier proteins aiming at focusing antibody responses to the peptides. However, both the synthetic peptides and the fusion proteins have failed to produce broadly protective responses [44–46]. One explanation could be that the MPER epitopes were not presented in the correct structural context in these constructs. Indeed, circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopy analysis showed that the solution conformation of T20, an MPER peptide carrying the 2F5 epitope, was predominantly random [44].
Crystallographic studies carried out with a range of peptides showed that the core 4E10 epitope adopted a helical conformation when complexed with the antibody [47]. The core 2F5 epitope contained a β-turn conformation enforced by the upstream 6-helix bundle structure of gp41 and the downstream 4E10 epitope [48]. Therefore, computational techniques were used to search and design a series of protein scaffolds, termed epitope scaffolds (ESs), which could accept grafting of the 2F5 and 4E10 epitopes and allow for preservation of their antibody-bound conformations. Ofek et al. identified five such scaffolds, designated ES1 to ES5, which could express the 2F5 epitope [49]. The resultant grafts exhibited nanomolar affinity for 2F5 and the epitope was shown to be in a conformation relatively similar to that of the peptide bound to 2F5. In another study, several ESs bearing the 4E10 epitope demonstrated higher affinities for 4E10 than the cognate peptide [50]. Crystal structures of the grafts showed a high degree of structural mimicry of 4E10-bound epitope conformation. In both cases, however, immunization of animals did not lead to broadly neutralizing sera although strong antibody responses to the grafts were observed [49–51]. Two mAbs isolated from mice immunized with the ES5-ES1 prime-boost group bound to an MPER peptide remarkably similarly as 2F5 [49], suggesting that at least 2F5-like antibodies binding to the core MPER epitope can be elicited. Notably, these two antibodies lacked long hydrophobic HCDR3s, which are a striking feature of 2F5 and 4E10 [52].
The discovery of the novel lipid-binding properties of 2F5 and 4E10 reveals additional complexity in their mechanisms of neutralization and provides more insight into MPER-targeted vaccine design. Both antibodies were found to significantly bind to a variety of purified lipids including phospholipids and to human cells [53–55]. Mutation of hydrophobic residues in 2F5 and 4E10 HCDR3s abrogated their lipid binding but had very little effects on binding of the antibodies to the MPER, suggesting the role of HCDR3s in antibody polyspecificity [56]. Another study also demonstrated that altering a hydrophobic patch on 2F5 HCDR3 did not affect binding of 2F5 to MPER peptides while a slight decrease in the hydrophobicity of 2F5 HCDR3 dramatically diminished the antibody neutralizing activity [57]. Further study suggested that 2F5 and 4E10 could associate initially with the viral membrane and subsequently capture the MPER [56, 58]. These results strongly indicate that for MPER-specific antibodies to be neutralizing, they probably need to engage not only amino acids of the peptide but also lipids on the virus membrane. Based on these findings, a combinatorial vaccine design approach was developed where MPER peptides and phospholipids were mixed as a formulation. The immunized animals induced antibodies that simultaneously bound to the MPER and adjacent lipids but inconsistently neutralized HIV-1 infection [59].
3.2. Gp120-based vaccine design
Gp120 contains most of the neutralizing epitopes. The CD4bs and the CoRbs are two functionally important conserved structures that can be targeted by antibody-mediated neutralizing responses. The recent identification of a panel of gp120-specific exceptionally potent bnAbs further validates the potential of gp120 as vaccine immunogens. The representative antibodies include VRC01 [24], which targets the CD4bs, PG9 and PG16 [23], which are directed against the conserved regions of variable loops of gp120 preferentially expressed on trimeric Envs, and the series of PGT antibodies [28], which bind to various novel epitopes on gp120. However, previous attempts to elicit broadly neutralizing antibodies using recombinant gp120 have failed likely partially because the highly flexible gp120 may present numerous conformations to the humoral immune system that are not found on the native viral spike and therefore, elicit antibodies that bind to recombinant gp120 but do not neutralize genetically diverse viruses [60].
Complexes of gp120 with sCD4 or CD4 mimetics (miniCD4) bind strongly to the HIV-1 coreceptor CCR5 and CD4-induced (CD4i) antibodies, which target the CoRbs [61, 62]. One of the gp120-sCD4 complexes elicited strong CD4i antibody responses in macaques that accounted for the control of SHIV challenge [63]. CD4i antibodies are abundant in patients with HIV-1 infection [64]. However, they generally do not or only weakly neutralize the virus as full-size antibody molecules due to steric occlusion of their epitopes [33, 65]. Further studies are needed to support the concept that gp120-sCD4 complexes can induce CD4i antibodies that are broadly neutralizing.
To present the CD4bs to the immune system and to reduce gp120 flexibility, structural and mutagenic approaches have been used to conformationally fix gp120 in CD4-bound states in the absence of CD4. These approaches include deletion of variable loops, introduction of inter-domain disulfide bonds, and substitution of the Phe43 cavity in the CD4bs [60, 66–68]. The stabilized gp120 generally had a significant increase in binding to sCD4 and CD4i antibodies compared with wild-type gp120. However, immunization with the stabilized gp120 showed only a trend of improvement in eliciting cross-reactive neutralizing antibodies relative to its wild-type counterpart [67–69]. One possible explanation is that the structural modifications hide or remove gp120 conformations recognized by some neutralizing antibodies. This line of reasoning is supported by the findings that interactions of some cross-reactive neutralizing antibodies against the CD4bs with the stabilized gp120 were abrogated [60, 66–68] and that a number of recently identified bnAbs targeted the conserved components of variable loops [23, 28], which were partially or completely deleted in the stabilized gp120.
We have recently demonstrated that CD4i antibodies could also induce conformational changes in gp120 and stabilize it in CD4-bound states without the need of variable loop deletions and mutagenic modifications [70]. We found that the CD4i antibodies m36 and m9 weakly enhanced CD4 binding to gp120 and that increasing the local concentrations of CD4i antibodies by co-expression with gp120 as single-chain fusion proteins resulted in gp120 conformations better recognized by CD4 and all gp120-specific bnAbs tested except the glycan-specific antibody 2G12. Our finding was further evidenced by cryo-electron microscopy analysis which showed that interactions with m36 and 17b lead to the same open conformations as those of trimeric Envs complexed with CD4 [71, 72]. Therefore, CD4i antibody–gp120 fusion proteins are a novel type of candidate HIV-1 vaccine immunogens with possibly better preserved neutralizing antibody epitopes compared to the above-described stabilized gp120.
