Summary:
De novo mutations of the neuronal sodium channel SCN8A have been identified in approximately 2% of individuals with epileptic encephalopathy. These missense mutations alter the biophysical properties of sodium channel Nav1.6 in ways that lead to neuronal hyperexcitability. We generated two mouse models carrying patient mutations N1768D and R1872W in order to characterize the effects on neuronal function in vivo. The conditional R1872W mutation is activated by expression of CRE recombinase, permitting characterization of the effects of the mutation on different classes of neurons and at different points in postnatal development. Preclinical drug testing in these mouse models provides support for several new therapies for this devastating disorder. Mutations of SCN8A that result in partial or complete loss-of-function are associated with intellectual disability and other disorders.
Keywords: SCN8A, encephalopathy, mutation, sodium channel, mouse model
Introduction:
The sodium channel gene SCN8A was first identified by molecular analysis of spontaneous neurological mutants in the mouse. Positional cloning and genome sequencing of mouse Scn8a from mutant lines identified two protein truncation mutations, a splice site mutation and a missense mutation, that were responsible for movement disorders ranging from tremor, ataxia, and dystonia to hind limb paralysis and lethality1-3. These spontaneous mouse mutations resulted in partial or complete loss of function of the sodium channel Nav1.6 and exhibited recessive inheritance and the absence of spontaneous tonic/clonic seizures. two sentences deleted here
The first human epileptogenic SCN8A mutation was described in 20124. Michael Hammer at the University of Arizona carried out complete genome sequencing of two parents, an affected child, and an unaffected sibling. A de novo A>G mutation was identified in the affected daughter, resulting in the SCN8A missense mutation p.Asn1768Asp (N1768D)4. To assess the pathogenicity of this mutation, we introduced it into a Nav1.6 cDNA and carried out functional evaluation in transfected ND7/23 cells. We observed a large increase in persistent current resulting in elevated firing of transfected neurons4. The causal role of the SCN8A mutation was confirmed in a knock-in mouse model that recapitulated epilepsy and sudden death5.
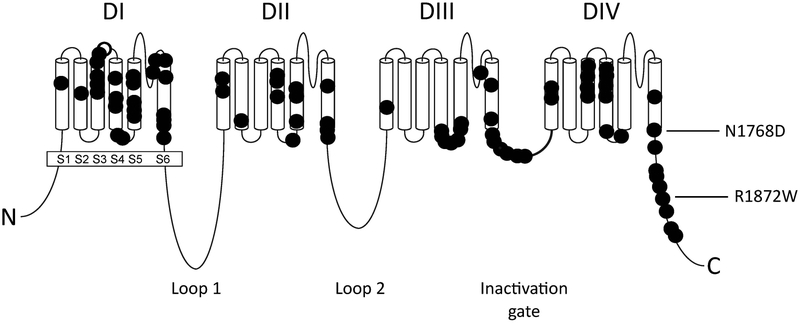
The important role of SCN8A in epileptic encephalopathy was quickly confirmed in additional patients. Today nearly 300 individuals with SCN8A encephalopathy have been reported in publications or described in databases maintained by family groups (e.g. www.SCN8A.net). Like the first case, the subsequently identified mutations arose de novo in the affected children, and result in missense substitutions of evolutionarily conserved amino acid residues that alter channel function6. The patient mutations are concentrated in transmembrane segments of the Nav1.6 channel protein, the inactivation gate and the cytoplasmic C-terminal domain (Figure 1).
Figure 1.
SCN8A mutations associated with epileptic encephalopathy. The protein structure of the four-domain sodium channel Nav1.6 is represented; each symbol represents a patient mutation. The positions of the two mutations studied in mouse models are indicated with arrows.
Functional studies of approximately 20 patient mutations have been carried out by expression in transfected cells, and 2 mutations have been studied in neurons of knock-in mice. None of the SCN8A encephalopathy mutations result in protein truncation or loss of channel function. Rather, these are missense mutations that alter the biophysical properties of the channel, such as altered voltage dependence of channel activation or inactivation, delayed channel inactivation, or increased resurgent or persistent current. Most mutations change more than one parameter, making it difficult to assign relative ‘severity’. The most common changes are a hyperpolarized shift in voltage dependence of activation, impaired channel inactivation, and elevated persistent current6. Each of these results in elevated neuronal firing, consistent with in vivo seizures.
