Abstract
The smallest independently folded antibody fragments, the domains, are emerging as promising scaffolds for candidate therapeutics and diagnostics that bind specifically targets of interest. The discovery of such binders is based on several technologies including structure-based design and generation of libraries of mutants displayed on phage or yeast, next-generation sequencing for diversity analysis, panning and screening of the libraries, affinity maturation of selected binders, and their expression, purification, and characterization for specific binding, function, and aggregation propensity. In this review, we describe these technologies as applied for the generation of engineered antibody domains (eAds), especially those derived from the human immunoglobulin heavy chain variable region (VH) and the second domain of IgG1 heavy chain constant region (CH2) as potential candidate therapeutics and diagnostics, and discuss examples of eAds against HIV-1 and cancer-related proteins.
Keywords: Antibody domains, human, library, phage display, scaffold, stability, therapeutics, yeast display
1. INTRODUCTION
Small-size binders based on human and nonhuman immunoglobulin (Ig) scaffolds have been extensively explored as promising candidate therapeutics and diagnostics [1–5]. They include a unique class of antibodies composed only of heavy chains (HCAbs) that are naturally formed in camels, dromedaries, and llamas. The variable regions of HCAbs are referred as VHHs and can recognize antigens as single-domain fragments. The Ig new antigen receptors (IgNARs) of sharks are also HCAbs and utilize single variable domains (VNARs) for binding to antigens [6]. In addition, there are non-antibody scaffolds such as the fibronectin type III domain (FN3), which has been extensively characterized and shown to be a robust scaffold for generating new binders [7]. Other non-antibody scaffolds include the Ig superfamily D1 domain of intercellular adhesion molecule 1 (ICAM-1) [8] and the Z1 domain from titin with the intracellular Ig fold [9] that have been recently engineered as potential scaffolds.
The heavy (VHs) and light (VLs) chain variable domains of human antibodies, known as domain antibodies (dAbs) in size ranging from 11 to 15 kDa, are potentially useful scaffolds for therapeutics development because of their human origin leading to no or reduced immunogenicity. Not only are dAbs much smaller than full-size antibodies, but also their paratopes are generally concentrated over smaller areas, which enables to target novel epitopes, e.g., the CD4-induced (CD4i) epitopes on the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) gp120, which are not accessible or only partially accessible to conventional antibodies or antibody fragments with paired light and heavy chain variable domains. However, most isolated human VHs and VLs are expressed generally in the form of insoluble inclusion bodies because of the exposed large hydrophobic patches which are otherwise buried at the VH/VL interface. It has been found that certain human VH and VL family members (e.g., HV3–23 and KV1–39) are more likely than others to be independently correctly folded and expressed as soluble proteins due to unique features in sequences and structures [10, 11]. They have been used as scaffolds to construct libraries for selection of dAbs targeting various human diseases. Interestingly, human antibody constant domain CH2 (CH3 for IgE and IgM) can be isolated as a monomeric soluble protein because it does not naturally involve any protein-protein interactions except weak carbohydrate interactions with the other CH2 domain. The CH2 domain has been extensively characterized as a scaffold to construct libraries for selection of small antibodies, which could have excellent properties such as prolonged half-lives and effector functions due to their potential ability to bind to Fc receptors [12]. Here, we use the term “engineered antibody domains (eAds)” to denote both variable and constant domains that are engineered to confer functions in addition to their native ones.
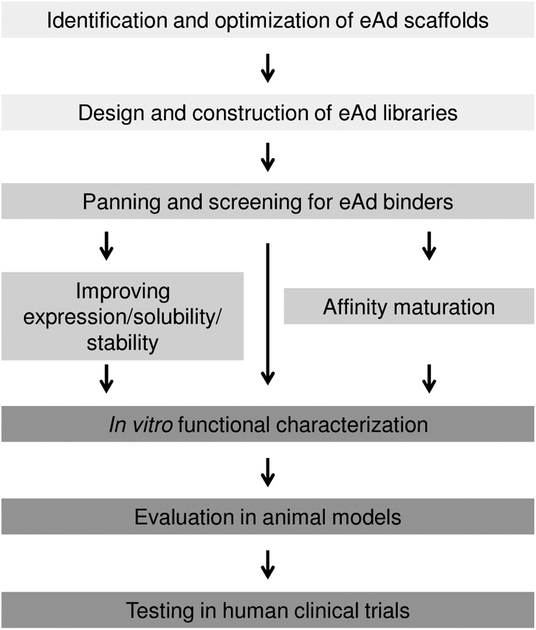
To facilitate the development of eAds, multiple approaches combining structure-based design, effective library technologies for generating eAds, panning and screening of the libraries, high-throughput eAd library sequencing, and improving biophysical properties are required that would lead to eAds with functionality and low aggregation propensity which could be suitable for evaluation in animal models and possible human testing of candidate eAds (Fig. 1). Here, we review current technologies for developing eAds, particularly those based on the VH and CH2 domains, and their usefulness, which we have already demonstrated by successful selection of high-affinity binders to viral [13–15] and cancer-related antigens [16].
Fig. (1).
A flow diagram indicating various stages and technologies involved in the development of eAds.
2. STRUCTURE-BASED DESIGN OF ANTIBODY DOMAIN SCAFFOLDS AND LIBRARIES
Conventional human antibodies have Y-shaped structures with two identical heavy chains paired with two identical light chains. The N-terminal domains of both heavy and light chains are variable and denoted as VH and VL, respectively. The other domains are constant; CH1 and CL are from the heavy and light chains, respectively, in the antigen-binding fragment (Fab) region, and CH2 and CH3 (and CH4 for IgE and IgM) in the fragment crystallizable (Fc) region. The antigen-binding sites are formed by six hypervariable regions, complementarity determining regions (CDRs), three on the VH and the other three on the VL domains. Interestingly, some animals produce unique HCAbs, which target antigens through a single variable domain displaying three or less CDRs. One example of HCAbs is the camelid IgG2 and IgG3 isotypes with size of about 85 kDa which unlike their IgG1, are heavy chain homodimers with a variable domain, designated as VHH, attached to two constant domains in each heavy chain [17]. The other example is the new antigen receptor antibodies (IgNARs) from shark with size of about 160 kDa that are composed of dimers of two heavy chains connected through disulfide bonds. Each of the chain contains a variable domain, designated as VNAR, and five constant domains [18].
The crystal structures of several VHHs and VNARs are known as free and complex forms, and are available in the Protein Data Bank [19]. Comparison of human VH structures with those of VHHs reveals that the framework hydrophobic residues in the VHs involved in the VH-VL interactions are mutated into hydrophilic residues in VHHs contributing to the stability of camelid and llama HCAbs [20]. In the IgNAR structures, VNARs do not have the C′ and C″ strands that normally contain CDR2s [6] which are similar to constant domains. Moreover, VHHs and VNARs encode CDR3s that are unusually longer and structurally more complex than those of human VHs. These conserved features dramatically increase the stability and diversity of VHH and VNAR repertoires, and define surface areas interacting with antigens resulting in novel paratopes that are different from those of conventional antibodies, indicating the molecular convergence between the HCAbs [21]. The molecular insights provided by HCAbs helped to develop single VH domain-based eAds, viz., camelization of human antibody domains and vice-versa for producing stable scaffolds [22, 23]. Remarkably, a soluble and refoldable human single VH antibody domain was previously identified as naturally occurring that originates from the VH3–23 germline, in which the framework hydrophobic residues involved in the VH-VL interface are retained and the only sequence changes from the corresponding human germline are confined to CDRs [24]. The structure of that VH domain antibody shows an unusual flipping of the side-chain of a framework residue (Trp47) into a cavity formed by the Gly35 of CDR1 that increases the hydrophilic surface. An eAd without the Ig intra-disulfide bond was recently developed using the human antibody VL domain binding to huntingtin (Htt) peptide [25].