HIV-1 gp120s are large proteins. They elicit various antibodies including non-neutralizing antibodies, some of which compete with cross-reactive neutralizing antibodies for binding to gp120 [9, 73]. Therefore, the use of native gp120 as vaccine immunogens could result in inefficient elicitation of antibodies to conserved neutralizing epitopes. An antigenecially resurfaced gp120, RSC3, was designed that specifically reacts with antibodies directed against the CD4bs [24]. Potent bnAbs including VRC01 and VRC02 were identified by using RSC3 as a probe for screening. RSC3 in soluble form or displayed on Chikungunya virus-like particles (VLPs) has been evaluated as vaccine immunogens (http://www.hivvaccineenterprise.org/conference/2011/sites/default/files/PL03.01,%20Boyington.pdf). The sera from immunized animals could only neutralize tier 1 HIV-1 isolates although they showed increased specificity to the CD4bs.
3.3. Env trimer-based vaccine design
PG9, PG16, and some PGT antibodies target quaternary structures of the Envs, indicating that Env trimers might be better able to induce such antibodies than monomeric Envs. However, cleaved (gp120/gp41)3 complexes are unstable and are difficult to manufacture. Recombinant gp140 proteins lacking the gp41 transmembrane segment can be partially trimerized but they are considered non-functional and when used in immunization, elicit relatively high levels of non-neutralizing antibodies [74]. An important task is, therefore, to reconstruct HIV-1 Env trimers that mimic native functional viral spikes. A previous study demonstrated that high-quality homologous HIV-1 Env trimers could be generated by fusing gp140 to T4 bacteriophage fibritin trimerization domains [75]. Recently, it was further reported that the same Env trimers could be produced in 293T cells with a yield sufficient for large-scale immunogenicity studies [76]. Interestingly, they recognized strongly only with CD4bs bnAbs but not with non-neutralizing CD4bs antibodies. They also showed detectable binding to PG9 and PG16 while the monomers did not react with the antibodies. Moreover, the trimers induced potent neutralizing antibody responses against diverse tier 1 and tier 2 viruses with titers substantially higher than those elicited by the corresponding gp120 monomers. This study provides proof-of-concept that suitably prepared Env trimers could accurately mimic the native viral spike and could be better vaccine immunogens than monomeric Envs.
Another method to produce functional Env trimers is to present them in situ on plasma membrane (e.g., on VLPs), assuming that the native environment may promote trimer stability [77]. However, non-functional Envs could also be co-expressed on the VLPs alongside the trimers. Interestingly, non-functional Envs appeared to be relatively enzyme-sensitive and could be selectively removed by endo H and chymotrypsin digestion [78].
3.4. Challenges and opportunities
Both traditional and rational vaccine design approaches have failed in the case of HIV-1. This could have been implicated by the unique features of the virus and its pathogenesis. HIV-1 displays enormous antigenic variability and high mutability, which rapidly develops viral variants resistant to antibodies generated by the immune system. It escapes humoral immunity by hiding functionally important structures (e.g., the CD4bs and CoRbs) while exposing immunogenic non-neutralizing epitopes (e.g., variable loops). It is therefore difficult for the immune system to initiate elicitation of HIV-1-blocking antibodies which must undergo extensive affinity maturation to be broadly neutralizing which could take years. In addition, HIV-1 progressively destroys the immune system by infecting and depleting CD4-expressing lymphocytes. Finally, HIV-1 establishes latency by integrating into the host chromosomes, and spreads not only in the form of cell-free viruses but also through cell-to-cell fusion. The fact that the natural immune responses in HIV-1-infected individuals cannot clear the infection indicates that effective HIV-1 vaccine design might not rely much on the native Envs as vaccine immunogens.
It was hypothesized (germline antibody hypothesis) that HIV-1 has evolved a strategy to reduce or eliminate the immunogenicity of the highly conserved epitopes of bnAbs by using ‘holes’ in the human germline B-cell receptor (BCR) repertoire (i.e., absent or very weak binding of germline antibodies to the bnAb epitopes that is not sufficient to initiate and/or maintain efficient immune responses) [79]. To test this hypothesis, germline-like antibodies corresponding most closely to b12, 2G12, and 2F5 were designed, produced and tested for binding to Envs [79]. The important finding is that they lacked measurable binding to Envs although the corresponding mature antibodies did. These results provided the first evidence that Env structures containing conserved vulnerable epitopes may not initiate humoral responses by binding to germline antibodies. Even if such responses are initiated by very weak binding, it is likely that they would be outcompeted by responses to structures containing epitopes of other antibodies that bind germline BCRs with much higher affinity/avidity. Recent studies with B cells expressing putative germline predecessors of bnAbs further supported and extended this finding. B cells carrying germline bnAb BCRs were not stimulated by infection-competent virions and by any of the tested vaccine candidates including soluble gp140 trimers and a multimerized, scaffolded epitope protein [80]. In another study, a large panel (n = 56) of recombinant Envs from clades A, B, and C was screened for binding to the germline predecessors of b12, NIH45–46 and 3BNC60, and for Env-induced B-cell activation [81]. Although the mature antibodies reacted with diverse Envs, the corresponding germline antibodies did not display Env reactivity. These results support the germline antibody hypothesis and suggest that a major challenge to elicit bnAbs is related to the lack of measurable Env binding to bnAb germline-like predecessors and therefore, using typical Envs may not initiate and/or maintain immune responses leading to elicitation of bnAbs [25, 55, 79–82]. These findings also suggest that components of effective immunogens need to engage low-affinity germline precursors and guide them through affinity maturation pathways resulting in the development of high-affinity broadly neutralizing antibodies. However, the maturation pathways leading to bnAbs are complex because of their high levels of somatic mutational diversification [55, 79, 83]. Moreover, it is uncertain what molecules initiate elicitation of HIV-1 bnAbs because every such antibody could be polyspecific if a sufficient amount of antigens are tested. Therefore, the rational approach based solely on the viral epitopes identified by analyzing the crystal structures of Env–bnAb complexes should be added by empirical approaches.