Approximately 1/3 of the patient mutations in SCN8A encephalopathy occur at recurrent positions, many of which are located in CpG islands that are sites of elevated mutation rate in the human genome7. Arginine residues such as 1617 and 1872 are the sites of recurrent mutations which can have dramatic effects on channel function. For example, loss of the positive charge at Arg1872 due to substitution of either of three neutral amino acids results in destabilization of the inactive state of the channel, resulting in elevated neuronal excitability8.
We have generated two in vivo mouse models expressing patient mutations, the original mutation Asn1768Asp (N1768D)5 and the recurrent mutation Arg1872Trp (R1872W)9. Both mice recapitulate the basic features of encephalopathy with spontaneous seizures and premature lethality. These mice enabled us to probe the effect of the mutations on firing patterns of different classes of mutations, secondary effects on expression of other genes, effects of Scn8a on cardiac function and responses to new drug therapies. The effects of strain background on seizure severity have also been examined. The features of the two mouse models are compared in Table 1.
Table 1.
Features of two mouse models of SCN8A encephalopathy. The N1768D mouse was generated by TALEN knock-in of the first described patient mutation7. The R1872W mouse carries a conditional allele of a recurrent human mutation that leads to lethality in early childhood11. Onset is the age at first observed seizure. Duration is the interval between first observed seizure and death.
| N1768D/+ | R1872W/+ | |
|---|---|---|
| patients | ambulatory, verbal, SUDEP at 15 yrs | nonambulatory, nonverbal |
| ephysiology | impaired inactivation | more severely impaired |
| mouse model: | Knock-in to Scn8a | conditional knock-in |
| onset | 2-4 months | 2 weeks |
| duration, onset to death | 1-2 week | 10 sec to 2 week depends on CRE |
| penetrance | 50-75% | 100% |
The N1768D mouse model of SCN8A encephalopathy.
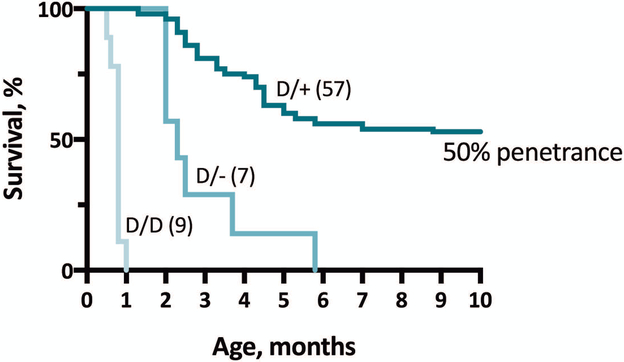
We used targeting with a TALEN endonuclease to knock-in the patient mutation SCN8A-N1768D into exon 26 of the mouse Scn8a gene10. Spontaneous seizures were observed in heterozygous adults beginning at 2 months of age. Mice are highly sensitive to gain-of-function mutations of Scn8a, and the time from first observed seizure to death varies from only one to four weeks depending on strain background (Figure 2).
Figure 2.
In vivo mouse model of the epileptic encephalopathy mutation SCN8A-N1768D. Knock-in of the patient mutation6 results in seizures and death in 50% of heterozygous D/+ animals. Combination with a null allele (D/−) or homozygosity of the mutant allele (D/D) exacerbates the phenotype, demonstrating partial protection by the wildtype allele in heterozygous animals9.
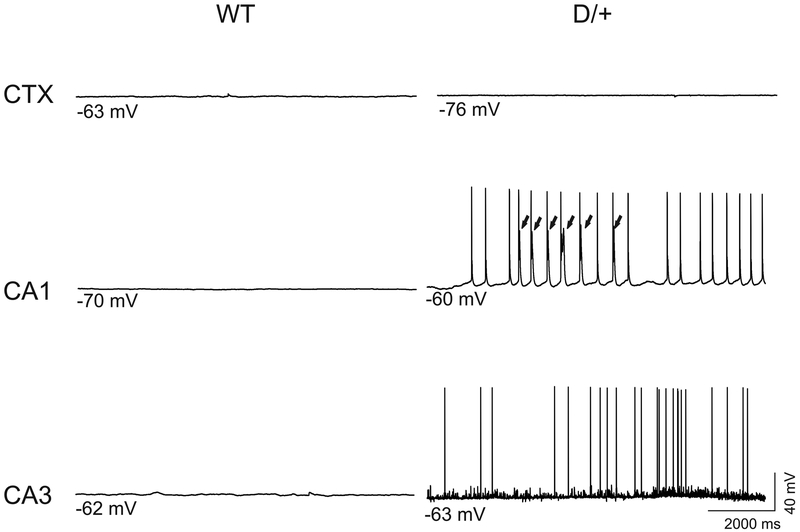
Recordings from hippocampal neurons revealed spontaneous firing in subsets of CA1 and CA3 neurons11 (Figure 3). Elevated activity was also observed in neurons from the medial entorhinal cortex that innervate the hippocampus12. Although Nav1.6 is expressed at a much lower level in cardiac myocytes, this expression is sufficent to generate cardiac arrythmia in the N1768D mice13,14.
