Abstract
Heterochiral DNA strand-displacement reactions enable sequence-specific interfacing of oligonucleotide enantiomers, making it possible to interface native d-nucleic acids with molecular circuits built using nuclease-resistant l-DNA. To date, all heterochiral reactions have relied on peptide nucleic acid (PNA), which places potential limits on the scope and utility of this approach. Herein, we now report heterochiral strand-displacement in the absence of PNA, instead utilizing chimeric d/l-DNA complexes to interface oligonucleotides of the opposite chirality. We show that these strand-displacement reactions can be easily integrated into multicomponent heterochiral circuits, are compatible with both DNA and RNA inputs, and can be engineered to function in serum-supplemented medium. We anticipate that these new reactions will lead to a wider application of heterochiral strand-displacement, especially in the design of biocompatible nucleic acid circuits that can reliably operate within living systems.
Keywords: strand-displacement reaction, l-DNA, peptide nucleic acid, heterochiral DNA circuit, microRNA
Graphical Abstract
Toehold-mediated strand-displacement has proven to be a powerful mechanism for programing DNA-based circuits and other nanodevices that are capable of autonomous decision-making and complex logic functions.1,2 While the predictability of Watson–Crick (WC) base pairing rules allows for precise control over the thermodynamic and kinetic parameters of strand-displacement reactions, it has also instilled a “homochiral” paradigm over the field. This is because oligonucleotides of opposite stereochemistry, d- versus l-(deoxy)ribose, are incapable of forming contiguous WC base pairs with each other.3,4 Therefore, it was presumed that the individual nucleic acid components of a strand-displacement reaction must have the same chirality. However, we recently introduced the concept of “heterochiral” DNA strand-displacement, wherein a strand of achiral peptide nucleic acid (PNA) is used to transfer sequence information between chiral d- and l-oligonucleotides.5 Heterochiral strand-displacement reactions now enable the construction of DNA nanotechnology having fully-integrated d- and l-oligonucleotide components, and open the door to interfacing living systems with molecular circuits and sensors composed of bio-orthogonal l-DNA.6 However, due to both synthetic challenges and high cost, PNA remains difficult for the average research laboratory to obtain in sufficient numbers and quantities to support an iterative design process. Furthermore, PNA suffers from poor water solubility, which also places limits on its length and nucleotide composition.7,8 Consequently, the scope and utility of heterochiral strand-displacement reactions and related nanotechnologies are potentially limited by their dependence on PNA. With this in mind, we now report two novel heterochiral strand-displacement reactions that enable sequence-specific interfacing of d- and l-oligonucleotides in the absence of PNA.
RESULTS AND DISCUSSION
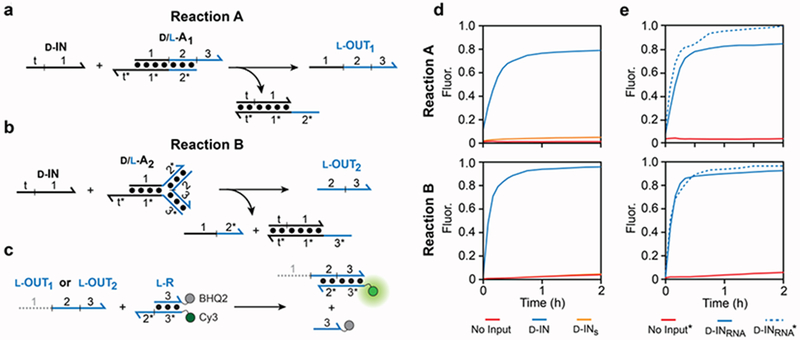
Previously reported heterochiral strand-displacement reactions employed pure d-DNA or pure l-DNA components, requiring an achiral PNA strand to serve as the intermediary between the two.5 However, we envisioned that the necessity for the PNA strand could be overcome by directly linking the d-DNA and l-DNA sequence domains within the same complex such that a homochiral strand-displacement reaction between a d-input and the d-DNA sequence domains triggers melting and release of the l-DNA sequence domain from the same complex (or vise versa), effectively yielding an output strand (or sequence domains) having the opposite chirality as the input. Herein, two such reactions are explored. Reaction A (Figure 1a) consists of a chimeric duplex (d/l-A1) comprising a portion of pure d-DNA (domain 1) and a portion of pure l-DNA (domains 2 and 3) directly linked by a typical 3′–5′ linkage. The reaction is initiated via binding of the d-DNA input (d-IN) to its toehold domain (t*), which resides on the