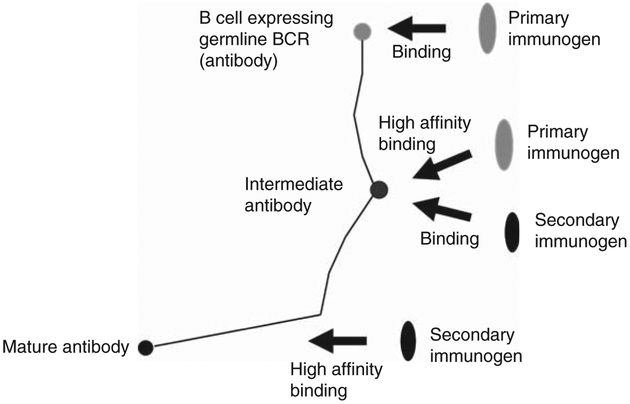
Because all known HIV-1 bnAbs are highly somatically mutated, we hypothesized that identifying antibodies that are intermediates in the pathways to maturation could help design conceptually novel vaccine immunogens [79, 83, 84]. Knowledge of the complete set of antibodies in an individual (antibodyome) is helping identify intermediate antibodies needed for design of such immunogens [27, 84]. We have also proposed a vaccination approach, where it is critical to identify primary immunogens that bind to the germlines of bnAbs and if needed, they should be combined with secondary immunogens that recognize intermediate and/or matured antibodies to guide the immune system through the long complex maturation pathways (Figure 1) [84]. Our approach has been widely discussed, modified, and expanded [85]. Attempts have been made to engineer Envs which are capable of interacting with putative germline predecessors or intermediates of bnAbs [27]. A recent study demonstrated that deglycosylation of Envs enhanced their binding to 2F5, 4E10, and their unmutated ancestor antibodies although the glycan-depleted Envs alone were unable to elicit neutralizing antibodies [86].
Figure 1. A cartoon schematically illustrating a vaccination approach using primary and secondary immunogens to guide antibody elicitation from germline antibodies through maturation pathways to bnAbs.
In this approach, B cells expressing germline BCRs are first activated by binding to a primary immunogen. The maturation pathways leading to mature bnAbs (denoted by black lines) are then directed for enhanced elicitation of bnAbs by interaction of the B cells expressing intermediate antibodies with secondary immunogens. The cartoon is modified from a previously published study [84].
4. Targeting HIV-1-infected cells for virus eradication
The recent report of possible cure of three HIV-1-infected patients after receiving bone marrow transplant has renewed hope for curing AIDS (http://www.aids2012.org/WebContent/File/AIDS2012_Media_Release_HIV_Cure_26_July_2012_EN.pdf) [16]. However, such treatment is not affordable to the vast majority of patients with HIV. In addition, the probability of finding a donor whose genetic type is a close match is low, especially when the immediate family members of patients are not available. Finally, the transplant poses potential safety risk to the recipients because they could inherit unknown diseases from the donors and transplant-related mortality is relatively high (up to 20%). It is therefore necessary to develop a safe, effective, affordable, and scalable cure for HIV. Here, we briefly describe possible mechanisms of HIV-1 latency and discuss the ‘activation–elimination’ strategy for clearance of the virus from its cellular reservoirs.
4.1. HIV-1 latency
The HIV-1 life cycle begins with the attachment of viral Envs to the primary receptor CD4 and a coreceptor, followed by virus–cell fusion and entry of the viral genome into target cells. After virus entry, the single-strand positive sense viral RNA undergoes reverse transcription, leading to the synthesis of double-strand DNA (dsDNA) [87]. The linear dsDNA is then transported into the nucleus and integrated into host cell chromosomes, allowing establishment of latency. The most well-defined HIV-1 latent reservoir is the memory CD4+ T cells. They can be directly infected or converted from activated CD4+ T cells that survive the infection [1–4]. Although CD4+ T cells constitute a relatively large population in peripheral blood and lymphoid tissues, there is a low total body load (< 107 cells) of latently infected resting CD4+ T cells with replication-competent integrated provirus [88]. HAART can reduce plasma virus to undetectable levels. However, the latent reservoir decays very slowly if at all during HAART with an estimated half-life of about 40 months, which translates to as long as 60 years for eradication solely of infected resting CD4+ T cells [89]. The situation is further complicated because additional reservoirs could exist including those in the densely packed lymphoid environments such as spleen, gut and lymph nodes, which requires that antiretroviral drugs must be able to penetrate into those tissues.
4.2. Activation of latent HIV-1
Current HIV-1 drugs target cellular receptors, viral Envs, or other factors (e.g., reverse-transcriptase and protease) essential for viral genome amplification and assembly. In the resting CD4+ T cells, however, there is only minimal transcription from the HIV-1 long terminal repeat (LTR) due to the absence of necessary host factors that are required for HIV-1 replication. Therefore, most proposed strategies involve activating the resting cells in some ways to induce expression from the HIV-1 genome so that the cells could be specifically targeted and killed.
Early forms of HIV activators are compounds that induce T cells to secrete lymphokines. For example, incubation with phytohemagglutinin (PHA) or phorbol myristate acetate (PMA) led to production of interleukin (IL)-2 and HIV in infected CD4+ T cells, and subsequently, death of the cells [90]. The T-cell activation correlated with induction of the transcription factor NF-κB, which binds to the enhancer regions of viral DNA resulting in an increase in HIV expression of up to 50-fold [91]. Later, it was found that some lymphokines such as IL-2 [92] and IL-7 [93] have direct effects on activation of HIV from latency. Other activators include CpG oligodeoxynucleotides (CpG ODNs), protein kinase C activators (PKCAs), histone deacetylase inhibitors (HDACIs), and methylation inhibitors [1]. CpG ODNs were shown to be able to stimulate HIV replication in ACH-2 cells by triggering NF-κB expression but their stimulating effects were not observed in Jurkat T cells [94]. The PKCA prostratin upregulated expression of viral products from latently infected cells, inhibited HIV-1 entry, and did not induce tumor formation [95]. Another PKCA, bryostatin, has recently been shown to be 1000-fold more potent in inducing latent HIV than prostratin [96]. Valproic acid (VPA), an HDACI, was capable of inducing expression of quiescent provirus, without fully activating cells or enhancing de novo infection [97]. A combination of VPA with the peptide entry inhibitor T20 accelerated clearance of resting CD4+ T cells in three of four patients [98]. Another HDACI, suberoylanilide hydroxamic acid or vorinostat, has also been tested in a translational clinical study. A single dose of vorinostat significantly increased (mean, by 4.8-fold) the expression of both HIV-1 RNA and biomarkers of cellular acetylation in cells isolated from all eight patients [99]. Vorinostat is the first HDACI approved by the US FDA for treatment of cutaneous T-cell lymphoma. These studies provide proof-of-concept that HDACIs could have potential for clinical use to activate latent HIV-1, which could be subsequently eradicated by HIV-1-specific therapeutics.
