Here, we report the draft genome sequence of Listeria innocua strain MEZLIS26, isolated from a healthy goat in Flagstaff, Eastern Cape Province, South Africa. The genome was sequenced using the Illumina MiSeq platform and had a length of 2,800,777 bp, with a G+C content of 37.4%, 2,755 coding DNA sequences (CDSs), 49 transfer RNAs (tRNAs), and 4 noncoding RNAs (ncRNAs).
ABSTRACT
Here, we report the draft genome sequence of Listeria innocua strain MEZLIS26, isolated from a healthy goat in Flagstaff, Eastern Cape Province, South Africa. The genome was sequenced using the Illumina MiSeq platform and had a length of 2,800,777 bp, with a G+C content of 37.4%, 2,755 coding DNA sequences (CDSs), 49 transfer RNAs (tRNAs), and 4 noncoding RNAs (ncRNAs).
ANNOUNCEMENT
Listeria spp. are small, motile, catalase-positive, non-spore-forming, rod-shaped, Gram-positive bacteria. The genus Listeria is currently known to consist of 20 species (1), of which L. monocytogenes is an important foodborne human pathogen causing serious epidemics and sporadic listeriosis (2, 3). Listeria spp. have been isolated from a wide variety of sources, and L. innocua is reported to be more commonly isolated than L. monocytogenes (4). L. innocua is a nonpathogenic surrogate species that is closely related to L. monocytogenes. Recently, atypical hemolytic L. innocua was reported to be virulent and can actively cross the intestinal epithelium and spread systemically to the liver and spleen, albeit to a lesser degree than L. monocytogenes (5). In addition to its clinical relevance (5–8) and similarity to L. monocytogenes, the genomes of L. innocua provide important information that helps understand the pathogenicity of L. monocytogenes. Limited data about the genome sequence of L. innocua are available. Here, we report the draft genome sequence of L. innocua strain MEZLIS26, isolated from a goat in Flagstaff, Eastern Cape, South Africa, in May 2018. The sample was collected in 10 ml of 0.1% buffered peptone water and incubated for 24 hours. Following enrichment in Listeria broth (Oxoid, England), the sample was streaked onto Listeria selective agar (Oxoid, England) and incubated at 37°C for 18 hours. A slant of the bacterial culture was shipped to North Carolina State University (NCSU) for further analysis as part of the GenomeTrakr project (9).
Colony PCR for the hemolysin (hly) gene was performed as previously described (10). An aliquot of overnight culture in brain heart infusion (BHI) broth was submitted to the Clinical Sciences Department at NCSU for matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis for further confirmation. DNA isolation was performed using a MasterPure DNA isolation kit (Lucigen, WI) according to the manufacturer’s protocol. Sequencing libraries were prepared using a Nextera XT library preparation kit (Illumina, CA). Sequencing was performed on the Illumina MiSeq platform using the v2 reagent kit, which yielded 250-bp paired-end (PE) reads.