Figure 3.
Spontaneous firing of hippocampal neurons in brain slices from N1768D mice13.
Secondary effects on gene expression were investigated by sequence analysis of transcripts from three brain regions, forebrain, cerebellum and hind brain15. Prior to seizure onset, we did not detect differences in transcript abundance between wildtype and mutant mice. Within 24 hours of the first observed seizure, there was elevation of transcripts associated with gliosis such as GFAP as well as neuropeptides previously associated with seizure susceptibility such as galanin and NPY15. We also observed a significant elevation of the neuropeptide NPW. These changes did not occur in mutant mice that did not develop seizures as a consequence of the incomplete penetrance of the N1768D mutation. We did not observe any compensatory up- or down-regulation of transcripts encoding other voltage-gated sodium channels, including Scn1a, Scn2a and Scn3a. The data suggest that there is not extensive remodeling of the brain via altered transcription prior to seizure onset.
The R1872W mouse model of SCN8A encephalopathy.
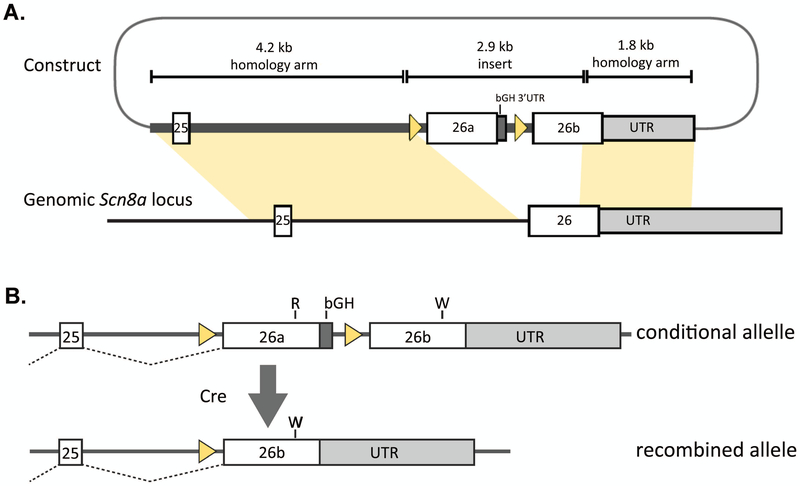
We used the same TALEN reagent to target exon 26 of mouse Scn8a to generate the R1872W mutant mouse line, but in this case the mutation is dependent on expression of CRE recombinase9. The targeting vector containing two copies of exon 26 is shown in Figure 4A. In the absence of CRE, the mice express a full diploid dose of Nav1.6 that contains wildtype exon 26a. Expression of CRE recombinase results in deletion of exon 26a and initiates expression of exon 26b containing the R1872W mutation (Figure 4B).
Figure 4.
The conditional mouse mutation SCN8A-R1872W. A. The targeting vector containing two copies of the final coding exon 26 of Scn8a was inserted into the endogenous gene. B. In the absence of CRE recombinase the wildtype exon 26a is expressed. Deletion of exon 26a by CRE initiates expression of the mutant exon 26b11.