We previously identified a phage-displayed HCAb by panning of a large human naive Fab library (size, approximately 1.5 × 1010) against a recombinant HIV-1 Env [15]. The VH domain of the HCAb, designated m0, was independently folded, solubly expressible, stable, and monomeric, and has become a promising scaffold for generating eAd libraries. m0 was also from the VH3–23 germline with unaltered hydrophobic framework residues at the VH-VL interface, and mutations in CDR1 and other framework regions compared to the germline. It has a CDR3 length of 18 amino acids. The mutations, particularly Ala to Asp at position 38 (IMGT numbering system) of the CDR1, Ala to Pro at position 55 of the framework, and those in the CDR3 might have contributed to the stability of m0. We used m0 for construction of a large-size (approximately 2.5 × 1010) phage-displayed eAd library by grafting natural CDR2s and CDR3s from five of our Fab libraries and randomly mutagenizing the CDR1. Panning of this library with an HIV-1 Env complexed with CD4 resulted in the identification of a very potent broadly cross-reactive eAd against HIV-1, m36, which neutralizes HIV-1 primary isolates from different clades with IC50s and IC90s in nM range [14].
We recently proposed that the human IgG1 constant domain CH2 could serve as a scaffold for construction of libraries containing diverse binders to various antigens [12]. CH2 is the smallest independently folded antibody domain that can be engineered to possess simultaneously antigen-binding sites and binding sites mediating effector and stability functions. We previously determined the crystal structure of a bacterially expressed monomeric human CH2 domain and found that the structure is almost identical to that of intact CH2 in Fc or IgG1, except for some disordered residues at the N terminus [26]. Based on the CH2 domain structure, we engineered a disulfide bond in addition to the native one resulting in a CH2 variant, m01, with increased stability [27]. We further generated a shortened form of CH2 domain, m01s, by removing seven residues at the N terminus that precede the stand A [28]. We found that shortening the N terminus significantly increases stability and reduces aggregation propensity of m01s without disrupting its conformation. We also designed libraries by grafting CDRs from our phage-displayed antibody libraries onto this improved CH2 scaffold and selected CH2-based bispecific eAds which showed binding to both antigens and neonatal Fc receptor (FcRn) [13]. We hypothesized that binding to the FcRn could increase antibody half-lives in vivo and binding to complements could lead to lysis of target cells. A recent pharmacokinetics study of CH2-based eAds showed relatively long serum half-lives, making them a unique scaffold suitable for development of targeted therapeutics [29]. Taken together, these studies suggest that eAds could be successfully developed from the human antibody VH domain- as well as the CH2 domain-based scaffolds by combining structure-based design and combinatorial library display and screening technologies as we described in the latter sections.
3. PHAGE-, YEAST-, AND MAMMALIAN CELL-BASED TECHNOLOGIES FOR EAD LIBRARY DISPLAY AND SELECTION
Since the invention of phage display technology by George P. Smith [30], numerous combinatory library display technologies have been developed and proven as powerful tools in the protein and antibody engineering field [31–34]. Due to its high efficacy and simplicity to perform, phage display has been broadly utilized in the isolation and engineering of antibody-based therapeutics and diagnostics development. Humira, the very first fully human antibody drug developed through phage display, has been approved by the US FDA and is on track to become the top selling biologics in the world, while numerous antibody-based therapeutics engineered using phage, yeast, mammalian cell, and other display technologies are at various clinical development stages.
Due to their small size and excellent biochemical and biophysical features, eAds have been extensively explored as candidates for the development of therapeutics and diagnostic reagents. Lately, naturally occurring single antibody domains from camel [17], shark [35], and lamprey [36, 37] were successfully displayed on phage or yeast and shown to be useful resources for the isolation and engineering of a wide range of eAd-based reagents for various biomedical applications. Similarly, for therapeutics development, fully human VH- and VL-based scaffolds have also been extensively explored and successfully validated using phage as a library display platform and engineering tool [15, 38].
Among all the other combinatory library display platforms, which were inspired by the invention of phage display, yeast display stands out as one of the most powerful protein engineering platforms. One of the main advantages the yeast display platform offers is its eukaryotic system providing very sophisticated protein folding and chaperones machinery, which allows efficient and consistent display of a variety of proteins; in return, this unique feature makes quantitative sorting of yeast libraries and highly efficient affinity maturation process possible. More importantly, a high-efficacy yeast electroporation protocol has been described recently [39], which enables construction of yeast display libraries with large size (up to 1010). This makes yeast display comparable to phage display system in terms of library size, and thus further simplifies the isolation of high affinity binders from naïve antibody libraries or other scaffold-based universal libraries. Previously, yeast display was used for the isolation of picomolar affinity binders from the variable lymphocyte receptor (VLR) library derived from lamprey [40]. We recently displayed human CH2-based library on yeast, and the concept of using CH2 domain as a scaffold for the isolation and engineering of CH2-based binders was fully validated [41].
Mammalian cells provide all necessary components for human antibody synthesis and processing under physiological conditions. Compared to the phage and yeast display systems where some human antibodies may not be expressed or correctly folded due to relatively modest protein folding machineries and other properties, mammalian cells could help retain the diversity of human antibody repertoires to a larger extent [42]. In addition, mammalian cells confer native and more consistent glycosylation which although rarely, can occur in the antigen-binding domains of antibodies and affect antibody functions. However, each mammalian cell can harbor multiple antibody species delivered by plasmid transfection, which requires many rounds of selection in order to enrich for cell clones with only a single antibody species, thus limiting the ability of mammalian cells to handle large libraries. Virus-mediated gene delivery systems have been vigorously explored to circumvent this drawback of mammalian cell display [42], which could eventually become a powerful tool for the development of novel candidate therapeutics and diagnostic reagents based on eAds.
4. GENERATION AND CHARACTERIZATION OF VARIABLE DOMAIN-BASED EAD LIBRARIES
While eAds selected from VHH, VNAR, and synthetic human VH and VL libraries may raise additional safety concerns (e.g., immunogenicity) when used in humans, libraries with fully human sequences are highly desirable in order to avoid or minimize potential adverse effects. Jirholt et al. [43] constructed an eAd library of 9 × 106 members by shuffling natural CDRs into a camelized human VH3–23 germline framework scaffold but no further characterization was described in terms of solubility and functionality of eAds from the library. As was previously reported [15], we accidentally identified a human VH-based scaffold by panning a large phage-displayed non-immune human Fab library against an HIV-1 Env gp120. One of the selected Fabs had a stop codon in the light chain but was still selected from the library and showed significant binding to the antigen as a heavy-chain only fragment. We cloned the VH domain of this antibody and found that it exhibited high levels of expression and high solubility. This completely natural VH, m0, belongs to the VH3–23 gene family and is diversified from the germline sequence by a number of somatic hypermutations. A homology-based model of m0 was developed using SWISS MODEL [44] and is shown in the Fig. 2A. m0 was used as a framework scaffold to construct a large phage-displayed human VH library m8l (size, approximately 2.5 × 1010) by grafting in vivo-formed heavy chain CDR2 (HCDR2) and CDR3 (HCDR3) repertoires from our other human antibody libraries and mutating four residues in the CDR1 (HCDR1). The library exhibited a high degree of variability and a large percentage of the tested VHs were expressed as soluble proteins in E. coli periplasm. The quality of the library was also validated by selection of VHs against different antigens. In contrast to the previously described library [43], there is no need to camelize the framework which is a part of naturally occurring antibodies in humans.