All above-described reagents non-specifically induce T-cell activation and therefore, could potentially be toxic. To identify activators specific for HIV-1, genes that could activate transcription from the HIV-1 LTR in an NF-κB-independent manner were screened [100]. The authors isolated an alternatively spliced form of the transcription factor Ets-1, ΔVIIEts-1, which reactivated latent HIV-1 in cells from patients on HAART without causing global T-cell activation. High-throughput screening also identified a new small molecule, AV6, which appeared to be specific and synergized with HDACIs in activating HIV transcription [101]. In another study, a cluster of cellular microRNAs were discovered that potently inhibited HIV-1 production in resting primary CD4+ T cells by targeting the 3′ ends of viral mRNA [102]. Their inhibitory activities could be neutralized by their corresponding 2′-O-methyl-oligoribonucleotide antisense microRNAs, resulting in increased HIV-1 production. Importantly, the antisense microRNAs did not affect HIV-1 expression in activated CD4+ T cells and cellular proliferation status, suggesting specific activation of latent virus.
4.3. Purging the reservoirs
Combinatorial treatment with HAART and activators has shown faster depletion of the latent reservoir than HAART alone [98], but methods to enhance killing of activated HIV-1-infected cells are still needed. Antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) are two conventional antibody Fc-mediated immune effector mechanisms for clearance of infected cells. Moldt et al. demonstrated that b12, when defucosylated, was 10-fold more efficient than fully fucosylated b12 in inhibiting viral replication and killing HIV-1-infected cells in an ADCC assay [103]. Such natural immune mechanisms are safe but whether they are sufficiently effective in eliminating the cellular reservoirs in vivo remains unclear and needs to be investigated. Cytotoxic T cells (CD8+ T cells) are another line of defense against intracellular pathogens. They recognize infected cells through T-cell receptors (TCRs), antibody-like proteins displayed on the cell surface for specific targeting of processed viral peptide antigens. To promote cytotoxic effects, CD8+ T cells have been engineered to express chimeric antigen receptors (CARs) with a transmembrane and an intracellular signaling domain involved in the cell activation and proliferation genetically fused to an antigen-binding domain such as an antibody fragment [104]. The modified T cells thus are able to specifically recognize and kill cells that the antigen-binding domain targets. CAR-mediated T-cell therapy appears to be successful in cancer treatment, having demonstrated promising clinical results [104]. This strategy is being tested for its efficacy in killing HIV-1-infected cells and very limited data have been currently publicly available.
Yet another extensively studied approach is to conjugate HIV-1 bnAbs with potent cytopathic toxins. The conjugates, termed immunotoxins, could specifically target the activated cells, which express the Envs on cell surfaces, and induce cell death by toxin-mediated cytopathic effects. Pastan, Berger and colleagues generated a recombinant immunotoxin, 3B3 (Fv)-PE38, which contained the scFv of an affinity-matured version (3B3) of the CD4bs antibody b12 and Pseudomonas aeruginosa exotoxin A (PE38) [105]. It not only potently inhibited spreading infection by all clinical HIV-1 isolates examined in peripheral blood mononuclear cells (PBMCs) and macrophages but also selectively killed infected cells. High-dose treatment of rhesus macaques with 3B3(Fv)-PE38 did not induce liver toxicity.
The clinical benefits that 3B3(Fv)-PE could confer in combination with HAART and activators to purge latent cells have been comprehensively discussed [106]. However, two major issues may limit the use of the same type of immunotoxins and need to be addressed. One is the generally high immunogenicity of protein-based cytotoxins derived from insects, bacteria, plants, or other nonhuman species. It has been observed in a Phase I clinical trial that sCD4-PE40, a fusion protein of sCD4 with Pseudomonas exotoxin PE40, elicited anti-PE40 antibodies in 58% of the recipients [107]. Based on the protocols from successful clinical use of immunotoxins in cancers, a short-term treatment, for example 1 – 2 weeks, has been recommended for such an anti-HIV immunotoxin that could be followed by products with toxins of different origins to promote cell killing efficacy and minimize antibody responses to toxins [106]. An alternative strategy is to use small molecule toxins, which are not or are less immunogenic and mediate high levels of cytotoxicity. The other issue is with the antibody arm of the conjugates, to which viral resistance could easily develop [9]. A recent study reported that VRC01, a bnAb that potently neutralized more than 90% of heterologous circulating HIV-1 isolates in vitro by mimicking CD4, was not sensitive to most virus strains in the slowly progressing HIV-1-infected donor, from which VRC01 was identified [108]. Treatment with bnAb–toxin conjugates could therefore leave some infected cells intact, resulting in a largely compromised therapeutic effect. One can speculate that certain non-neutralizing mAbs targeting conserved epitopes on HIV-1 Envs could be used. Such antibodies might not exert selection pressure on the virus which induces escape mutations on the viral Envs but only experimentation can prove it.
Another potential issue with the use of HIV-1 mAbs for virus eradication is that they might not recognize well the Envs expressed on the surface of infected cells. HIV-1 could exhibit differential exposure and/or conformations of epitopes on viral and infected cell membranes [109]. However, almost all known mAbs are selected by screening for binding to recombinant HIV-1 Envs or cell-free viruses. Therefore, HIV-1 mAbs should be evaluated for their ability to bind infected cell-membrane-associated Envs before being used as components of immunotoxins.
The fact that HIV-1 requires CD4 for entry and cell-to-cell transmission prompts us to suggest reconsideration using sCD4 as a component to specifically target potentially all infected cells. The possible reasons for the lack of clinical benefit in the sCD4-PE40 clinical trial remain unclear [106] and should not diminish the enthusiasm for, and tempo of, further research efforts. CD4 also interacts with the class II major histocompatibility complex (MHCII) molecules, which raises safety concerns especially when sCD4 is conjugated with toxic molecules. In addition, both four-domain (D1–4, ~ 50 kDa) and two-domain (D1D2, ~ 25 kDa) sCD4 derivatives are relatively large so they may have difficulty penetrating into the densely packed lymphoid tissues such as gut and lymph nodes, in which HIV replication mostly occurs in patients on HAART [110].