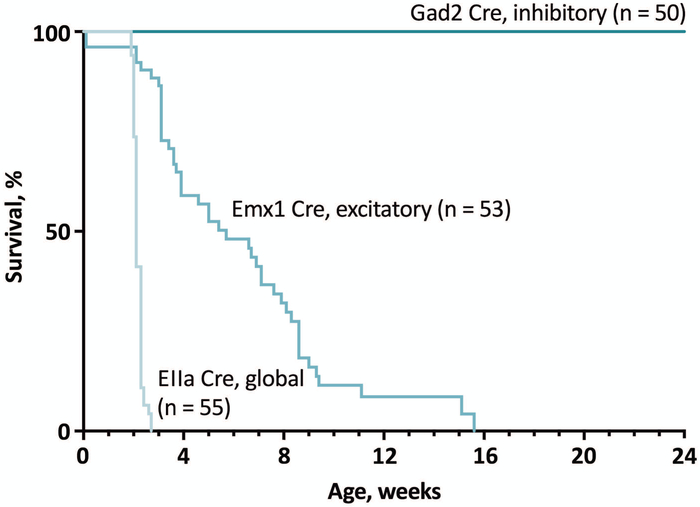
We observed dramatic results of global activation of the mutant channel early in development by EIIA-CRE, with sudden onset of seizures at 14 days of age and death within 24 hours of the first observed seizures (Figure 5). Activation of R1872W specifically in excitatory neurons of the forebrain was sufficient to initiate seizures and death (Figure 5). Activation of R1872W exclusively in inhibitory neurons using GAD2 CRE or DLX5a CRE was not pathogenic (Figure 5). Expression of the mutant channel in the adult by tamoxifen treatment of mice expressing the CAG-CRE-ER also resulted in 100% penetrance of spontaneous seizures and sudden death, demonstrating the continued sensitivity of adult neurons and an apparent cell-autonomous effect of the mutant channel in the mature CNS9.
Figure 5.
Predominant role of excitatory neurons in SCN8A encephalopathy in the conditional R1872W mouse. Global pre-implantation activation of the mutant allele by EIIA-CRE results in seizure onset at 2 weeks of age followed rapidly by lethality. Activation of the mutant allele in excitatory forebrain neurons by Emx1 CRE also results in 100% penetrance of seizures and death, approximately 4 weeks later than the EIIA-CRE. In contrast, activation by the GAD2 CRE in inhibitory neurons had no detectable pathogenicity11.
These results are consistent with the major role of Nav1.6 in the initiation of action potentials at the AIS of excitatory neurons, and the elevated excitability of neurons expressing the mutant channel. This is in striking contrast with the Dravet Syndrome mouse, in which inhibitory neurons appear to be the primary target responsible for seizure initiation due to reduced activity of Scn1a (Ref.16).
Survival is approximately 4 weeks longer in mice expressing the Emx1-CRE compared with the ubiquitously expressed EIIA-CRE. This may be explained by the expression of the EIIA-CRE in many cells that do not express Emx1-CRE, including neurons in the brain stem that regulate respiration and cardiac function, as well as cardiac myocytes that express Nav1.6 at a low level13,14.
Pre-clinical drug testing in the SCN8A mutant mice.
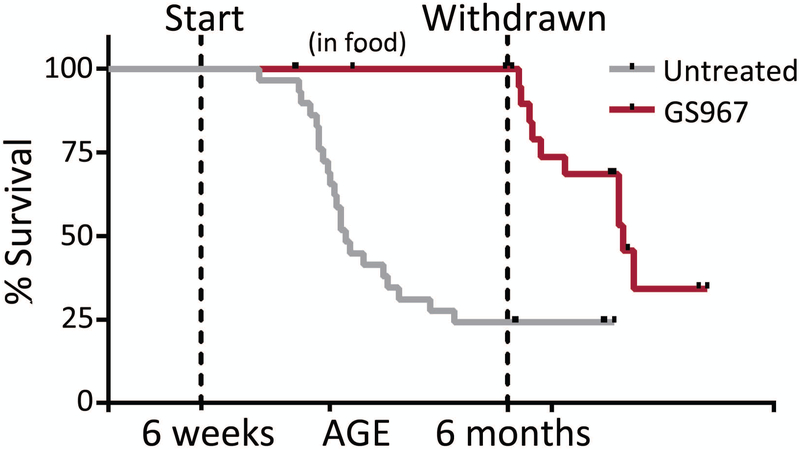
Existing anti-epileptic drugs are inadequate for seizure control in the majority of children with epileptic encephalopathy. The availability of mouse mutants expressing patient mutations has made it possible to evaluate new therapeutic approaches. The polycyclic sodium channel modulator GS967/Prax330 acts to reduce persistent current17. Nav1.6 contributes to persistent current in CNS neurons, as demonstrated by the reduced persistent current in hippocampal CA1 neurons18, cerebellar granule cells19, and trigeminal mesencephalic neurons20 from Scn8a null mice. Administration of GS967/Prax330 in the food to Scn8a N1768D/+ mice rescues lethality and reduces the incidence of spontaneous seizures by more than 50% 21 (Figure 6). The compound is also effective in mice with mutations of Scn1a and Scn2a 17,22, possibly through its effect on wildtype Nav1.6. Phase 1 clinical trials of this compound are in progress.