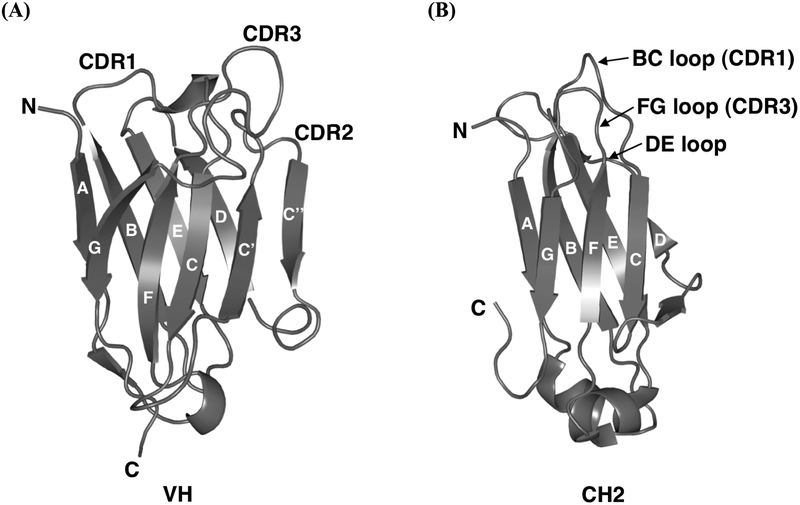
Fig. (2).
Ribbon diagrams illustrating (A) a model of the human antibody VH domain, m0, and (B) the crystal structure of the human IgG1 CH2 domain, which have been used as scaffolds for eAd library construction. The β-strands are labeled through A-G, having the same topology and similar structures except there are two additional strands, C′ and C″, in the VH domain. The CDRs or loop regions between those β-strands are at the same end of the barrel. The CDR1–3 in VH and BC, DE, and FG loops in CH2 are marked.
To further increase the library diversity, we developed a second-generation library m9l (size, approximately 1 × 1010) by grafting light chain CDR3 (LCDR3) repertoires into the CDR1 of library m8l [45]. Our design is based on the observation that in general, LCDR3 is significantly more diverse than HCDR1 not only in sequences but also in lengths. In addition, there could be a need to compensate the loss of antigen-interacting surface contributed by the hypervariable loops of VLs. We speculated that grafting of non-cognate LCDR3 would lead to novel paratopes, resulting that the library could be more suited for selection of antibodies to some antigens or epitopes of the same antigen than the existing libraries based on conventional designs. In line with this speculation, our results showed efficient selection of different antibodies from m9l against the human cancer-related antigens, insulin-like growth factor II (IGF-II) and IGF-I receptor (IGF-IR), compared to those selected from the first-generation library m8l [45]. However, it appears that we need to pay a price for grafting non-cognate LCDR3 into HCDR1 because soluble expression of VHs randomly selected from m9l is somewhat lower than that from library m8l. It is to be expected that the use of noncognate grafting could result in misfolding and aggregation of some VHs. However, most of the antigen-selected VHs from m9l are soluble and expressed at high levels. We found that they contain a variety of LCDR3s and HCDR2s originated from different germlines, and HCDR3s of varying lengths.
5. GENERATION AND CHARACTERIZATION OF CONSTANT DOMAIN-BASED EAD LIBRARIES
Compared to a human VH domain, the CH2 domain is also composed of similar antiparallel β-strands, A-G, forming two sheets and interspersed with loops including BC loop (CDR1), DE loop, and FG loop (CDR3), but lacking C′ C″ loop (CDR2) due to the absence of C′ and C″ strands in CH2 (Fig. 2B). This suggests that the CH2 scaffold could sustain diversification of these loops for the development of libraries containing CH2 domain-based binders to antigens. Additionally, CH2 binders could be engineered to retain some effector functions as rendered by the constant domains of IgGs. To this end, we constructed a large (size, approximately 5 × 1010) CH2-based library [46], in which we mutated all residues randomly in the BC and FG loops into four frequently occurring residues, tyrosine, alanine, aspartic acid, and serine. A glycine residue was added at the C-terminal end of each loop to increase flexibility to accommodate the possible changes required for stability and antigen recognition of binders. To test whether potentially useful binders could be selected from the CH2-based library, we used an HIV-1 gp120 fused with human two-domain soluble CD4 (gp120Bal–sCD4) as a panning antigen. A number of positive clones were successfully selected. The highest affinity binder, m1a1, recognized specifically a highly conserved CD4i epitope and inhibited to various extents seven of nine HIV-1 isolates from different clades tested. These results thus provide a proof of concept for using the CH2 domain as a robust scaffold to construct eAd libraries.
Recently, we described a novel strategy based on multi-step PCR to construct a library using m01s, a shortened CH2 domain with an N-terminal seven residues deletion, as a scaffold [13]. The multi-step PCR resulted in the precise grafting of human antibody HCDR3s onto the loop FG of m01s. Using this HCDR3 grafting method and limited mutagenesis of loops BC and DE, we generated a phage-displayed eAd library m01sl. After panning this library against an HIV-1 membrane proximal external region (MPER) peptide, sp62, we selected and characterized one of the binders, m2a1, which exhibited relatively lower propensity for aggregation. We found that m2a1 interacted non-competitively with sp62 and FcRn, suggesting that CH2-based eAds could confer both antigen-binding activity and effector functions of full-size antibodies. A major drawback for some applications of small-size binders is their short half-lives in serum (in the order of minutes) partially due to lack of FcRn binding. m2a1 and the wild-type CH2 bind to FcRn in a pH-dependent manner, which may account for an extended half-life (approximately 10 hours) of human CH2 in mice [29]. These findings have significant implications for the stability and related pharmacokinetics of constant domain-based eAds in vivo.
6. GENERATION OF SOLUBLE MONOMERIC FC AS A SCAFFOLD FOR EAD LIBRARIES
In our efforts to develop soluble and functional Fc monomers (mFcs) that could exert their therapeutic effects with prolonged half-lives, we used structure-based rational protein design and multiple screening methods [47]. Briefly, we constructed a phage display library (size, approximately 1.3 × 109) randomly mutating several residues that are located at the CH3 dimer interface of human IgG1 Fc. The library was panned against protein G resulting in a library enriched for soluble and well-folded mFcs. The resultant library was further panned against human FcRn for five rounds; in each round, a buffer at pH6.0 was used for washing and a buffer at pH7.4 was used for elution to select pH-dependent binders. Finally, we selected three mFc proteins by screening the final enriched library using non-reducing SDS-PAGE and western blots of the soluble periplasmic fraction of E. coli. Two of the three selected mFcs each contains seven mutations, whereas the other mFc contains six substitutions in the CH3 dimer interface. The mutations were partially reversed and the resultant proteins were analyzed by size exclusion chromatography for their monomeric states. We found that only four specific mutations are essential to produce mFcs, albeit at a lower expression level.
The mFcs are promising tools to explore FcRn binding and for the development of fusion proteins. The mFcs themselves could serve as novel antibody scaffolds for construction of libraries containing diverse binders to a variety of antigens. It has been reported that IgG1 Fc can be engineered to acquire antigen binding properties [48]. Importantly, the larger exposed surface areas available on mFcs, compared to the wild-type Fc dimer, provide more sites for protein engineering to design point mutations and CDR grafting onto mFc frameworks, which could enable novel modes of interactions with antigens. It is possible that binders selected from mFc scaffold-based libraries, which have molecular masses of about 27 kDa, similar to that of an antibody single-chain variable fragment (scFv), could possess much longer in vivo half-lives.