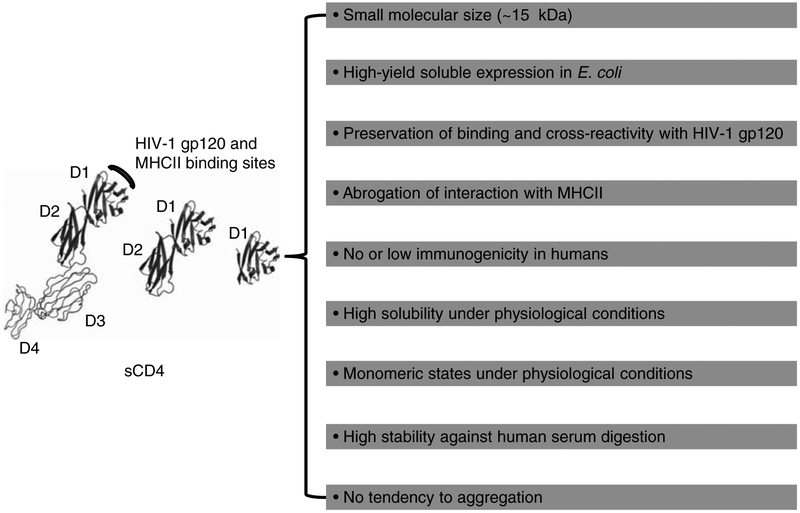
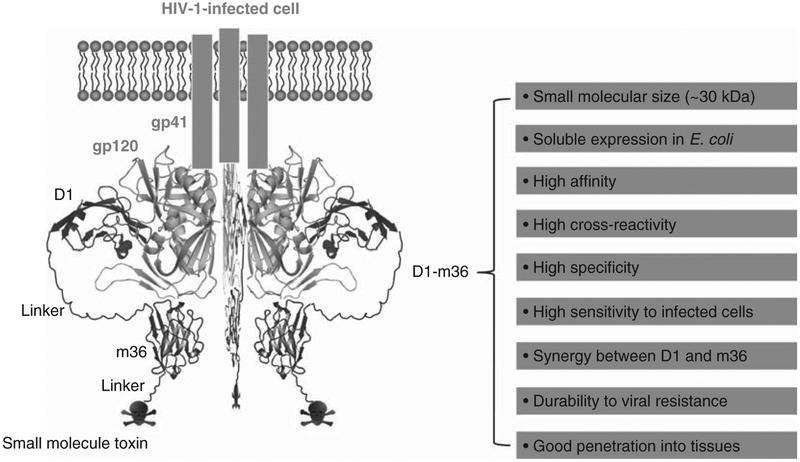
We have generated highly soluble, stable single-domain human sCD4 (D1, ~ 15 kDa) that shows significantly increased neutralizing activity and greatly decreased non-specific binding to human blood cell lines BJAB (B cells expressing MHCII) and SUPT1 (T cells) compared to D1D2 [111]. Therefore, the D1 mutant could be superior to D1D2 and D1–4 for conjugation with toxins if the mutagenesis does not introduce immunogenicity in humans (Figure 2). We previously identified an eAd m36 (~ 15 kDa) that targets the CoRbs on gp120, potently neutralizes HIV-1 isolates from different clades, and synergizes with D1D2 in neutralizing the virus [33]. Moreover, m36 did not bind to BJAB and SUPT1 cells (Chen W et al., unpublished data). We therefore hypothesize that the D1–m36–small molecule toxin combination could have relatively small size, high avidity, cross-reactivity, specificity, and sensitivity to low-level cell-surface expression of gp120, which all together could lead to efficient eradication of HIV-1 when combined with HAART and agents that activate latently infected cells (Figure 3).
Figure 2. Excellent properties that engineered single human CD4 domain D1 could exhibit.
The X-ray crystal structures of four-domain (D1–4), two-domain (D1D2) and single-domain (D1) sCD4 were adapted from Protein Data Bank (PDB) entry 1WIO. Note that the D1 structure was obtained as part of the D1D2; D1 has not been crystallized yet.
Figure 3. Schematic representation of specific targeting of potentially all HIV-1-infected cells by D1–m36–small molecule toxin conjugates.
The X-ray crystal structures of HIV-1 gp120 and D1 were adapted from PDB entry 1GC1. The m36 structure is derived from homology modeling using SWISS-MODEL (http://swissmodel.expasy.org/) based on the crystal structure of a human VH domain (PDB entry 1T2J). m36 was manually docked into the CoRbs of gp120 according to the crystal structure of D1D2–gp120–17b complex (PDB entry 1GC1). Favorable properties that the D1–m36 combination could exhibit are shown on the right.
5. Expert opinion
The ultimate goal of AIDS research is to develop an effective vaccine for those who are potentially at risk of HIV infection and to cure patients who are already infected. Identifying novel potent bnAbs and understanding their mechanisms of action can undoubtedly have implications for AIDS vaccine development and finding a cure. The very weak or lack of measurable binding of the bnAb germline predecessors to HIV-1 Envs is an important recent finding which urges rethinking concerning the decade-long efforts in HIV vaccine design using matured bnAbs as templates [79, 84]. If extensive somatic diversification is indeed a prerequisite for anti-HIV-1 antibodies to be broadly neutralizing, design of immunogens that can not only bind to germline BCR predecessors of bnAbs but also guide them through maturation pathways becomes an urgent task (Figure 1). The advent of high-throughput next-generation sequencing technologies, e.g., the Roche 454 sequencing platform (http://www.454.com), allows for exploration of almost whole antibody repertoires generated by the immune system at any given time points, accelerating identification of broadly neutralizing antibody maturation pathways [27]. However, the extreme complexity of the maturation pathways may again ‘pour cold water’ on our pursuit of a protective HIV-1 vaccine.