d-DNA portion of the chimeric duplex (d/l-A1). The predicted melting temperature of the l-DNA portion of d/l-A1 (domain 2) is sufficiently low (~30 °C) such that subsequent invasion of the input strand through domain 1 results in spontaneous melting of the complex and release of the output strand (L-OUT1) containing the l-DNA sequence domains (2 and 3) (Figure S1a). In Reaction B (Figure 1b), the same basic principles are applied to a three-way junction (d/l-A2) consisting of one pure d-DNA arm and two pure l-DNA arms. As before, the reaction is initiated via binding of the d-DNA input (d-IN) to its toehold domain (t*) on the d-DNA arm. Because the melting temperatures of domains 2 and 3 are independently low (Figure S1b), separation of the two strands within domain 1 by the invading input (d-IN) results in disassembly of the three-way junction and release of the l-DNA output (l-OUT2). While Reaction A (i.e. duplex d/l-A1) benefits from a relatively simple design, the advantage of Reaction B (i.e. complex d/l-A2) is that it releases a pure l-DNA output. Importantly, for both Reactions A and B, release of their respective output strands Importantly, for both Reactions A and B, release of their leads to the “activation” of domain 2, allowing it to serve as an input for downstream strand-displacement reactions having l-DNA components. Given that efficient strand-displacement can be achieved with DNA toeholds having as little as 4 nucleotides9, the length of domain 2, which must remain short enough to allow for thermodynamic melting, is not expected to significantly limit the design of downstream circuit components.
Figure 1.
PNA-independent heterochiral strand displacement reactions. (a,b) Schematic illustration of heterochiral strand-displacement Reaction A (a) and B (b). D-DNA (black) and L-DNA (blue) are distinguished by color and are depicted as lines with the half-arrow indicting the 3′ end. (c) Fluorescent reporter strategy. (d,e) Fluorescent monitoring (Cy3) of heterochiral circuits. All reaction mixtures containing 100 nM of either d/l-A1 (top) or d/l-A2 (bottom), 300 nM l-R, 300 mM NaCl, and 10 mM TRIS (pH 7.6) were carried out at 37 °C. Reactions were initiated with either 150 nM of the indicated DNA input (d) or 1 μM RNA input (e). The asterisk indicated the presence of 0.1 mg/mL HeLa cell RNA extracts. Sequences of all strands are listed in Table S1.
In order to test the above designs, we assembled a heterochiral circuit wherein release of the l-DNA output strand generates a fluorescent signal through a second reaction with a fluorescent reporter complex (L-R) (Figure 1c). In the absence of PNA, the entire heterochiral circuit was easily prepared using a common solid-phase DNA synthesizer employing well-established protocols and commercial reagents (see Supplementary Information). It is important to emphasize that the sequences of domains 2 and 3 are identical in both chimeric complexes (d/l-A1 and d/l-A2), allowing the same reporter complex (L-R) to be used to monitor both reactions. Furthermore, l-R is the identical reporter complex previously used to monitor PNA-dependent reactions.5 As shown in Figure 1d, addition of d-IN to a reaction mixture containing either d/l-A1 or d/l-A2, along with l-R, resulted in the rapid generation of a fluorescent signal corresponding to production of the l-output strand. Reaction A proceeded somewhat more slowly than Reaction B (1.5 × 104 M−1 s−1 and 2.9 × 104 M−1 s−1, respectively). Nevertheless, both reactions were considerably faster than the PNA-dependent heterochiral strand-displacement reactions reported previously (9.6 × 102 M−1 s−1),5 highlighting another potential advantage of the current approach. As expected, no significant fluorescent signal was observed for either circuit in the absence of an input. An input strand with a scrambled toehold domain (d-INs) also failed to activate an appreciable fluorescent signal. Together, the above results confirm that Reactions A and B behave as intended, and can be easily substituted for a PNA-dependent reaction in a strand-displacement cascade.