7. HIGH-THROUGHPUT 454 SEQUENCING FOR CHARACTERIZATION OF EAD LIBRARIES
Size and sequence diversities are thought to be key elements in the high-quality libraries. In the past decades, library diversity was estimated mainly by standard Sanger sequencing of only a limited number of randomly selected antibody clones due to the relatively high sequencing costs and the requirement of preparation of plasmid DNAs or PCR products for every single clone [3]. This situation has recently changed with the advent of next-generation sequencing (NGS) technologies [49, 50], a representative of which is the Roche 454 sequencing platform. The average length of antibody variable domains is compatible with a single read length obtainable from 454 sequencing method, 400–450 bp, which makes it possible to reveal the entire sequence of the domain without any sequence assembly process. We recently performed 454 sequencing of antibody variable domains from human cord blood and adult IgM libraries. Analysis of the sequences has provided valuable insights into the breadth of antibody repertoires expressed and B cell diversity including germline gene usage, junctional diversity, and somatic mutations [51, 52]. Although highly efficient, NGS technologies, particularly the 454 antibody sequencing, pose significant challenges for identifying sequencing errors [53]. The IMGT/HighV-QUEST is a very useful tool for the analysis of 454 antibody sequencing data and can handle some sequencing errors, mainly insertion and deletion [54]. However, certain types of errors such as point mutations could not be corrected, particularly those in the CDRs, unless several repeated sequencing reactions are performed or possible inferences are made from clonally related sequences sharing some repeated mutations which could be distinguished from errors. It could be easier to detect errors in framework regions because they are highly conserved, and their corresponding germline sequences are known, which may allow the evaluation of sequence quality to a certain extent.
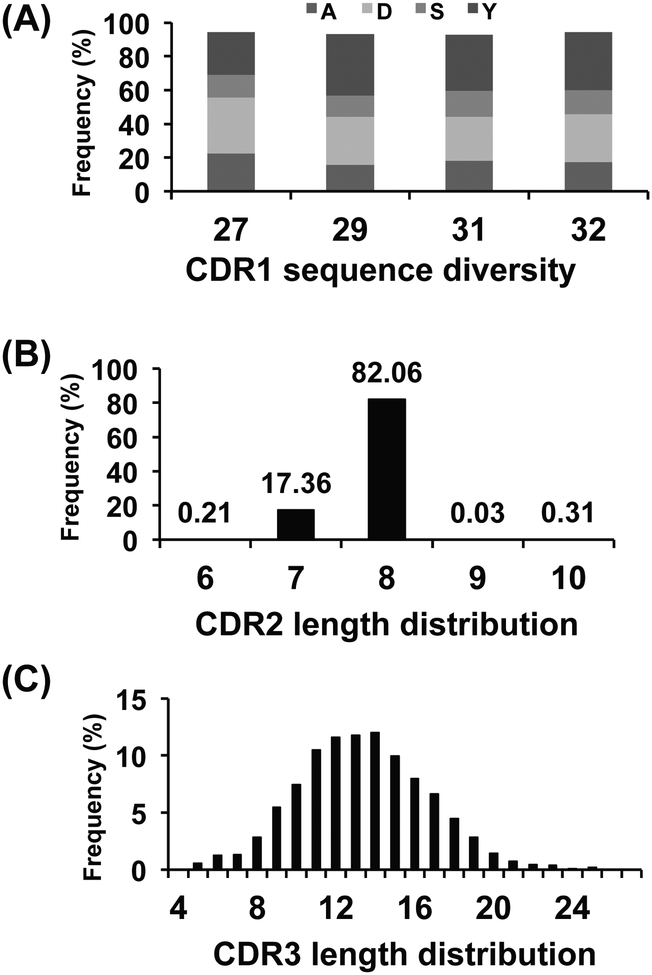
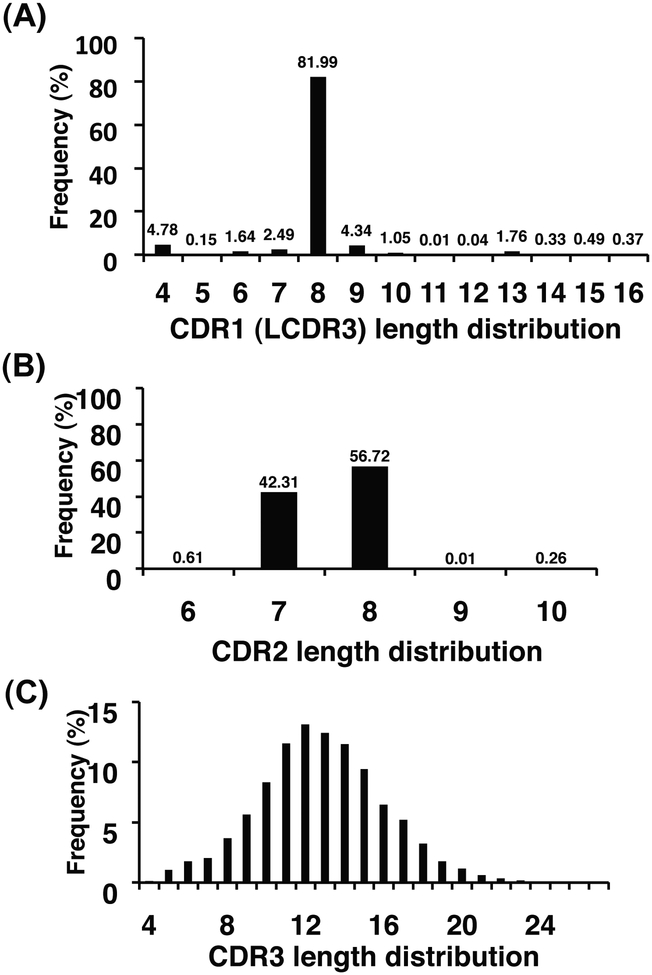
We used high-throughput 454 sequencing method to characterize the sequence diversity of the two VH-based libraries m8l and m9l (Table 1). Although there are limitations due to sampling depth, PCR efficiency, and sequencing errors, we obtained relatively larger datasets compared to our previous random sequence characterizations [15, 45]. The unique numbers of eAds in Table 1 represent the total numbers of eAd amino acid sequences which are functionally productive, without stop codons and/or frame shifts. For library m8l, sequence diversity arising from the amino acid compositions of CDR1, and the lengths of CDR2 and CDR3 are shown in Fig. 3. Frequency distribution of site-directed mutations to tyrosine, alanine, aspartic acid, and serine in the CDR1 showed the dominance of aspartic acid and tyrosine residues at the average values of 29% and 32%, respectively, at the specific positions of CDR1 (Fig. 3A). CDR2 and CDR3 length distributions are shown in Fig. 3B and Fig. 3C respectively. Library m9l was constructed by replacing the CDR1s of library m8l with a library of human antibody LCDR3 to increase diversity [45]. The length distribution of LCDR3s grafted onto the CDR1 is shown in Fig. 4A, in which 82% of the eAds have a CDR1 (LCDR3) length of 8 amino acids. Of note is that most of the natural HCDR1s have 8 amino acids in lengths which might be optimized to connect across two β-sheets in the VH domains. We found 11,518 unique CDR1 (LCDR3) sequences at the amino acid level which contributed to the diversity of library m9l. CDR2 and CDR3 length distributions are shown in Fig. 4B and 4C, respectively. Both CDR2 and CDR3 length distributions of the two libraries were found to be similar to those observed for natural Ig repertoires [51]. Though the lengths of CDR2 are mostly restricted to 7 and 8 amino acids up to 99% in each library, we found numerous unique CDR2 sequences in the eAd libraries (Table 1). The productive CDR3s are the result of unique V-D-J rearrangements which largely determine the diversity of the eAd libraries. The lengths of HCDR3s from the two libraries ranged from 4 to 27 amino acids (Fig. 3C and 4C).