Figure 6.
Rescue of Scn8aN1768D mice by GS967/Prax330. The polycyclic drug GS967/Prax330 18 was incorporated into pelleted food and fed to the heterozygous mutant mice from the age of 1.5 months (pre-seizure onset) to 6 months. The mice were protected against seizures and neurotoxicity until removal of the drug19. p<0.0001 (Log rank). Disease penetrance was higher than in Figure 2; this represents experimental variability within the same mouse line.
A series of compounds with preferential effects on Nav1.6 developed by Xenon Pharmaceuticals are in Phase 1 clinical trials. previous sentence modified Antisense oligonucleotides that reduce transcript abundance are a potential third route to protection against gain-of-function mutations in SCN8A 23; this approach is potentially applicable to SCN8A encephalaphy and testable in the mutant mice.
Summary of mouse models.
Investigation of two mouse models has clearly demonstrated that gain-of-function mutations of SCN8A are sufficient to generate severe seizures and a lethal phentype. Neurons from mutant mice exhibit hyperactivity, with greater severity and earlier lethality in the R1872W mutant. These mice have been used for pre-clinical evaluation of new drugs, accelerating progress to clinical trials. Analysis of the conditional R1872W mutant mouse demonstrated the specific role of excitatory neurons in SCN8A encephalopathy, in contrast to the key role of inhibitory neurons in Dravet Syndrome. The susceptibility of the adult nervous system to activation of the R1872W allele observed in the conditional mouse model indicates that life-long treatment will be required to protect against the effects of gain-of-function SCN8A alleles in the CNS.
The expanding spectrum of SCN8A disorders.
The addition of SCN8A into gene testing panels and the increase in exome sequencing is resulting in the association of SCN8A with a broader spectrum of clinical conditions24-27, and this growth is likely to continue. Protein truncation mutations and missense mutations resulting in complete loss of channel activity have been identified in individuals with intellectual disability unacompanied by seizures26,28. Partial loss of channel activity was identified in a family with the SCN8A mutation that segregated with isolated myoclonus27. We anticipate the future discovery of many more mutations in patients with movement disorders like those seen in the spontaneous mouse Scn8a mutants, such as ataxia, dystonia and essential tremor29.
*De novo GOF mutations in SCN8A encephalopathy alter the biophysical properties of sodium channel Nav1.6 and result in neuronal hyperexcitability.
*Functional effects of pathogenic mutations include impaired channel inactivation, premature channel opening, and elevated persistent current.
*Mouse lines expressing the SCN8A patient mutations p.Asn1768Asp and p.Arg1872Trp recapitulate the human disorder with spontaneous seizures and premature lethality.
*Conditional expression of p.Arg1872Trp in excitatory forebrain neurons causes seizures and premature lethality but expression in inhibitory neurons is nonpathogenic.
*Loss-of-function variants of SCN8A are less severe clinically, resulting in isolated intellectual disability, myoclonus, or mild seizure disorders.
Acknowledgements:
Our work on SCN8A has been supported by NIH R01 NS034509 and by the Dravet Syndrome Foundation, the Wishes for Elliott Foundation and the Cute Syndrome Foundation. I thank Dr. Jacy Wagnon for helpful discussions.
Footnotes
Ethical publication: I confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Disclosure: The author has no conflict of interest to disclose.