As enantiomers, d-DNA and l-DNA have the same physical and chemical properties, yet l-DNA is completely resistant to degradation by cellular nucleases.10 This makes l-DNA an ideal nucleic acid analog for engineering bio-stable DNA-based nanodevices that interact with and operate within living systems.11–13 With this application in mind, we first tested whether heterochiral circuits comprising Reactions A and B could tolerate an RNA version of d-IN (d-INRNA), which has the identical sequence to microRNA-155. MicroRNAs represent an important class of disease biomarkers that often serve as inputs for diagnostic and/or therapeutic DNA-based nanodevices.14 As shown in Figure 1e, addition of d-INRNA to a reaction mixture containing either d/l-A1 or d/l-A2, along with l-R, resulted in the rapid activation of a fluorescent response, although a higher concentration of the RNA input (d-INRNA) was needed to obtain a signal equivalent to the DNA input (d-IN) at 150 nM (Figure S2). Similar results were obtained in the presence of a large excess of non-specific RNA from HeLa cells (0.1 mg/mL), demonstrating the specificity of these designs.
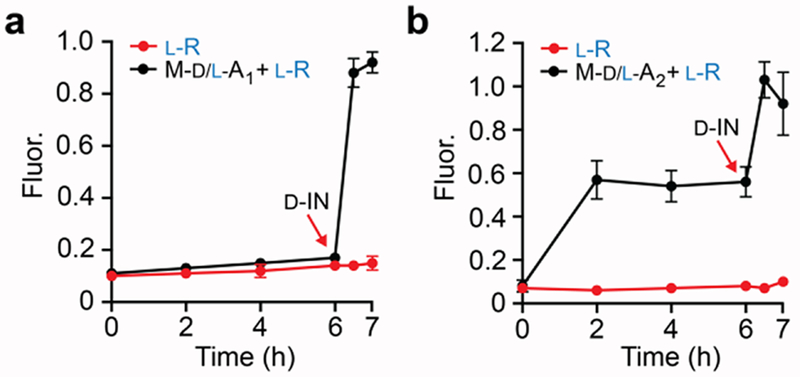
Finally, we examined the behavior of Reactions A and B in cell medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Nuclease degradation of the d-DNA portion of either chimeric complex (d/l-A1 or d/l-A2) will result in the release of the l-output and activation of the reporter complex l-R (i.e. circuit leakage). Indeed, incubation of both heterochiral circuits (d/l-A1/l-R or d/l-A2/l-R) in serum-supplemented medium resulted in significant circuit leakage after 6 hours and failed to further activate upon addition of the input (Figure S3a,c), indicating that the d-DNA domains were degraded. It is worth noting, however, that Reaction A components (d/l-A1/l-R) were still more stable under these conditions than a version of the circuit comprised entirely of d-DNA (Figure S3b). In order to increase the overall stability of d/l-A1 and d/l-A2, we replaced the d-DNA portions of both chimeric complexes with d-2′-O-methyl (2′OMe) ribonucleotides (Figure S4), which have increased stability against nuclease degradation (Figure S5).15 As expected, the 2′OMe versions of d/l-A1 and d/l-A2 (M-d/l-A1 and M-d/l-A2, respectively) improved the performance of their corresponding heterochiral circuits in serum-supplemented DMEM as compared to their all DNA counterparts (Figure 2). Most notably, the 2′OMe version of Reaction A showed a ~5-fold enhancement in fluorescence upon addition of input (d-IN) following the 6 hour incubation in serum-supplemented medium (Figure 2a). Reaction B components could also be activated by input following the 6-hour incubation in medium, however, the background signal for this system was considerably higher than Reaction A despite the 2′OMe modifications. While further optimization is needed, the above results demonstrate that the heterochiral strand-displacement reactions described herein can be made compatible with complex biological environments.
Figure 2.
Monitoring of Reaction A (a) and Reaction B (b) in DMEM supplemented with 10% FBS. 2′OMe-modified versions of d/l-A1 and d/l-A2 (M-d/l-A1 and M-d/l-A2, respectively) were employed in these experiments. The concentration of circuit components was identical to those described in Figure 1 and reactions were carried out at 37 °C. d-IN (1 μM) was added after 6 hours.