Given the continued development of exceptionally potent HIV-1 inhibitors as a complement to the highly successful HARRT and the increased understanding of HIV-1 pathogenesis and latency, the time has come for consideration of how we could give the patients complete relief from the disease. The century-old ‘magic bullet’ concept based on mAbs [112] has achieved great success in therapy of cancers including lymphoma [113] and leukemia [114]. The CAR concept could also hold promise for cancer therapy [104]. These strategies may be more advantageous for HIV-1 because the targeted molecules are encoded by the virus, thereby minimizing adverse effects encountered in cancer treatment where normal cells expressing low levels of target antigens can be affected. The central challenge of HIV-1 eradication, however, is the absent expression of viral Envs on the latently infected cell surfaces and the extremely high variability of the Envs. The HDACI vorinostat, which has been clinically used for treatment of T-cell lymphoma [99], and new activators that are being developed, are promising as agents to activate the latent virus. The highly conserved CD4bs on gp120 is essential for host cell recognition and infection by the virus, representing a major vulnerability of HIV-1. sCD4, theoretically, is most appropriate for specific targeting of the vulnerability, providing that HIV-1 can easily escape the known bnAbs including those closely mimicking CD4 (Figure 2). The ability of sCD4 to bind to MHCII-expressing cells requires serious consideration but the interaction could be eliminated by decreasing the size of sCD4 to a single domain D1 and mutagenesis without losing cross-reactivity with HIV-1 gp120 [111]. Another issue with sCD4 is that at low concentrations, it could enhance HIV-1 infectivity presumably because it helps expose the CoRbs, the other functionally conserved element of HIV-1 gp120, and initiates further conformational changes in gp41 that ultimately lead to virus–cell fusion. Under HARRT, however, the enhancement effect of sCD4 is likely to be marginal if any. It could be reduced or eliminated by combining sCD4 with CD4i antibodies (e.g., m36 and 17b), which target the CoRbs and synergize with sCD4 in binding and neutralizing HIV-1 (Figure 3). Based on the considerations outlined above, we suggest that sCD4–CD4i antibody fusion proteins, in particular D1–m36 which is small and does not or only very weakly bind to human blood cells, are promising as ‘navigation systems’ of the ‘magic bullets, ‘ the ‘CARs, ‘ or naturally occurring killer cells for HIV-1 eradication.
Footnotes
Declaration of interest
This project was supported by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health (NIH) and the Intramural Research Program of the NIH, National Cancer Institute. The authors have no other competing interests to declare.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Smith MZ, Wightman F, Lewin SR. HIV reservoirs and strategies for eradication. Curr HIV AIDS Rep 2012;9:5–15 [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, Fauci AS. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS 2012;26:1261–8 [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Autran B, Berkhout B, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012;12:607–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer S, Josefsson L, Coffin JM. HIV reservoirs and the possibility of a cure for HIV infection. J Intern Med 2011;270:550–60 [DOI] [PubMed] [Google Scholar]
- 5.Barbaro G, Scozzafava A, Mastrolorenzo A, et al. Highly active antiretroviral therapy: current state of the art, new agents and their pharmacological interactions useful for improving therapeutic outcome. Curr Pharm Des 2005;11:1805–43 [DOI] [PubMed] [Google Scholar]
- 6.Rusconi S, Scozzafava A, Mastrolorenzo A, et al. An update in the development of HIV entry inhibitors. Curr Top Med Chem 2007;7:1273–89 [DOI] [PubMed] [Google Scholar]
- 7.Dimitrov DS, Marks JD. Therapeutic antibodies: current state and future trends–is a paradigm change coming soon? Methods Mol Biol 2009;525:1–27, xiii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrov DS. Therapeutic proteins. Methods Mol Biol 2012;899:1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Dimitrov DS. Monoclonal antibody-based candidate therapeutics against HIV type 1. AIDS Res Hum Retroviruses 2012;28:425–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong R, Chen W, Dimitrov DS. Candidate antibody-based therapeutics against HIV-1. BioDrugs 2012;26:143–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ada GL. Human vaccines. Dev Biol Stand 1994;82:181–8 [PubMed] [Google Scholar]
- 12.Adkins JC, Wagstaff AJ. Recombinant hepatitis B vaccine: a review of its immunogenicity and protective efficacy against hepatitis B. BioDrugs 1998;10:137–58 [DOI] [PubMed] [Google Scholar]
- 13.Li G, Chen W, Yan W, et al. Comparison of immune responses against foot-and-mouth disease virus induced by fusion proteins using the swine IgG heavy chain constant region or beta-galactosidase as a carrier of immunogenic epitopes. Virology 2004;328:274–81 [DOI] [PubMed] [Google Scholar]
- 14.Bonsignori M, Alam SM, Liao HX, et al. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol 2012;20:532–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009;361:2209–20. [DOI] [PubMed] [Google Scholar]; •• First report of modest efficacy of an AIDS vaccine in humans.
- 16.Hutter G, Nowak D, Mossner M, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 2009;360:692–8 [DOI] [PubMed] [Google Scholar]; •• First report of a possible functional cure of AIDS by bone marrow transplant.
- 17.Chen W, Dimitrov DS. Human monoclonal antibodies and engineered antibody domains as HIV-1 entry inhibitors. Curr Opin HIV AIDS 2009;4:112–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton DR, Poignard P, Stanfield RL, et al. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 2012;337:183–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roben P, Moore JP, Thali M, et al. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 1994;68:4821–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchacher A, Predl R, Strutzenberger K, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 1994;10:359–69 [DOI] [PubMed] [Google Scholar]
- 21.Purtscher M, Trkola A, Gruber G, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 1994;10:1651–8 [DOI] [PubMed] [Google Scholar]
- 22.Stiegler G, Kunert R, Purtscher M, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 2001;17:1757–65 [DOI] [PubMed] [Google Scholar]
- 23.Walker LM, Phogat SK, Chan-Hui PY, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009;326:285–9 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes novel potent HIV-1 bnAbs PG9 and PG16.
- 24.Wu X, Yang ZY, Li Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010;329:856–61 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes novel potent HIV-1 bnAbs VRC01–03.
- 25.Scheid JF, Mouquet H, Ueberheide B, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011;333:1633–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diskin R, Scheid JF, Marcovecchio PM, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 2011;334:1289–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011;333:1593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011;477:466–70 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a series of PGT antibodies as novel potent HIV-1 bnAbs.
- 29.Huang J, Ofek G, Laub L, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 2012;491:406–12 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a novel MPER-specific bnAb that is not autoreactive.