Table 1.
454 antibody sequencing reveals high sequence diversity of eAd libraries generated by engineering of CDRs or loops.
| eAd libraries | Numbers of unique eAds | Numbers of unique CDRs | ||
|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | ||
| m81a | 7,770 | 956 | 2,398 | 6,731 |
| m91b | 26,197 | 11,518 | 6,140 | 21,553 |
| m01slc | 37,818 | 28,626d | 2,463e | 15,645f |
A human VH library based on m0 scaffold and grafting of in vivo-formed HCDR2s and HCDR3s from our other human antibody libraries onto their respective CDRs, and mutating four residues in the CDR1 of m0 [15].
A second-generation human VH library constructed by grafting of LCDR3s from our other human antibody libraries into the CDR1 of library m8l [45].
A human IgG1 CH2 domain-based eAd library using m01s as a framework scaffold in which the FG loop was replaced by the HCDR3s from library m8l, and mutations were intro duced to BC and DE loops by site-directed mutagenesis [13].
BC loop;
DE loop;
FG loop
Fig. (3).
Analysis of sequence diversity of the VH-based library m8l by 454 sequencing. (A) Frequencies of site-directed mutations to A, D, S or Y at specific positions in the CDR1. (B) CDR2 length distribution. (C) CDR3 length distribution.
Fig. (4).
Analysis of sequence diversity of the VH-based library m9l by 454 sequencing. (A) CDR1 (LCDR3) length distribution. (B) CDR2 length distribution. (C) CDR3 length distribution.
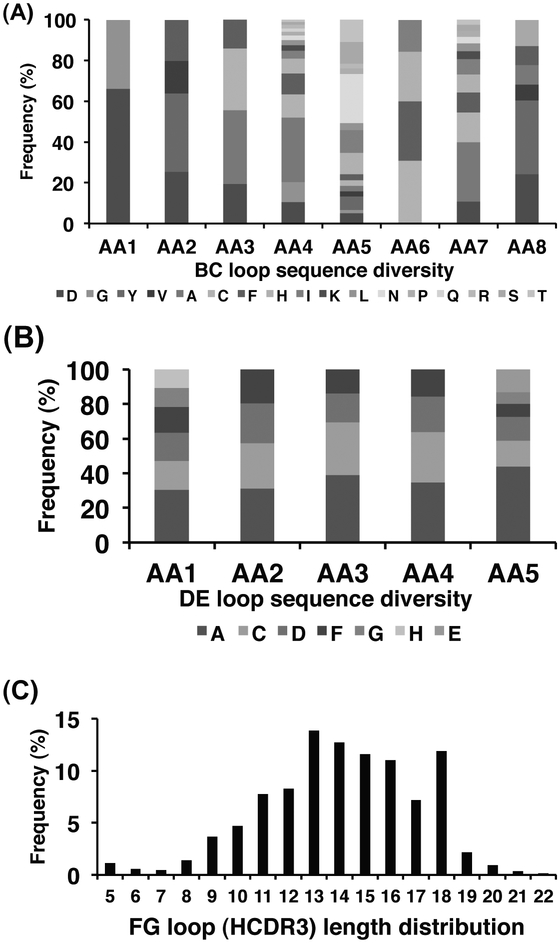
We created several eAd libraries based on the antibody CH2 domain as a scaffold [13, 46]. Here, we described the sequence diversity analysis of a CH2-based eAd library m01sl as determined by 454 sequencing. m01s, the truncated CH2 domain, was used as a framework scaffold where the FG loop was replaced by the HCDR3s from library m8l. In addition, eight residues in the BC loop and five residues in the DE loop were randomized by using degenerate primers [13]. Relatively larger amino acid composition diversity was observed in the BC loop (Fig. 5A) compared to the DE loop (Fig. 5B). The CDR3 lengths range from 5 to 22 amino acids (Fig. 5C). A large number of unique sequences found in the VH- and CH2-based eAds libraries further indicate high sequence diversity of the libraries (Table 1).
Fig. (5).
Analysis of sequence diversity of the CH2-based library m01sl by 454 sequencing. (A) Frequencies of site-directed mutations at eight residues in the BC loop. (B) Frequencies of site-directed mutations at five residues in the DE loop. (C) FG loop (HCDR3) length distribution. AA, amino acid.
The knowledge of diversity of eAd libraries provided by 454 antibody sequencing can also be useful in improving the biophysical properties of eAds [55], finding binders without screening [56], and understanding the maturation pathways of antibodies in response to antigens [57].
8. IDENTIFICATION OF EAD-BASED BINDERS TO EPITOPES THAT ARE NOT ACCESSIBLE OR ONLY PARTIALLY ACCESSIBLE TO LARGE-SIZE ANTIBODIES
Like libraries based on conventional antibody formats (e.g., Fab and scFv), large highly diverse eAd libraries derived from natural or synthetic sources could yield antibodies against virtually any antigens. Importantly, eAd libraries are a valuable source of antibodies targeting cryptic epitopes and antigens in obstructed locations where efficient penetration is critical for successful treatment. The small size of eAds leads to relatively good penetration into tissues and the ability to bind into cavities or active sites of protein targets which may not be accessible to larger-size antibodies and fragments. In the following two sections, we discussed two appropriate examples where the use of eAd libraries is shown to be beneficial for selection of binders against size-restricted epitopes on the surface of protein molecules.
8.1. Identification of eAds Targeting Sterically Restricted (Hidden) Epitopes
EAds show enhanced access to sterically restricted areas on the surfaces of some molecules. HIV-1 Env gp120 is an example of such molecules. HIV-1 entry is triggered by binding of the gp120 to the cellular receptor CD4. The binding induces conformational changes in gp120 leading to exposure of the coreceptor-binding site (CoRbs) [58]. Therefore, the CD4-binding site (CD4bs) and the CoRbs on gp120 are attractive targets for antibody therapy. Because the CoRbs is located toward the target cell membrane, the small space between gp120 and the target cell membrane is not sufficient to accommodate large-size antibodies; however, it is accessible to small-size antibody fragments or domains. A previous study clearly demonstrated that generally, the size of the CoRbs-directed neutralizing antibodies in different forms (IgG, Fab and scFv) is inversely correlated with their ability to neutralize the virus [59].
We successfully identified a number of eAds from library m8l that target the CoRbs of HIV-1 gp120 [14]. One of the highest affinity binders, m36, had a very high yield of soluble protein (about 70 mg/L from bacterial shaking culture), was monomeric in PBS (pH7.4), and showed a Tm value of 67°C. It potently neutralized genetically diverse HIV-1 isolates in vitro [14]. In a humanized NOD/SCID/γcnull mouse model, m36.4, an affinity-matured version of m36, provided sterilizing protection of four of six animals against intrasplenical challenge with high-titer HIV-1 (>1000 TCID50s) while extensive infection was detected in all four control animals (Zhang et al., unpublished work) [60]. Increasing antibody size by joining m36 to cargo proteins led to a complete blockade of neutralization suggesting the existence of steric occlusion with the highly conserved functionally important structure that the antibody targets. m36 is the first reported human eAd against HIV-1 and represents a novel class of potent, broadly cross-reactive HIV-1 inhibitors based on human eAds.