References
- 1.Burgess DL, Kohrman DC, Galt J, et al. Mutation of a new sodium channel gene, Scn8a, in the mouse mutant ‘motor endplate disease’. Nature Genetics 1995;10:461–465, 1995. [DOI] [PubMed] [Google Scholar]
- 2.Kohrman DC, Harris JB, Meisler MH. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. Unusual splicing due to a minor class AT-AC intron. J Biol Chem 1996a;271:17576–17581. [DOI] [PubMed] [Google Scholar]
- 3.Kohrman DC, Smith MR, Goldin AL, et al. A missense mutation in the sodium channel gene Scn8a is responsible for cerebellar ataxia in the mouse mutant jolting. J. Neurosci.1996b;16:5993–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veeramah KR, O’Brien JE, Meisler MH, et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am J Hum Genet 2012;90:502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagnon JL, Korn MJ, Parent R, et al. Convulsive seizures and SUDEP in a mouse model of SCN8A epileptic encephalopathy. Hum Mol Genet 2015;24:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meisler MH, Helman G, Hammer MF, et al. SCN8A Encephalopathy: Research Progress and Prospects. Epilepsia 2016;57:1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagnon JL, Meisler MH. Recurrent and Non-Recurrent Mutations of SCN8A in Epileptic Encephalopathy. Front Neurol 2015;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagnon JL, Barker BS, Hounshell JA, et al. Pathogenic mechanism of recurrent mutations of SCN8A in epileptic encephalopathy. Ann Clin Transl Neurol 2016;3:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunton-Stasyshyn RKA, Wagnon JL, Wengert ER, et al. Prominent role of forebrain excitatory neurons in SCN8A encephalopathy. BRAIN 2019;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JM, Meisler MH. Modeling human epilepsy by TALEN targeting of mouse sodium channel Scn8a. Genesis 2014;52:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Santiago LF, Yuan Y., Wagnon JL, et al. Neuronal hyperexcitability in a mouse model of SCN8A encephalopathy. Proc. Nat’l. Acad. Sci. 2017;114:2383–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ottolini M, Barker BS, Gaykema RP, et al. Aberrant Sodium Channel Currents and Hyperexcitability of Medial Entorhinal Cortex Neurons in a Mouse Model of SCN8A Encephalopathy. J. Neurosci. 2017;37:7643–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noujaim SF, Kaur K, Milstein M, et al. A null mutation of the neuronal sodium channel NaV1.6 disrupts action potential propagation and excitation-contraction coupling in the mouse heart. FASEB J. 2012; 26:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frasier CR, Wagnon JL, Bao Y, et al. Cardiac arrhythmia in a mouse model of SCN8A Epileptic Encephalopathy. Proc. Nat’l. Acad. Sci. 2016113:12838–12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprissler RS, Wagnon JL, Bunton-Stasyshyn RK, et al. Altered gene expression profile in a mouse model of Scn8a epilepsy reveals reactive astrocytosis in response to seizures. Exptl. Neurol. 2016;288:134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu FH, Mantegazza M, Westenbroek RE et al. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006; 9:1142–1149 [DOI] [PubMed] [Google Scholar]
- 17.Anderson LL, Thompson CH, Hawkins NA, et al. Antiepileptic activity of preferential inhibitors of persistent sodium current. Epilepsia. 2014;55:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Royeck M, Horstmann MT, Remy S, et al. Role of axonal NaV1.6 sodium channels in action potential initiation of CA1 pyramidal neurons. J Neurophysiol. 2008. 100:2361–2380. [DOI] [PubMed] [Google Scholar]
- 19.Osorio N, Cathala L, Meisler MH et al. Persistent Nav1.6 current at axon initial segments tunes spike timing of cerebellar granule cells. J Physiol. 2010. 588(Pt 4):651–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enomoto A, Han JM, Hsiao CF, et al. Sodium currents in mesencephalic trigeminal neurons from Nav1.6 null mice. J Neurophysiol 2007. 98:710–719. [DOI] [PubMed] [Google Scholar]
- 21.Baker EM, Thompson CH, Hawkins NA, et al. The novel sodium channel modulator GS-458967 (GS967) is an effective treatment in a mouse model of SCN8A encephalopathy. Epilepsia 2018;59:1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson LL, Hawkins NA, Thompson CH, et al. Unexpected Efficacy of a Novel Sodium Channel Modulator in Dravet Syndrome. Sci Rep. 2017;7:1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roovers J, De Jonghe P, Weckhuysen S. The therapeutic potential of RNA regulation in neurological disorders. Expert Opin Ther Targets. 2018; 22:1017–1028. [DOI] [PubMed] [Google Scholar]
- 24.Larsen J, Carvill GL, Gardella E, et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology 2015;84:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardella E, Becker F, Moller RS, et al. Benign infantile seizures and paroxysmal dyskinesia caused by an SCN8A mutation. Ann Neurol 2016;79:428–436. [DOI] [PubMed] [Google Scholar]
- 26.Wagnon JL, Barker BS, Ottolini M, et al. Loss-of-function mutations of SCN8A in intellectual disability without seizures. Neurology Genetics 2017;3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagnon JL, Mencacci NE, Barker BS, et al. Partial loss-of-function of sodium channel SCN8A in familial isolated myoclonus. Human Mutation 2018. 39:965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trudeau MM, Dalton JC, Day JW, et al. Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia and mental retardation. J Med Genet. 2006;43:527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharkey LM, Jones JM, Hedera P, et al. Parkinsonism Relat Disord. Evaluation of SCN8A as a candidate gene for autosomal dominant essential tremor. 2009; 15:321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]