CONCLUSION
In summary, we have successfully demonstrated heterochiral strand-displacement in the absence of PNA, instead utilizing chimeric d/l-DNA complexes to interface oligonucleotides of the opposite chirality. To the best of our knowledge, this is the first time chimeric d/l-oligonucleotides have been used in a strand-displacement scheme. We note that while the reactions presented herein convert d-inputs into l-outputs, the same design principles can be applied to the reverse reaction (i.e. converting l-inputs into d-outputs). By lifting restrictions previously imposed on the system by PNA, this work is expected to lead to a wider application of heterochiral strand-displacement reactions in the design of DNA/RNA nanotechnology. Importantly, the heterochiral circuits described herein can be engineered to be stable in harsh biological environments and are compatible with RNA inputs, thereby further expanding the toolkit for interfacing endogenous nucleic acids signals (e.g. microRNAs and mRNAs) with molecular circuits and other nanodevices composed of nuclease-resistant l-DNA/RNA. Towards this goal, it will be important to examine the operation of these new designs in living cells.
Supplementary Material
Acknowledgments
Funding
J.T.S is a CPRIT Scholar of Cancer Research supported by the Cancer Prevention and Research Institute of Texas (RR150038). Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R21EB027855.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Materials and Methods. Supplementary Figures S1–6 and Table S1. (PDF)
Conflict of Interest
The authors declare no competing financial interests.
REFERENCES
- (1).Simmel FC; Yurke B; Singh HR Principles and applications of nucleic acid strand displacement reactions. Chem. Rev 2019, 119, 6326–6369. [DOI] [PubMed] [Google Scholar]
- (2).Zhang DY; Seelig G Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem 2011, 3, 103–113. [DOI] [PubMed] [Google Scholar]
- (3).Garbesi A; Capobianco ML; Colonna FP; Tondelli L; Arcamone F; Manzini G; Hilbers CW; Aelen JME; Blommers MJJ l-DNAs as potential antimessenger oligonucleotides: A reassessment. Nucleic Acids Res. 1993, 21, 4159–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hoehlig K; Bethge L; Klussmann S Stereospecificity of oligonucleotide interactions revisited: No evidence for heterochiral hybridization and ribozyme/DNAzyme activity. PLoS ONE [Online] 2015, 10, e0115328 https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0115328 (accessed August 15, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kabza AM; Young BE; Sczepanski JT Heterochiral DNA strand-displacement circuits. J. Am. Chem. Soc 2017, 139, 17715–17718. [DOI] [PubMed] [Google Scholar]
- (6).Li J; Green AA; Yan H; Fan C Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem 2017, 9, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Braasch DA; Corey DR Synthesis, Analysis, purification, and intracellular delivery of peptide nucleic acids. Methods 2001, 23, 97–107. [DOI] [PubMed] [Google Scholar]
- (8).Sahu B; Sacui I; Rapireddy S; Zanotti KJ; Bahal R; Armitage BA; Ly DH Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing γ-peptide nucleic acids with superior hybridization properties and water solubility. J. Org. Chem 2011, 76, 5614–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhang DY; Winfree E Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc 2009, 131, 17303–17314. [DOI] [PubMed] [Google Scholar]
- (10).Hauser NC; Martinez R; Jacob A; Rupp S; Hoheisel JD; Matysiak S Utilising the left-helical conformation of L-DNA for analysing different marker types on a single universal microarray platform. Nucleic Acids Res. 2006, 34, 5101–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhong W; Sczepanski JT A Mirror image fluorogenic aptamer sensor for live-cell imaging of microRNAs. ACS Sens. 2019, 4, 566–570. [DOI] [PubMed] [Google Scholar]
- (12).Ke G; Wang C; Ge Y; Zheng N; Zhu Z; Yang CJ l-DNA Molecular beacon: a safe, stable, and accurate intracellular nano-thermometer for temperature sensing in living cells. J. Am. Chem. Soc 2012, 134, 18908–18911. [DOI] [PubMed] [Google Scholar]
- (13).Cui L; Peng R; Fu T; Zhang X; Wu C; Chen H; Liang H; Yang CJ; Tan W Biostable L-DNAzyme for sensing of metal ions in biological systems. Anal. Chem 2016, 88, 1850–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cheng Y; Dong L; Zhang J; Zhao Y; Li Z Recent advances in microRNA detection. Analyst 2018, 143, 1758–1774. [DOI] [PubMed] [Google Scholar]
- (15).Groves B; Chen Y-J; Zurla C; Pochekailov S; Kirschman JL; Santangelo PJ; Seelig G Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol 2016, 11, 287–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.