8.2. Identification of eAds Binding to Non-Overlapping Epitopes
Soluble ligands have been linked to the pathogenesis of many diseases including cancer and immune disorders. Several monoclonal antibodies (mAbs) have been approved by the US FDA to treat ligand-related diseases. Although the approved antibodies block the interactions of ligands with corresponding receptors, they do not remove the ligands and actually increase their half-lives due to the long half-lives of the antibodies. Since antibody binding is a reversible process, the dissociation of antibodies will release ligands which continue to function. We hypothesized that antibodies targeting non-overlapping epitopes on the same ligand molecule could cross-link the ligand and form large immune complexes, which can strongly bind to Fc gamma receptor (FcγR)-expressing effector cells due to avidity effects of multivalent antibody Fc leading to internalization of the complexes. However, many soluble ligands are so small (e.g., IGF-II, approximately 7 kDa) that binding of conventional blocking antibodies typically leaves only a small ligand surface area, which may not be accessible to non-competing antibodies with large antigen-binding domains. Because of the small size and relatively small antigen-binding sites of eAds, it is more likely to select non-competing antibodies from eAd libraries.
We previously identified an IGF-II-specific antibody (m610) from one of our Fab libraries, which potently inhibits cancer cell growth in vitro [61] and in a mouse model with implanted human bone marrow [62]. m610.27, an affinity-matured version of m610, was selected by constructing, panning, and screening a light chain-shuffled library of m610. However, all our previous efforts to select antibodies non-competing with m610.27 in binding to IGF-II from our Fab and scFv libraries failed until we used our eAd library m8l, from which we successfully selected m630.3 [16]. We then generated a bispecific antibody m660 by fusing scFv m610.27 and eAd m630.3 to the N termini of human IgG1 heavy and light chain constant regions, respectively. As expected, large complexes of m660 were formed in the presence of IGF-II that bound to FcγRII-expressing BJAB cells much more efficiently than the monospecific antibody-IGF-II complexes. The m660-IGF-II complexes but not the m610.27-IGF-II and m630.3-IGF-II complexes were phagocytosed by phorbol 12-myristate 13-acetate (PMA)-stimulated macrophage-like U937 cells. A mixture of m610.27 and m630.3 exhibited similar properties. To our knowledge, these mAbs are the first reported to target nonoverlapping epitopes on a cancer-related ligand, and could represent a novel class of candidate therapeutics against cancer. Our approach could be also used to down-regulate other disease-related soluble ligands.
9. CHARACTERIZATION OF EAD STABILITY AND AGGREGATION PROPENSITY
Here, we review current characterization methods in examining protein stability, soluble aggregation, and unfolding of proteins as applicable to eAds. Proteins are dynamic molecules and undergo conformational changes under slight variations in physiochemical conditions. Protein monomers can unfold partially or completely if physicochemical conditions cause a shift in the equilibrium. During the process, two or more of such nonnative monomers might tend to aggregate. Monitoring the degree of antibody aggregation and more importantly, understanding how to avoid it is pivotal to ensure quality of antibody therapeutics.
Protein unfolding studies use intrinsic or extrinsic fluorescence spectroscopy method to characterize changes in overall fluorescence spectrum of antibodies induced by temperature ramp or denaturants, for example, guanidine hydrochloride and urea. Proteins with aromatic amino acids are intrinsically fluorescent when excited with UV light. The excitation and emission wavelengths of the three aromatic amino acids are: 280/348 nm for tryptophan, 274/303 nm for tyrosine, and 257/282 nm for phenylalanine. Compared with tyrosine and phenylalanine, tryptophan has greater absorptivity, higher quantum yield, and resonance energy transfer. The fluorescence spectrum of tryptophan usually reflects that of a protein containing the three amino acids. The amino acids may or may not be exposed on surface of a folded protein. During the unfolding process, residues embedded within a folded protein may become exposed and the change will be reflected on the change in the overall fluorescence of the protein. Also, the fluorescence spectrum of tryptophan is solvent dependent; when the polarity of solvent decreases, the spectrum shifts to shorter wavelengths and its intensity increases. For example, tryptophan in the hydrophobic environment of a folded protein displays maximum fluorescence at 330 nm, while tryptophan exposed to water has maximum fluorescence at 350 nm.
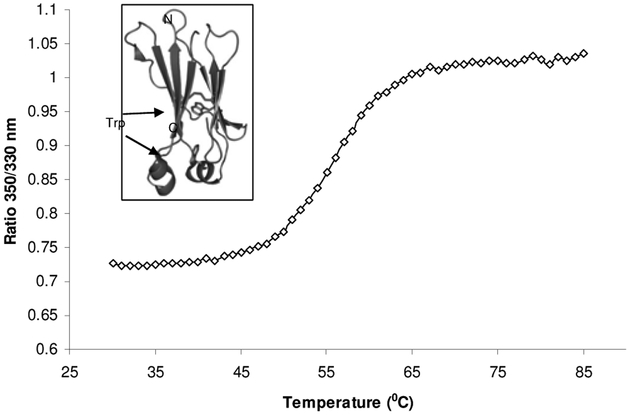
The shift of tryptophan fluorescence has been applied to study conformational states of proteins. Multiple instruments utilizing these unique features of tryptophan are available. We used Avacta Optim1000 instrument, which combines function of spectrofluorometer and static light scattering, to record the spectrum of temperature induced unfolding of a recombinant CH2 domain of human IgG1. Approximately 9 μl of purified CH2 protein at 0.2 mg/ml was used. The sample temperature was raised at the rate of one degree per min ute and held for 1 minute at each temperature, and was plotted against the ratio of 350 nm/330 nm as indicated in Fig. 6. There are two tryptophan residues in the CH2 domain and both are buried inside as found in the crystal structure (inset of Fig. 6). One tryptophan residue is found normally packed against the disulfide bond at the core of Ig domain connecting the two β-sheets. The other tryptophan residue is at one of the short helix and closer to the C-terminal end.
Fig. (6).
Temperature induced unfolding of the recombinant human IgG1 CH2 domain. The graph was obtained using Avacta Optim1000 instrument. The ratio of 350 nm/330 nm indicated that the two tryptophan residues in CH2 were exposed as the protein unfolded at 50–60 °C. The unfolding transition thermal mid points (Tm) was calculated to be 56.2 °C. The two tryptophan residues in the CH2 domain are buried inside the folded protein as seen in the crystal structure [26], inset of the figure.
Small proteins such as eAds may have few tryptophan residues and each in its local environment gives out different emission spectrum. In such case, intrinsic fluorescence may not reflect change of protein structures. Under these circumstances, examining fluorescence spectrum with the assistance of extrinsic probes will be helpful. For this, 1-anilinonaphtalene-8-sulfonate (ANS) (excitation wavelength, 370 nm; emission wavelength, 480 nm) or Thioflavin T (ThT) is used as a fluorescent probe. ANS has negligible weak green fluorescence in aqueous environment, but exhibits intense blue fluorescence when binding to proteins and membranes. ANS generally binds to hydrophobic sites in proteins. As more ANS probes gain access to exposed hydrophobic pockets, a significant increase in fluorescence intensity is observed. In the meantime, a blue shift of emission wavelength maximum accompanies the binding. The fluorescence intensity of ANS also changes when the pH of protein solution changes. At pH3.5, it exhibits high intensity and at pH7.5, the intensity decreases by 50 folds and the emission wavelength also shifts by 10–20 nm. Alternatively, ThT is a benzothiazole yellow dye that exhibits enhanced fluorescence upon binding to β sheet-rich structures, such as in amyloid aggregates. The fluorescence intensity of ThT is quantitatively proportional to the concentrations of soluble aggregates. Due to this property and its lack of appreciable binding to monomer proteins, ThT is frequently used to study soluble aggregation of proteins. Thus, the combination of fluorescence probes, such as ANS and ThT, with fluorescence spectroscopy has been proven to be a powerful tool to study protein folding, unfolding, and aggregation. The analyses require only a small amount of proteins, and various buffer conditions can be tested for formulation studies.