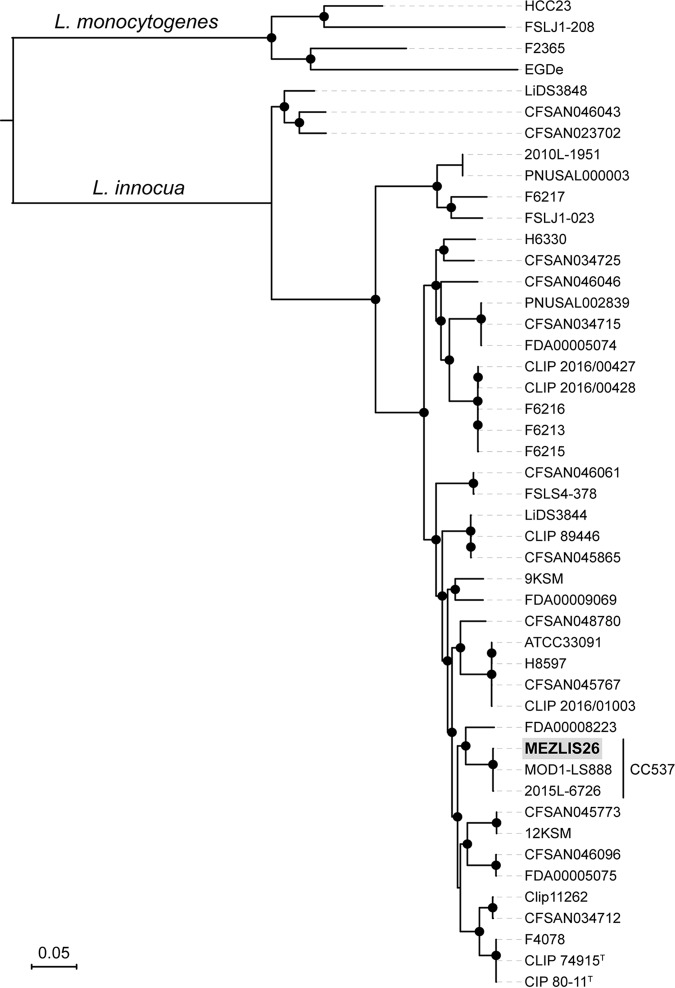
A total of 1,294 Mb (or ∼1.3 Gb) raw data reads were generated, and a total of 1.191 Mb (or ∼1.2 Gb) cleaned reads were obtained using Trim Galore, a Perl wrapper for Cutadapt (11), and FastQC (12) using the functions –paired, –phred33, –clip_R1 11, –clip_R2 11, –three_prime_clip_R1 3, and –three_prime_clip_R2 3. The N50 value of the cleaned sequence reads was 234 bp. Sequences were assembled using Unicycler version 0.4.7 (13) into 12 contigs of at least 200 nucleotides (nt) long, using default parameters with the addition of the –min_fasta_length 200 parameter. Assembly quality was assessed using QUAST (13), yielding a total of 2,800,777 bp, with a G+C content of 37.4%, an N50 value of 1,410,057 bp, and an L50 value of 1. Prokka version 1.13 (14) was used for annotation, indicating that the genome contained 2,755 coding DNA sequences (CDSs) and 49 tRNA, 1 transfer-messenger RNA (tmRNA), and 3 rRNA genes. The average nucleotide identity BLAST against L. innocua Clip11262 (GenBank accession number NC_003212) was of 98.73%, confirming species identity (15). To better understand the phylogenetic placement of isolate MEZLIS26, a maximum likelihood phylogeny was inferred from the core genome alignment of 42 L. innocua and 4 L. monocytogenes public genomes (5) using Parsnp, implemented in Harvest suite v.1.1.2 (16) and visualized with iTol v.4.2 (17). Isolate MEZLIS26 clustered within clonal complex CC537 (nonhemolytic L. innocua) together with isolates MOD1-LS888 and 2015L-6726 (SRA accession numbers SRR1481929 and SRR2915359, respectively), isolated from food in the United States (Fig. 1).
FIG 1.
Phylogenetic positioning of isolate MEZLIS26 (highlighted in gray) within L. innocua. Representative genomes of L. monocytogenes were used as the outgroup. The maximum likelihood phylogeny was inferred from 642,408 core genome SNPs. Black circles represent bootstrap branch support values higher than 90% based on 1,000 replicates.
Data availability.
This whole-genome sequencing project has been deposited at DDBJ/ENA/GenBank under the BioProject number PRJNA514279 (BioSample accession number SAMN11604718 and GenBank accession number AADHQU000000000). The version described in this paper is the first version, AADHQU010000000. The sequences have been submitted to the Sequence Read Archive (SRA) under the accession numbers SRX5806851 and SRR9029426. All isolates used in this study are also publicly available in https://bigsdb.pasteur.fr/listeria/.
ACKNOWLEDGMENTS
The whole-genome sequencing work is supported by the National Institutes of Health/Food and Drug Administration under award number 1U18FD006780-01.