- 30.Zhu Z, Qin HR, Chen W, et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J Virol 2011;85:11401–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abela IA, Berlinger L, Schanz M, et al. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog 2012;8:e1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimitrov DS, Willey RL, Sato H, et al. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol 1993;67:2182–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W, Zhu Z, Feng Y, et al. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc Natl Acad Sci USA 2008;105:17121–6 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the first potent, broadly neutralizing human eAd against HIV-1.
- 34.Forsman A, Beirnaert E, Aasa-Chapman MM, et al. Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)-neutralizing properties and high affinity for HIV-1 gp120. J Virol 2008;82:12069–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy LE, Quigley AF, Strokappe NM, et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med 2012;209:1091–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun M, Pace C, Yu J, et al. Rational design of a novel bispecific HIV-1-neutralizing antibody, Ibalizumabm36 [abstracts:436]. 18th Conference on Retroviruses and Opportunistic Infections; 2012 [Google Scholar]
- 37.Pegu A, Yang Z, Chen X, et al. VRC01 provides sterilizing protection to non human primates from mucosal SHIV challenges. J Immunol 2011;186:155.11 [Google Scholar]
- 38.Zhang C, Zheng J, Chen W, et al. Protective immunity: eAd inhibit in vivo HIV infection in humanized mice and can be expressed by lentiviral vectors to reprogam cells to secrete broadly neutralizing anti-HIV antibodies [abstracts:379]. 18th Conference on Retroviruses and Opportunistic Infections; 2011 [Google Scholar]
- 39.Klein F, Halper-Stromberg A, Horwitz JA, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balazs AB, Chen J, Hong CM, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 2012;481:81–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray ES, Madiga MC, Moore PL, et al. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J Virol 2009;83:11265–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen X, Parks RJ, Montefiori DC, et al. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol 2009;83:3617–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang MY, Wang Y, Mankowski MK, et al. Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: possible role of the prolonged half-life of the immunogen. Vaccine 2009;27:857–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joyce JG, Hurni WM, Bogusky MJ, et al. Enhancement of alpha -helicity in the HIV-1 inhibitory peptide DP178 leads to an increased affinity for human monoclonal antibody 2F5 but does not elicit neutralizing responses in vitro. Implications for vaccine design. J Biol Chem 2002;277:45811–20 [DOI] [PubMed] [Google Scholar]
- 45.Zhou M, Kostoula I, Brill B, et al. Prime boost vaccination approaches with different conjugates of a new HIV-1 gp41 epitope encompassing the membrane proximal external region induce neutralizing antibodies in mice. Vaccine 2012;30:1911–16 [DOI] [PubMed] [Google Scholar]
- 46.Dennison SM, Sutherland LL, Jaeger FH, et al. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One 2011;6:e27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardoso RM, Zwick MB, Stanfield RL, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 2005;22:163–73 [DOI] [PubMed] [Google Scholar]
- 48.Ofek G, Tang M, Sambor A, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 2004;78:10724–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ofek G, Guenaga FJ, Schief WR, et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl AcadSci USA 2010;107:17880–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correia BE, Ban YE, Holmes MA, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 2010;18:1116–26 [DOI] [PubMed] [Google Scholar]
- 51.Guenaga J, Dosenovic P, Ofek G, et al. Heterologous epitope-scaffold prime: boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant. PLoS One 2011;6:e16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burton DR. Scaffolding to build a rational vaccine design strategy. Proc Natl Acad Sci USA 2010;107:17859–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haynes BF, Fleming J, St Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005;308:1906–8 [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Martinez S, Lorizate M, Katinger H, et al. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res Hum Retroviruses 2006;22:998–1006 [DOI] [PubMed] [Google Scholar]
- 55.Xiao XD, Chen WZ, Feng Y, et al. Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses Basel 2009;1:802–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; • First attemtp to elucidate maturation pathways of HIV-1 bnAbs in the context of their germline predecessors.
- 56.Alam SM, Morelli M, Dennison SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA 2009;106:20234–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ofek G, McKee K, Yang Y, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol 2010;84:2955–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim M, Sun ZY, Rand KD, et al. Antibody mechanics on a membrane-bound HIV segment essential for GP41-targeted viral neutralization. Nat Struct Mol Biol 2011;18:1235–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matyas GR, Wieczorek L, Beck Z, et al. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS 2009;23:2069–77 [DOI] [PubMed] [Google Scholar]
- 60.Xiang SH, Kwong PD, Gupta R, et al. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol 2002;76:9888–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin G, Burke B, Thai R, et al. Stabilization of HIV-1 envelope in the CD4-bound conformation through specific cross-linking of a CD4 mimetic. J Biol Chem 2011;286:21706–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fouts TR, Tuskan R, Godfrey K, et al. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J Virol 2000;74:11427–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeVico A, Fouts T, Lewis GK, et al. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc Natl Acad Sci USA 2007;104:17477–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Decker JM, Bibollet-Ruche F, Wei X, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 2005;201:1407–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Labrijn AF, Poignard P, Raja A, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 2003;77:10557–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou T, Xu L, Dey B, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 2007;445:732–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dey B, Svehla K, Xu L, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog 2009;5:e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dey B, Pancera M, Svehla K, et al. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol 2007;81:5579–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y, McKee K, Tran K, et al. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site.J Biol Chem 2012;287:5673–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W, Feng Y, Wang Y, et al. Fusion proteins of HIV-1 envelope glycoprotein gp120 with CD4-induced antibodies showed enhanced binding to CD4 and CD4 binding site antibodies. Biochem Biophys Res Commun 2012;425:931–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tran EE, Borgnia MJ, Kuybeda O, et al. Structural mechanism of Trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 2012;8:e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meyerson JR, Tran EEH, Kuybeda O, et al. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc Natl Acad Sci USA In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen W, Zhu Z, Liao H, et al. Cross-reactive human IgM-derived monoclonal antibodies that bind to HIV-1 envelope glycoproteins. Viruses 2010;2:547–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des 2007;13:213–27 [DOI] [PubMed] [Google Scholar]
- 75.Nkolola JP, Peng H, Settembre EC, et al. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J Virol 2010;84:3270–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kovacs JM, Nkolola JP, Peng H, et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci USA 2012;109:12111–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang L, Song Y, Li X, et al. HIV-1 Virus-Like Particles Produced by Stably Transfected Drosophila S2 Cells: a Desirable Vaccine Component. J Virol 2012;86:7662–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tong T, Crooks ET, Osawa K, et al. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol 2012;86:3574–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao X, Chen W, Feng Y, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun 2009;390:404–9 [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First report of lack of interaction of HIV-1 bnAb predecessors with Envs.