Circular Dichroism (CD) method utilizes the chiral nature of amino acids. When circularly polarized light passes through a protein sample, the speeds between right and left polarizations differ (cL ≠ cR) as well as their wavelengths (λL ≠ λR) and the extent to which they are absorbed (εL≠εR). CD readout is the difference (Δε ≡ εL- εR). Furthermore, secondary structures of proteins will also impart a distinct CD to its respective molecules. Antibody domains consist of multiple β sheet structures. With α helix and β sheet of proteins having their CD spectral signature, CD has its distinct advantage for stability studies of these antibody domains. Although CD gives less specific information on structure than crystallography and NMR spectroscopy, it is a quick method that only requires a small amount of proteins. When combined with sample conditions such as solvent conditions, temperature, pH, salinity, and the presence of various cofactors, it may provide much structural information on antibody domains.
Dynamic light scattering (DLS) is also called photon correlation spectroscopy, PCS. It measures Brownian motion and relates this to the size of particles. If particles or molecules are illuminated with a laser, the intensity of scattered light fluctuates at a rate that is dependent upon the size of particles, as smaller particles are “kicked” further by solvent molecules and move more rapidly. Analysis of these intensity fluctuations yields the velocity of Brownian motion and hence particle size using the Stokes-Einstein relationship. The measurement is non-invasive, and the same sample could be measured for multiple times after being subjected to high temperature or other stress. In a monodisperse sample, the average particle diameter could be measured. Protein samples are often polydisperse systems with multiple sizes of particles. Particularly in aggregation test, many populations of particles with large size may exist, and DLS method is one of the choices to trace the change during protein aggregation. Many newly designed DLS instruments incorporate in situ temperature control, and allow monitoring of particle size in relation to temperature changes.
In contrast to DLS, static light scattering (SLS) measures time-averaged intensity of scattered light. The intensity of light scattered over a period, e.g., 30 seconds, is accumulated for a number of concentrations of samples. This time averaging eliminates the inherent fluctuations in the signal. Usually a series of known concentrations of protein samples are measured for scattering intensity. The Rayleigh equation is applied to estimate the molecular weights of proteins. Similar to DLS method, SLS measurement is non-invasive and thus, is appropriate for stress or aggregation tests.
CONCLUSIONS
We discussed discovery of eAds and their potential applications as candidate therapeutics and diagnostics, which involves identification of eAd scaffolds and several cutting-edge technologies for structure-based design, generating eAd libraries, display platforms, high-throughput sequencing, computational methods to improve biophysical properties, and functional evaluation and testing. We also briefly described sequence diversity of VH- and CH2-based eAd libraries using 454 sequencing analysis, which corroborates quality and as representative at large of these libraries. We further demonstrated the usefulness of eAd libraries in the selection of binders against HIV-1 and cancer-related proteins. Finally, eAds possess characteristic structural and functional features, mainly small size, stability, and ability to target epitopes non-accessible to conventional antibodies that would make them useful for future research and biotechnological applications.
ACKNOWLEDGEMENTS
We thank Drs. B. Haynes, H. Liao, C. Broder and T. Fouts for reagents. We thank Laboratory of Molecular Technology and Advanced Biomedical Computing Center of SAIC-Frederick Inc. for providing Roche 454 sequencing service and support. We are grateful to Marie-Paule Lefranc and Eltaf Alamyar at the IMGT® for providing access to IMGT/HighV-QUEST. We thank Ms. Maria G. Singarayan for constructing PostgreSQL database and developing JAVA standalone applications. This project was supported by the Intramural AIDS Targeted Antiviral Program (IATAP) of the National Institutes of Health (NIH) and the U.S.-China Program for Biomedical Research Cooperation.
Footnotes
CONFLICT OF INTEREST
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, the Gates Foundation, and the Federal funds from the NIH, National Cancer Institute, under Contract No. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U. S. Government. The authors have no other competing interests to declare.
REFERENCES
- [1].Stijlemans B, Conrath K, Cortez-Retamozo V, et al. Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J Biol Chem 2004; 279: 1256–61. [DOI] [PubMed] [Google Scholar]
- [2].Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 2005; 23: 1126–36. [DOI] [PubMed] [Google Scholar]
- [3].de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact 2011; 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wesolowski J, Alzogaray V, Reyelt J, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol 2009; 198: 157–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Holt LJ, Herring C, Jespers LS, Woolven BP, Tomlinson IM. Domain antibodies: proteins for therapy. Trends Biotechnol 2003; 21: 484–90. [DOI] [PubMed] [Google Scholar]
- [6].Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004; 305: 1770–3. [DOI] [PubMed] [Google Scholar]
- [7].Bloom L, Calabro V. FN3: a new protein scaffold reaches the clinic. Drug Discov Today 2009; 14: 949–55. [DOI] [PubMed] [Google Scholar]
- [8].Owens RM, Gu X, Shin M, Springer TA, Jin MM. Engineering of single Ig superfamily domain of intercellular adhesion molecule 1 (ICAM-1) for native fold and function. J Biol Chem 2010; 285: 15906–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bruning M, Barsukov I, Franke B, et al. The intracellular Ig fold: a robust protein scaffold for the engineering of molecular recognition. Protein Eng Des Sel 2012; 25: 205–12. [DOI] [PubMed] [Google Scholar]
- [10].Ewert S, Huber T, Honegger A, Pluckthun A. Biophysical properties of human antibody variable domains. J Mol Biol 2003; 325: 531–53. [DOI] [PubMed] [Google Scholar]
- [11].Ewert S, Cambillau C, Conrath K, Pluckthun A. Biophysical properties of camelid V(HH) domains compared to those of human V(H)3 domains. Biochemistry 2002; 41: 3628–36. [DOI] [PubMed] [Google Scholar]
- [12].Dimitrov DS. Engineered CH2 domains (nanoantibodies). MAbs 2009; 1: 26–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gong R, Wang Y, Ying T, Dimitrov DS. Bispecific Engineered Antibody Domains (Nanoantibodies) That Interact Noncompetitively with an HIV-1 Neutralizing Epitope and FcRn. PLoS ONE 2012; 7: e42288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen W, Zhu Z, Feng Y, Dimitrov DS. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc Natl Acad Sci USA 2008; 105: 17121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol 2008; 382: 779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen W, Feng Y, Zhao Q, Zhu Z, Dimitrov DS. Human Monoclonal Antibodies Targeting Nonoverlapping Epitopes on Insulin-like Growth Factor II as a Novel Type of Candidate Cancer Therapeutics. Mol Cancer Ther 2012; 11: 1400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hamers-Casterman C, Atarhouch T, Muyldermans S, et al. Naturally occurring antibodies devoid of light chains. Nature 1993; 363: 446–8. [DOI] [PubMed] [Google Scholar]
- [18].Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995; 374: 168–73. [DOI] [PubMed] [Google Scholar]
- [19].Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 2007; 35: D301–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Engineering 1994; 7: 1129–35. [DOI] [PubMed] [Google Scholar]