We thank the South African National Research Foundation for supporting this research through the Thuthuka Funding Instrument (grant number TTK170411226583) and the Swedish Research Council (VR) through grant number 2016-02606. We thank Lyndy Harden and Siddhartha Thakur from Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University (Raleigh, NC). We thank the Genome Trakr Network and the Whole Genome Sequencing Program for foodborne pathogen traceback and the Center for Food Safety and Applied Nutrition (CFSAN), U.S. Food and Drug Administration (FDA), for support in the whole-genome sequencing (WGS) of the strain MEZLIS26 as part of the US Food and Drug Administration's WGS surveillance effort.
M.E.E.Z. conceived, coordinated, and supervised the research project, isolated the strain, prepared and wrote the manuscript, and submitted the strain for WGS. R.A.H. and A.M. conducted the phylogenetic analysis. R.A.H. and A.M. contributed to manuscript writing. N.N., O.T.Z., M.L., and J.D.J. reviewed the manuscript. M.E.E.Z. critically revised the manuscript. All authors approved the final version of the manuscript.
REFERENCES
- 1.Leclercq A, Moura A, Vales G, Tessaud-Rita N, Aguilhon C, Lecuit M. 2019. Listeria thailandensis sp. nov. Int J Syst Evol Microbiol 69:74–81. doi: 10.1099/ijsem.0.003097. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman S, Maculloch B, Batz M. 2015. Economic burden of major foodborne illnesses acquired in the United States. United States Department of Agriculture, Economic Research Service, Washington, DC. [Google Scholar]
- 3.Buchrieser C, Rusniok C, The Listeria Consortium, Kunst F, Cossart P, Glaser P. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol 35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 4.Petran RL, Swanson KM. 1993. Simultaneous growth of Listeria monocytogenes and Listeria innocua. J Food Prot 56:616–618. doi: 10.4315/0362-028X-56.7.616. [DOI] [PubMed] [Google Scholar]
- 5.Moura A, Disson O, Lavina M, Thouvenot P, Huang L, Leclercq A, Fredriksson-Ahomaa M, Eshwar AK, Stephan R, Lecuit M. 2019. Atypical hemolytic Listeria innocua isolates are virulent, albeit less than Listeria monocytogenes. Infect Immun 87:e00758-18. doi: 10.1128/IAI.00758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favaro M, Sarmati L, Sancesario G, Fontana C. 2014. First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep 1. doi: 10.1099/jmmcr.0.003103. [DOI] [Google Scholar]
- 7.Perrin M, Bemer M, Delamare C. 2003. Fatal case of Listeria innocua bacteremia. J Clin Microbiol 41:5308–5309. doi: 10.1128/jcm.41.11.5308-5309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocha P, Dalmasso A, Grattarola C, Casalone C, Del Piero F, Bottero MT, Capucchio MT. 2013. Atypical cerebral listeriosis associated with Listeria innocua in a beef bull. Res Vet Sci 94:111–114. doi: 10.1016/j.rvsc.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Stevens EL, Timme R, Brown EW, Allard MW, Strain E, Bunning K, Musser S. 2017. The public health impact of a publically available, environmental database of microbial genomes. Front Microbiol 8:808. doi: 10.3389/fmicb.2017.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A, Grover S, Batish VK. 2015. Exploring specific primers targeted against different genes for a multiplex PCR for detection of Listeria monocytogenes. 3 Biotech 5:261–269. doi: 10.1007/s13205-014-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 12.Wingett SW, Andrews S. 2018. FastQ screen: a tool for multi-genome mapping and quality control. Version 2. F1000Res 7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 15.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 16.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome sequencing project has been deposited at DDBJ/ENA/GenBank under the BioProject number PRJNA514279 (BioSample accession number SAMN11604718 and GenBank accession number AADHQU000000000). The version described in this paper is the first version, AADHQU010000000. The sequences have been submitted to the Sequence Read Archive (SRA) under the accession numbers SRX5806851 and SRR9029426. All isolates used in this study are also publicly available in https://bigsdb.pasteur.fr/listeria/.