- 80.Ota T, Doyle-Cooper C, Cooper AB, et al. Anti-HIV B Cell Lines as Candidate Vaccine Biosensors. J Immunol 2012;189:4816–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoot SJ, McGuire A, Cohen KW, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen W, Streaker ED, Russ DE, et al. Characterization of germline antibody libraries from human umbilical cord blood and selection of monoclonal antibodies to viral envelope glycoproteins: implications for mechanisms of immune evasion and design of vaccine immunogens. Biochem Biophys Res Commun 2012;417:1164–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiao X, Feng Y, Chen W, et al. Guiding the immune system through complex maturation pathways: a novel multi-immunogen approach for elicitation of broadly neutralizing antibodies. Retrovirology 2009;6:P35 [Google Scholar]
- 84.Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs 2010;2:347–56 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes a novel approach to elicit bnAbs against HIV-1.
- 85.Haynes BF, Kelsoe G, Harrison SC, et al. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol 2012;30:423–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma BJ, Alam SM, Go EP, et al. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog 2011;7:e1002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abbas W, Herbein G. Molecular understanding of HIV-1 Latency. Adv Virol 2012;2012:574967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997;387:183–8 [DOI] [PubMed] [Google Scholar]; • Quantification of latently HIV-1-infected resting CD4+ T cells in humans.
- 89.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 1999;5:512–17 [DOI] [PubMed] [Google Scholar]; • Determination of the decay half-life of latent HIV-1 reservoirs during HAART.
- 90.Zagury D, Bernard J, Leonard R, et al. Long-term cultures of HTLV-III–infected T cells: a model of cytopathology of T-cell depletion in AIDS. Science 1986;231:850–3 [DOI] [PubMed] [Google Scholar]
- 91.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 1987;326:711–13 [DOI] [PubMed] [Google Scholar]
- 92.Chun TW, Engel D, Mizell SB, et al. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med 1999;5:651–5 [DOI] [PubMed] [Google Scholar]
- 93.Scripture-Adams DD, Brooks DG, Korin YD, et al. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol 2002;76:13077–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheller C, Ullrich A, McPherson K, et al. CpG oligodeoxynucleotides activate HIV replication in latently infected human T cells. J Biol Chem 2004;279:21897–902 [DOI] [PubMed] [Google Scholar]
- 95.Hezareh M Prostratin as a new therapeutic agent targeting HIV viral reservoirs. Drug News Perspect 2005;18:496–500 [DOI] [PubMed] [Google Scholar]
- 96.Dechristopher BA, Loy BA, Marsden MD, et al. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat Chem 2012;4:705–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ylisastigui L, Archin NM, Lehrman G, et al. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 2004;18:1101–8 [DOI] [PubMed] [Google Scholar]
- 98.Lehrman G, Hogue IB, Palmer S, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 2005;366:549–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Archin NM, Liberty AL, Kashuba AD, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012;487:482–5 [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows that a clinically used anti-cancer drug is capable of activating latent HIV-1.
- 100.Yang HC, Shen L, Siliciano RF, et al. Isolation of a cellular factor that can reactivate latent HIV-1 without T cell activation. Proc Natl Acad Sci USA 2009;106:6321–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Micheva-Viteva S, Kobayashi Y, Edelstein LC, et al. High-throughput screening uncovers a compound that activates latent HIV-1 and acts cooperatively with a histone deacetylase (HDAC) inhibitor. J Biol Chem 2011;286:21083–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang J, Wang F, Argyris E, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 2007;13:1241–7 [DOI] [PubMed] [Google Scholar]
- 103.Moldt B, Shibata-Koyama M, Rakasz EG, et al. A nonfucosylated variant of the anti-HIV-1 monoclonal antibody b12 has enhanced FcgammaRIIIa-mediated antiviral activity in vitro but does not improve protection against mucosal SHIV challenge in macaques. J Virol 2012;86:6189–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Curran KJ, Pegram HJ, Brentjens RJ. Chimeric antigen receptors for T cell immunotherapy: current understanding and future directions. J Gene Med 2012;14:405–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kennedy PE, Bera TK, Wang QC, et al. Anti-HIV-1 immunotoxin 3B3(Fv)-PE38: enhanced potency against clinical isolates in human PBMCs and macrophages, and negligible hepatotoxicity in macaques. J Leukoc Biol 2006;80:1175–82 [DOI] [PubMed] [Google Scholar]
- 106.Berger EA, Pastan I. Immunotoxin complementation of HAART to deplete persisting HIV-infected cell reservoirs. PLoS Pathog 2010;6:e1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davey RT Jr, Boenning CM, Herpin BR, et al. Use of recombinant soluble CD4 Pseudomonas exotoxin, a novel immunotoxin, for treatment of persons infected with human immunodeficiency virus. J Infect Dis 1994;170:1180–8 [DOI] [PubMed] [Google Scholar]
- 108.Wu X, Wang C, O’Dell S, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol 2012;86:5844–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Steckbeck JD, Sun C, Sturgeon TJ, et al. Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes. PLoS ONE 2010;5:e15261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chun TW, Nickle DC, Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis 2008;197:714–20 [DOI] [PubMed] [Google Scholar]
- 111.Chen W, Feng Y, Gong R, et al. Engineered single human CD4 domains as potent HIV-1 inhibitors and components of vaccine immunogens. J Virol 2011;85:9395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first successful design and generation of bacterially solubly expressible, stable single human CD4 domains with high and broad neutralizing activity.
- 112.Adler MJ, Dimitrov DS. Therapeutic antibodies against cancer. Hematol Oncol Clin North Am 2012;26:447–81.vii [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duvic M, Talpur R. Optimizing denileukin diftitox (Ontak) therapy. Future Oncol 2008;4:457–69 [DOI] [PubMed] [Google Scholar]
- 114.Stasi R, Evangelista ML, Buccisano F, et al. Gemtuzumab ozogamicin in the treatment of acute myeloid leukemia. Cancer Treat Rev 2008;34:49–60 [DOI] [PubMed] [Google Scholar]