- [21].Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol 2011; 9: e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dottorini T, Vaughan CK, Walsh MA, LoSurdo P, Sollazzo M. Crystal structure of a human VH: requirements for maintaining a monomeric fragment. Biochemistry 2004; 43: 622–8. [DOI] [PubMed] [Google Scholar]
- [23].Conrath K, Vincke C, Stijlemans B, et al. Antigen binding and solubility effects upon the veneering of a camel VHH in framework-2 to mimic a VH. J Mol Biol 2005; 350: 112–25. [DOI] [PubMed] [Google Scholar]
- [24].Jespers L, Schon O, James LC, Veprintsev D, Winter G. Crystal structure of HEL4, a soluble, refoldable human V(H) single domain with a germ-line scaffold. J Mol Biol 2004; 337: 893–903. [DOI] [PubMed] [Google Scholar]
- [25].Schiefner A, Chatwell L, Korner J, et al. A disulfide-free single-domain V(L) intrabody with blocking activity towards huntingtin reveals a novel mode of epitope recognition. J Mol Biol 2011; 414: 337–55. [DOI] [PubMed] [Google Scholar]
- [26].Prabakaran P, Vu BK, Gan J, Feng Y, Dimitrov DS, Ji X. Structure of an isolated unglycosylated antibody C(H)2 domain. Acta Crystallogr D Biol Crystallogr 2008; 64: 1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gong R, Vu BK, Feng Y, et al. Engineered human antibody constant domains with increased stability. J Biol Chem 2009; 284: 14203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gong R, Wang Y, Feng Y, Zhao Q, Dimitrov DS. Shortened engineered human antibody CH2 domains: increased stability and binding to the human neonatal Fc receptor. J Biol Chem 2011; 286: 27288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gehlsen K, Gong R, Bramhill D, et al. Pharmacokinetics of engineered human monomeric and dimeric CH2 domains. MAbs 2012; 4: 466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Smith GP. Filamentous Fusion Phage - Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science 1985; 228: 1315–7. [DOI] [PubMed] [Google Scholar]
- [31].Beerli RR, Bauer M, Buser RB, et al. Isolation of human monoclonal antibodies by mammalian cell display. Proc Natl Acad Sci USA 2008; 105: 14336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 1997; 15: 553–7. [DOI] [PubMed] [Google Scholar]
- [33].Francisco JA, Earhart CF, Georgiou G. Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci USA 1992; 89: 2713–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA 1997; 94: 4937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dooley H, Flajnik MF, Porter AJ. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol Immunol 2003; 40: 25–33. [DOI] [PubMed] [Google Scholar]
- [36].Suzuki T, Shin T, Fujiyama A, Kohara Y, Kasahara M. Hagfish leukocytes express a paired receptor family with a variable domain resembling those of antigen receptors. J Immunol 2005; 174: 2885–91. [DOI] [PubMed] [Google Scholar]
- [37].Xu G, Tasumi S, Pancer Z. Yeast surface display of lamprey variable lymphocyte receptors. Methods Mol Biol 2011; 748: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Davies J, Riechmann L. Antibody Vh Domains as Small Recognition Units. Bio-Technology 1995; 13: 475–9. [DOI] [PubMed] [Google Scholar]
- [39].Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Engineering Design & Selection 2010; 23: 155–9. [DOI] [PubMed] [Google Scholar]
- [40].Tasumi S, Velikovsky CA, Xu G, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA 2009; 106: 12891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhao Q, Zhu Z, Dimitrov DS. Yeast display of engineered antibody domains. Methods Mol Biol 2012; 899: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Beerli RR, Rader C. Mining human antibody repertoires. MAbs 2010; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jirholt P, Ohlin M, Borrebaeck CA, Soderlind E. Exploiting sequence space: shuffling in vivo formed complementarity determining regions into a master framework. Gene 1998; 215: 471–6. [DOI] [PubMed] [Google Scholar]
- [44].Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006; 22: 195–201. [DOI] [PubMed] [Google Scholar]
- [45].Chen W, Zhu Z, Feng Y, Dimitrov DS. A large human domain antibody library combining heavy and light chain CDR3 diversity. Mol Immunol 2010; 47: 912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xiao X, Feng Y, Vu BK, Ishima R, Dimitrov DS. A large library based on a novel (CH2) scaffold: identification of HIV-1 inhibitors. Biochem Biophys Res Commun 2009; 387: 387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ying T, Chen W, Gong R, Feng Y, Dimitrov DS. Soluble monomeric IgG1 Fc. J Biol Chem 2012; 287: 19399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wozniak-Knopp G, Bartl S, Bauer A, et al. Introducing antigen-binding sites in structural loops of immunoglobulin constant domains: Fc fragments with engineered HER2/neu-binding sites and antibody properties. Protein Eng Des Sel 2010; 23: 289–97. [DOI] [PubMed] [Google Scholar]
- [49].Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs 2010; 2: 347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Persson MA. Twenty years of combinatorial antibody libraries, but how well do they mimic the immunoglobulin repertoire? Proc Natl Acad Sci USA 2009; 106: 20137–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Prabakaran P, Chen W, Singarayan MG, et al. Expressed antibody repertoires in human cord blood cells: 454 sequencing and IMGT/HighV-QUEST analysis of germline gene usage, junctional diversity, and somatic mutations. Immunogenetics 2012; 64: 337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Prabakaran P, Zhu Z, Chen W, et al. Origin, diversity, and maturation of human antiviral antibodies analyzed by high-throughput sequencing. Front Microbiol 2012; 3: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Prabakaran P, Streaker E, Chen W, Dimitrov DS. 454 antibody sequencing - error characterization and correction. BMC Res Notes 2011; 4: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Alamyar E, Giudicelli V, Duroux P, Lefranc MP. IMGT/HighVQUEST: A High-Throughput System and Web Portal for the Analysis of Rearranged Nucleotide Sequences of Antigen Receptors In: Journées Ouvertes en Biologie, Informatique et Mathématiques. Montpellier, France; 2010. pp. P63. [Google Scholar]
- [55].Dudgeon K, Famm K, Christ D. Sequence determinants of protein aggregation in human VH domains. Protein Eng Des Sel 2009; 22: 217–20. [DOI] [PubMed] [Google Scholar]
- [56].Reddy ST, Ge X, Miklos AE, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol 2010; 28: 965–9. [DOI] [PubMed] [Google Scholar]
- [57].Xiao X, Chen W, Feng Y, Dimitrov DS. Maturation Pathways of Cross-Reactive HIV-1 Neutralizing Antibodies. Viruses 2009; 1: 802–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998; 393: 648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Labrijn AF, Poignard P, Raja A, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 2003; 77: 10557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chen W, Xiao X, Wang Y, Zhu Z, Dimitrov DS. Bifunctional fusion proteins of the human engineered antibody domain m36 with human soluble CD4 are potent inhibitors of diverse HIV-1 isolates. Antiviral Res 2010; 88: 107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feng Y, Zhu Z, Xiao X, Choudhry V, Barrett JC, Dimitrov DS. Novel human monoclonal antibodies to insulin-like growth factor (IGF)-II that potently inhibit the IGF receptor type I signal transduction function. Mol Cancer Ther 2006; 5: 114–20. [DOI] [PubMed] [Google Scholar]
- [62].Kimura T, Kuwata T, Ashimine S, et al. Targeting of bone-derived insulin-like growth factor-II by a human neutralizing antibody suppresses the growth of prostate cancer cells in a human bone environment. Clin Cancer Res 2010; 16: 121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]